Xanthohumol Alters Gut Microbiota Metabolism and Bile Acid Dynamics in Gastrointestinal Simulation Models of Eubiotic and Dysbiotic States
Abstract
1. Introduction
2. Results
2.1. Gut Microbiota-Derived Metabolism of XN Varies by Colon Compartment and Time
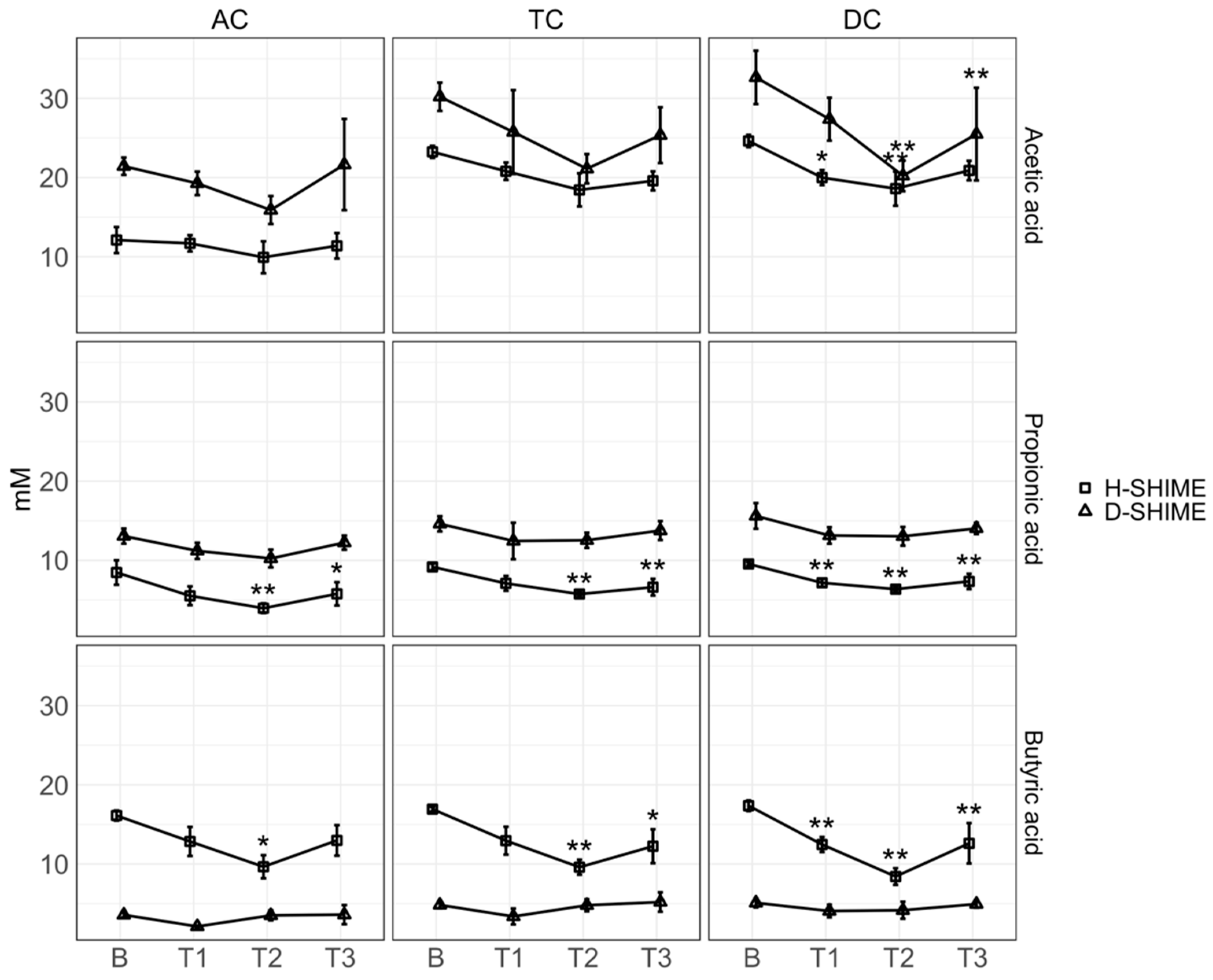
2.2. XN Reduces Short-Chain Fatty Acid Concentration
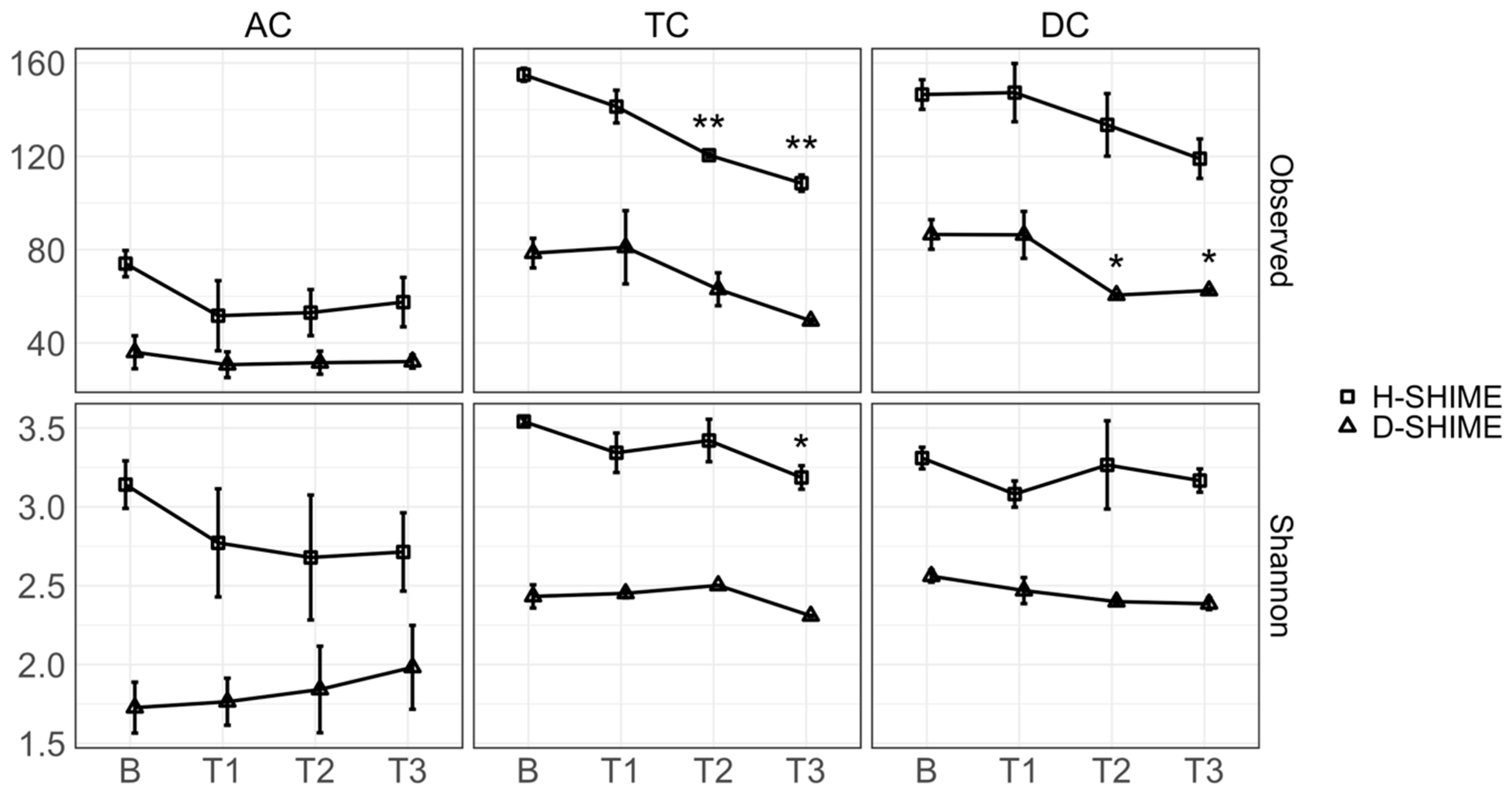
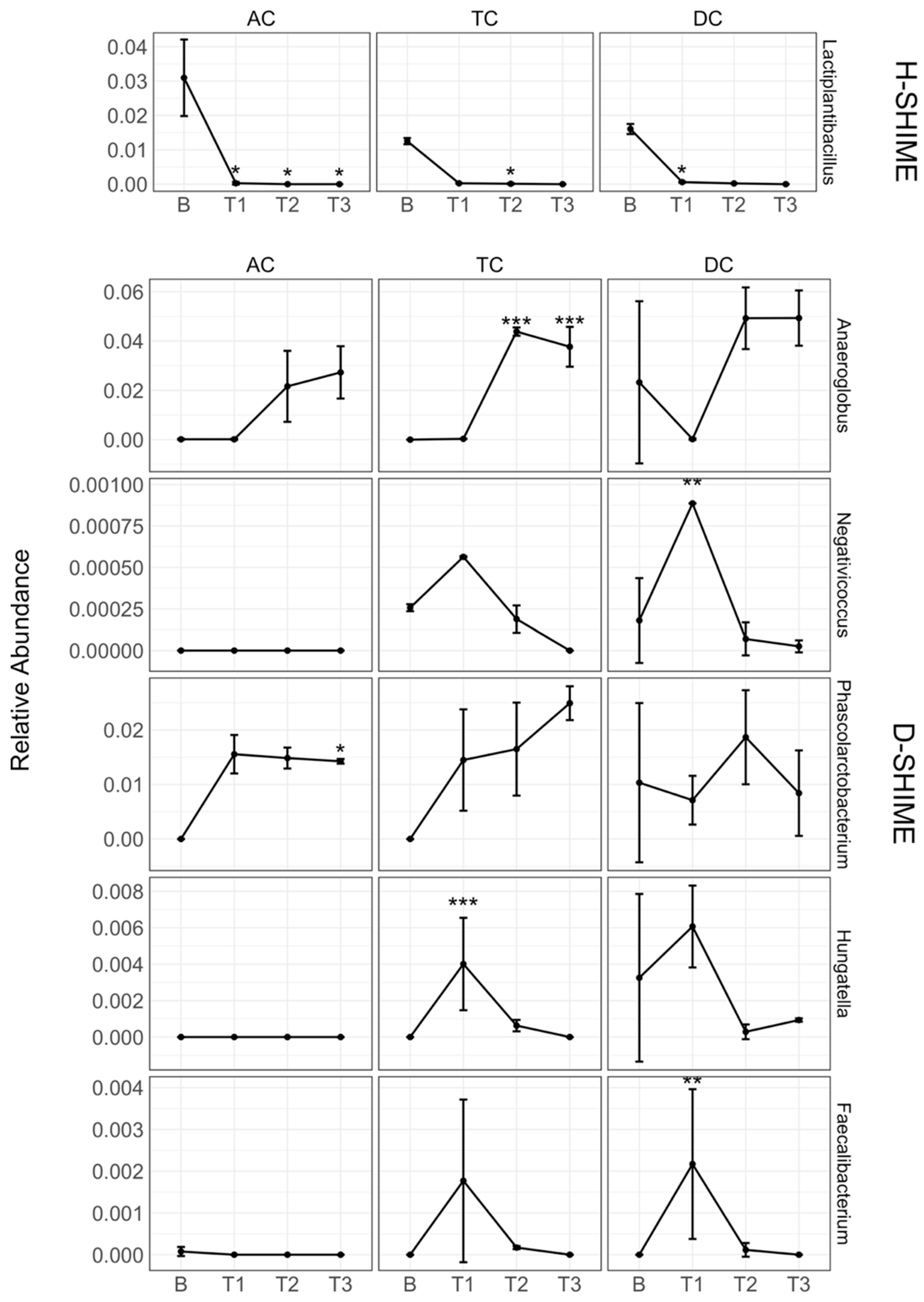
2.3. XN Alters Gut Microbiota Community Structure
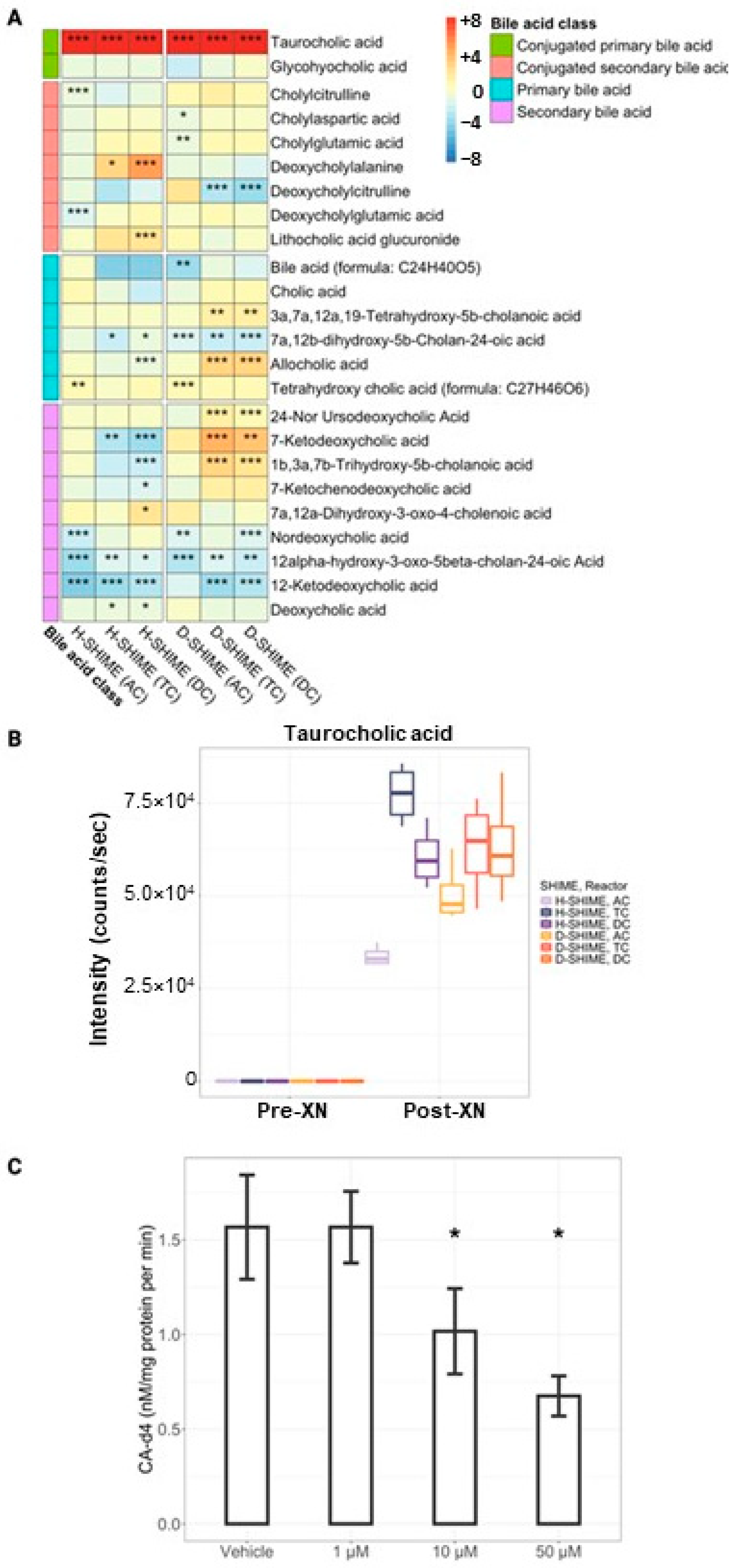
2.4. XN Supplementation Inhibits Bile Acid Metabolism Within the Digestive Meta-Metabolome
2.5. XN Inhibits Bile Salt Hydrolase Activity
3. Discussion
4. Materials and Methods
4.1. Fecal Microbiota Sample Collection
4.2. Long-Term Colonic Incubation
4.3. Experimental Design and Dosage Information
4.4. Quantification of XN Microbial Metabolites
4.5. Quantification of Short-Chain Fatty Acids
4.6. Bacterial 16S rRNA Gene Sequencing, Data Management, and ASV Normalization
4.7. Metabolomics
4.8. Bile Salt Hydrolase (BSH) Activity
4.9. LC-SRM-MS/MS Analysis of Cholic Acid-d4 Generation
4.10. Statistical Analysis
4.11. Data and Code Availability
