FGFR2 Might Be a Promising Therapeutic Target for Some Solid Tumors: Analysis of 1312 Cancers with FGFR2 Abnormalities
Abstract
1. Introduction
2. Results
2.1. Patient Background of Cases with FGFR2 Gene Abnormalities
2.2. Distribution of FGFR2 Genetic Abnormalities
2.3. Treatment with FGFR Inhibitors
3. Discussion
4. Materials and Methods
4.1. Extraction of Subject
4.2. Extraction of Genetic Abnormalities
4.3. Patients’ Clinicopathologic Analysis
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kuroda, K.; Yashiro, M.; Miki, Y.; Sera, T.; Yamamoto, Y.; Sugimoto, A.; Nishimura, S.; Kushiyama, S.; Togano, S.; Okuno, T.; et al. Circulating Tumor Cells with FGFR2 Expression Might Be Useful to Identify Patients with Existing FGFR2-overexpressing Tumor. Cancer Sci. 2020, 111, 4500–4509. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Otsuki, S.; Ohsawa, H.; Hirano, A.; Kazuno, H.; Yamashita, S.; Egami, K.; Shibata, Y.; Yamamiya, I.; Yamashita, F.; et al. Discovery of Futibatinib: The First Covalent FGFR Kinase Inhibitor in Clinical Use. ACS Med. Chem. Lett. 2023, 14, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Sootome, H.; Fujita, H.; Ito, K.; Ochiiwa, H.; Fujioka, Y.; Ito, K.; Miura, A.; Sagara, T.; Ito, S.; Ohsawa, H.; et al. Futibatinib Is a Novel Irreversible FGFR 1–4 Inhibitor That Shows Selective Antitumor Activity against FGFR-Deregulated Tumors. Cancer Res. 2020, 80, 4986–4997. [Google Scholar] [CrossRef]
- Goyal, L.; Meric-Bernstam, F.; Hollebecque, A.; Valle, J.W.; Morizane, C.; Karasic, T.B.; Abrams, T.A.; Furuse, J.; Kelley, R.K.; Cassier, P.A.; et al. Futibatinib for FGFR2-Rearranged Intrahepatic Cholangiocarcinoma. N. Engl. J. Med. 2023, 388, 228–239. [Google Scholar] [CrossRef]
- Silverman, I.M.; Hollebecque, A.; Friboulet, L.; Owens, S.; Newton, R.C.; Zhen, H.; Féliz, L.; Zecchetto, C.; Melisi, D.; Burn, T.C. Clinicogenomic Analysis of FGFR2-Rearranged Cholangiocarcinoma Identifies Correlates of Response and Mechanisms of Resistance to Pemigatinib. Cancer Discov. 2021, 11, 326–339. [Google Scholar] [CrossRef]
