Copy Number Alteration Profiling from Plasma cfDNA WES in Advanced NSCLC
Abstract
1. Introduction
2. Results
2.1. Sample Characteristics
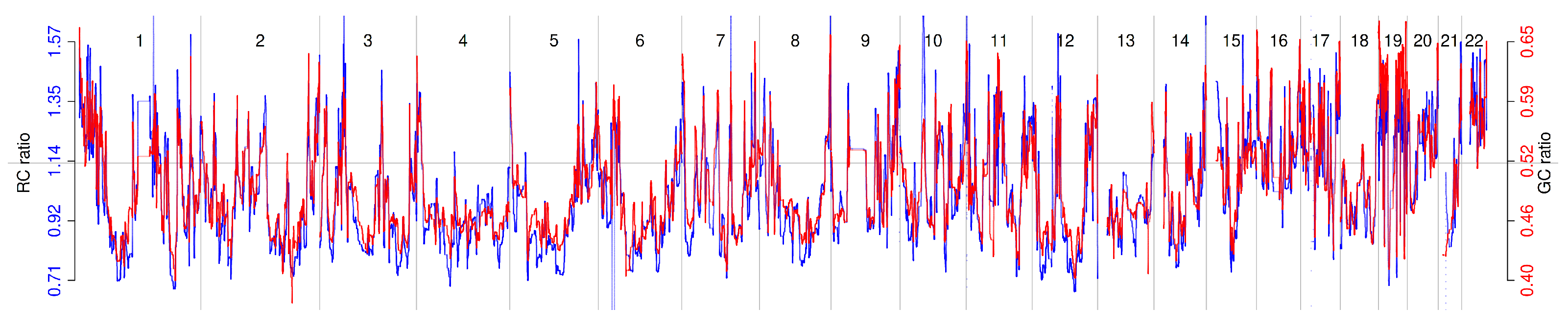
2.2. Relationship Between Read Count Patterns and GC Content
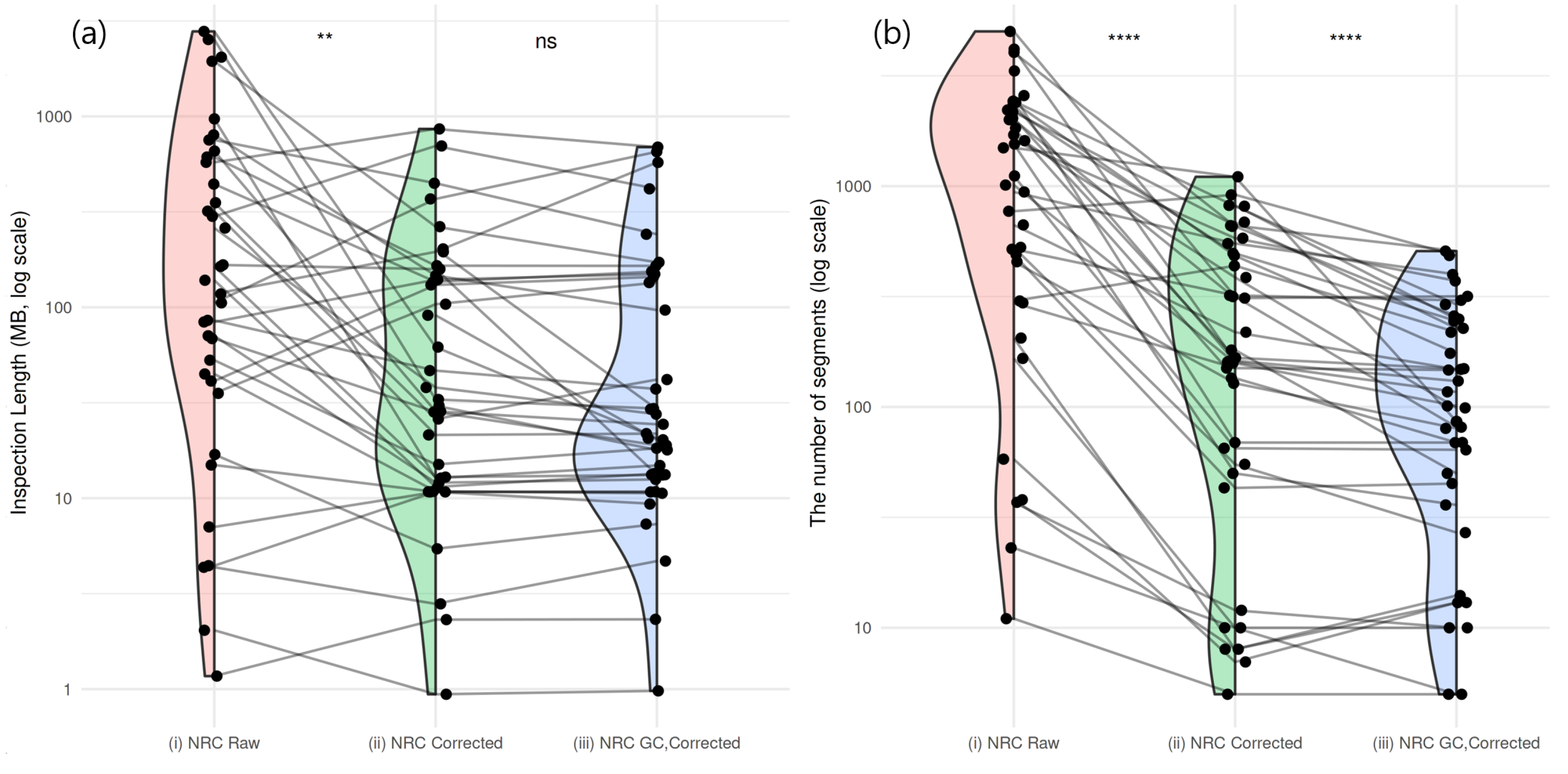
2.3. Residual GC Bias Correction in Normal Subjects
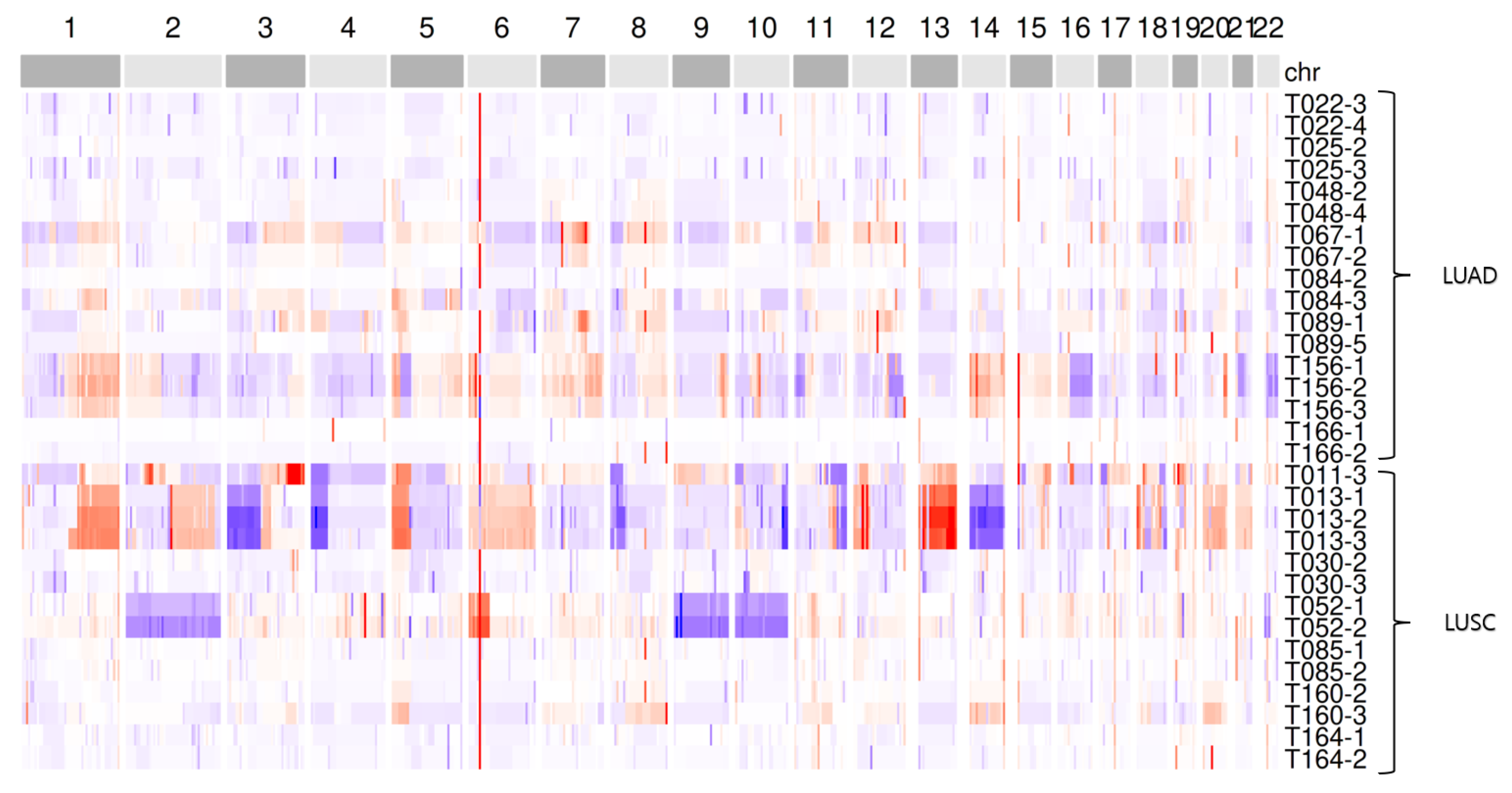
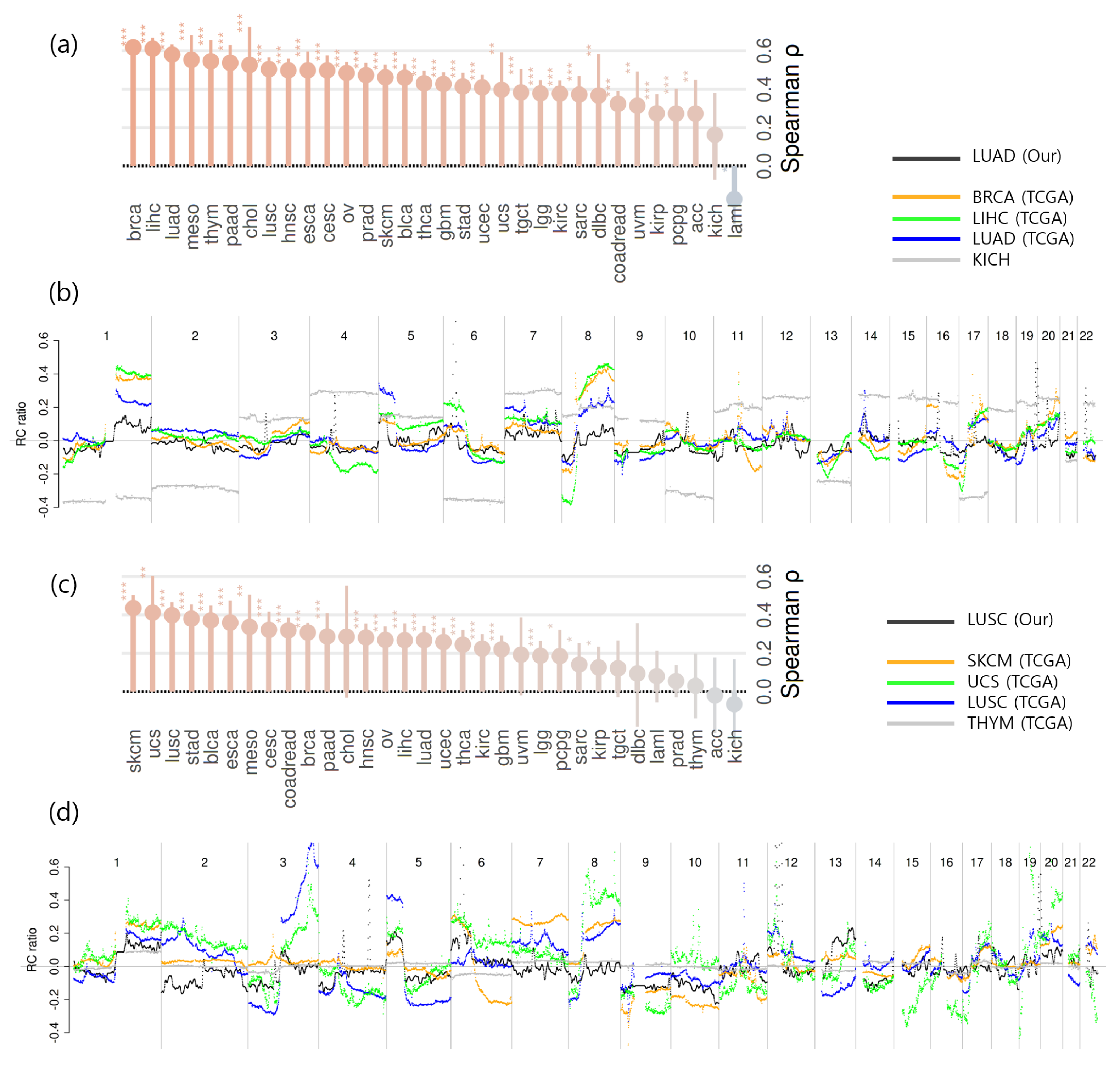
2.4. CNA Detection and Comparison of Aggregated CNA Profiles Between Cancer Types
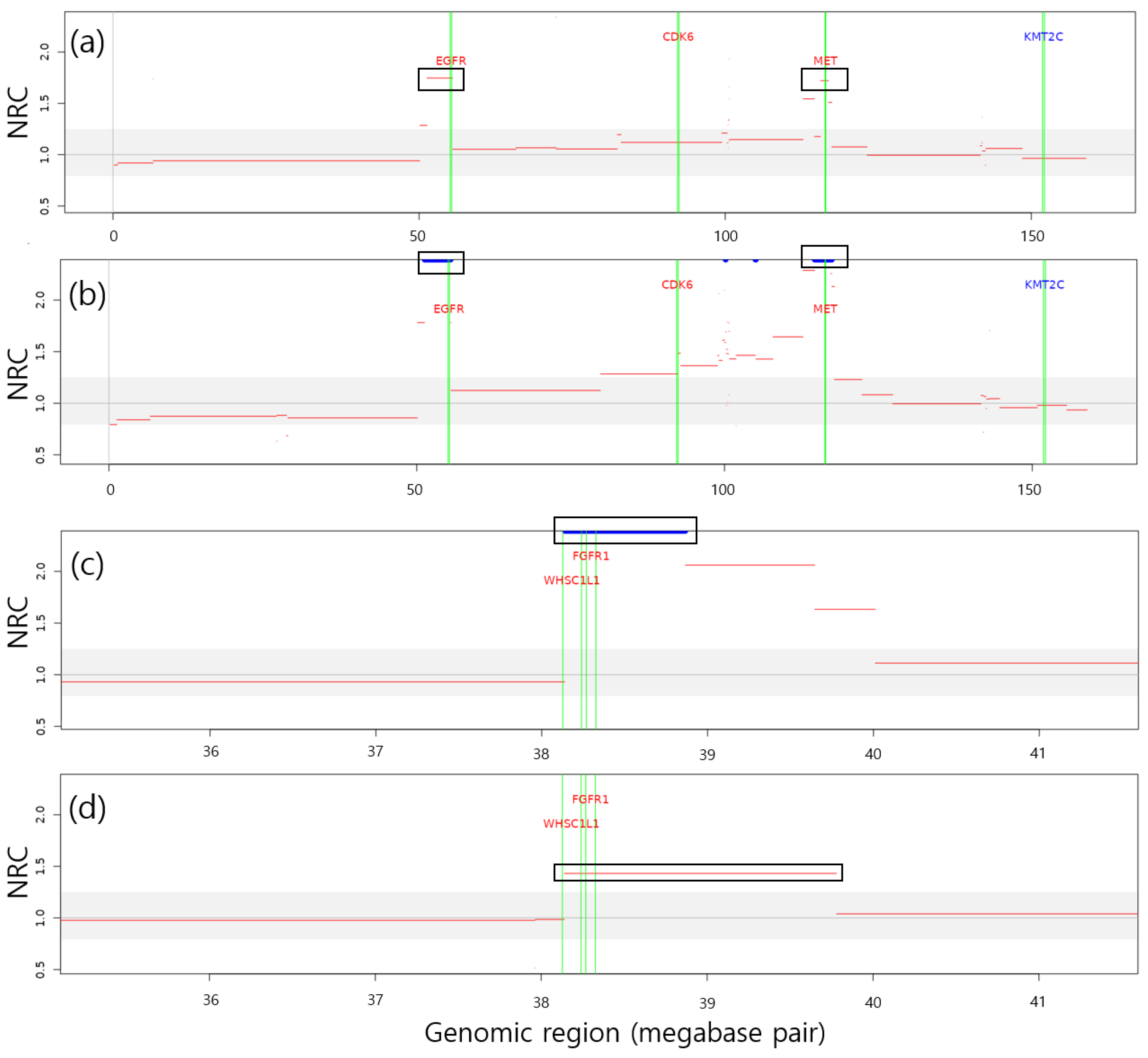
2.5. Identification of LUAD- and LUSC-Related Genes Within CNA Regions
2.6. Effect of GC Bias Correction on CNA Detection Performance
3. Discussion
3.1. Improvement of CNA Detection Performance
3.2. Enhancing Detection Sensitivity and Threshold Adaptation
3.3. Challenges in Detecting CNAs from cfDNA WES
3.4. Clinical Relevance of CNA-Affected Genes in LUAD and LUSC
3.5. Limitations in Linking CNA Signals to Clinical Outcomes
4. Materials and Methods
4.1. Patients, Samples, and WES Procedures
4.2. Raw Copy Number Ratio Quantification and Common Fluctuation Control
4.3. GC Bias Correction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CBS | Circular binary segmentation |
| cfDNA | Cell-free DNA |
| CNA | Copy number alteration |
| CNV | Copy number variation |
| ctDNA | Circulating tumor DNA |