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Dietz, B.M.; Kang, Y.-H.; Liu, G.; Eggler, A.L.; Yao, P.; Chadwick, L.R.; Pauli, G.F.; Farnsworth, N.R.; Mesecar, A.D.; van Breemen, R.B.; et al. Xanthohumol Isolated from Humulus Lupulus Inhibits Menadione-Induced DNA Damage through Induction of Quinone Reductase. Chem. Res. Toxicol. 2005, 18, 1296–1305. [Google Scholar] [CrossRef]
- Colgate, E.C.; Miranda, C.L.; Stevens, J.F.; Bray, T.M.; Ho, E. Xanthohumol, a Prenylflavonoid Derived from Hops Induces Apoptosis and Inhibits NF-kappaB Activation in Prostate Epithelial Cells. Cancer Lett. 2007, 246, 201–209. [Google Scholar] [CrossRef]
- Paraiso, I.L.; Tran, T.Q.; Magana, A.A.; Kundu, P.; Choi, J.; Maier, C.S.; Bobe, G.; Raber, J.; Kioussi, C.; Stevens, J.F. Xanthohumol Ameliorates Diet-Induced Liver Dysfunction via Farnesoid X Receptor-Dependent and Independent Signaling. Front. Pharmacol. 2021, 12, 643857. [Google Scholar] [CrossRef]
- Zhang, Y.; Bobe, G.; Revel, J.S.; Rodrigues, R.R.; Sharpton, T.J.; Fantacone, M.L.; Raslan, K.; Miranda, C.L.; Lowry, M.B.; Blakemore, P.R.; et al. Improvements in Metabolic Syndrome by Xanthohumol Derivatives Are Linked to Altered Gut Microbiota and Bile Acid Metabolism. Mol. Nutr. Food Res. 2020, 64, 1900789. [Google Scholar] [CrossRef]
- Vicente de Andrade Silva, G.; Demaman Arend, G.; Antonio Ferreira Zielinski, A.; Di Luccio, M.; Ambrosi, A. Xanthohumol Properties and Strategies for Extraction from Hops and Brewery Residues: A Review. Food Chem. 2023, 404, 134629. [Google Scholar] [CrossRef] [PubMed]