- Morizane, C.; Ueno, M.; Ioka, T.; Tajika, M.; Ikeda, M.; Yamaguchi, K.; Hara, H.; Yabusaki, H.; Miyamoto, A.; Iwasa, S.; et al. Tasurgratinib in Patients with Cholangiocarcinoma or Gastric Cancer: Expansion Part of the First-in-human Phase I Study. Cancer Sci. 2025, 116, 192–203. [Google Scholar] [CrossRef]
- Kawakami, H. New Therapeutic Target Molecules for Gastric and Gastroesophageal Junction Cancer. Int. J. Clin. Oncol. 2024, 29, 1228–1236. [Google Scholar] [CrossRef]
- Hanssens, C.; Mouna, O.; Meyers, M.; Hendlisz, A. State-of-the-Art and Trends in Fibroblast Growth Factor Receptor-Directed Therapies in Gastro-Intestinal Malignancies. Curr. Opin. Oncol. 2024, 36, 320–325. [Google Scholar] [CrossRef]
- Helsten, T.; Elkin, S.; Arthur, E.; Tomson, B.N.; Carter, J.; Kurzrock, R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin. Cancer Res. 2016, 22, 259–267. [Google Scholar] [CrossRef]
- Krook, M.A.; Reeser, J.W.; Ernst, G.; Barker, H.; Wilberding, M.; Li, G.; Chen, H.-Z.; Roychowdhury, S. Fibroblast Growth Factor Receptors in Cancer: Genetic Alterations, Diagnostics, Therapeutic Targets and Mechanisms of Resistance. Br. J. Cancer 2021, 124, 880–892. [Google Scholar] [CrossRef]
- Nakazawa, K.; Yashiro, M.; Hirakawa, K. Keratinocyte Growth Factor Produced by Gastric Fibroblasts Specifically Stimulates Proliferation of Cancer Cells from Scirrhous Gastric Carcinoma. Cancer Res. 2003, 63, 8848–8852. [Google Scholar]
- Nakamura, K.; Yashiro, M.; Matsuoka, T.; Tendo, M.; Shimizu, T.; Miwa, A.; Hirakawa, K. A Novel Molecular Targeting Compound as K-SamII/FGF-R2 Phosphorylation Inhibitor, Ki23057, for Scirrhous Gastric Cancer. Gastroenterology 2006, 131, 1530–1541. [Google Scholar] [CrossRef]
- Yashiro, M.; Shinto, O.; Nakamura, K.; Tendo, M.; Matsuoka, T.; Matsuzaki, T.; Kaizaki, R.; Miwa, A.; Hirakawa, K. Synergistic Antitumor Effects of FGFR2 Inhibitor with 5-Fluorouracil on Scirrhous Gastric Carcinoma. Int. J. Cancer 2010, 126, 1004–1016. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, L.; Park, H.; Chhim, S.; Tanphanich, M.; Yashiro, M.; Kim, K.J. Monoclonal Antibodies to Fibroblast Growth Factor Receptor 2 Effectively Inhibit Growth of Gastric Tumor Xenografts. Clin. Cancer Res. 2010, 16, 5750–5758. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, H.; Yashiro, M.; Fukuoka, T.; Hasegawa, T.; Morisaki, T.; Kasashima, H.; Masuda, G.; Noda, S.; Hirakawa, K. Diffuse-Type Gastric Cancer Cells Switch Their Driver Pathways from FGFR2 Signaling to SDF1/CXCR4 Axis in Hypoxic Tumor Microenvironments. Carcinogenesis 2015, 36, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Yashiro, M.; Kuroda, K.; Masuda, G.; Okuno, T.; Miki, Y.; Yamamoto, Y.; Sera, T.; Sugimoto, A.; Kushiyama, S.; Nishimura, S.; et al. Clinical Difference between Fibroblast Growth Factor Receptor 2 Subclass, Type IIIb and Type IIIc, in Gastric Cancer. Sci. Rep. 2021, 11, 4698. [Google Scholar] [CrossRef]
- Kohno, T.; Kato, M.; Kohsaka, S.; Sudo, T.; Tamai, I.; Shiraishi, Y.; Okuma, Y.; Ogasawara, D.; Suzuki, T.; Yoshida, T.; et al. C-CAT: The National Datacenter for Cancer Genomic Medicine in Japan. Cancer Discov. 2022, 12, 2509–2515. [Google Scholar] [CrossRef]
- Mukai, Y.; Ueno, H. Establishment and Implementation of Cancer Genomic Medicine in Japan. Cancer Sci. 2021, 112, 970–977. [Google Scholar] [CrossRef]
- Nishikubo, H.; Kawabata, K.; Kanei, S.; Aoyama, R.; Ma, D.; Sano, T.; Imanishi, D.; Sakuma, T.; Maruo, K.; Fan, C.; et al. Multi-Cancer Genome Profiling for Neurotrophic Tropomyosin Receptor Kinase (NTRK) Fusion Genes: Analysis of Profiling Database of 88,688 Tumors. Cancers 2025, 17, 2250. [Google Scholar] [CrossRef]
- Jeske, Y.W.; Ali, S.; Byron, S.A.; Gao, F.; Mannel, R.S.; Ghebre, R.G.; DiSilvestro, P.A.; Lele, S.B.; Pearl, M.L.; Schmidt, A.P.; et al. FGFR2 Mutations Are Associated with Poor Outcomes in Endometrioid Endometrial Cancer: An NRG Oncology/Gynecologic Oncology Group Study. Gynecol. Oncol. 2017, 145, 366–373. [Google Scholar] [CrossRef]
