Validation of GDAP1 and HECW2 as Epigenetic Markers of Alcohol Use Disorder in Blood and Brain
Abstract
1. Introduction
2. Results
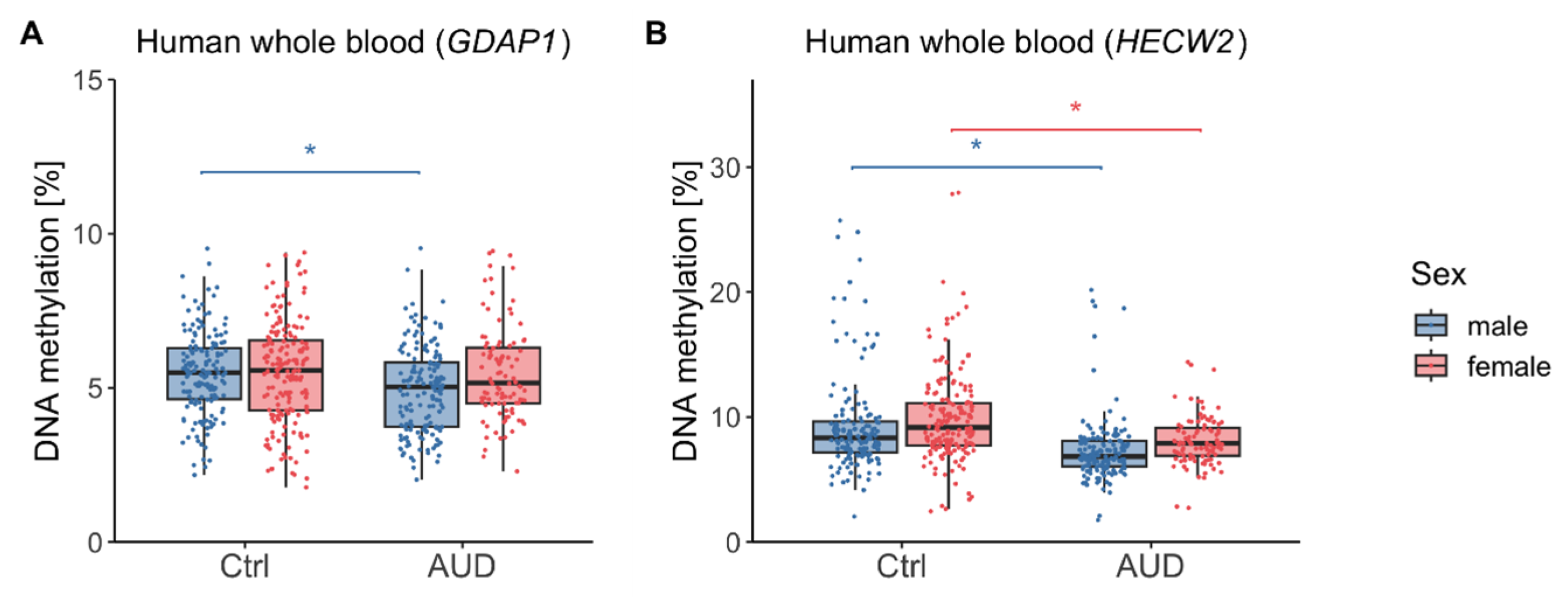
2.1. DNA methylation (DNAm) in Human Whole Blood
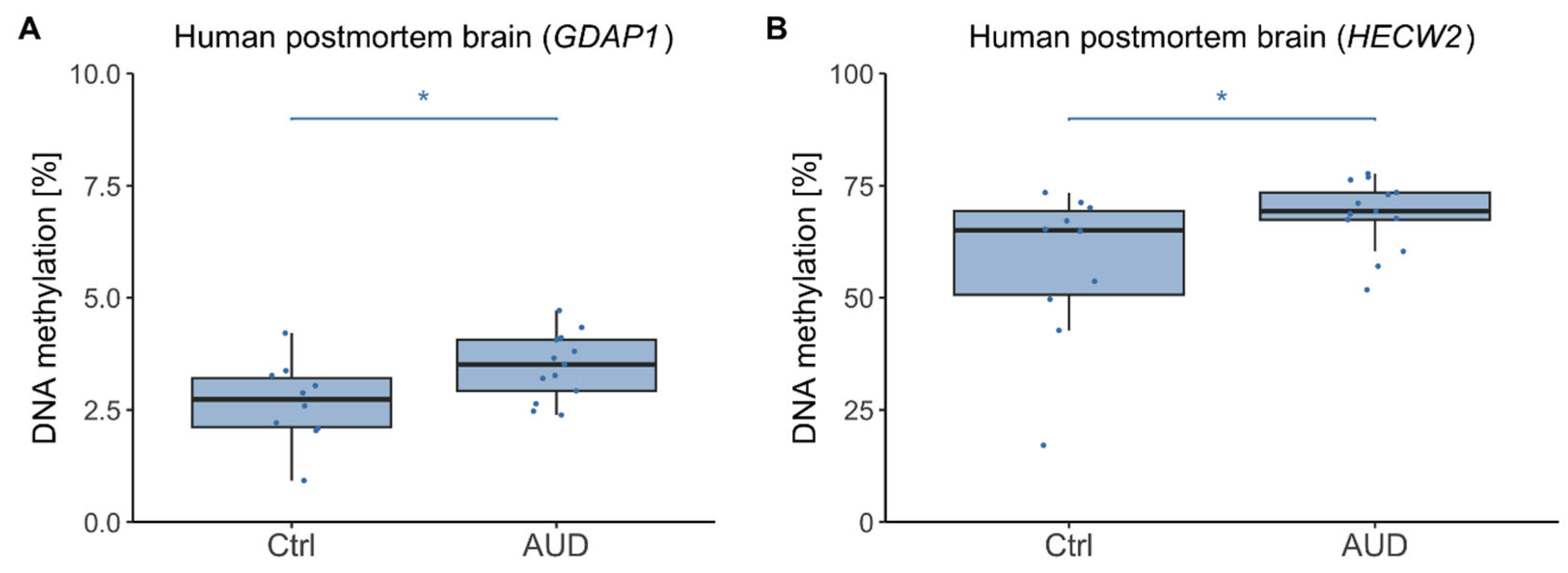
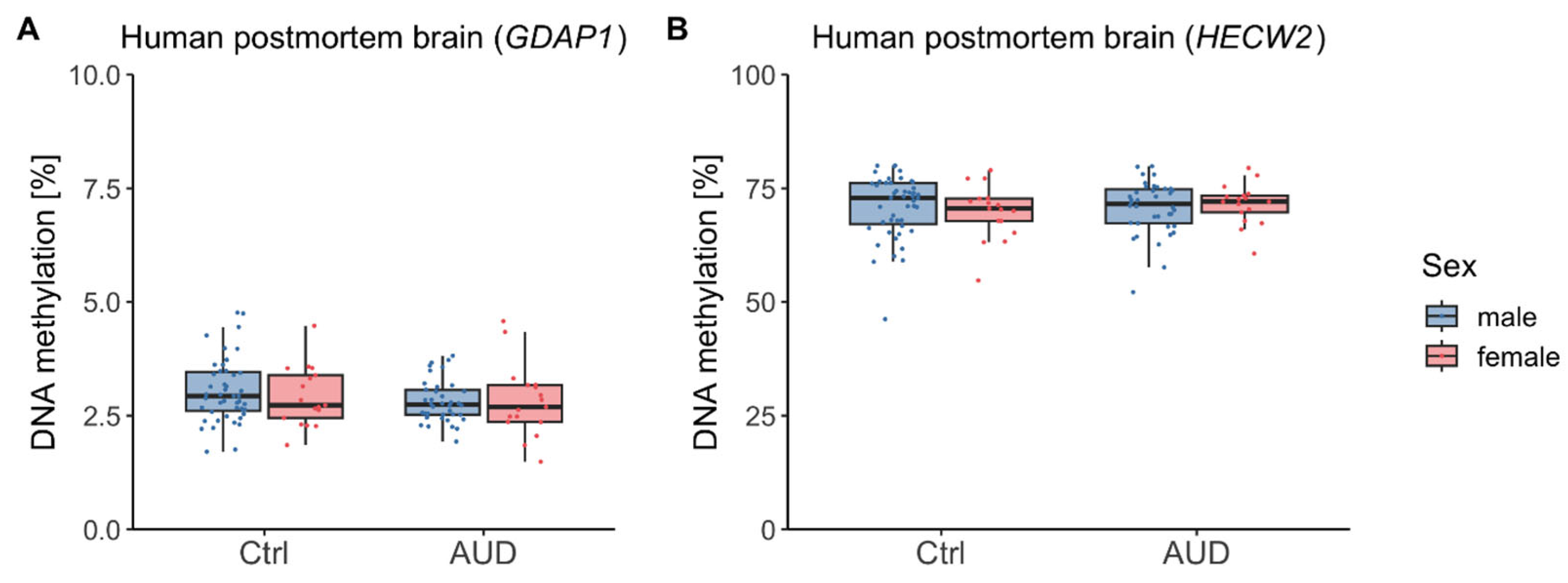
2.2. DNAm in Human Postmortem Brain
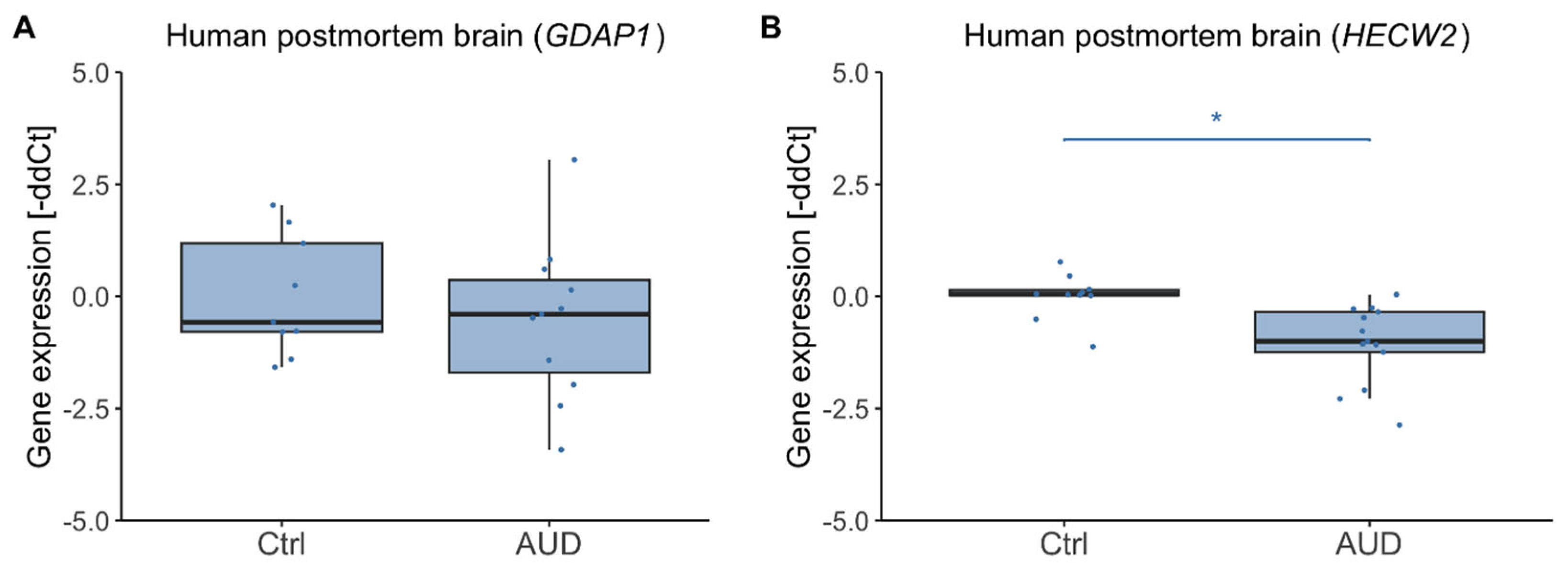
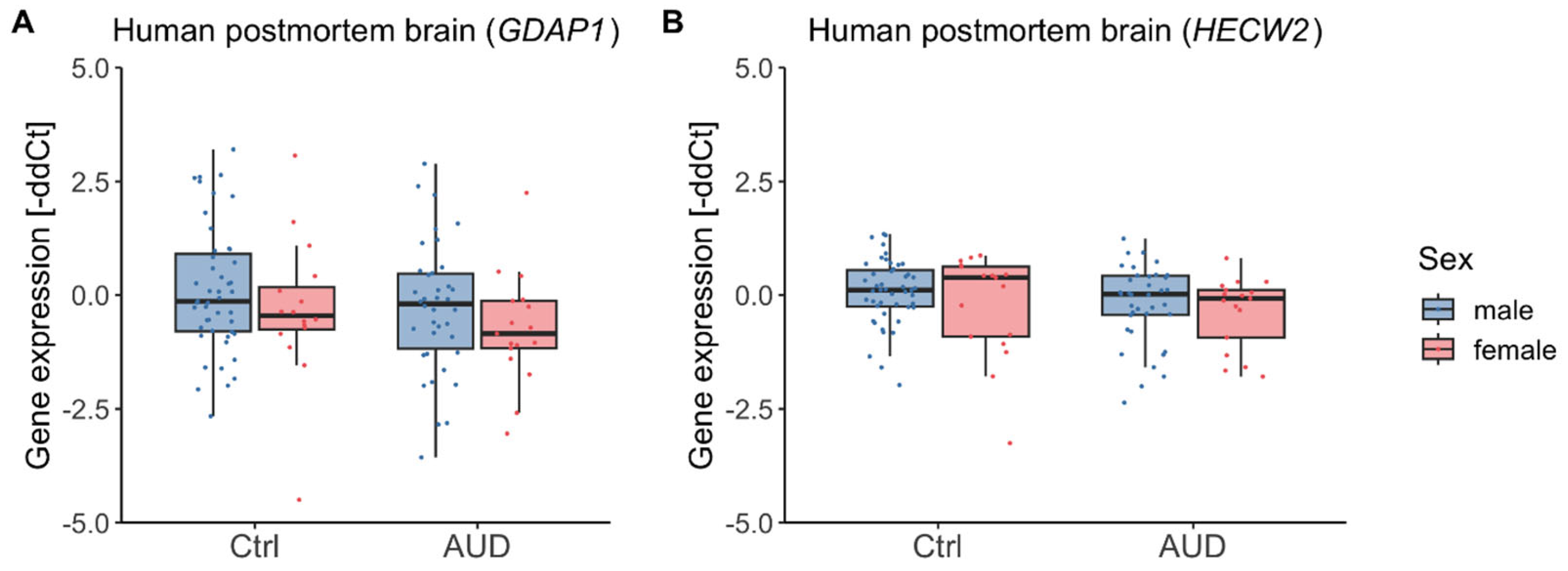
2.3. Gene Expression in Human Postmortem Brain
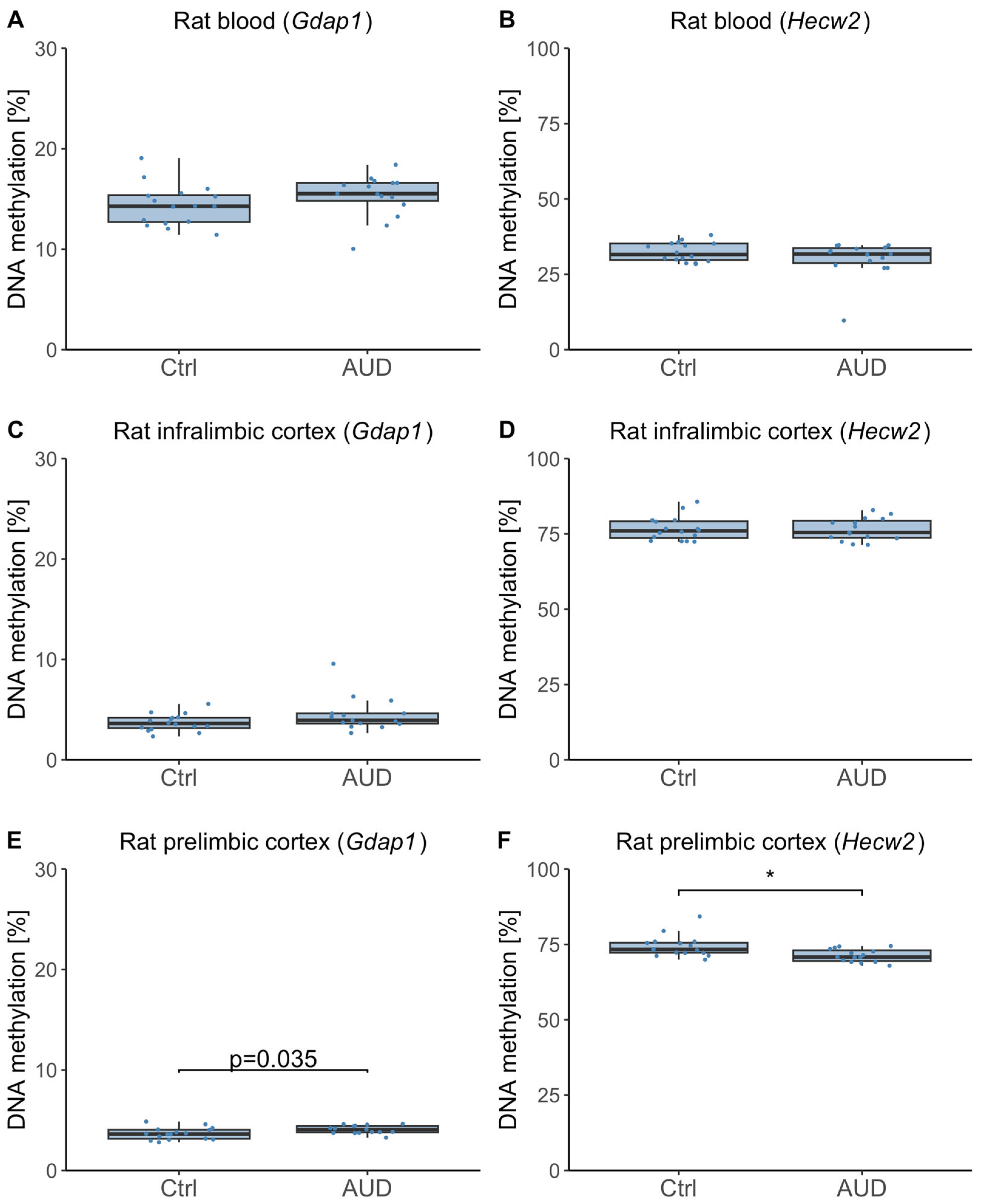
2.4. DNAm in Whole Blood and Brain Samples from an Alcohol Use Disorder (AUD) Animal Model
3. Discussion
3.1. DNAm Findings in Human Whole Blood
3.2. Postmortem Brain Tissue and Directionality of DNAm Effects
3.3. Insights from the Rat Model
3.4. Limitations
3.5. Conclusions and Outlook
4. Materials and Methods
4.1. Study Samples
4.1.1. Human Whole Blood Samples
4.1.2. Human Postmortem Brain Samples
4.1.3. Induction of Alcohol Dependence in Rats
4.2. DNAm Analysis
4.2.1. Human Whole Blood Samples
4.2.2. Human Postmortem Brain Samples
4.2.3. Rat Blood and Brain Samples
4.3. Gene Expression Analysis
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUD | Alcohol use disorder |
| DNAm | DNA methylation |
| DALYs | Disability-adjusted life years |
| EWAS | epigenome-wide association study |
| GDAP1 | Ganglioside-induced Differentiation Associated Protein 1 |
| HECW2 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| SD | Standard deviation |
| BMI | Body mass index |
| Ctrl | Controls |
| PMI | Postmortem interval |
| DSM-IV | Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition |
| CIE | Chronic intermittent ethanol exposure |
| qRT-PCR | Quantitative real-time PCR |
| RIN | RNA integrity number |
| GLS | Generalized least squares |
References