| ddPCR | Droplet digital polymerase chain reaction |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| NRC | Normalized read count |
| NSCLC | Non-small cell lung cancer |
| PD | Progressive disease |
| PFS | Progression-free survival |
| QC | Quality control |
| RC | Read count |
| SNV | Single-nucleotide variant |
| TCGA | The Cancer Genome Atlas |
| TF | Tumor fraction |
| TI wavelet transform | Translation-invariant wavelet transform |
| WES | Whole-exome sequencing |
| WGS | Whole-genome sequencing |
References
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef]
- Koboldt, D.C. Best Practices for Variant Calling in Clinical Sequencing. Genome Med. 2020, 12, 91. [Google Scholar] [CrossRef]
- Mandelker, D.; Ceyhan-Birsoy, O. Evolving Significance of Tumor-Normal Sequencing in Cancer Care. Trends Cancer 2020, 6, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Choi, C.M.; Lee, S.H.; Lee, S.; Jeong, M.K. Read Count Patterns and Detection of Cancerous Copy Number Alterations in Plasma Cell-Free DNA Whole Exome Sequencing Data for Advanced Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2022, 23, 12932. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404, Erratum in Cancer Discov. 2012, 2, 960.. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Campbell, J.D.; Alexandrov, A.; Kim, J.; Wala, J.; Berger, A.H.; Pedamallu, C.S.; Shukla, S.A.; Guo, G.; Brooks, A.N.; et al. Distinct Patterns of Somatic Genome Alterations in Lung Adenocarcinomas and Squamous Cell Carcinomas. Nat. Genet. 2016, 48, 607–616. [Google Scholar] [CrossRef]
