Abstract
Litchi is an important subtropical fruit, highly valued by consumers for its vibrant color and distinctive flavor. B-box (BBX) proteins, which are zinc finger transcription factors, play a crucial role in regulating plant growth, development, and stress responses. Nevertheless, the specific function of BBX genes in the development and coloration of litchi fruit remains inadequately understood. In this study, 21 LcBBX genes (designated as LcBBX1-LcBBX21) were identified within the litchi genome. These genes were categorized into five sub-families based on phylogenetic analysis and were found to be unevenly distributed across 12 chromosomes. Promoter analysis revealed a rich presence of light-responsive elements, such as the G-box, and abscisic acid (ABA) responsive elements, including ABRE, within the promoter regions of LcBBX genes. Protein–protein interaction predictions indicated that the majority of LcBBX genes have the potential to interact with the light-responsive factor HY5. Transcriptome analysis and qRT-PCR results demonstrated that LcBBX genes exhibit tissue-specific expression patterns. Notably, most LcBBX genes were highly expressed prior to fruit coloration, whereas LcBBX4 and LcBBX10 were upregulated during the fruit coloration phase. Furthermore, LcBBX1/4/6/7/15/19 were upregulated in response to light following the removal of shading. The findings suggest that LcBBX4 may directly regulate anthocyanin biosynthesis in litchi pericarp. This study provides critical insights into the molecular mechanisms underlying litchi fruit development and coloration.
1. Introduction
Transcription factors (TFs) can regulate various processes in plant growth and development, as well as responses to biotic or abiotic stresses, by activating or inhibiting the expression of their target genes [1]. Zinc finger transcription factors are among the largest TF families in plants and play a crucial role in regulating diverse biological functions [2]. BBX proteins are a class of zinc finger transcription factors containing conserved B-box domains, which can interact with DNA, RNA, or proteins to precisely regulate plant growth and development [3]. BBX proteins exhibit typical structural features: they usually contain one or two B-box motifs at the N-terminus (each approximately 40 amino acid residues in length), and some members also have a CCT motif in the C-terminal region [4]. Based on the consensus sequence and spacing characteristics of zinc-binding residues, B-box domains can be divided into two types, namely B-box1 (C-X2-C-X7-8-C-X2-D-X-A-X-L-C-X2-C-D-X3-H) and B-box2 (C-X2-C-X3- P-X4-C-X2-D-X3-L-C-X2-C-D-X3-H) [5]. The CCT domain contains 42–43 amino acid residues, and previous studies have confirmed that this domain plays a key role in the DNA-binding ability, transcriptional activity, and nuclear localization function of BBX proteins [6,7]. According to the differences in the composition of B-box and CCT domains, the BBX protein family is usually divided into 5 subfamilies [8,9].
As an important class of regulatory factors in plants, BBX proteins play critical roles in multiple processes of plant growth and development, including the regulation of plant flowering, seedling photomorphogenesis, pigment accumulation, and abiotic stress response. CONSTANS (AtBBX1) is the first identified BBX protein in Arabidopsis thaliana; under long-day conditions, it can activate the expression of the FLOWERING LOCUS (FT) gene to regulate the photoperiodic flowering process [10]. In the flowering regulatory pathway, different BBX proteins exhibit distinct functions: AtBBX6 and AtBBX24 can promote plant flowering [11,12], while AtBBX4, AtBBX5, and AtBBX7 exert flowering-inhibiting effects [13,14,15]. In addition, under red light conditions, AtBBX11 interacts with PHYTOCHROME-INTERACTING FACTOR 3 (PIF3), thereby promoting photomorphogenesis [16]. In contrast, AtBBX30 and AtBBX32 are negative regulators of photomorphogenesis, and their expression at the transcriptional level is negatively regulated by ELONGATED HYPOCOTYL 5 (HY5) [17].
BBX genes have been confirmed to be involved in regulating anthocyanin biosynthesis in multiple species, including pear [18], apple [19], cherry [20], strawberry [21], and grape [22]. In Arabidopsis thaliana, AtBBX21/22/23 positively regulate anthocyanin accumulation [23,24,25], while AtBBX24/25 exert negative regulation [26,27]; under UV-B irradiation, AtBBX31 interacts with HY5 to significantly promote anthocyanin accumulation [28]. In contrast, AtBBX32 can negatively regulate anthocyanin biosynthesis by inhibiting the activity of the BBX21-HY5 complex [29]. In addition, BBX proteins also play important regulatory roles in abiotic stress responses. For example, heterologous expression of AtBBX29 in sugarcane can significantly increase proline accumulation and antioxidant enzyme activity in sugarcane, thereby enhancing sugarcane’s drought resistance and anti-aging ability [30]; overexpression of CmBBX24 in chrysanthemum markedly improves tolerance to freezing and drought stresses [31]; heterologous expression of apple MdBBX10 in Arabidopsis significantly increases seed germination rate and seedling root length, while enhancing tolerance to drought and salt stresses [32].
Litchi (Litchi chinensis Sonn.), an evergreen tree of the genus Litchi (Sapindaceae), is an important subtropical fruit. The reddening of litchi pericarp is mainly driven by anthocyanin accumulation [33]. R2R3-MYB, bHLH, and WD40 are core transcription factor families that regulate anthocyanin synthesis [34]. Among them, the LcMYB1 transcription factor in litchi has been confirmed to positively regulate the biosynthesis of pericarp anthocyanins [35,36,37,38]. Transcription factors such as NAC [39], bZIP [40], and ERF [41] have also been successively identified and proven to be involved in anthocyanin accumulation in litchi fruit. However, whether BBX transcription factors regulate anthocyanin accumulation in litchi pericarp remains unclear. To clarify the regulatory mechanism of BBX genes on anthocyanin biosynthesis in litchi pericarp, this study conducted a systematic genome-wide identification and analysis of the litchi BBX gene family, including analyses of physicochemical properties, phylogenetic and evolutionary relationships, chromosomal localization, collinearity, conserved domain characteristics, gene structure, promoter cis-acting element prediction, and protein–protein interaction (PPI) network prediction of LcBBX genes. Meanwhile, we comparatively analyzed the expression patterns of litchi BBX family members across different tissues, pericarp developmental stages (pre-coloration, coloration, ripening), and corresponding light conditions, screened candidate LcBBX genes potentially regulating litchi pericarp coloration, and provided a theoretical basis for further exploring the function of LcBBX genes in anthocyanin biosynthesis and pericarp coloration.
2. Results
2.1. Identification and Physicochemical Properties of LcBBX Genes
In this study, we identified 21 BBX family members from the litchi genome and designated them LcBBX1 to LcBBX21 based on their chromosomal order. Physicochemical analysis revealed that the amino acid lengths of LcBBX proteins range from 132 aa (LcBBX16) to 499 aa (LcBBX19), with molecular weights varying between 15.21 and 56.19 kDa. The predicted isoelectric points ranged from 4.44 (LcBBX5) to 9.67 (LcBBX7). Based on instability index predictions, only LcBBX4, LcBBX11, and LcBBX21 were classified as stable proteins (instability index < 40), whereas the remaining members were unstable. The results of subcellular localization prediction showed that all LcBBX proteins are localized in the nucleus, except that LcBBX2 is localized in chloroplasts and LcBBX14 is localized in the cytoplasm (Table S2).