- Pollock, P.M.; Gartside, M.G.; Dejeza, L.C.; Powell, M.A.; Mallon, M.A.; Davies, H.; Mohammadi, M.; Futreal, P.A.; Stratton, M.R.; Trent, J.M.; et al. Frequent Activating FGFR2 Mutations in Endometrial Carcinomas Parallel Germline Mutations Associated with Craniosynostosis and Skeletal Dysplasia Syndromes. Oncogene 2007, 26, 7158–7162. [Google Scholar] [CrossRef]
- Hyung, S.; Han, B.; Jung, J.; Kim, S.T.; Hong, J.Y.; Park, S.H.; Zang, D.Y.; Park, J.O.; Park, Y.S.; Kim, K.-M.; et al. Incidence of FGFR2 Amplification and FGFR2 Fusion in Patients with Metastatic Cancer Using Clinical Sequencing. J. Oncol. 2022, 2022, 9714570. [Google Scholar] [CrossRef]
- Hu, Z.I.; Ross, J.S.; Pavlick, D.; Hsiehchen, D. Age and Sex Affects the Frequency and Mutation Type of FGFR2 Alterations in Cholangiocarcinoma. JCO Oncol. Adv. 2024, 1, e2400027. [Google Scholar] [CrossRef]
- Borad, M.J.; Gores, G.J.; Roberts, L.R. Fibroblast Growth Factor Receptor 2 Fusions as a Target for Treating Cholangiocarcinoma. Curr. Opin. Gastroenterol. 2015, 31, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Dutt, A.; Salvesen, H.B.; Chen, T.-H.; Ramos, A.H.; Onofrio, R.C.; Hatton, C.; Nicoletti, R.; Winckler, W.; Grewal, R.; Hanna, M.; et al. Drug-Sensitive FGFR2 Mutations in Endometrial Carcinoma. Proc. Natl. Acad. Sci. USA 2008, 105, 8713–8717. [Google Scholar] [CrossRef] [PubMed]
- Sunami, K.; Ichikawa, H.; Kubo, T.; Kato, M.; Fujiwara, Y.; Shimomura, A.; Koyama, T.; Kakishima, H.; Kitami, M.; Matsushita, H.; et al. Feasibility and Utility of a Panel Testing for 114 Cancer-Associated Genes in a Clinical Setting: A Hospital-Based Study. Cancer Sci. 2019, 110, 1480–1490. [Google Scholar] [CrossRef]
- Yatabe, Y.; Sunami, K.; Goto, K.; Nishio, K.; Aragane, N.; Ikeda, S.; Inoue, A.; Kinoshita, I.; Kimura, H.; Sakamoto, T.; et al. Multiplex Gene-Panel Testing for Lung Cancer Patients. In Pathology International; Blackwell Publishing: Oxford, UK, 2020; pp. 921–931. [Google Scholar] [CrossRef]
- Woodhouse, R.; Li, M.; Hughes, J.; Delfosse, D.; Skoletsky, J.; Ma, P.; Meng, W.; Dewal, N.; Milbury, C.; Clark, T.; et al. Clinical and Analytical Validation of Foundation One Liquid CDx, a Novel 324-Gene CfDNA-Based Comprehensive Genomic Profiling Assay for Cancers of Solid Tumor Origin. PLoS ONE 2020, 15, e0237802. [Google Scholar] [CrossRef]