- World Health Organization. Global Status Report on Alcohol and Health and Treatment of Substance Use Disorders; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Agabio, R.; Pisanu, C.; Luigi Gessa, G.; Franconi, F. Sex differences in alcohol use disorder. Curr. Med. Chem. 2017, 24, 2661–2670. [Google Scholar] [CrossRef]
- Verhulst, B.; Neale, M.C.; Kendler, K.S. The heritability of alcohol use disorders: A meta-analysis of twin and adoption studies. Psychol. Med. 2015, 45, 1061–1072. [Google Scholar] [CrossRef]
- Zilberman, M.L.; Tavares, H.; Blume, S.B.; El-Guebaly, N. Substance use disorders: Sex differences and psychiatric comorbidities. Can. J. Psychiatry 2003, 48, 5–13. [Google Scholar] [CrossRef]
- Nieratschker, V.; Batra, A.; Fallgatter, A.J. Genetics and epigenetics of alcohol dependence. J. Mol. Psychiatry 2013, 1, 11. [Google Scholar] [CrossRef]
- Wedemeyer, F.; Kaminski, J.A.; Zillich, L.; Hall, A.S.; Friedel, E.; Witt, S.H. Prospects of Genetics and Epigenetics of Alcohol Use Disorder. Curr. Addict. Rep. 2020, 7, 446–452. [Google Scholar] [CrossRef]
- Bird, A. The essentials of DNA methylation. Cell 1992, 70, 5–8. [Google Scholar] [CrossRef]
- Berkel, T.D.; Pandey, S.C. Emerging role of epigenetic mechanisms in alcohol addiction. Alcohol. Clin. Exp. Res. 2017, 41, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Gelernter, J. Human genetics and epigenetics of alcohol use disorder. J. Clin. Investig. 2024, 134, e172885. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Marioni, R.E.; Hedman, Å.K.; Pfeiffer, L.; Tsai, P.-C.; Reynolds, L.M.; Just, A.C.; Duan, Q.; Boer, C.G.; Tanaka, T. A DNA methylation biomarker of alcohol consumption. Mol. Psychiatry 2018, 23, 422–433. [Google Scholar] [CrossRef]