2.2. Phylogenetic Analysis of LcBBX Genes
To elucidate the evolutionary relationships among the BBX genes of litchi, Arabidopsis, rice, pear, and pineapple, we constructed a phylogenetic tree using the BBX protein sequences from Arabidopsis (32), rice (30), pear (25), pineapple (19), and litchi (21). The cluster analysis revealed that the LcBBX genes could be classified into five distinct subfamilies (Group I–V), containing 4, 3, 2, 7, and 5 members, respectively. Previous studies have demonstrated that both AtBBX21 and AtBBX22 act as positive regulators of anthocyanin biosynthesis in Arabidopsis [23,24]. Among them, LcBBX8 clustered closely with AtBBX21, and LcBBX4 grouped with AtBBX22, suggesting that these BBX proteins may share a common evolutionary origin and could perform similar biological roles in litchi (Figure 1).
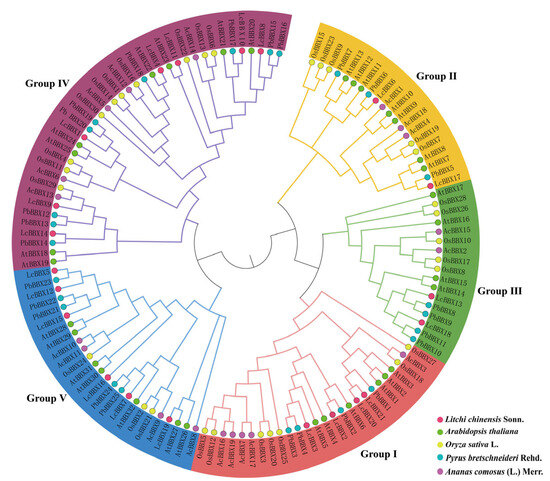
Figure 1.
Phylogenetic relationships of the BBX gene family in litchi, Arabidopsis, rice, pear, and pineapple. Different coloured branches represent different subfamilies. The red, green, yellow, blue, and purple circular shapes represent the BBX genes of litchi, Arabidopsis, rice, pear, and pineapple.
2.3. Conserved Domains, Gene Structure, and Conserved Motifs of the LcBBX Genes
Conserved domain analysis showed that all 21 LcBBXs in litchi contain the B-box domain, among which members of subfamilies Group I/II/III also contain the CCT domain (Figure 2B). Gene structure analysis revealed that the number of introns in LcBBX genes shows subfamily-specific characteristics. In subfamilies Group Ⅰ/Ⅲ/Ⅳ, all members contain only 1 intron except for LcBBX7/16, which have no introns, while LcBBX members in subfamily Group Ⅱ contain multiple introns (4–5 introns) (Figure 2C).
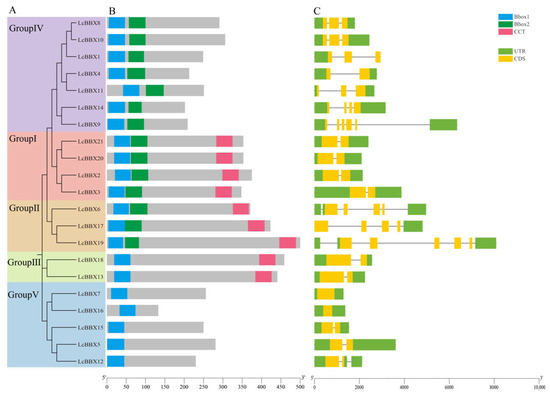
Figure 2.
Characterization of LcBBXs. (A) neighbor-joining tree analysis and its grouping of LcBBXs; (B) conserved structural domains of LcBBXs. The three conserved structural domains are represented by boxes of three colors; (C) gene structure of LcBBXs. Green and yellow boxes indicate UTR and CDS.
In the analysis of conserved motifs in litchi BBX protein sequences, 10 distinct conserved motifs were identified (Figure S1). Combined analysis of the conserved motif results (Figure S1B) and the conserved domain results (Figure 2B) revealed that Motif1 corresponds to the Bbox1 domain, Motif3 corresponds to the Bbox2 domain, and Motif2 corresponds to the CCT domain. Additionally, Motif5/6/7/8 are exclusively present in LcBBX20/21, Motif9 is only found in LcBBX5/12/15, and Motif10 exists solely in LcBBX13/18.
2.4. Chromosomal Localization and Collinearity Analysis of LcBBX Genes
The 21 LcBBX genes were unevenly distributed across 12 litchi chromosomes. Among them, chromosomes 1 and 12 each contained 3 LcBBX genes; chromosomes 2, 5, 8, 14, and 15 each had 2 LcBBX members; and chromosomes 3, 4, 9, 10, and 11 each harbored 1 LcBBX gene. Further observation revealed that most LcBBX genes were located in the gene-dense regions of the chromosomes (Figure 3).
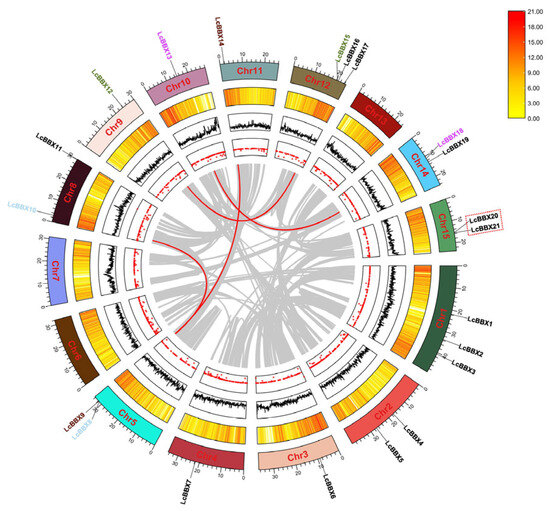
Figure 3.
Intraspecific collinearity analysis of the LcBBX gene family members. The segmental duplicated gene pairs are linked by red lines, and the tandem duplicated gene pairs are marked with red dashed lines. The figure from outside to inside shows gene chromosome localization, gene density, genomic GC ratio, and genomic gap statistics.
To explore the expansion mechanism of the LcBBX gene family in litchi, we performed an intraspecific collinearity analysis (Figure 3). The results showed that there were 4 segmental duplication events in the litchi LcBBX family, involving LcBBX8-LcBBX10, LcBBX9-LcBBX14, LcBBX12-LcBBX15, and LcBBX13-LcBBX18. In addition, LcBBX20 and LcBBX21 are a pair of tandem duplicated genes, with a sequence similarity of approximately 99.16%.
In addition, to further explore the evolutionary characteristics of the litchi BBX gene family, this study analyzed the collinearity relationships between litchi and related species (longan, rambutan), important fruit trees (apple, pineapple), and model plants (Arabidopsis, rice) (Figure 4). Among them, longan and rambutan, which belong to the same Sapindaceae family as litchi, share 23 and 26 homologous BBX genes with litchi, respectively (Figure 4A); apple and Arabidopsis, as dicotyledonous plants like litchi, share 41 and 25 homologous BBX genes with litchi, respectively (Figure 4B); pineapple and rice are monocotyledonous plants, and they have relatively fewer homologous BBX genes with litchi, with only 10 and 4, respectively (Figure 4C).
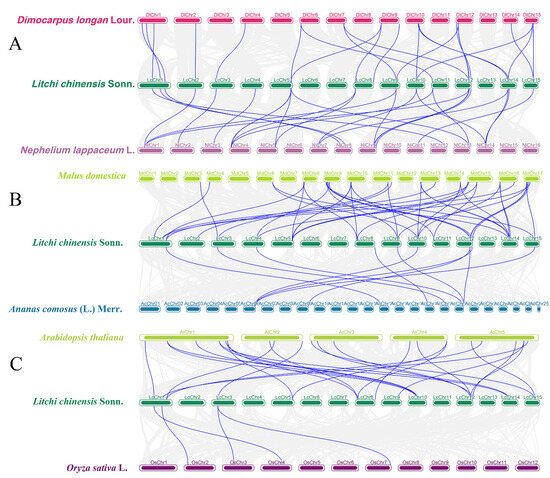
Figure 4.