- Harikumar, K.B.; Kunnumakkara, A.B.; Ahn, K.S.; Anand, P.; Krishnan, S.; Guha, S.; Aggarwal, B.B. Modification of the Cysteine Residues in IκBα Kinase and NF-κB (P65) by Xanthohumol Leads to Suppression of NF-κB–Regulated Gene Products and Potentiation of Apoptosis in Leukemia Cells. Blood 2009, 113, 2003–2013. [Google Scholar] [CrossRef]
- Lee, I.-S.; Lim, J.; Gal, J.; Kang, J.C.; Kim, H.J.; Kang, B.Y.; Choi, H.J. Anti-Inflammatory Activity of Xanthohumol Involves Heme Oxygenase-1 Induction via NRF2-ARE Signaling in Microglial BV2 Cells. Neurochem. Int. 2011, 58, 153–160. [Google Scholar] [CrossRef]
- Krajka-Kuźniak, V.; Paluszczak, J.; Baer-Dubowska, W. Xanthohumol Induces Phase II Enzymes via Nrf2 in Human Hepatocytes in Vitro. Toxicol. Vitr. 2013, 27, 149–156. [Google Scholar] [CrossRef]
- Luo, Y.; Eggler, A.L.; Liu, D.; Liu, G.; Mesecar, A.D.; van Breemen, R.B. Sites of Alkylation of Human Keap1 by Natural Chemoprevention Agents. J. Am. Soc. Spectrom. 2007, 18, 2226–2232. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.L.; Elias, V.D.; Hay, J.J.; Choi, J.; Reed, R.L.; Stevens, J.F. Xanthohumol Improves Dysfunctional Glucose and Lipid Metabolism in Diet-Induced Obese C57BL/6J Mice. Arch. Biochem. Biophys. 