- Lohoff, F.W.; Clarke, T.-K.; Kaminsky, Z.A.; Walker, R.M.; Bermingham, M.L.; Jung, J.; Morris, S.W.; Rosoff, D.; Campbell, A.; Barbu, M. Epigenome-wide association study of alcohol consumption in N = 8161 individuals and relevance to alcohol use disorder pathophysiology: Identification of the cystine/glutamate transporter SLC7A11 as a top target. Mol. Psychiatry 2022, 27, 1754–1764. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Montalvo-Ortiz, J.L.; Zhang, X.; Southwick, S.M.; Krystal, J.H.; Pietrzak, R.H.; Gelernter, J. Epigenome-wide DNA methylation association analysis identified novel loci in peripheral cells for alcohol consumption among European American male veterans. Alcohol. Clin. Exp. Res. 2019, 43, 2111–2121. [Google Scholar] [CrossRef]
- Ruggeri, B.; Nymberg, C.; Vuoksimaa, E.; Lourdusamy, A.; Wong, C.P.; Carvalho, F.M.; Jia, T.; Cattrell, A.; Macare, C.; Banaschewski, T. Association of protein phosphatase PPM1G with alcohol use disorder and brain activity during behavioral control in a genome-wide methylation analysis. Am. J. Psychiatry 2015, 172, 543–552. [Google Scholar] [CrossRef]
- Brückmann, C.; Islam, S.A.; MacIsaac, J.L.; Morin, A.M.; Karle, K.N.; Di Santo, A.; Wüst, R.; Lang, I.; Batra, A.; Kobor, M.S. DNA methylation signatures of chronic alcohol dependence in purified CD3+ T-cells of patients undergoing alcohol treatment. Sci. Rep. 2017, 7, 6605. [Google Scholar] [CrossRef]
- Witt, S.H.; Frank, J.; Frischknecht, U.; Treutlein, J.; Streit, F.; Foo, J.C.; Sirignano, L.; Dukal, H.; Degenhardt, F.; Koopmann, A. Acute alcohol withdrawal and recovery in men lead to profound changes in DNA methylation profiles: A longitudinal clinical study. Addiction 2020, 115, 2034–2044. [Google Scholar] [CrossRef]
- Zillich, L.; Frank, J.; Streit, F.; Friske, M.M.; Foo, J.C.; Sirignano, L.; Heilmann-Heimbach, S.; Dukal, H.; Degenhardt, F.; Hoffmann, P. Epigenome-wide association study of alcohol use disorder in five brain regions. Neuropsychopharmacology 2022, 47, 832–839. [Google Scholar] [CrossRef]