Collinearity analysis of the BBX gene family across multiple species. (A) Collinearity analysis between litchi, longan, and rambutan. (B) Collinearity analysis between litchi, apple, and pineapple. (C) Collinearity analysis between litchi, Arabidopsis, and rice. Gray lines represent the collinear within litchi and other plant genomes, blue lines emphasize the pairs of syntenic BBX genes.
2.5. Analysis of cis-Acting Elements in the Promoters of LcBBX Genes
Analyzing and predicting the type, number, and location of cis-acting elements in the promoter of a target gene is of great significance for revealing the molecular basis of how genes are precisely regulated. The cis-acting elements in the promoter region of litchi LcBBX genes were predicted and analyzed using the PlantCARE database. The results showed that the promoters of all members of the LcBBX family contained a variety of cis-acting elements, mainly involving elements related to light response, hormone response, stress response, and growth and development regulation (Figure 5). Among these, the light response elements had the largest number of types and quantities, with the G-Box element accounting for the highest proportion (84 in total); these light response elements were unevenly distributed in the LcBBX promoter sequences (Figure S2), indicating that the expression of litchi BBX family genes may be regulated by light signals.
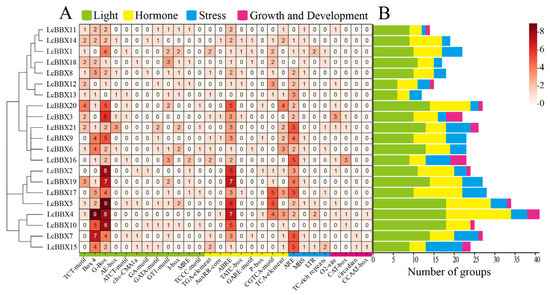
Figure 5.
Statistics of cis-acting elements of promoters in LcBBX genes. The green, yellow, blue, and pink modules represent light-responsive elements, hormone-responsive elements, stress-responsive elements, and growth and development elements. (A) number of each element in LcBBXs; (B) number of elements per group in LcBBXs.
In the promoter sequences of LcBBX genes, a variety of cis-acting elements related to endogenous hormone responses were also predicted, including auxin-responsive cis-acting elements (TGA-element and AuxRR-core), ABA-responsive cis-acting element (ABRE), gibberellin-responsive cis-acting elements (TATC-box, GARE-motif, and P-box), methyl jasmonate-responsive cis-acting element (CGTCA-motif), and salicylic acid-responsive cis-acting element (TCA-element) (Figure 5). Among them, except for LcBBX13, the promoter regions of all other members contain at least one ABRE element, indicating that ABA plays an important role in the transcriptional regulation of LcBBX.
A large number of stress-responsive cis-acting elements were also identified in the promoter sequences of LcBBX genes. Among them, the anaerobic induction element (ARE) was present in the promoter sequences of 19 LcBBX genes; the drought-induced response element (MBS) was found in 13 LcBBX genes; the low-temperature-induced response element (LTR) existed in 4 LcBBX genes; and the defense and stress response element (TC-rich repeats) was detected in 6 LcBBX genes. In addition, elements such as the zein metabolism regulatory element (O2-site), meristem expression regulatory element (CAT-box), and circadian rhythm regulatory element (circadian) were also present in the promoter sequences of multiple LcBBX genes. These results indicate that LcBBXs play important roles in plant growth and development as well as in responses to external stresses.
2.6. GO Enrichment Annotation and Protein–Protein Interaction Network Prediction of LcBBX Genes
GO functional enrichment analysis was performed on 21 LcBBX genes (Figure S3), which could be classified into three categories: 16 biological processes, 4 molecular functions, and 1 cellular component (Figure S3). In the biological process category, LcBBX genes were significantly enriched in terms related to "response to light" and "post-embryonic development", indicating that LcBBX genes may be involved in plant light signal response and growth and development regulation. In the molecular function category, the functions of LcBBX genes were mainly associated with "DNA-binding transcription factor activity", which can bind to the transcriptional cis-regulatory regions of DNA molecules, thereby activating or inhibiting the expression of downstream structural genes. In the cellular component category, LcBBX genes were enriched in the nucleus, suggesting that they may be involved in transcriptional regulation processes within the nucleus.
The results of protein interaction prediction showed that there are numerous interaction relationships among members of the litchi BBX genes, with the number of interactions ranging from 1 to 10. Among them, LcBBX19 may interact with 10 other LcBBX proteins. The light response factor HY5 and its homologous protein HY5 HOMOLOG (HYH) may interact with 12 members of the litchi LcBBX genes, and these 12 members (except LcBBX21) all belong to the Group IV and Group V subfamilies. GATA2 of the GATA transcription factor family, as a positive regulator of photomorphogenesis, only interacts with BBX genes of the Group IV subfamily (Figure 6). These findings provide an important reference for further analyzing the molecular regulatory network of BBX genes in litchi pericarp.
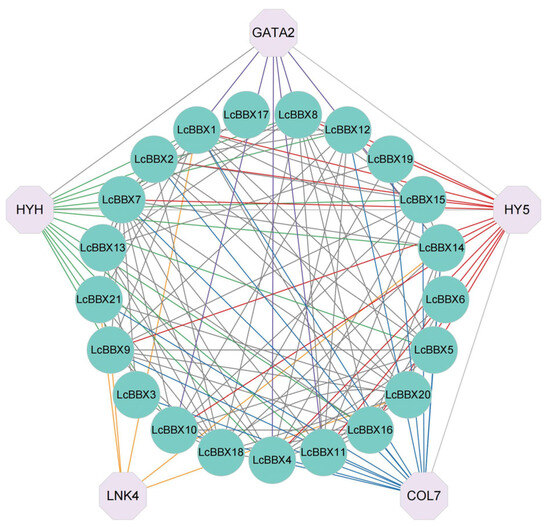
Figure 6.
Protein–protein interaction network of LcBBX proteins. The circle represents LcBBXs, and the octagon represents genes that interact with LcBBXs. Each gene that interacts with LcBBX is connected using lines of different colors. The red lines indicate LcBBXs that interact with HY5; the green lines indicate that LcBBXs interact with HYH; the purple lines indicate that LcBBXs interact with GATA2.
2.7. Prediction of miRNA Targets for Members of the LcBBX Genes
Predicting the miRNA targets of LcBBX family members is helpful for further studying the interaction between miRNAs and LcBBXs, as well as their functions in biological processes. The results showed that 16 litchi BBX genes have miRNA target binding sites (Table S3). Among them: LcBBX5/9/14/16 have miRNA target sites in both the CDS region and UTR region; LcBBX6/7/8/11/13/15/17/18/19/20 have one or more miRNA target sites in the CDS region; LcBBX10 has miRNA target sites only in the UTR region.
2.8. Tissue-Specific Expression of LcBBX Gene Family Members
To explore the potential functions of LcBBX genes and their roles in growth and development, this study analyzed the expression patterns of litchi BBX genes in different tissues and developmental processes. The results showed that the gene expression characteristics of litchi LcBBX exhibited obvious tissue specificity: LcBBX17/19 were specifically highly expressed in male flowers, while all LcBBX genes maintained low transcriptional levels in female flowers; LcBBX6/21/18/9/1/20/13/4/8 showed high transcriptional levels in leaves; In fruits, LcBBX3/14/6/21/18/9/1/20/12/5 were highly expressed in the pericarp; LcBBX10/8/5 had high transcriptional levels in the aril. Only LcBBX11 was highly expressed in small fruits and seeds (Figure 7, Table S4). These findings suggest that LcBBX may play a more important role in pericarp development and color formation in litchi.
