Comparative Analysis of Mucosa-Associated and Luminal Gut Microbiota in Pediatric Ulcerative Colitis
Abstract
1. Introduction
2. Results
2.1. Cohort and Sample Characteristics
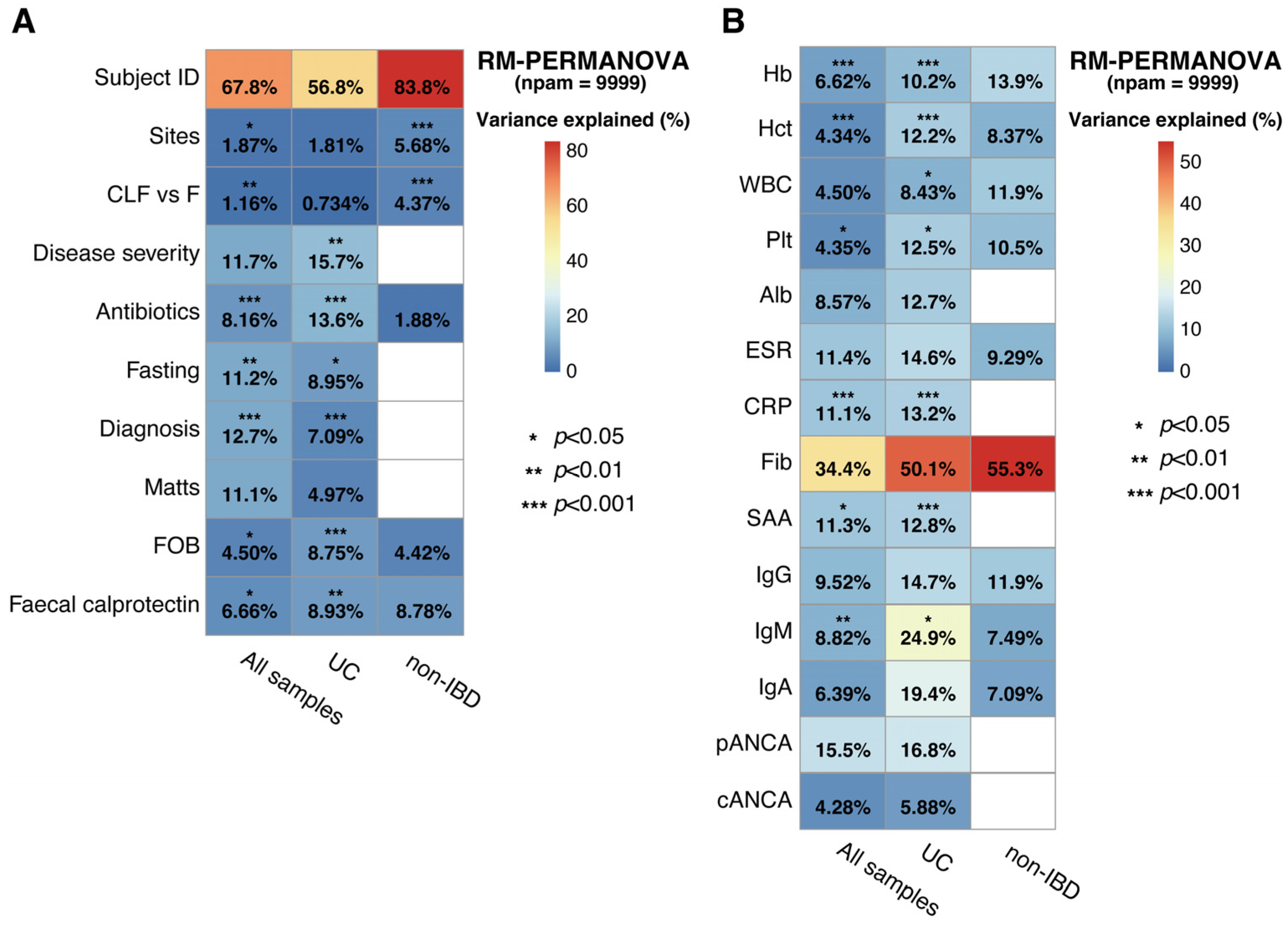
2.2. Association of Host Factors with Gut Microbial Taxonomic Variation in UC and Non-IBD
2.3. Association of the Specific Microbial Taxa with Clinical Metadata
2.4. Homogenous Distribution of Microbiota in MAM Along the Colon
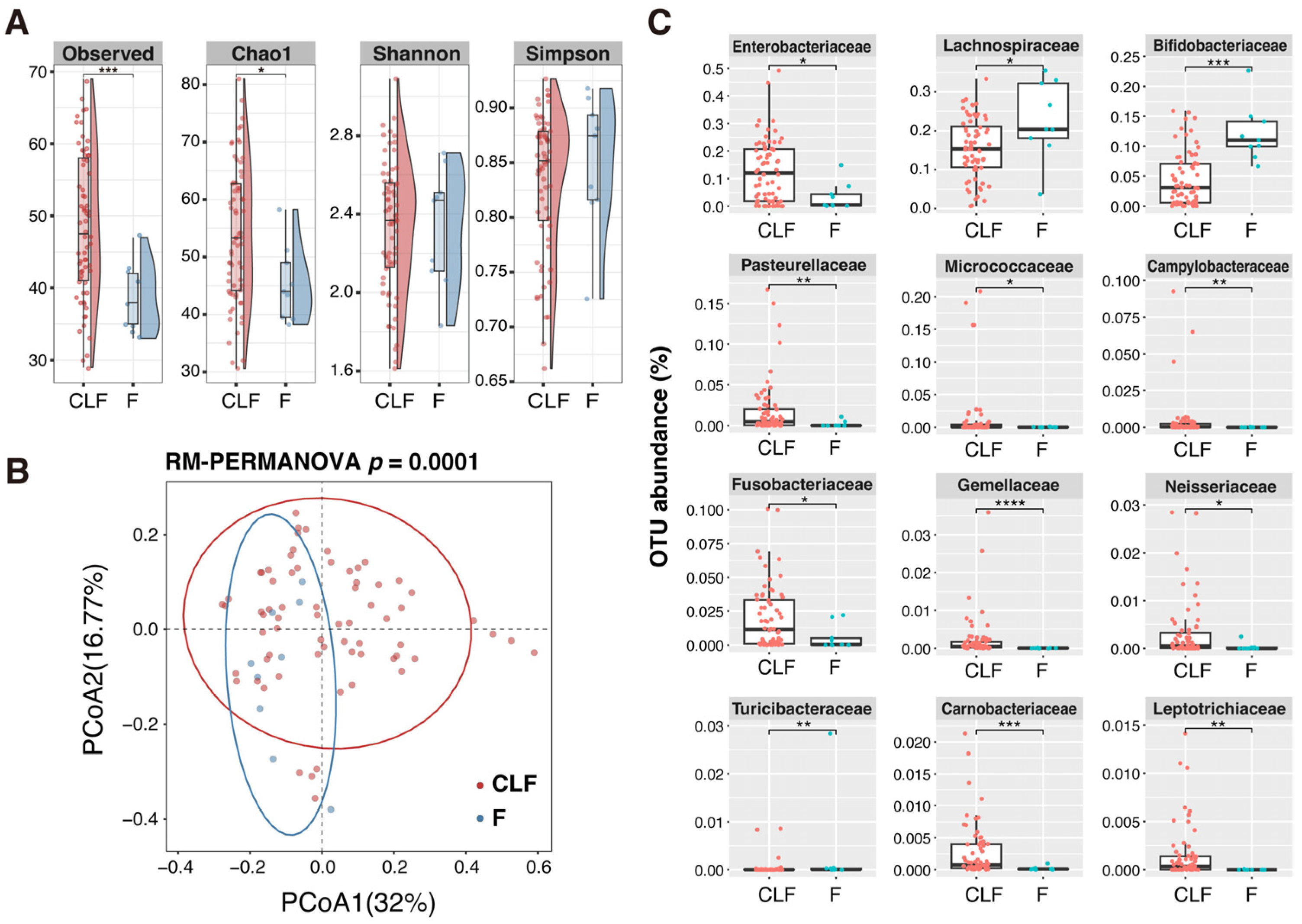