- Szatkiewicz, J.P.; Wang, W.; Sullivan, P.F.; Wang, W.; Sun, W. Improving Detection of Copy-Number Variation by Simultaneous Bias Correction and Read-Depth Segmentation. Nucleic Acids Res. 2013, 41, 1519–1532. [Google Scholar] [CrossRef]
- Bos, M.K.; Angus, L.; Nasserinejad, K.; Jager, A.; Jansen, M.P.; Martens, J.W.; Sleijfer, S. Whole Exome Sequencing of Cell-Free DNA—A Systematic Review and Bayesian Individual Patient Data Meta-Analysis. Cancer Treat. Rev. 2020, 83, 101951. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive Genomic Characterization of Squamous Cell Lung Cancers. Nature 2012, 489, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, S.; Yasui, K.; Iizasa, T.; Imoto, I.; Fujisawa, T.; Inazawa, J. TERC Identified as a Probable Target within the 3q26 Amplicon That Is Detected Frequently in Non-Small Cell Lung Cancers. Clin. Cancer Res. 2003, 9, 4705–4713. [Google Scholar]
- Pelosi, G.; Curto, B.D.; Nicholson, A.G.; Manzotti, M.; Spaggiari, L.; Maisonneuve, P.; Pasini, F.; Iannucci, A. 3q26 Amplification and Polysomy of Chromosome 3 in Squamous Cell Lesions of the Lung: A Fluorescence In Situ Hybridization Study. Clin. Cancer Res. 2007, 13, 1995–2004. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.J.; Watanabe, H.; Mermel, C.H.; Yu, S.; Perner, S.; Verhaak, R.G.; Kim, S.Y.; Wardwell, L.; Tamayo, P.; Gat-Viks, I.; et al. SOX2 Is an Amplified Lineage-Survival Oncogene in Lung and Esophageal Squamous Cell Carcinomas. Nat. Genet. 2009, 41, 1238–1242. [Google Scholar] [CrossRef]
- Kang, J.U.; Koo, S.H.; Kwon, K.C.; Park, J.W.; Kim, J.M. Gain at Chromosomal Region 5p15.33, Containing TERT, Is the Most Frequent Genetic Event in Early Stages of Non-Small Cell Lung Cancer. Cancer Genet. Cytogenet. 2008, 182, 1–11. [Google Scholar] [CrossRef]
- Zhu, C.Q.; Cutz, J.C.; Liu, N.; Lau, D.; Shepherd, F.A.; Squire, J.A.; Tsao, M.S. Amplification of Telomerase (hTERT) Gene Is a Poor Prognostic Marker in Non-Small-Cell Lung Cancer. Br. J. Cancer 2006, 94, 1452–1459. [Google Scholar] [CrossRef]
- Yang, H.; Wen, L.; Zhao, C.; Li, X.; Shan, C.; Liu, D.; Hong, W.; Zhou, Z.; Zhou, C.; Cai, L.; et al. EGFR Amplification Is a Putative Resistance Mechanism for NSCLC–LM Patients with TKI Therapy and Is Associated with Poor Outcome. Front. Oncol. 2022, 12, 902664. [Google Scholar] [CrossRef]
- Yoshimura, K.; Inoue, Y.; Inui, N.; Karayama, M.; Yasui, H.; Hozumi, H.; Suzuki, Y.; Furuhashi, K.; Fujisawa, T.; Enomoto, N.; et al. MET Amplification and Efficacy of Nivolumab in Patients With NSCLC. JTO Clin. Res. Rep. 2021, 2, 100239. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Q.; Zeng, X.; Wang, M.; Dong, S.; Yang, B.; Tu, X.; Wei, T.; Xie, W.; Zhang, C.; et al. MET Amplification Attenuates Lung Tumor Response to Immunotherapy by Inhibiting STING. Cancer Discov. 2021, 11, 2726–2737. [Google Scholar] [CrossRef]
- Dutt, A.; Ramos, A.H.; Hammerman, P.S.; Mermel, C.; Cho, J.; Sharifnia, T.; Chande, A.; Tanaka, K.E.; Stransky, N.; Greulich, H.; et al. Inhibitor-Sensitive FGFR1 Amplification in Human Non-Small Cell Lung Cancer. PLoS ONE 2011, 6, e20351. [Google Scholar] [CrossRef]
- Yuan, G.; Flores, N.M.; Hausmann, S.; Lofgren, S.M.; Kharchenko, V.; Angulo-Ibanez, M.; Sengupta, D.; Lu, X.; Czaban, I.; Azhibek, D.; et al. Elevated NSD3 Histone Methylation Activity Drives Squamous Cell Lung Cancer. Nature 2021, 590, 504–508. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Liu, S.; Wu, X.; Marti, T.M.; Dorn, P.; Schmid, R.A.; Peng, R.W.; Shu, Y. Dissecting the Immunological Profiles in NSD3-Amplified LUSC through Integrative Multi-Scale Analyses. Cancers 2022, 14, 4997. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhou, X.; Li, H.; Yin, T.; Jiang, Y. Prognosis of Immunotherapy for Non-Small Cell Lung Cancer with CDKN2A Loss of Function. J. Thorac. Dis. 2024, 16, 507–515. [Google Scholar] [CrossRef]