- Zillich, L.; Poisel, E.; Frank, J.; Foo, J.C.; Friske, M.M.; Streit, F.; Sirignano, L.; Heilmann-Heimbach, S.; Heimbach, A.; Hoffmann, P. Multi-omics signatures of alcohol use disorder in the dorsal and ventral striatum. Transl. Psychiatry 2022, 12, 190. [Google Scholar] [CrossRef] [PubMed]
- Lohoff, F.W.; Roy, A.; Jung, J.; Longley, M.; Rosoff, D.B.; Luo, A.; O’connell, E.; Sorcher, J.L.; Sun, H.; Schwandt, M. Epigenome-wide association study and multi-tissue replication of individuals with alcohol use disorder: Evidence for abnormal glucocorticoid signaling pathway gene regulation. Mol. Psychiatry 2021, 26, 2224–2237. [Google Scholar] [CrossRef]
- Andrade-Brito, D.E.; Núñez-Ríos, D.L.; Martínez-Magaña, J.J.; Nagamatsu, S.T.; Rompala, G.; Zillich, L.; Witt, S.H.; Clark, S.L.; Lattig, M.C.; Montalvo-Ortiz, J.L. Neuronal-specific methylome and hydroxymethylome analysis reveal significant loci associated with alcohol use disorder. Front. Genet. 2024, 15, 1345410. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.L.; Chan, R.F.; Zhao, M.; Xie, L.Y.; Copeland, W.E.; Penninx, B.W.; Aberg, K.A.; van den Oord, E.J. Dual methylation and hydroxymethylation study of alcohol use disorder. Addict. Biol. 2022, 27, e13114. [Google Scholar] [CrossRef] [PubMed]
- Longley, M.J.; Lee, J.; Jung, J.; Lohoff, F.W. Epigenetics of alcohol use disorder—A review of recent advances in DNA methylation profiling. Addict. Biol. 2021, 26, e13006. [Google Scholar] [CrossRef]
- Philibert, R.A.; Penaluna, B.; White, T.; Shires, S.; Gunter, T.; Liesveld, J.; Erwin, C.; Hollenbeck, N.; Osborn, T. A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics 2014, 9, 1212–1219. [Google Scholar] [CrossRef] [PubMed]