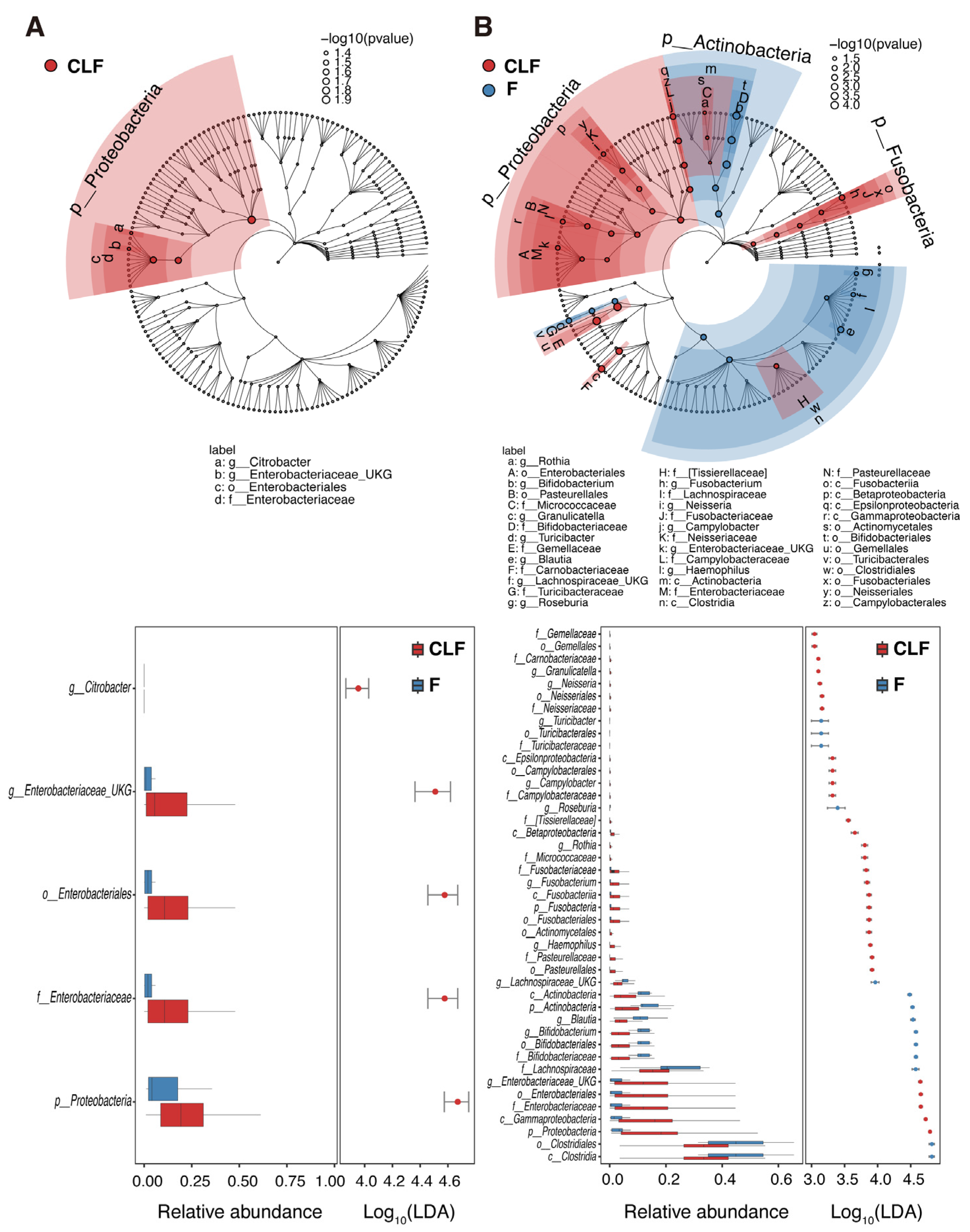
2.5. Microbial Community Structure of MAM and Feces in Pediatric UC and Non-IBD Patients
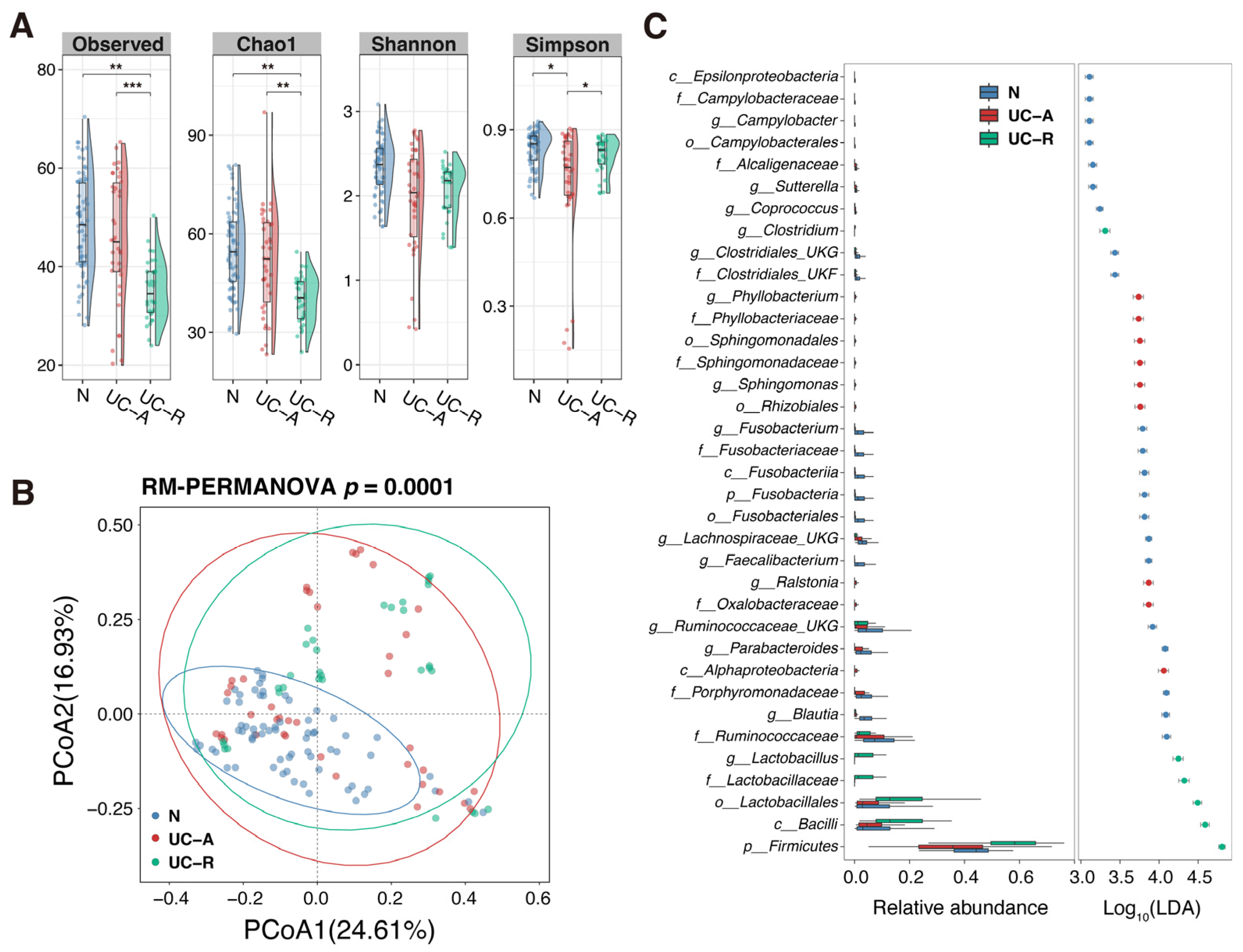
2.6. Alteration in Community Structure of MAM Depending on UC Disease Activity
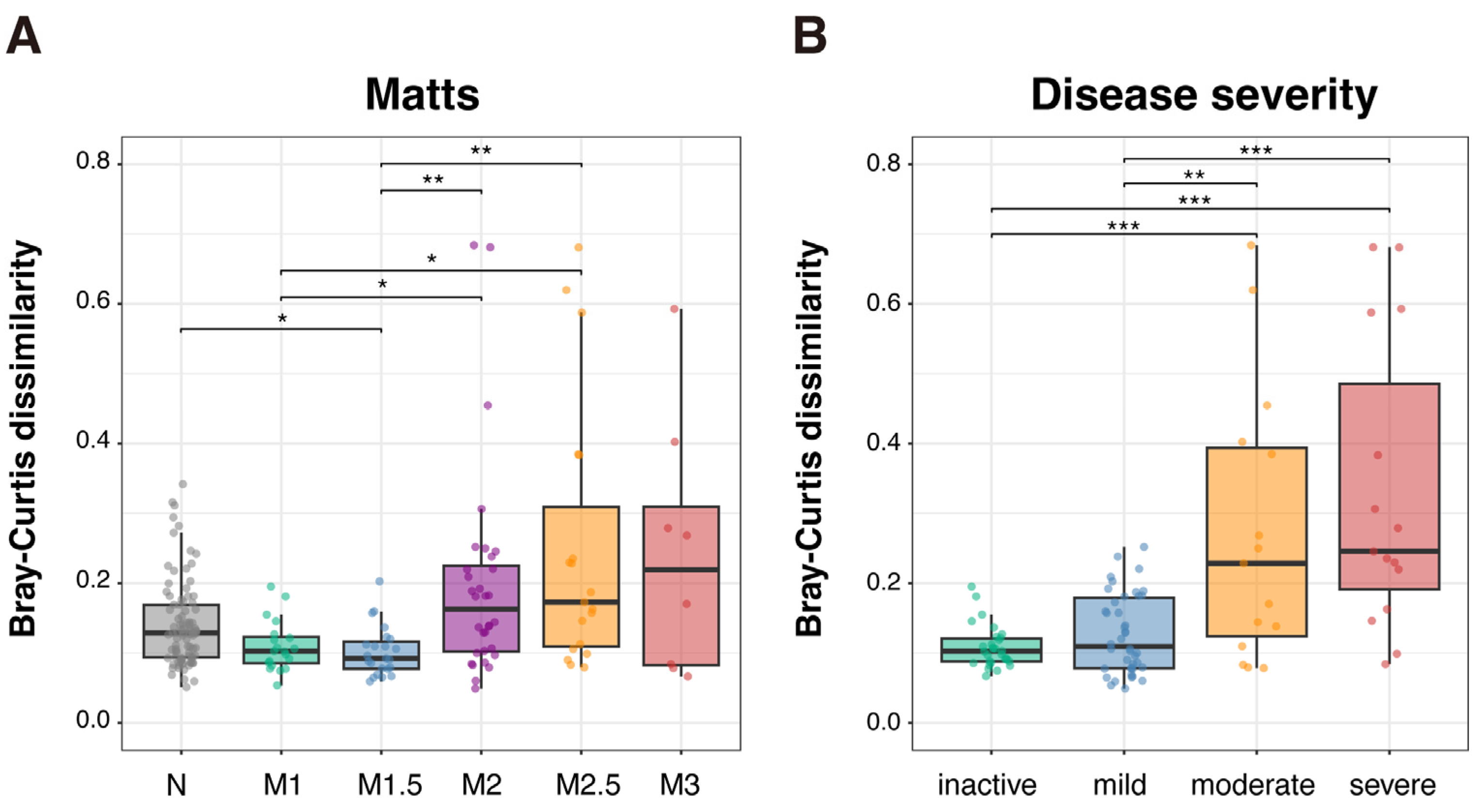
2.7. Profiling of MAM Community Structure According to UC Disease Severity
2.8. Profiling of MAM Community Structure According to Mucosal Inflammation
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. Colon Lavage Collection
4.3. DNA Preparation
4.4. 16S rRNA Gene Analysis
4.5. Statistical Analysis
5. Conclusions
- (i)
- The compositions of MAM were similar to fecal microbiota in pediatric UC patients, while they were significantly different in non-IBD, which might have resulted from the dysfunction of the mucous layer to separate MAM and luminal microbiota.
- (ii)
- MAM in non-IBD showed higher α-diversity compared with feces, while only Enterobacteriaceae was enriched in MAM in pediatric UC.
- (iii)
- Aggravation in UC disease severity or mucosal inflammation promotes inter-regional difference among the colonic MAMs, leading to the enrichment in microbes that usually reside in the oral cavity, oropharynx, upper intestine, or that have environmental origins.
- (iv)
- Specific microbes in MAMs may be associated with inflammatory markers and humoral immune response, including autoantibodies.