- Watson, A.S.; Krause, H.B.; Elliott, A.; Farrell, A.; Liu, S.V.; Ma, P.C.; VanderWalde, A.; Sledge, G.W.; Spetzler, D.; Schenk, E.L.; et al. Use of Oncogene Overlap by Tissue-Based Next-Generation Sequencing to Explore the Mutational Landscape and Survival Impact of HER2, KRAS and MET Copy-Number Gain in Nonsmall Cell Lung Cancer. Clin. Lung Cancer 2024, 25, 712–722.e1. [Google Scholar] [CrossRef]
- Knudsen, E.S.; Nambiar, R.; Rosario, S.R.; Smiraglia, D.J.; Goodrich, D.W.; Witkiewicz, A.K. Pan-Cancer Molecular Analysis of the RB Tumor Suppressor Pathway. Commun. Biol. 2020, 3, 158. [Google Scholar] [CrossRef] [PubMed]
- Goel, S.; DeCristo, M.J.; Watt, A.C.; BrinJones, H.; Sceneay, J.; Li, B.B.; Khan, N.; Ubellacker, J.M.; Xie, S.; Metzger-Filho, O.; et al. CDK4/6 Inhibition Triggers Anti-Tumour Immunity. Nature 2017, 548, 471–475. [Google Scholar] [CrossRef]
- Weir, B.A.; Woo, M.S.; Getz, G.; Perner, S.; Ding, L.; Beroukhim, R.; Lin, W.M.; Province, M.A.; Kraja, A.; Johnson, L.A.; et al. Characterizing the Cancer Genome in Lung Adenocarcinoma. Nature 2007, 450, 893–898. [Google Scholar] [CrossRef]
- Goglia, A.G.; Alshalalfa, M.; Khan, A.; Isakov, D.R.; Hougen, H.Y.; Swami, N.; Kannikal, J.; Mcbride, S.M.; Gomez, D.R.; Punnen, S.; et al. Pan-Cancer Genomic Analysis Reveals FOXA1 Amplification Is Associated with Adverse Outcomes in Non–Small Cell Lung, Prostate, and Breast Cancers. JNCI J. Natl. Cancer Inst. 2025, 117, 188–197. [Google Scholar] [CrossRef]