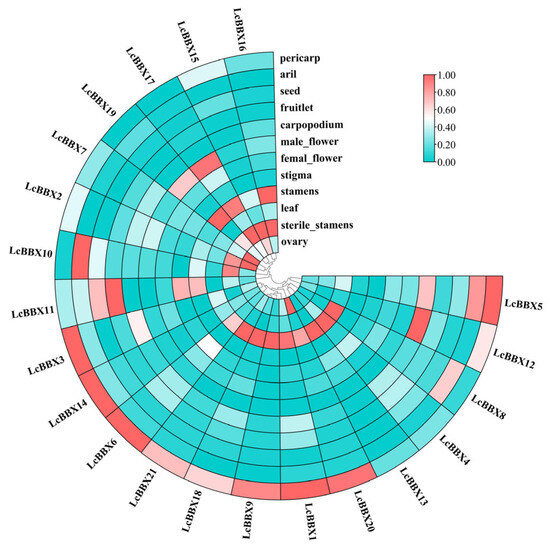
Figure 7.
The expression profiles of litchi BBX genes in different tissues. A heatmap shows the relative expression levels in pericarp, aril (flesh), seed, fruitlet, carpopodium, male flower, female flower, stigma, stamens, leaf, sterile stamens, and ovary.
To further reveal the potential functions of LcBBX in the process of litchi fruit development, this study analyzed the gene expression characteristics of LcBBX genes at different fruit development stages. The results showed that most LcBBX genes (LcBBX15/14/3/9/2/13/7/1/16/20) had high transcriptional levels before pericarp coloration, while LcBBX5/8/12/17/18 had relatively high expression levels during the fruit coloration stage. At the fruit ripening stage, LcBBX21/6/19/10/4 were highly expressed, followed by slightly lower expression of genes such as LcBBX13/7/1/16. The expression levels of LcBBX4/10/19 gradually increased with pericarp development and ripening, which was consistent with the accumulation trend of anthocyanins in the pericarp, suggesting that they may be involved in the regulation of anthocyanin biosynthesis (Figure 8, Table S5). The expression patterns of LcBBX genes exhibit distinct stage-specificity and complementary characteristics, providing important clues for investigating the functions of LcBBX genes during fruit developmental stages.
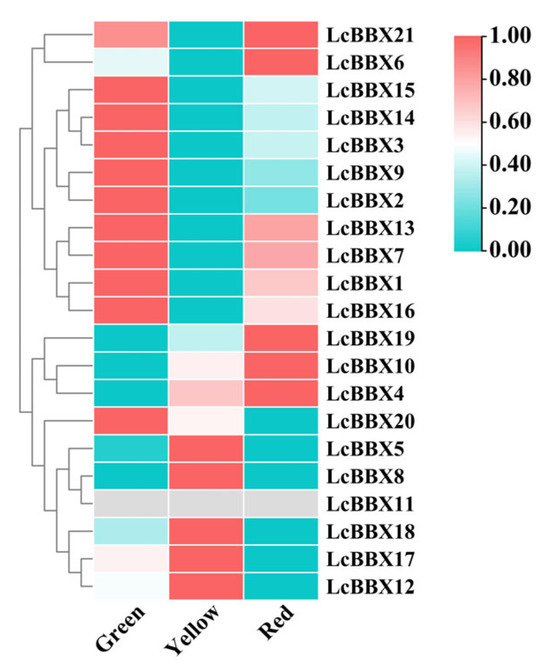
Figure 8.
The expression profiles of LcBBX genes at different stages of pericarp coloring. The three developmental stages include green pericarp, yellow pericarp, and red pericarp.
2.9. qRT-PCR Analysis of Expression Patterns of LcBBX Genes in Litchi Pericarp at Different Developmental Stages
qRT-PCR was used to further analyze the gene expression characteristics of litchi fruit at three stages: pre-coloration (50 DAFB), coloration (50 DAFB), and ripening (70 DAFB). The results showed that most LcBBX genes (LcBBX1/2/3/9/14/15/16/21) had high expression levels before pericarp coloration, decreased expression during the coloration stage, and then increased again at the ripening stage. This pattern was similar to the transcriptome data of different developmental stages (Figure 8). In contrast, the expression levels of LcBBX4/5/6/13 increased during the pericarp coloration stage. In addition, LcBBX10/11/12/17 were highly expressed during the pericarp coloration and ripening stages, while LcBBX8 was highly expressed at the pericarp ripening stage (Figure 9). In summary, the expression levels of most LcBBX genes change during the developmental stages of litchi pericarp, suggesting that they may directly or indirectly participate in the anthocyanin accumulation process in litchi pericarp.
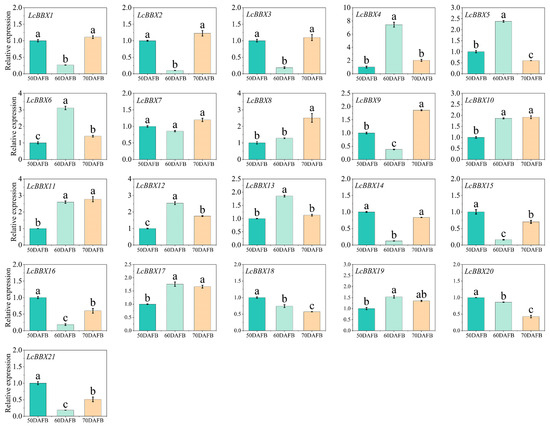
Figure 9.
Expression patterns of LcBBX genes in different stages of pericarp development, namely the pre-coloration stage (50 DAFB), coloration stage (60 DAFB), and ripening stage (70 DAFB) of litchi pericarp. Data are means ± SEM of three biological replicates. Different letters mean significant differences at the 0.05 level.
2.10. qRT-PCR Analysis of Expression Patterns of LcBBX Genes in Litchi Pericarp Under Light Induction
Previous studies have shown that light can induce the biosynthesis of anthocyanins in litchi pericarp [42]. The qRT-PCR results revealed that after de-shading of litchi fruits, the expression levels of multiple LcBBX genes changed significantly with light exposure time: the expression levels of LcBBX1/7/15 increased on the first day after light recovery; LcBBX4/5/6/8/19 were upregulated on the third day after light recovery. In contrast, the expression levels of LcBBX3/9/12/14/18 decreased after light recovery (Figure 10). In conclusion, LcBBX genes exhibit diverse expression patterns in litchi pericarp during the light stage after de-shading. This differential expression characteristic suggests that they may play different regulatory roles in the light response of litchi pericarp and the subsequent anthocyanin accumulation, providing a basis for further analyzing the functions of LcBBX genes in processes such as light signal transduction and anthocyanin biosynthesis in litchi pericarp.
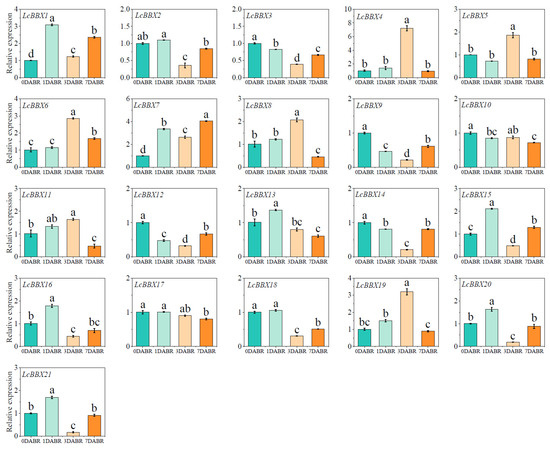
Figure 10.
Expression patterns of LcBBX genes in litchi pericarp with the removal of shading treatment. DABR, days after bag removal. Data are means ± SEM of three biological replicates. Different letters mean significant differences at the 0.05 level.