- Brückmann, C.; Di Santo, A.; Karle, K.N.; Batra, A.; Nieratschker, V. Validation of differential GDAP1 DNA methylation in alcohol dependence and its potential function as a biomarker for disease severity and therapy outcome. Epigenetics 2016, 11, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Kawecka, E.; Plättner, H.; Ederer, L.; Niemann, K.; Pasche, S.; Zimmermann, M.; Edelmann, S.; Nieratschker, V. GDAP1 Is Dysregulated at DNA Methylation and H3K4me3 Levels in Alcohol Use Disorder. Int. J. Mol. Sci. 2025, 26, 1623. [Google Scholar] [CrossRef]
- Bertholet, A.; Delerue, T.; Millet, A.; Moulis, M.; David, C.; Daloyau, M.; Arnauné-Pelloquin, L.; Davezac, N.; Mils, V.; Miquel, M. Mitochondrial fusion/fission dynamics in neurodegeneration and neuronal plasticity. Neurobiol. Dis. 2016, 90, 3–19. [Google Scholar] [CrossRef]
- Shen, H.; Kou, Q.; Shao, L.; Zhang, J.; Li, F. E3 ubiquitin ligase HECW2: A promising target for tumour therapy. Cancer Cell Int. 2024, 24, 374. [Google Scholar] [CrossRef] [PubMed]
- Dugué, P.A.; Wilson, R.; Lehne, B.; Jayasekara, H.; Wang, X.; Jung, C.H.; Joo, J.E.; Makalic, E.; Schmidt, D.F.; Baglietto, L. Alcohol consumption is associated with widespread changes in blood DNA methylation: Analysis of cross-sectional and longitudinal data. Addict. Biol. 2021, 26, e12855. [Google Scholar] [CrossRef]
- Mundorf, A.; Rommel, S.; Verheyen, M.; Mergia, E.; Peters, M.; Freund, N. Cigarette smoke exposure has region-specific effects on GDAP1 expression in mouse hippocampus. Psychiatry Res. 2020, 289, 112979. [Google Scholar] [CrossRef]
- Friske, M.M.; Torrico, E.C.; Haas, M.J.; Borruto, A.M.; Giannone, F.; Hade, A.-C.; Yu, Y.; Gao, L.; Sutherland, G.T.; Hitzemann, R. A systematic review and meta-analysis on the transcriptomic signatures in alcohol use disorder. Mol. Psychiatry 2024, 30, 310–326. [Google Scholar] [CrossRef]