- (v)
- We identified nine potential microbial markers to discriminate pediatric UC from non-IBD patients.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Alb | Albumin |
| CLF | Colonic lavage fluid |
| cANCA | Cytoplasmic antineutrophil cytoplasmic antibodies |
| CD | Crohn’s disease |
| CRP | C-reactive protein |
| ESR | Erythrocyte sedimentation rate |
| F | Feces |
| FDR | False discovery rate |
| Fib | Fibrinogen |
| FOB | Fecal occult blood |
| Hct | Hematocrit |
| Hb | Hemoglobin |
| IBD | Inflammatory bowel disease |
| LEfSe | Linear discriminant analysis effect size |
| LDA | Linear discriminant analysis |
| MaAsLin2 | Association analysis by linear model coefficients 2 |
| MAM | Mucosa-associated microbiota |
| OTU | Operational taxonomic unit |
| pANCA | Peripheral antineutrophil cytoplasmic antibodies |
| PCoA | Principal coordinate analysis |
| PERMANOVA | Permutational multivariate analysis of variance |
| Plt | Platelet count |
| PUCAI | Pediatric Ulcerative Colitis Activity Index |
| ROC | Receiver operating characteristic |
| RM-PERMANOVA | Repeated measures PERMANOVA |
| SAA | Serum amyloid A protein |
| UC | Ulcerative colitis |
| WBC | White blood cell count |
References
- Guan, Q.A. comprehensive review and update on the pathogenesis of inflammatory bowel disease. J. Immunol. Res. 2019, 2019, 7247238. [Google Scholar] [CrossRef]
- Liu, J.Z.; van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R.; Takahashi, A.; Ripke, S.; Lee, J.C.; Jostins, L.; Shah, T.; et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Ishige, T.; Tomomasa, T.; Hatori, R.; Tatsuki, M.; Igarashi, Y.; Sekine, K.; Arakawa, H. Temporal trend of pediatric inflammatory bowel disease: Analysis of national registry data 2004 to 2013 in Japan. J. Pediatr. Gastroenterol. Nutr. 2017, 65, e80–e82. [Google Scholar] [CrossRef]
- Sýkora, J.; Pomahačová, R.; Kreslová, M.; Cvalínová, D.; Štych, P.; Schwarz, J. Current global trends in the incidence of pediatric-onset inflammatory bowel disease. World J. Gastroenterol. 2018, 24, 2741–2763. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Griffiths, A.; Markowitz, J.; Wilson, D.C.; Turner, D.; Russell, R.K.; Fell, J.; Ruemmele, F.M.; Walters, T.; Sherlock, M.; et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: The Paris classification. Inflamm. Bowel Dis. 2011, 17, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Malaty, H.M.; Abraham, B.P.; Mehta, S.; Garnett, E.A.; Ferry, G.D. The natural history of ulcerative colitis in a pediatric population: A follow-up population-based cohort study. Clin. Exp. Gastroenterol. 2013, 6, 77–83. [Google Scholar] [CrossRef]
- Zakerska-Banaszak, O.; Tomczak, H.; Gabryel, M.; Baturo, A.; Wolko, L.; Michalak, M.; Malinsla, N.; Mankowska-Wierzbicka, D.; Eder, P.; Dobrowolska, A.; et al. Dysbiosis of gut microbiota in Polish patients with ulcerative colitis: A pilot study. Sci. Rep. 2021, 11, 2166. [Google Scholar] [CrossRef]
- Nishida, A.; Inoue, R.; Inatomi, O.; Bamba, S.; Naito, Y.; Andoh, A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 2018, 11, 1–10. [Google Scholar] [CrossRef]
- Frank, D.N.; Robertson, C.E.; Hamm, C.M.; Kpadeh, Z.; Zhang, T.; Chen, H.; Zhu, W.; Sartor, R.B.; Boedeker, E.C.; Harpaz, N.; et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 2011, 17, 179–184. [Google Scholar] [CrossRef]
- Casén, C.; Vebø, H.C.; Sekelja, M.; Hegge, F.T.; Karlsson, M.K.; Ciemniejewska, E.; Dzankovic, S.; Føyland, C.; Nestestog, R.; Engstrand, L.; et al. Deviations in human gut microbiota: A novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment. Pharmacol. Ther. 2015, 42, 71–83. [Google Scholar] [CrossRef]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef]
- Juge, N. Relationship between mucosa-associated gut microbiota and human diseases. Biochem. Soc. Trans. 2022, 50, 1225–1236. [Google Scholar] [CrossRef]
- Swidsinski, A.; Ladhoff, A.; Pernthaler, A.; Swidsinski, S.; Loening-Baucke, V.; Ortner, M.; Weber, J.; Hoffmann, U.; Schreiber, S.; Dietel, M.; et al. Mucosal flora in inflammatory bowel disease. Gastroenterology 2002, 122, 44–54. [Google Scholar] [CrossRef]
- Seksik, P.; Rigottier-Gois, L.; Gramet, G.; Sutren, M.; Pochart, P.; Marteau, P.; Jian, R.; Doré, J. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut 2003, 52, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, E.; Taida, T.; Kawasumi, M.; Ohkusa, T.; Sato, N.; Ohno, H. Analysis of colonic mucosa-associated microbiota using endoscopically collected lavage. Sci. Rep. 2022, 12, 1758. [Google Scholar] [CrossRef] [PubMed]
- Ringel, Y.; Maharshak, N.; Ringel-Kulka, T.; Wolber, E.A.; Sartor, R.B.; Carroll, I.M. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes 2015, 6, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Carstens, A.; Roos, A.; Andreasson, A.; Magnuson, A.; Agréus, L.; Halfvarson, J.; Engstrand, L. Differential clustering of fecal and mucosa-associated microbiota in ‘healthy’ individuals. J. Dig. Dis. 2018, 19, 745–752. [Google Scholar] [CrossRef]