- Humar, M.; Kern, I.; Vlacic, G.; Hadzic, V.; Cufer, T. Insulin-like Growth Factor 1 Receptor Expression in Advanced Non-Small-Cell Lung Cancer and Its Impact on Overall Survival. Radiol. Oncol. 2017, 51, 195–202. [Google Scholar] [CrossRef]
- Huang, L.N.; Wang, D.S.; Chen, Y.Q.; Li, W.; Hu, F.D.; Gong, B.L.; Zhao, C.L.; Jia, W. Meta-Analysis for Cyclin E in Lung Cancer Survival. Clin. Chim. Acta 2012, 413, 663–668. [Google Scholar] [CrossRef]
- Li, H.; Van Der Merwe, P.A.; Sivakumar, S. Biomarkers of Response to PD-1 Pathway Blockade. Br. J. Cancer 2022, 126, 1663–1675. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.H.; Jun, H.J.; Choi, Y.; Kim, J.C.; Jang, H.; Park, S.M.; Kwon, O.; Choi, C.M.; Kim, S.J.; Choi, J.G.; et al. Investigating Treatment Response and Immune Profile in Association with Pattern Identification in NSCLC Patients Scheduled for Immune Checkpoint Inhibitor Monotherapy (HARMONY Study): A Protocol for a Prospective Observational Study. Med. Case Rep. Study Protoc. 2022, 3, e0247. [Google Scholar] [CrossRef]
- Jang, H.; Lee, H. Multiresolution Correction of GC Bias and Application to Identification of Copy Number Alterations. Bioinformatics 2019, 35, 3890–3897. [Google Scholar] [CrossRef] [PubMed]