3. Discussion
At present, in addition to the MBW complex, transcription factors such as NAC [39], bZIP [40], and ERF [41] have been successively identified and confirmed to be involved in anthocyanin accumulation in litchi fruits. The BBX transcription factor plays a crucial role in regulating plant growth and development, and it may also play an important part in litchi fruit development and color formation. In this study, 21 LcBBX genes were screened from the litchi genome, and they were unevenly distributed on 12 chromosomes (Figure 3). In terms of gene number, BBX family members in litchi are fewer than in Arabidopsis thaliana (32) [3] and rice (30) [43], but higher than in pineapple (19) [44] and Chinese chestnut (18) [45], indicating that the distribution of BBX genes varies significantly among different plant species. Generally, gene duplication events such as segmental duplication and tandem duplication are the core driving forces for plant genome evolution and environmental adaptation [46]. The results of collinearity analysis showed that there were 4 pairs of segmentally duplicated genes and 1 pair of tandemly duplicated genes in the LcBBX gene family (Figure 3), which might be an important pathway for the expansion of the litchi LcBBX gene family. Collinearity analysis with other plant species revealed that the number of BBX homologous genes between litchi and other dicotyledonous plants was significantly higher than that between monocotyledonous plants. This suggests that dicotyledonous and monocotyledonous plants have undergone different gene duplication events during their evolutionary process, leading to the differentiation of BBX genes between the two types of plants (Figure 4). Phylogenetic tree analysis divided the 21 LcBBX family genes into 5 subfamilies, and members of the same subfamily had a consistent combination of conserved domains. This classification pattern is highly consistent with that in Arabidopsis and rice, implying the evolutionary conservation of the BBX gene family classification across different plants (Figure 1). Meanwhile, the three types of conserved domains (B-box1, B-box2, and CCT) contained in LcBBX genes have highly similar sequences, which further confirms the stability of the conserved domains of the LcBBX family during evolution and provides a structural basis for the subsequent analysis of their functional conservation (Figure 2 and Figure S1).
Cis-acting elements in the promoter region play a crucial regulatory role in the functional diversity of genes [47], and they are mainly classified into four categories: light-responsive elements, hormone-responsive elements, stress-responsive elements, and growth and development elements. Among the cis-acting elements in the promoters of LcBBX genes, the light-responsive element G-box is the most abundant (Figure 5). Previous studies have confirmed that the transcription factor HY5 can regulate the expression of downstream genes by binding to the G-box in their promoter regions [48], and BBX proteins can serve as rate-limiting cofactors for HY5 [49]. In Arabidopsis thaliana, HY5 can interact with AtBBX21/22/23 to collectively regulate plant photomorphogenesis [23,24,25]. Meanwhile, GO functional enrichment analysis showed that most LcBBX family members are significantly enriched in light response-related terms, suggesting that they may be involved in the light signal response process of litchi (Figure S3). Among the hormone-responsive elements, the ABA-responsive element ABRE is the most numerous and widely distributed in the promoters of LcBBX genes (Figure 5 and Figure S2). Additionally, existing studies have verified that ABA can promote anthocyanin accumulation in litchi pericarp [50]. These findings indicate that LcBBX genes may respond to both light and ABA signals, and participate in the integration of light and ABA signal pathways by regulating the expression of downstream structural genes, thereby modulating the biosynthesis of anthocyanins in litchi fruits. The results of protein–protein interaction prediction revealed that there are numerous interaction relationships among members of the LcBBX gene family, and they may exert regulatory functions synergistically by forming protein complexes; LcBBX proteins can also interact with light-responsive factors such as HY5, HYH, and GATA2 (Figure 6). It can be concluded that LcBBX genes play important roles in responding to light and ABA signals and regulating anthocyanin biosynthesis.
BBX genes are involved in regulating anthocyanin biosynthesis in various plant species, and their functions are often associated with light response [20,51,52]. In this study, the expression patterns of LcBBX family members exhibited distinct tissue-specificity, among which 10 LcBBX genes (LcBBX1/3/5/6/9/12/14/18/20/21) showed high expression levels in the pericarp (Figure 7). Most LcBBX genes were highly expressed before pericarp coloration. Furthermore, the expression levels of LcBBX4/10/19 gradually increased with the development of the pericarp, which was almost consistent with the accumulation trend of anthocyanins in the pericarp (Figure 8). Similarly, in Rubus chingii, RcBBX genes also displayed differential expression patterns in roots, stems, leaves, flowers, and fruits, and most RcBBX genes were highly expressed before pericarp coloration during fruit development [53]. This shows a certain similarity to the expression pattern of LcBBX genes in litchi, suggesting that the function of BBX genes in regulating fruit development and anthocyanin synthesis processes has a certain evolutionary conservation. qRT-PCR results showed that as the coloration process of litchi pericarp progressed, the expression levels of genes such as LcBBX4/5/6/10 increased significantly (Figure 9). Meanwhile, multiple LcBBX genes changed obviously after the shading treatment was removed: the expression levels of LcBBX1/4/5/6/7/8/15/19 increased significantly within 1–3 days after light recovery, while the expression levels of LcBBX9/12/14/18 decreased significantly after light recovery (Figure 10). It can be seen that LcBBX genes have obvious light-responsive characteristics and may play a key regulatory role in the light-induced anthocyanin biosynthesis process in litchi pericarp.
4. Materials and Methods
4.1. Plant Materials and Sources of Public Data
The ‘Feizixiao’ litchi cultivar was cultivated at the Litchi and Longan Experimental Base, Institute of South Subtropical Crops, Chinese Academy of Tropical Agricultural Sciences. For the purpose of litchi pericarp sampling at various developmental stages, samples were collected at 50 days after full bloom (DAFB, pre-coloration stage), 60 DAFB (coloration stage), and 70 DAFB (ripening stage). At each developmental stage, three trees were selected as biological replicates, and 10 fruits were uniformly collected from different orientations of each tree. The pericarps were then peeled, immediately flash-frozen in liquid nitrogen, and preserved. For the sampling of litchi fruits under shading treatment, at 42 DAFB, three litchi trees exhibiting consistent growth vigor and normal fruit set were randomly selected, and 10 branches per tree were subjected to shading treatment. The shading treatment concluded at 63 DAFB, and pericarp samples were collected at 0 days after shading removal, as well as at 1, 3, and 7 days post-unshading. At each time point, 10 shaded fruits were randomly harvested from each of the three trees, and the fruits collected from each individual tree were stored appropriately.
In addition, transcriptome data of different litchi tissues were obtained from the accession number PRJNA747875 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA747875 (accessed on 4 March 2024)), including pericarp, aril (flesh), seed, fruitlet, carpopodium, male flower, female flower, stigma, stamens, leaf, sterile stamens, and ovary. Transcriptome data of the litchi fruit coloration process were derived from the accession number PRJNA261000 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA261000 (accessed on 8 March 2024)), covering the green (pre-coloration), yellow (coloration stage), and red (ripening stage).
4.2. Identification of LcBBX Gene Family Members
Litchi genome information, including DNA, GFF, CDS, and protein files, was downloaded from SapBase (http://www.sapindaceae.com/Download.html (accessed on 17 October 2024)). Arabidopsis thaliana genome data were downloaded from TAIR (https://www.arabidopsis.org/ (accessed on 17 October 2024)). Protein sequences of AtBBX family members were downloaded from the PlantTFDB database (https://planttfdb.gao-lab.org/ (accessed on 17 October 2024)) as query sequences, and the BLAST tool in TBtools software (version 2.363) was used to search for homologous sequences in the litchi protein database. In addition, the hidden Markov model (HMM) of the BBX zinc finger domain (PF00643) was downloaded from the InterPro database (https://www.ebi.ac.uk/interpro/entry/pfam/PF00643/ (accessed on 21 October 2024)), and the HMMER software package (version 3.0) was used for homologous sequence analysis (threshold of 1 × 10−5). Using AtBBX proteins as query sequences, TBtools v2.363 was then employed to search for putative LcBBX proteins (e value < 1 × 10−5 and coverage ≥ 50%). The search results of HMMER and BLAST were combined as candidate members of the litchi BBX family. The conserved domains in protein sequences were analyzed using the Web CD-search tool of NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 23 October 2024)), and protein sequences without the B-box conserved domain were excluded to finally obtain the members of the litchi BBX gene family. The ExPASy website (https://web.expasy.org/protparam/ (accessed on 27 October 2024)) was used to analyze parameters such as relative molecular weight, number of amino acids, isoelectric point, and instability index. The WoLF PSORT website (https://wolfpsort.hgc.jp/ (accessed on 27 October 2024)) was used to predict subcellular localization.