- Watanabe, K.; Ogawa, M.; Shinozaki-Ushiku, A.; Tsutsumi, S.; Tatsuno, K.; Aburatani, H.; Kage, H.; Oda, K. Real-World Data Analysis of Genomic Alterations Detected by a Dual DNA–RNA Comprehensive Genomic Profiling Test. Cancer Sci. 2025, 116, 1984–1995. [Google Scholar] [CrossRef]

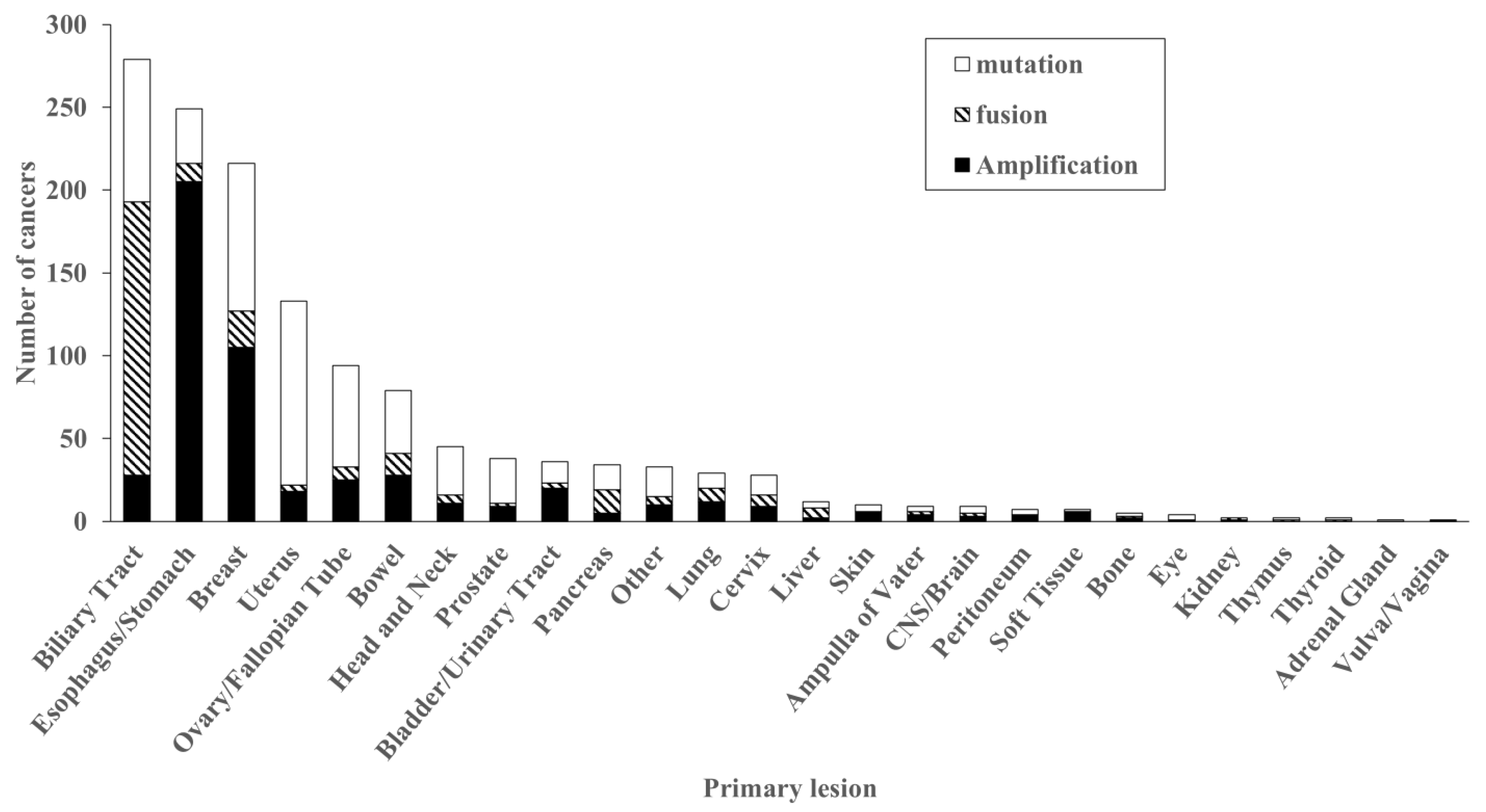
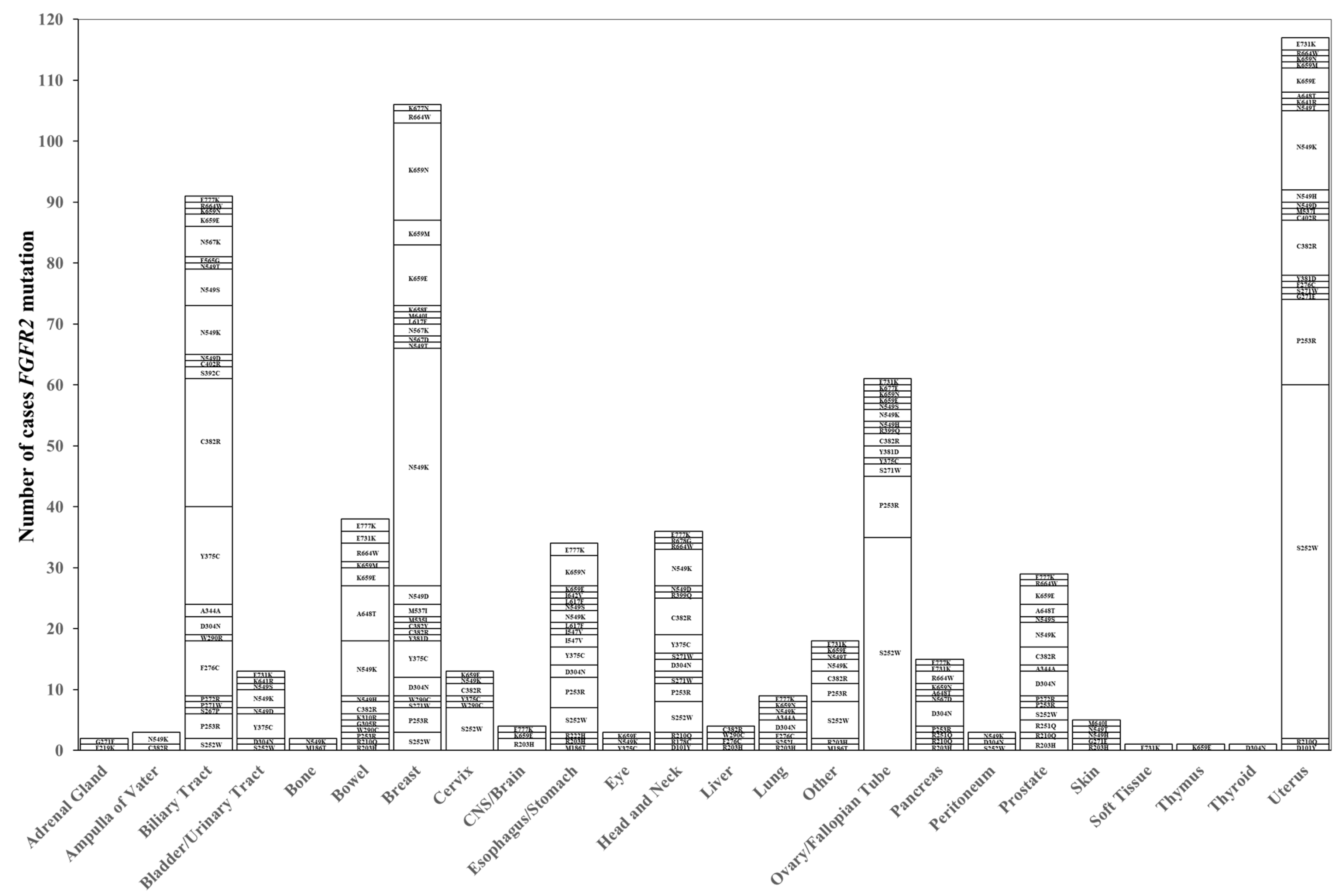
| Patient and Samples | FGFR2 Alteration (n = 1312) | FGFR2 Wild (n = 99,919) | p-Value | |
|---|---|---|---|---|
| Gender | Male (n = 50,437) | 513 (39.1%) | 49,924 (50.0%) | |
| Female (n = 50,790) | 799 (60.9%) | 49,991 (50.0%) | <0.001 | |
| Age | <60 (n = 38,129) | 539 (41.1%) | 37,406 (37.4%) | |
| 60 ≦ (n = 63,103) | 773 (58.9%) | 62,514 (62.6%) | <0.01 | |
| Smoking | Positive (n = 43,945) | 476 (36.3%) | 43,469 (43.5%) | |
| Negative (n = 51,599) | 763 (58.2%) | 50,836 (50.9%) | <0.001 | |
| Alcohol | Positive (n = 11,627) | 138 (10.5%) | 11,489 (11.5%) | |
| Negative (n = 77,339) | 1031 (78.6%) | 76,308 (76.4%) | <0.001 | |
| Assay sample | DNA assay (n = 96,494) | 1261 (96.1%) | 95,233 (95.3%) | |
| RNA assay (n = 4737) | 51 (3.9%) | 4686 (4.7%) | N.S. | |
| Sample type | Tissue (n = 83,485) | 1126 (85.8%) | 82,359 (82.4%) | |
| Blood (n = 17,746) | 186 (14.2%) | 17,560 (17.6%) | <0.001 | |
| Sampling site | Primary tumor (n = 57,323) | 797 (60.7%) | 56,526 (56.6%) | |
| Metastatic tumor (n = 25,777) | 326 (24.8%) | 25,451 (25.5%) | <0.001 | |
| Organ | Fusion Partner (Number of Cases) |
|---|---|
| Ampulla of Vater (n = 2) | CCSER2 (1), RBM2 (1) |
| Biliary Tract (n = 165) | BICC1 (29), TACC2 (9), AHCYL1 (6), CCDC6 (5), VCL (5), CTNNA3 (6), KIAA1217 (6), PAWR (5), KIAA1598 (4), SHROOM3 (4), INA (3), TACC1 (3), AFF3 (2), ANKRD28 (2), BEND3 (2), CASP7 (2), CCAR1 (2), CLIP1 (2), COL16A1 (2), DDX21 (2), NOL4 (2), NRBF2 (2), POC1B (2), RBFOX2 (2), SORBS1 (2), SORBS2 (2), SYNPO2 (2), A1CF (1), ALS2CR12 (1), ARHGAP21 (1), ATAD2 (1), BAIAP2L1 (1), CBY1 (1), CCDC147 (1), CCDC171 (1), CCSER2 (1), CCT7 (1), CD2AP (1), CLASP2 (1), CROCC (1), CUX1 (1), DNAH8 (1), DSP (1), EBF3 (1), EEA1 (1), FOXP1 (1), GPHN (1), GPX3 (1), GRB2 (1), HDX (1), HOOK1 (1), KIAA1210 (1), MITF (1), MYLK (1), NELL2 (1), NRAP (1), PAH (1), PDE4DIP (1), PKD2L1 (1), POLDIP3 (1), PPP1R21 (1), PRDM16 (1), RASAL2 (1), RMND1 (1), SEC63 (1), 14-Sep (1), SLMAP (1), TRA2B (1), TRIM54 (1), TTC28 (1), TULP3 (1), TXLNA (1), UBE2K (1), WAC (1), WARS1 (1), WDR65 (1), ZMYM4 (1) |
| Bladder/Urinary Tract (n = 3) | TACC2 (2), CCDC6 (1) |
| Bone (n = 1) | ATE1 (1) |
| Bowel (n = 12) | TACC2 (2), ATE1 (1), CIT (1), FOXP1 (1), GLS (1), KIAA1217 (1), POC1B (1), POF1B (1), PTBP3 (1), STK31 (1), TBC1D1 (1) |
| Breast (n = 22) | ATE1 (3), C10orf88 (1), CCDC6 (1), CCSER2 (1), CLIP1 (1), CPXM2 (1), CTNNA3 (1), FAM160B1 (1), GAB2 (1), KIAA1598 (1), KIF11 (1), MPP7 (1), PRDX3 (1), PTPRE (1), RBM20 (1), RGS10 (1), SLMAP (1), TACC2 (1), VCL (1) |
| Cervix (n = 7) | TACC2 (2), ATE1 (1), BICC1 (1), CASP7 (1), FARS2 (1), PRKN (1) |
| CNS/Brain (n = 2) | SOGA1 (1), UGP2 (1) |
| Esophagus/Stomach (n = 11) | TACC2 (3), TRIM44 (2), ATE1 (2), DMBT1 (1), LAMP1 (1), PPAPDC1A (1), SKIL (1) |
| Head and Neck (n = 5) | CCDC102A (1), FILIP1 (1), FOXP1 (1), KIAA1217 (1), PBLD (1) |
| Kidney (n = 1) | KIFC1 (1) |
| Liver (n = 6) | BICC1 (3), AMTN (1), CCDC6 (1), KIAA1217 (1) |
| Lung (n = 8) | CIT (1), CTNNA3 (1), EEA1 (1), FOXP4 (1), MCC (1), NAV2 (1), NRBF2 (1), TACC2 (1) |
| Other (n = 5) | BICC1 (2), ACPP (1), CCDC6 (1), YPEL5 (1) |
| Ovary/Fallopian Tube (n = 8) | C10orf11 (1), DLG5 (1), KIAA1217 (1), KIF24 (1), RASEF (1), TRIM23 (1), TRIM8 (1), ZMYND11 (1) |
| Pancreas (n = 14) | SYCP1 (2), CAT (1), CGNL1 (1), CIT (1), FOXP1 (1), FRMD4A (1), KIAA1598 (1), POLDIP3 (1), TACC2 (1), TBC1D1 (1), TFCP2 (1), TMEM132B (1), TXLNA (1) |
| Prostate (n = 2) | CTNNA3 (1), TACC2 (1) |
| Thymus (n = 1) | KIAA1217 (1) |
| Thyroid (n = 1) | TACC2 (1) |
| Uterus (n = 4) | ACTN1 (1), ATE1 (1), CCDC6 (1), PPP2R5C (1) |
| Therapeutic Response of FGFR2 Inhibitors | FGFR2 Alteration Type | ||||
|---|---|---|---|---|---|