- Auta, J.; Zhang, H.; Pandey, S.C.; Guidotti, A. Chronic alcohol exposure differentially alters one-carbon metabolism in rat liver and brain. Alcohol. Clin. Exp. Res. 2017, 41, 1105–1111. [Google Scholar] [CrossRef]
- Cruise, T.M.; Kotlo, K.; Malovic, E.; Pandey, S.C. Advances in DNA, histone, and RNA methylation mechanisms in the pathophysiology of alcohol use disorder. Adv. Drug Alcohol Res. 2023, 3, 10871. [Google Scholar] [CrossRef]
- Heilig, M.; Barbier, E.; Johnstone, A.; Tapocik, J.; Meinhardt, M.; Pfarr, S.; Wahlestedt, C.; Sommer, W. Reprogramming of mPFC transcriptome and function in alcohol dependence. Genes Brain Behav. 2017, 16, 86–100. [Google Scholar] [CrossRef]
- Cui, H.Z.; Sun, M.Z.; Wang, R.Z.; Li, C.Y.; Huang, Y.X.; Huang, Q.J.; Qiao, X.M. DNA methylation in the medial prefrontal cortex regulates alcohol-related behavior in rats. Yi Chuan 2020, 42, 112–125. [Google Scholar]
- Barbier, E.; Tapocik, J.D.; Juergens, N.; Pitcairn, C.; Borich, A.; Schank, J.R.; Sun, H.; Schuebel, K.; Zhou, Z.; Yuan, Q. DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J. Neurosci. 2015, 35, 6153–6164. [Google Scholar] [CrossRef]
- Domi, A.; Cadeddu, D.; Lucente, E.; Gobbo, F.; Edvardsson, C.; Petrella, M.; Jerlhag, E.; Ericson, M.; Söderpalm, B.; Adermark, L. Pre-and postsynaptic signatures in the prelimbic cortex associated with “alcohol use disorder” in the rat. Neuropsychopharmacology 2024, 49, 1851–1860. [Google Scholar] [CrossRef]
- Jarczak, J.; Miszczak, M.; Radwanska, K. Is DNA methylation in the brain a mechanism of alcohol use disorder? Front. Behav. Neurosci. 2023, 17, 957203. [Google Scholar] [CrossRef] [PubMed]
- Soyka, M.; Preuss, U.; Koller, G.; Zill, P.; Bondy, B. Dopamine D 4 receptor gene polymorphism and extraversion revisited: Results from the Munich gene bank project for alcoholism. J. Psychiatr. Res. 2002, 36, 429–435. [Google Scholar] [CrossRef]