4.3. Construction of the Phylogenetic Tree of LcBBX Genes
MEGA11 software (version 11.0.13) was used to construct the phylogenetic tree of the BBX gene family members from litchi, Arabidopsis, rice, pear, and pineapple. ClustalW was used for multiple sequence alignment of all protein members. The neighbor-joining (NJ) method was adopted, with 1000 bootstrap replicates for reliability testing; other parameters were set to default, and the phylogenetic tree was plotted.
4.4. Analysis of Conserved Domains, Gene Structure, and Conserved Motifs of LcBBX Genes
Protein sequences of litchi BBX genes were submitted to the CD-search Tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 23 October 2024)) for conserved domain prediction, and were visualized using the Gene Structure View tool in TBtools software (version 2.363). Using the litchi genome GFF file, the Gene Structure View tool in TBtools software was used to analyze and visualize the gene structure of litchi BBX genes. The MEME website (https://meme-suite.org/meme/ (accessed on 3 December 2024)) was used to analyze protein conserved motifs, with the number of motifs set to 10 (other parameters are default values), and were visualized using the Gene Structure View tool in TBtools software.
4.5. Chromosomal Localization and Collinearity Analysis of LcBBX Genes
Gene files and genome annotation files of litchi, as well as those of longan and rambutan (Sapindaceae), Arabidopsis and apple (dicotyledons), and pineapple and rice (monocotyledons), were used. The One Step MCScanX tool in TBtools software was employed to visualize intraspecific collinearity of litchi and interspecific collinearity between litchi and other species, so as to analyze the evolutionary relationship of BBX genes. Data of longan and rambutan were obtained from SapBase (http://www.sapindaceae.com/Download.html (accessed on 6 August 2024)); data of Arabidopsis were from the TAIR database (https://www.arabidopsis.org/ (accessed on 17 October 2024)); data of rice and apple were from the Ensembl database (http://plants.ensembl.org/index.html (accessed on 13 March 2025)); and pineapple data were from the Ananas Genomic Database (http://pineapple.zhangjisenlab.cn/pineapple/html/download.html (accessed on 13 March 2025)).
4.6. Analysis of cis-Acting Elements in LcBBX Gene Promoters
The Gtf/Gff3 Sequences Extract tool in TBtools software v2.363 was used to extract the 2000 bp gene sequence upstream of the start codon of LcBBX genes as the promoter region. Promoter sequences were submitted to the Plant CARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 12 March 2025)) for cis-acting element prediction, and the results were visualized using the Simple BioSequence Viewer tool in TBtools software v2.363.
4.7. GO Enrichment and Protein–Protein Interaction Prediction of LcBBX Genes
Protein sequences of LcBBX genes were submitted to the online STRING website (https://cn.string-db.org/) for protein–protein interaction prediction of litchi BBX proteins. The Functional Enrichment Visualization tool on this website was used to perform GO enrichment analysis of LcBBX genes.
4.8. Prediction of miRNA Targets of LcBBX Genes
Litchi miRNA annotation files were downloaded from SapBase (http://www.sapindaceae.com/sRNA/litchi-miRNA-list.html (accessed on 14 March 2025)). The psRNATarget website (https://www.zhaolab.org/psRNATarget/ (accessed on 14 March 2025)) was used to predict microRNA (miRNA) target binding sites in the coding sequence (CDS) and untranslated region (UTR) of LcBBX genes, with the expected value set to ≤4.5 and other parameters set to default.
4.9. Quantitative Real-Time PCR (qRT-PCR) Analysis of LcBBX Genes at Different Pericarp Developmental Stages
Total RNA was isolated utilizing the Fast Universal Plant RNA Extraction Kit (Huayueyang, Beijing, China). The purity of the RNA, indicated by OD260/280 and OD260/230 ratios, was assessed with a NanoDrop UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and RNA integrity was examined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA). Reverse transcription of the total RNA to synthesize cDNA for quantitative real-time PCR (qRT-PCR) analysis was conducted using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA). qRT-PCR primers were designed with the Batch q-PCR Primer Design plugin in TBtools software version 2.363 (Table S1). The MaActin gene (GenBank accession number: HQ615689) served as the internal reference [50]. qRT-PCR was executed on a QuantStudio 1 Real-Time PCR system following the manufacturer’s protocol for the ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). The thermal cycling conditions were as follows: initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Each reaction was performed in three technical replicates. The relative expression levels of LcBBXs were calculated using the 2−∆∆Ct method [54]. Data visualization was carried out using Origin 2024 software.
4.10. Statistical Analysis
The qRT-PCR data underwent a one-way analysis of variance (ANOVA) using SPSS (version 23.0), followed by Tukey’s multiple comparison test to conduct statistical analyses. Prior to performing the ANOVA tests, the assumptions of variance homogeneity and normality were meticulously verified to ensure the validity of the statistical conclusions [55]. Each experiment comprised three independent biological replicates, and the qRT-PCR results are reported as the mean ± standard error (SE) of these replicates. Statistical significance was determined at a threshold of p < 0.05, with different letters indicating significant differences among the samples.
5. Conclusions
In this study, 21 LcBBX genes (LcBBX1-LcBBX21) were identified from the litchi genome, which are unevenly distributed across 12 chromosomes. Based on phylogenetic relationships, these LcBBX members can be divided into 5 subfamilies (Group I to V). The promoter regions of LcBBX genes are rich in the light-responsive element G-box and the ABA-responsive element ABRE. Protein interaction prediction revealed that most LcBBX proteins can interact with HY5, suggesting that LcBBX may be involved in the regulation of physiological processes by integrating light and ABA signals. In terms of gene expression characteristics, LcBBX genes exhibit distinct tissue-specific expression patterns. Most members are highly expressed before pericarp coloration; in addition, LcBBX4/10 are upregulated during the fruit coloration stage. After the removal of shading treatment, the expressions of LcBBX1/4/6/7/15/19 are upregulated in response to light. In conclusion, LcBBX4 may directly participate in the regulation of anthocyanin biosynthesis in litchi pericarp. This study clarifies the basic characteristics and functional potential of the LcBBX family in litchi, and provides an important reference for subsequent functional verification of LcBBX genes and research on the transcriptional regulatory mechanism of litchi anthocyanin biosynthesis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms262210834/s1.