- Chen, L.; Wang, W.; Zhou, R.; Ng, S.C.; Li, J.; Huang, M.; Zhou, F.; Wang, X.; Shen, B.; Kamm, M.A.; et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine 2014, 93, e51. [Google Scholar] [CrossRef]
- Altomare, A.; Putignani, L.; Del Chierico, F.; Cocca, S.; Angeletti, S.; Ciccozzi, M.; Tripiciano, C.; Piccola, B.D.; Cicala, M.; Guarino, M.P.L. Gut mucosal-associated microbiota better discloses inflammatory bowel disease differential patterns than faecal microbiota. Dig. Liver Dis. 2019, 51, 648–656. [Google Scholar] [CrossRef]
- Tong, M.; Li, X.; Parfrey, L.W.; Roth, B.; Ippoliti, A.; Wei, B.; Borneman, J.; McGovern, D.P.B.; Frank, D.N.; Li, E.; et al. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS ONE 2013, 8, e80702. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.P.; Schippa, S.; Zamboni, I.; Penta, M.; Chiarini, F.; Seganti, L.; Osborn, J.; Falconieri, P.; Borrelli, O.; Cucchiara, S. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 2006, 55, 1760–1767. [Google Scholar] [CrossRef] [PubMed]
- Alipour, M.; Zaidi, D.; Valcheva, R.; Jovel, J.; Martínez, I.; Sergi, C.; Walter, J.; Mason, A.L.; Ka-Shu Wong, G.; Gieleman, L.A.; et al. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J. Crohn’s Colitis 2016, 10, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Jalanka, J.; Cheng, J.; Hiippala, K.; Ritari, J.; Salojärvi, J.; Ruuska, T.; Kalliomäki, M.; Satokari, R. Colonic mucosal microbiota and association of bacterial taxa with the expression of host antimicrobial peptides in pediatric ulcerative colitis. Int. J. Mol. Sci. 2020, 21, 6044. [Google Scholar] [CrossRef]
- Sjöberg, F.; Barkman, C.; Nookaew, I.; Östman, S.; Adlerberth, I.; Saalman, R.; Wold, A.E. Low-complexity microbiota in the duodenum of children with newly diagnosed ulcerative colitis. PLoS ONE 2017, 12, e0186178. [Google Scholar] [CrossRef]
- Turner, D.; Ruemmele, F.M.; Orlanski-Meyer, E.; Griffiths, A.M.; de Carpi, J.M.; Bronsky, J.; Veres, G.; Aloi, M.; Strisciuglio, C.; Braegger, C.P.; et al. Management of paediatric ulcerative colitis, Part 1: Ambulatory care-an evidence-based guideline from European Crohn’s and Colitis Organization and European Society of Paediatric Gastroenterology, Hepatology and Nutrition. J. Pediatr. Gastroenterol. Nutr. 2018, 67, 257–291. [Google Scholar] [CrossRef]
- Turner, D.; Ruemmele, F.M.; Orlanski-Meyer, E.; Griffiths, A.M.; de Carpi, J.M.; Bronsky, J.; Veres, G.; Aloi, M.; Strisciuglio, C.; Braegger, C.P.; et al. Management of paediatric ulcerative colitis, Part 2: Acute severe colitis-an evidence-based consensus guideline From the European Crohn’s and Colitis Organization and the European Society of Paediatric Gastroenterology, Hepatology and Nutrition. J. Pediatr. Gastroenterol. Nutr. 2018, 67, 292–310. [Google Scholar] [CrossRef]
- Hirano, A.; Umeno, J.; Okamoto, Y.; Shibata, H.; Ogura, Y.; Moriyama, T.; Torisu, T.; Fujioka, S.; Fuyuno, Y.; Kawarabayasi, Y.; et al. Comparison of the microbial community structure between inflamed and non-inflamed sites in patients with ulcerative colitis. J. Gastroenterol. Hepatol. 2018, 33, 1590–1597. [Google Scholar] [CrossRef]
- Nishihara, Y.; Ogino, H.; Tanaka, M.; Ihara, E.; Fukaura, K.; Nishioka, K.; Chinen, T.; Tanaka, Y.; Nakayama, J.; Kang, D.; et al. Mucosa-associated gut microbiota reflects clinical course of ulcerative colitis. Sci. Rep. 2021, 11, 13743. [Google Scholar] [CrossRef]
- Shah, R.; Cope, J.L.; Nagy-Szakal, D.; Dowd, S.; Versalovic, J.; Hollister, E.B.; Kellermayer, R. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes 2016, 7, 384–396. [Google Scholar] [CrossRef]
- Kurokawa, K.; Itoh, T.; Kuwahara, T.; Oshima, K.; Toh, H.; Toyoda, A.; Takami, H.; Morita, H.; Sharma, V.K.; Srivastava, T.P. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007, 14, 169–181. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; May, P.; Laczny, C.C.; Lebrun, L.A.; Bellora, C.; Krishna, A.; Wampach, L.; Schnieder, J.G.; Hogan, A.; de Beaufort, C.; et al. Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat. Microbiol. 2016, 2, 16180, Erratum in Nat. Microbiol. 2017, 2, 16227. [Google Scholar] [CrossRef]
- Kashiwagi, S.; Naito, Y.; Inoue, R.; Takagi, T.; Nakano, T.; Inada, Y.; Fukui, A.; Katada, K.; Mizushima, K.; Kamada, K.; et al. Mucosa-associated microbiota in the gastrointestinal tract of healthy Japanese subjects. Digestion 2020, 10, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Malmborg, P.; Grahnquist, L.; Ideström, M.; Lindholm, J.; Befrits, R.; Björk, J.; Montgomery, S.; Hildebrand, H. Presentation and progression of childhood-onset inflammatory bowel disease in Northern Stockholm County. Inflamm. Bowel Dis. 2015, 21, 1098–1108. [Google Scholar] [CrossRef] [PubMed]