- Zill, P.; Baghai, T.; Zwanzger, P.; Schüle, C.; Eser, D.; Rupprecht, R.; Möller, H.; Bondy, B.; Ackenheil, M. SNP and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene provide evidence for association with major depression. Mol. Psychiatry 2004, 9, 1030–1036. [Google Scholar] [CrossRef]
- Sutherland, G.; Sheedy, D.; Stevens, J.; McCrossin, T.; Smith, C.; van Roijen, M.; Kril, J. The NSW brain tissue resource centre: Banking for alcohol and major neuropsychiatric disorders research. Alcohol 2016, 52, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Harper, C.; Kril, J.; Daly, J. Does a “moderate” alcohol intake damage the brain? J. Neurol. Neurosurg. Psychiatry 1988, 51, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, M.W.; Sommer, W.H. Postdependent state in rats as a model for medication development in alcoholism. Addict. Biol. 2015, 20, 1–21. [Google Scholar] [CrossRef]
- Hermann, D.; Hirth, N.; Reimold, M.; Batra, A.; Smolka, M.N.; Hoffmann, S.; Kiefer, F.; Noori, H.R.; Sommer, W.H.; Reischl, G. Low μ-opioid receptor status in alcohol dependence identified by combined positron emission tomography and post-mortem brain analysis. Neuropsychopharmacology 2017, 42, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, M.W.; Hansson, A.C.; Perreau-Lenz, S.; Bauder-Wenz, C.; Stählin, O.; Heilig, M.; Harper, C.; Drescher, K.U.; Spanagel, R.; Sommer, W.H. Rescue of infralimbic mGluR2 deficit restores control over drug-seeking behavior in alcohol dependence. J. Neurosci. 2013, 33, 2794–2806. [Google Scholar] [CrossRef] [PubMed]