Author Contributions
Conceptualization, T.L. and Y.W.; methodology and validation, Y.C. and W.S.; formal analysis and investigation, H.Z. and Y.C.; resources, H.Z. and Y.C.; data curation, T.L. and Y.C.; writing—original draft preparation, T.L. and Y.W.; writing—review and editing, Y.C. and Y.W.; visualization, Y.C.; supervision, Y.W. and H.Z.; project administration, Y.W. and H.Z.; funding acquisition, H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The project was funded by the major science and technology project of the major science and technology project of Hainan province (ZDKJ2021006) and the priming scientific research foundation of Hainan University [KYQD (ZR)-20090].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article and Supplementary Materials, further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gong, W.; Shen, Y.-P.; Ma, L.-G.; Pan, Y.; Du, Y.-L.; Wang, D.-H.; Yang, J.-Y.; Hu, L.-D.; Liu, X.-F.; Dong, C.-X.; et al. Genome-Wide ORFeome Cloning and Analysis of Arabidopsis Transcription Factor Genes. Plant Physiol. 2004, 135, 773–782. [Google Scholar] [CrossRef]
- Takatsuji, H. Zinc-Finger Transcription Factors in Plants. Cell. Mol. Life Sci. 1998, 54, 582–596. [Google Scholar] [CrossRef]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.-H. The Arabidopsis B-Box Zinc Finger Family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The BBX Family of Plant Transcription Factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Crocco, C.D.; Botto, J.F. BBX Proteins in Green Plants: Insights into Their Evolution, Structure, Feature and Functional Diversification. Gene 2013, 531, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis Circadian Clock Protein, TOC1, Is a DNA-Binding Transcription Factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef]
- Jang, S.; Marchal, V.; Panigrahi, K.C.S.; Wenkel, S.; Soppe, W.; Deng, X.-W.; Valverde, F.; Coupland, G. Arabidopsis COP1 Shapes the Temporal Pattern of CO Accumulation Conferring a Photoperiodic Flowering Response. EMBO J. 2008, 27, 1277–1288. [Google Scholar] [CrossRef]
- Ming, M.; Yi, M.; Sun, K.; Zu, A.; Zhang, J.; Fu, F.; Cao, F.; Yang, X. Genome-Wide Identification and Expression Analysis of the Ginkgo Biloba B-Box Gene Family in Response to Hormone Treatments, Flavonoid Levels, and Water Stress. Int. J. Mol. Sci. 2025, 26, 8427. [Google Scholar] [CrossRef]
- Yu, L.; Lyu, Z.; Liu, H.; Zhang, G.; He, C.; Zhang, J. Insights into the Evolutionary Origin and Expansion of the BBX Gene Family. Plant Biotechnol. Rep. 2022, 16, 205–214. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Shen, Y.; Chang, H.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The Flowering Time Regulator CONSTANS Is Recruited to the FLOWERING LOCUS T Promoter via a Unique Cis-element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef]
- Hassidim, M.; Harir, Y.; Yakir, E.; Kron, I.; Green, R.M. Over-Expression of CONSTANS-LIKE 5 Can Induce Flowering in Short-Day Grown Arabidopsis. Planta 2009, 230, 481–491. [Google Scholar] [CrossRef]
- Li, F.; Sun, J.; Wang, D.; Bai, S.; Clarke, A.K.; Holm, M. The B-Box Family Gene STO (BBX24) in Arabidopsis Thaliana Regulates Flowering Time in Different Pathways. PLoS ONE 2014, 9, e87544. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wang, Z. Overexpression of COL9, a CONSTANS-LIKE Gene, Delays Flowering by Reducing Expression of CO and FT in Arabidopsis Thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Carvallo, M.; Hamilton, E.E.; Preuss, S.; Kay, S.A. Arabidopsis B-BOX32 Interacts with CONSTANS-LIKE3 to Regulate Flowering. Proc. Natl. Acad. Sci. USA 2017, 114, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, Y. The Arabidopsis thaliana CONSTANS -LIKE 4 (COL4)—A Modulator of Flowering Time. Front. Plant Sci. 2019, 10, 651. [Google Scholar] [CrossRef]
- Song, Z.; Heng, Y.; Bian, Y.; Xiao, Y.; Liu, J.; Zhao, X.; Jiang, Y.; Deng, X.W.; Xu, D. BBX11 Promotes Red Light-Mediated Photomorphogenic Development by Modulating phyB-PIF4 Signaling. aBIOTECH 2021, 2, 117–130. [Google Scholar] [CrossRef]
- Heng, Y.; Lin, F.; Jiang, Y.; Ding, M.; Yan, T.; Lan, H.; Zhou, H.; Zhao, X.; Xu, D.; Deng, X.W. B-Box Containing Proteins BBX30 and BBX31, Acting Downstream of HY5, Negatively Regulate Photomorphogenesis in Arabidopsis. Plant Physiol. 2019, 180, 497–508. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box Protein, Positively Regulates Light-induced Anthocyanin Accumulation by Activating MYB10 in Red Pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef]
- Bai, S.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. An Apple B-Box Protein, MdCOL11, Is Involved in UV-B- and Temperature-Induced Anthocyanin Biosynthesis. Planta 2014, 240, 1051–1062. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, Y.; Sun, Y.; Zhang, X.; Du, B.; Turupu, M.; Yao, Q.; Gai, S.; Tong, S.; Huang, J.; et al. Two B-Box Proteins, PavBBX6/9, Positively Regulate Light-Induced Anthocyanin Accumulation in Sweet Cherry. Plant Physiol. 2023, 192, 2030–2048. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Yue, M.; Jiang, L.; Zhang, N.; Luo, Y.; Chen, Q.; Zhang, Y.; Wang, Y.; Li, M.; et al. FaMYB5 Interacts with FaBBX24 to Regulate Anthocyanin and Proanthocyanidin Biosynthesis in Strawberry (Fragaria × Ananassa). Int. J. Mol. Sci. 2023, 24, 12185. [Google Scholar] [CrossRef]
- Liu, W.; Tang, R.; Zhang, Y.; Liu, X.; Gao, Y.; Dai, Z.; Li, S.; Wu, B.; Wang, L. Genome-Wide Identification of B-Box Proteins and VvBBX44 Involved in Light-Induced Anthocyanin Biosynthesis in Grape (Vitis vinifera L.). Planta 2021, 253, 114. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, Y.; Li, J.; Lin, F.; Holm, M.; Deng, X.W. BBX21, an Arabidopsis B-Box Protein, Directly Activates HY5 and Is Targeted by COP1 for 26S Proteasome-Mediated Degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655–7660. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Johansson, H.; Hettiarachchi, C.; Irigoyen, M.L.; Desai, M.; Rubio, V.; Holm, M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-Box Protein Involved in Light-Dependent Development and Gene Expression, Undergoes COP1-Mediated Ubiquitination. Plant Cell 2008, 20, 2324–2338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huai, J.; Shang, F.; Xu, G.; Tang, W.; Jing, Y.; Lin, R. A PIF1/PIF3-HY5-BBX23 Transcription Factor Cascade Affects Photomorphogenesis. Plant Physiol. 2017, 174, 2487–2500. [Google Scholar] [CrossRef]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-Box Proteins Regulate Photomorphogenesis by Oppositely Modulating HY5 through Their Diverse C-Terminal Domains. Plant Physiol. 2018, 176, 2963–2976. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX Protein BBX25 Interacts with HY5, Negatively Regulating BBX22 Expression to Suppress Seedling Photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef]