| Amplification (n = 8) | Fusion (n = 138) | Mutation (n = 2) | Amplification and Fusion (n = 4) | Mutation and Fusion (n = 2) | |
| CR (n = 0) | 0 | 0 | 0 | 0 | 0 |
| PR (n = 21) | 1 | 20 | 0 | 0 | 0 |
| SD (n = 27) | 0 | 24 | 1 | 1 | 1 |
| PD (n = 14) | 1 | 12 | 0 | 1 | 0 |
| NE (n = 23) | 4 | 18 | 0 | 1 | 0 |
| ORR; CR + PR (n = 21) | 1 | 20 | 0 | 0 | 0 |
| DCR; CR + PR + SD (n = 48) | 1 | 44 | 1 | 1 | 1 |
| Cancer Type | DCR (n = 48) | PD + NE (n = 37) |
|---|---|---|
| Ampulla of vater (n = 2) | 1 | 1 |
| Biliary tract (n = 73) | 44 | 29 |
| Esophagus/Stomach (n = 3) | 0 | 3 |
| Liver (n = 3) | 1 | 2 |
| Other (n = 2) | 2 | 0 |
| Pancreas (n = 2) | 0 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishikubo, H.; Ma, D.; Sano, T.; Imanishi, D.; Sakuma, T.; Fan, C.; Yamamoto, Y.; Yamamori, M.; Yashiro, M. FGFR2 Might Be a Promising Therapeutic Target for Some Solid Tumors: Analysis of 1312 Cancers with FGFR2 Abnormalities. Int. J. Mol. Sci. 2025, 26, 10777. https://doi.org/10.3390/ijms262110777
Nishikubo H, Ma D, Sano T, Imanishi D, Sakuma T, Fan C, Yamamoto Y, Yamamori M, Yashiro M. FGFR2 Might Be a Promising Therapeutic Target for Some Solid Tumors: Analysis of 1312 Cancers with FGFR2 Abnormalities. International Journal of Molecular Sciences. 2025; 26(21):10777. https://doi.org/10.3390/ijms262110777
Chicago/Turabian StyleNishikubo, Hinano, Dongheng Ma, Tomoya Sano, Daiki Imanishi, Takashi Sakuma, Canfeng Fan, Yurie Yamamoto, Motohiro Yamamori, and Masakazu Yashiro. 2025. "FGFR2 Might Be a Promising Therapeutic Target for Some Solid Tumors: Analysis of 1312 Cancers with FGFR2 Abnormalities" International Journal of Molecular Sciences 26, no. 21: 10777. https://doi.org/10.3390/ijms262110777
APA StyleNishikubo, H., Ma, D., Sano, T., Imanishi, D., Sakuma, T., Fan, C., Yamamoto, Y., Yamamori, M., & Yashiro, M. (2025). FGFR2 Might Be a Promising Therapeutic Target for Some Solid Tumors: Analysis of 1312 Cancers with FGFR2 Abnormalities. International Journal of Molecular Sciences, 26(21), 10777. https://doi.org/10.3390/ijms262110777