2016, 599, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.L.; Johnson, L.A.; de Montgolfier, O.; Elias, V.D.; Ullrich, L.S.; Hay, J.J.; Paraiso, I.L.; Choi, J.; Reed, R.L.; Revel, J.S.; et al. Non-Estrogenic Xanthohumol Derivatives Mitigate Insulin Resistance and Cognitive Impairment in High-Fat Diet-Induced Obese Mice. Sci Rep 2018, 8, 613. [Google Scholar] [CrossRef]
- Legette, L.; Karnpracha, C.; Reed, R.L.; Choi, J.; Bobe, G.; Christensen, J.M.; Rodriguez-Proteau, R.; Purnell, J.Q.; Stevens, J.F. Human Pharmacokinetics of Xanthohumol, an Antihyperglycemic Flavonoid from Hops. Mol. Nutr. Food Res. 2014, 58, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Legette, L.; Ma, L.; Reed, R.L.; Miranda, C.L.; Christensen, J.M.; Rodriguez-Proteau, R.; Stevens, J.F. Pharmacokinetics of Xanthohumol and Metabolites in Rats after Oral and Intravenous Administration. Mol. Nutr. Food Res. 2012, 56, 466–474. [Google Scholar] [CrossRef]
- Logan, I.E.; Shulzhenko, N.; Sharpton, T.J.; Bobe, G.; Liu, K.; Nuss, S.; Jones, M.L.; Miranda, C.L.; Vasquez-Perez, S.; Pennington, J.M.; et al. Xanthohumol Requires the Intestinal Microbiota to Improve Glucose Metabolism in Diet-Induced Obese Mice. Mol. Nutr. Food Res. 2021, 65, 2100389. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Li, J.; Li, Y.; Zhao, F.; Wang, J.; Ye, P.; Zeng, Z. Xanthohumol Ameliorates Dextran Sodium Sulfate-Induced Colitis in Mice by Inhibiting of NF-κB Signaling Pathways and Modulating Intestinal Microbiota. Eur. J. Nutr. 2024, 64, 21. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-N.; Han, B.; Zhang, M.-Q.; Chai, N.-N.; Yu, F.-L.; Qi, W.-H.; Tian, M.-Y.; Sun, D.-Z.; Huang, Y.; Song, Q.-X.; et al. Therapeutic Effects and Mechanisms of Isoxanthohumol on DSS-Induced Colitis: Regulating T Cell Development, Restoring Gut Microbiota, and Improving Metabolic Disorders. Inflammopharmacol 2024, 32, 1983–1998. [Google Scholar] [CrossRef]
- Cermak, P.; Olsovska, J.; Mikyska, A.; Dusek, M.; Kadleckova, Z.; Vanicek, J.; Nyc, O.; Sigler, K.; Bostikova, V.; Bostik, P. Strong Antimicrobial Activity of Xanthohumol and Other Derivatives from Hops (Humulus lupulus L.) on Gut Anaerobic Bacteria. APMIS 2017, 125, 1033–1038. [Google Scholar] [CrossRef]
- Paraiso, I.L.; Plagmann, L.S.; Yang, L.; Zielke, R.; Gombart, A.F.; Maier, C.S.; Sikora, A.E.; Blakemore, P.R.; Stevens, J.F. Reductive Metabolism of Xanthohumol and 8-Prenylnaringenin by the Intestinal Bacterium Eubacterium Ramulus. Mol. Nutr. Food Res. 2019, 63, 1800923. [Google Scholar] [CrossRef]