- Yadav, A.; Bakshi, S.; Yadukrishnan, P.; Lingwan, M.; Dolde, U.; Wenkel, S.; Masakapalli, S.K.; Datta, S. The B-Box-Containing MicroProtein miP1a/BBX31 Regulates Photomorphogenesis and UV-B Protection. Plant Physiol. 2019, 179, 1876–1892. [Google Scholar] [CrossRef]
- Holtan, H.E.; Bandong, S.; Marion, C.M.; Adam, L.; Tiwari, S.; Shen, Y.; Maloof, J.N.; Maszle, D.R.; Ohto, M.; Preuss, S.; et al. BBX32, an Arabidopsis B-Box Protein, Functions in Light Signaling by Suppressing HY5-Regulated Gene Expression and Interacting with STH2/BBX21. Plant Physiol. 2011, 156, 2109–2123. [Google Scholar] [CrossRef]
- Xiang, J.; Li, M.; Li, Y.; Liu, Y.; Wei, L.; Zheng, T.; Wu, J.; Yu, Y.; Cheng, J. Overexpression of Grapevine VyTRXy Improves Drought Tolerance by Maintaining Photosynthesis and Enhancing the Antioxidant and Osmolyte Capacity of Plants. Int. J. Mol. Sci. 2023, 24, 16388. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, C.; Xu, Y.; Wei, Q.; Imtiaz, M.; Lan, H.; Gao, S.; Cheng, L.; Wang, M.; Fei, Z.; et al. A Zinc Finger Protein Regulates Flowering Time and Abiotic Stress Tolerance in Chrysanthemum by Modulating Gibberellin Biosynthesis. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Yuan, L.; Sun, Q.; Zhang, S.; Wang, X. A B-Box Zinc Finger Protein, MdBBX10, Enhanced Salt and Drought Stresses Tolerance in Arabidopsis. Plant Mol. Biol. 2019, 99, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-J.; Lai, B.; Zhao, J.-T.; Qin, Y.-H.; He, J.-M.; Huang, X.-M.; Wang, H.-C.; Hu, G.-B. Sequence Differences in LcFGRT4 Alleles Are Responsible for the Diverse Anthocyanin Composition in the Pericarp of Litchi chinensis. Mol. Breed. 2016, 36, 93. [Google Scholar] [CrossRef]
- Jaakola, L. New Insights into the Regulation of Anthocyanin Biosynthesis in Fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Li, Z.; Song, Y.; Zhu, H.; Lin, S.; Huang, R.; Jiang, Y.; Duan, X. LcNAC13 Physically Interacts with LcR1MYB1 to Coregulate Anthocyanin Biosynthesis-Related Genes during Litchi Fruit Ripening. Biomolecules 2019, 9, 135. [Google Scholar] [CrossRef]
- Li, S.; Qin, Y.; Jing, S.; Wang, D.; Zhang, Z.; Qin, Y.; Hu, G.; Zhao, J. Metabolome and Transcriptome Analyses Reveal the Molecular Mechanisms of LcMYB1 Regulating Anthocyanin Accumulation in Litchi Hairy Roots. Plant Physiol. Biochem. 2023, 200, 107749. [Google Scholar] [CrossRef]
- Lai, B.; Li, X.-J.; Hu, B.; Qin, Y.-H.; Huang, X.-M.; Wang, H.-C.; Hu, G.-B. LcMYB1 Is a Key Determinant of Differential Anthocyanin Accumulation among Genotypes, Tissues, Developmental Phases and ABA and Light Stimuli in Litchi Chinensis. PLoS ONE 2014, 9, e86293. [Google Scholar] [CrossRef]
- Lai, B.; Du, L.-N.; Liu, R.; Hu, B.; Su, W.-B.; Qin, Y.-H.; Zhao, J.-T.; Wang, H.-C.; Hu, G.-B. Two LcbHLH Transcription Factors Interacting with LcMYB1 in Regulating Late Structural Genes of Anthocyanin Biosynthesis in Nicotiana and Litchi Chinensis during Anthocyanin Accumulation. Front. Plant Sci. 2016, 7, 166. [Google Scholar] [CrossRef]
- Zou, S.-C.; Zhuo, M.-G.; Abbas, F.; Hu, G.-B.; Wang, H.-C.; Huang, X.-M. Transcription Factor LcNAC002 Coregulates Chlorophyll Degradation and Anthocyanin Biosynthesis in Litchi. Plant Physiol. 2023, 192, 1913–1927. [Google Scholar] [CrossRef]
- Hu, B.; Lai, B.; Wang, D.; Li, J.; Chen, L.; Qin, Y.; Wang, H.; Qin, Y.; Hu, G.; Zhao, J. Three LcABFs Are Involved in the Regulation of Chlorophyll Degradation and Anthocyanin Biosynthesis During Fruit Ripening in Litchi Chinensis. Plant Cell Physiol. 2019, 60, 448–461. [Google Scholar] [CrossRef]
- Zhuo, M.-G.; Wang, T.-Y.; Huang, X.-M.; Hu, G.-B.; Zhou, B.-Y.; Wang, H.-C.; Abbas, F. ERF Transcription Factors Govern Anthocyanin Biosynthesis in Litchi Pericarp by Modulating the Expression of Anthocyanin Biosynthesis Genes. Sci. Hortic. 2024, 337, 113464. [Google Scholar] [CrossRef]
- Zhang, H.-N.; Li, W.-C.; Wang, H.-C.; Shi, S.-Y.; Shu, B.; Liu, L.-Q.; Wei, Y.-Z.; Xie, J.-H. Transcriptome Profiling of Light-Regulated Anthocyanin Biosynthesis in the Pericarp of Litchi. Front. Plant Sci. 2016, 7, 963. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The Rice B-Box Zinc Finger Gene Family: Genomic Identification, Characterization, Expression Profiling and Diurnal Analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef]
- Ouyang, Y.; Pan, X.; Wei, Y.; Wang, J.; Xu, X.; He, Y.; Zhang, X.; Li, Z.; Zhang, H. Genome-Wide Identification and Characterization of the BBX Gene Family in Pineapple Reveals That Candidate Genes Are Involved in Floral Induction and Flowering. Genomics 2022, 114, 110397. [Google Scholar] [CrossRef]
- Yu, L.; Wang, D.; Huang, R.; Cao, F.; Guo, C.; Zhang, J. Genome-Wide Identification, Characterization and Expression Profile Analysis of BBX Gene Family in Chinese Chestnut (Castanea Mollissima). Plant Biotechnol. Rep. 2024, 18, 129–142. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene Duplication and Evolution in Recurring Polyploidization–Diploidization Cycles in Plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-Regulatory Elements: Molecular Mechanisms and Evolutionary Processes Underlying Divergence. Nat. Rev. Genet. 2012, 13, 59–69. [Google Scholar] [CrossRef]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of Transcription Factor HY5 Genomic Binding Sites Revealed Its Hierarchical Role in Light Regulation of Development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef]
- Bursch, K.; Toledo-Ortiz, G.; Pireyre, M.; Lohr, M.; Braatz, C.; Johansson, H. Identification of BBX Proteins as Rate-Limiting Cofactors of HY5. Nat. Plants 2020, 6, 921–928. [Google Scholar] [CrossRef]
- Wei, Y.-Z.; Hu, F.-C.; Hu, G.-B.; Li, X.-J.; Huang, X.-M.; Wang, H.-C. Differential Expression of Anthocyanin Biosynthetic Genes in Relation to Anthocyanin Accumulation in the Pericarp of Litchi Chinensis Sonn. PLoS ONE 2011, 6, e19455. [Google Scholar] [CrossRef]
- Plunkett, B.J.; Henry-Kirk, R.; Friend, A.; Diack, R.; Helbig, S.; Mouhu, K.; Tomes, S.; Dare, A.P.; Espley, R.V.; Putterill, J.; et al. Apple B-Box Factors Regulate Light-Responsive Anthocyanin Biosynthesis Genes. Sci. Rep. 2019, 9, 17762. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Tian, S.; Hao, W.; Du, L. Two B-Box Proteins, MaBBX20 and MaBBX51, Coordinate Light-Induced Anthocyanin Biosynthesis in Grape Hyacinth. Int. J. Mol. Sci. 2022, 23, 5678. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, G.; Chen, J.; Ying, Y.; Yao, L.; Li, X.; Teixeira Da Silva, J.A.; Yu, Z. Role of Rubus Chingii BBX Gene Family in Anthocyanin Accumulation during Fruit Ripening. Front. Plant Sci. 2024, 15, 1427359. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).