- Sommer, J.U.; Schmitt, A.; Heck, M.; Schaeffer, E.L.; Fendt, M.; Zink, M.; Nieselt, K.; Symons, S.; Petroianu, G.; Lex, A. Differential expression of presynaptic genes in a rat model of postnatal hypoxia: Relevance to schizophrenia. Eur. Arch. Psychiatry Clin. Neurosci. 2010, 260, 81–89. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar][Green Version]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkat, D.; Team, R.C. nlme: Linear and Nonlinear Mixed Effects Models. R Package Version 2018, 3, 1. [Google Scholar][Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiegand, A.; Friske, M.; Edelmann, S.; Bender, A.; Fischer, L.; Zill, P.; Koller, G.; Bakalkin, G.; Sommer, W.H.; Hansson, A.C.; et al. Validation of GDAP1 and HECW2 as Epigenetic Markers of Alcohol Use Disorder in Blood and Brain. Int. J. Mol. Sci. 2025, 26, 10840. https://doi.org/10.3390/ijms262210840
Wiegand A, Friske M, Edelmann S, Bender A, Fischer L, Zill P, Koller G, Bakalkin G, Sommer WH, Hansson AC, et al. Validation of GDAP1 and HECW2 as Epigenetic Markers of Alcohol Use Disorder in Blood and Brain. International Journal of Molecular Sciences. 2025; 26(22):10840. https://doi.org/10.3390/ijms262210840
Chicago/Turabian StyleWiegand, Ariane, Marion Friske, Susanne Edelmann, Annika Bender, Lea Fischer, Peter Zill, Gabriele Koller, Georgy Bakalkin, Wolfgang H. Sommer, Anita C. Hansson, and et al. 2025. "Validation of GDAP1 and HECW2 as Epigenetic Markers of Alcohol Use Disorder in Blood and Brain" International Journal of Molecular Sciences 26, no. 22: 10840. https://doi.org/10.3390/ijms262210840
APA StyleWiegand, A., Friske, M., Edelmann, S., Bender, A., Fischer, L., Zill, P., Koller, G., Bakalkin, G., Sommer, W. H., Hansson, A. C., & Nieratschker, V. (2025). Validation of GDAP1 and HECW2 as Epigenetic Markers of Alcohol Use Disorder in Blood and Brain. International Journal of Molecular Sciences, 26(22), 10840. https://doi.org/10.3390/ijms262210840