- Xi, R.; Lee, S.; Xia, Y.; Kim, T.M.; Park, P.J. Copy Number Analysis of Whole-Genome Data Using BIC-seq2 and Its Application to Detection of Cancer Susceptibility Variants. Nucleic Acids Res. 2016, 44, 6274–6286. [Google Scholar] [CrossRef] [PubMed]
- Olshen, A.B.; Venkatraman, E.S.; Lucito, R.; Wigler, M. Circular Binary Segmentation for the Analysis of Array-Based DNA Copy Number Data. Biostatistics 2004, 5, 557–572. [Google Scholar] [CrossRef]
- Venkatraman, E.S.; Olshen, A.B. A Faster Circular Binary Segmentation Algorithm for the Analysis of Array CGH Data. Bioinformatics 2007, 23, 657–663. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, H.; Jeong, M.-K. Copy Number Alteration Profiling from Plasma cfDNA WES in Advanced NSCLC. Int. J. Mol. Sci. 2025, 26, 11111. https://doi.org/10.3390/ijms262211111
Jang H, Jeong M-K. Copy Number Alteration Profiling from Plasma cfDNA WES in Advanced NSCLC. International Journal of Molecular Sciences. 2025; 26(22):11111. https://doi.org/10.3390/ijms262211111
Chicago/Turabian StyleJang, Ho, and Mi-Kyung Jeong. 2025. "Copy Number Alteration Profiling from Plasma cfDNA WES in Advanced NSCLC" International Journal of Molecular Sciences 26, no. 22: 11111. https://doi.org/10.3390/ijms262211111
APA StyleJang, H., & Jeong, M.-K. (2025). Copy Number Alteration Profiling from Plasma cfDNA WES in Advanced NSCLC. International Journal of Molecular Sciences, 26(22), 11111. https://doi.org/10.3390/ijms262211111