- Van Limbergen, J.; Russell, R.K.; Drummond, H.E.; Aldhous, M.C.; Round, N.K.; Nimmo, E.R.; Smith, L.; Gillett, P.M.; McGrogan, P.; Weaver, L.T. Definition of phenotypic characteristics of childhood-onset inflammatory bowel disease. Gastroenterology 2008, 135, 1114–1122. [Google Scholar] [CrossRef]
- Bourgonje, A.R.; Roo-Brand, G.; Lisotto, P.; Sadabad, M.S.; Reitsema, R.D.; de Goffau, M.C.; Faber, K.N.; Dijkstra, G.; Harmsen, H.J.M. Patients with inflammatory bowel disease show IgG immune responses towards specific intestinal bacterial genera. Front. Immunol. 2022, 13, 842911. [Google Scholar] [CrossRef]
- Luo, Z.; Li, M.; Wu, Y.; Meng, Z.; Martin, L.; Zhang, L.; Ogunrinde, E.; Zhou, Z.; Qin, S.; Wan, Z.; et al. Systemic translocation of Staphylococcus drives autoantibody production in HIV disease. Microbiome 2019, 7, 25. [Google Scholar] [CrossRef]
- Smitka, K.; Prochazkova, P.; Roubalova, R.; Dvorak, J.; Papezova, H.; Hill, M.; Pokorny, J.; Kittnar, O.; Bilej, M.; Tlaskalva-Hogenova, H. Current aspects of the role of autoantibodies directed against appetite-regulating hormones and the gut microbiome in eating disorders. Front. Endocrinol. 2021, 12, 613983. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knight, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef]
- Schirmer, M.; Denson, L.; Vlamakis, H.; Franzosa, E.A.; Thomas, S.; Gotman, N.M.; Rufo, P.; Baker, S.S.; Sauer, C.; Markowitz, J.; et al. Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe 2018, 24, 600–610.e4. [Google Scholar] [CrossRef]
- Somineni, H.K.; Kugathasan, S. The microbiome in patients with inflammatory diseases. Clin. Gastroenterol. Hepatol. 2019, 17, 243–255. [Google Scholar] [CrossRef]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, S.; Nagao-Kitamoto, H.; Jiao, Y.; Gillilland, M.G., III; Hayashi, A.; Imai, J.; Sugihara, K.; Miyoshi, M.; Brazil, J.C.; Kuffa, P.; et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell 2020, 182, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Nagao-Kitamoto, H.; Leslie, J.L.; Kitamoto, S.; Jin, C.; Thomsson, K.A.; Gillilland, M.G., III; Kuffa, P.; Goto, Y.; Jenq, R.R.; Ishii, C.; et al. Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat. Med. 2020, 26, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, S.; Nagao-Kitamoto, H.; Hein, R.; Schmidt, T.M.; Kamada, N. The bacterial connection between the oral cavity and the gut diseases. J. Dent. Res. 2020, 99, 1021–1029. [Google Scholar] [CrossRef]
- Imhann, F.; Bonder, M.J.; Vila, A.V.; Fu, J.; Mujagic, Z.; Vork, L.; Tigchelaar, E.F.; Jankipersadsing, S.A.; Cenit, M.C.; Harmsen, H.J.M.; et al. Proton pump inhibitors affect the gut microbiome. Gut 2016, 65, 740–748. [Google Scholar] [CrossRef]
- Brito, F.; de Barros, F.C.; Zaltman, C.; Carvalho, A.T.P.; de Vasconcellos Carneiro, A.J.; Fischer, R.G.; Gustafsson, A.; de Silva Figueredo, C.M. Prevalence of periodontitis and DMFT index in patients with Crohn’s disease and ulcerative colitis. J. Clin. Periodontol. 2008, 35, 555–560. [Google Scholar] [CrossRef]
- Nishino, K.; Nishida, A.; Inoue, R.; Kawada, Y.; Ohno, M.; Sakai, S.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Kawahara, M.; et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 2018, 53, 95–106. [Google Scholar] [CrossRef]
- Seishima, J.; Iida, N.; Kitamura, K.; Yutani, M.; Wang, Z.; Seki, A.; Yamashita, T.; Sakai, Y.; Honda, M.; Yamashita, T.; et al. Gut-derived Enterococcus faecium from ulcerative colitis patients promotes colitis in a genetically susceptible mouse host. Genome Biol. 2019, 20, 252. [Google Scholar] [CrossRef]
- Watanabe, Y.; Nagai, F.; Morotomi, M. Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl. Environ. Microbiol. 2012, 78, 511–518. [Google Scholar] [CrossRef]
- Ikeyama, N.; Murakami, T.; Toyoda, A.; Mori, H.; Iino, T.; Ohkuma, M.; Sakamoto, M. Microbial interaction between the succinate-utilizing bacterium Phascolarctobacterium faecium and the gut commensal Bacteroides thetaiotaomicron. Microbiologyopen 2020, 9, e1111. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.H.; Ma, X.; Zhao, J.C.; Wang, X.; Gao, C.Q. Succinate metabolism and its regulation of host-microbe interactions. Gut Microbes 2023, 15, 2190300. [Google Scholar] [CrossRef]
- Littlewood-Evans, A.; Sarret, S.; Apfel, V.; Loesle, P.; Dawson, J.; Zhang, J.; Muller, A.; Tigani, B.; Kneuer, R.; Patel, S.; et al. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J. Exp. Med. 2016, 213, 1655–1662. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, H.; Wang, X.; Lu, Y.; Yang, C.; Yang, P. Coinfection with Fusobacterium nucleatum can enhance the attachment and invasion of Porphyromonas gingivalis or Aggregatibacter actinomycetemcomitans to human gingival epithelial cells. Arch. Oral Biol. 2015, 60, 1387–1393. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Inagaki, S.; Kimizuka, R.; Okuda, K.; Hosaka, Y.; Nakagawa, T.; Ishihara, K. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 2008, 54, 349–355. [Google Scholar] [CrossRef]
- Sakanaka, A.; Kuboniwa, M.; Shimma, S.; Alghamdi, S.A.; Mayumi, S.; Lamont, R.J.; Fukusaki, E.; Amano, A. Fusobacterium nucleatum metabolically integrates commensals and pathogens in oral biofilms. mSystems 2022, 7, e0017022. [Google Scholar] [CrossRef]
- Yamaguchi-Kuroda, Y.; Kikuchi, Y.; Kokubu, E.; Ishihara, K. Porphyromonas gingivalis diffusible signaling molecules enhance Fusobacterium nucleatum biofilm formation via gene expression modulation. J. Oral Microbiol. 2023, 15, 2165001. [Google Scholar] [CrossRef]
- Han, Y.W. Fusobacterium nucleatum: A commensal-turned pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Fan, Z.; Tang, P.; Li, C.; Yang, Q.; Xu, Y.; Su, C.; Li, L. Fusobacterium nucleatum and its associated systemic diseases: Epidemiologic studies and possible mechanisms. J. Oral Microbiol. 2022, 15, 2145729. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Baik, J.E.; Lagana, S.M.; Han, R.P.; Raab, W.J.; Sahoo, D.; Dalerba, P.; Wang, T.C.; Han, Y.W. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep. 2019, 20, e47638. [Google Scholar] [CrossRef]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M.; et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef]
- Garrett, W.S. The gut microbiota and colon cancer. Science 2019, 364, 1133–1135. [Google Scholar] [CrossRef] [PubMed]
- Koliarakis, I.; Messaritakis, I.; Nikolouzakis, T.K.; Hamilos, G.; Souglakos, J.; Tsiaoussis, J. Oral bacteria and intestinal dysbiosis in colorectal cancer. Int. J. Mol. Sci. 2019, 20, 4146. [Google Scholar] [CrossRef]
- Wang, N.; Fang, J.-Y. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 2023, 31, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, N.; Nakayama-Imaohji, H.; Emmanuel, M.; Tada, A.; Kondo, T.; Kondo, S.; Kusaka, T.; Kuwahara, T. Reactivity of autologous serum IgG to gut microbes in pediatric ulcerative colitis. Int. J. Mol. Sci. 2025, 26, 8196. [Google Scholar] [CrossRef] [PubMed]
- Hotta, F.; Eguchi, H.; Nakayama-Imaohji, H.; Kuwahara, T.; Tada, A.; Yagi, H.; Shimomura, Y.; Kusaka, S. Microbiome analysis of contact lens care solutions and tear fluids of contact lens wearers: Possible involvement of streptococcal antigens in allergic symptoms related to contact lens wear. Int. J. Mol. Med. 2020, 46, 1367–1376. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]



| UC | Non-IBD | |
|---|---|---|
| No. patient | 19 | 19 |
| %Male | 52.6 | 57.9 |
| Age (years) * | 12.7 ± 3.1 | 10.1 ± 3.6 |
| Disease Activity | ||
| Active | 10 | − |
| Remission | 9 | − |
| Disease type | ||
| Proctitis | 2 | − |
| Left-sided colitis | 0 | − |
| Extensive colitis | 0 | − |
| Pancolitis | 17 | − |
| No. CLF sample | ||
| Ascending colon | 18 | 19 |
| Transverse colon | 17 | 15 |
| Sigmoid colon | 19 | 18 |
| Rectum | 18 | 16 |
| No. fecal sample | 14 | 9 |
| Matts score | ||
| 1 | 23 | − |
| 2 | 34 | − |
| 3−4 | 15 | − |
| Stool test | ||
| %FOB | 36.0 | 30.6 |
| Calprotectin (μg/g) ** | 1515 (376.5−3997.5) | 49 (13.4−230.8) |
| Therapies | ||
| Immunosuppressants | 10 | 0 |
| Biologics | 7 | 0 |
| Antibiotics | 4 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondo, T.; Kondo, S.; Nakayama-Imaohji, H.; Tada, A.; Tabassum, N.; Munyeshyaka, E.; Koyano, K.; Nakamura, S.; Kusaka, T.; Kuwahara, T. Comparative Analysis of Mucosa-Associated and Luminal Gut Microbiota in Pediatric Ulcerative Colitis. Int. J. Mol. Sci. 2025, 26, 10775. https://doi.org/10.3390/ijms262110775
Kondo T, Kondo S, Nakayama-Imaohji H, Tada A, Tabassum N, Munyeshyaka E, Koyano K, Nakamura S, Kusaka T, Kuwahara T. Comparative Analysis of Mucosa-Associated and Luminal Gut Microbiota in Pediatric Ulcerative Colitis. International Journal of Molecular Sciences. 2025; 26(21):10775. https://doi.org/10.3390/ijms262110775
Chicago/Turabian StyleKondo, Takeo, Sonoko Kondo, Haruyuki Nakayama-Imaohji, Ayano Tada, Nafisa Tabassum, Emmanuel Munyeshyaka, Kosuke Koyano, Shinji Nakamura, Takashi Kusaka, and Tomomi Kuwahara. 2025. "Comparative Analysis of Mucosa-Associated and Luminal Gut Microbiota in Pediatric Ulcerative Colitis" International Journal of Molecular Sciences 26, no. 21: 10775. https://doi.org/10.3390/ijms262110775
APA StyleKondo, T., Kondo, S., Nakayama-Imaohji, H., Tada, A., Tabassum, N., Munyeshyaka, E., Koyano, K., Nakamura, S., Kusaka, T., & Kuwahara, T. (2025). Comparative Analysis of Mucosa-Associated and Luminal Gut Microbiota in Pediatric Ulcerative Colitis. International Journal of Molecular Sciences, 26(21), 10775. https://doi.org/10.3390/ijms262110775