- Possemiers, S.; Rabot, S.; Espín, J.C.; Bruneau, A.; Philippe, C.; González-Sarrías, A.; Heyerick, A.; Tomás-Barberán, F.A.; De Keukeleire, D.; Verstraete, W. Eubacterium Limosum Activates Isoxanthohumol from Hops (Humulus lupulus L.) into the Potent Phytoestrogen 8-Prenylnaringenin In Vitro and in Rat Intestine. J. Nutr. 2008, 138, 1310–1316. [Google Scholar] [CrossRef]
- Molly, K.; Woestyne, M.V.; Smet, I.D.; Verstraete, W. Validation of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) Reactor Using Microorganism-Associated Activities. Microb. Ecol. Health Dis. 1994, 7, 191–200. [Google Scholar] [CrossRef]
- Van de Wiele, T.; Van den Abbeele, P.; Ossieur, W.; Possemiers, S.; Marzorati, M. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Springer: Cham, Switzerland, 2015; pp. 305–317. [Google Scholar]
- Jamieson, P.E.; Smart, E.B.; Bouranis, J.A.; Choi, J.; Danczak, R.E.; Wong, C.P.; Paraiso, I.L.; Maier, C.S.; Ho, E.; Sharpton, T.J.; et al. Gut Enterotype-Dependent Modulation of Gut Microbiota and Their Metabolism in Response to Xanthohumol Supplementation in Healthy Adults. Gut Microbes 2024, 16, 2315633. [Google Scholar] [CrossRef]
- Quinn, R.A.; Melnik, A.V.; Vrbanac, A.; Fu, T.; Patras, K.A.; Christy, M.P.; Bodai, Z.; Belda-Ferre, P.; Tripathi, A.; Chung, L.K.; et al. Global Chemical Effects of the Microbiome Include New Bile-Acid Conjugations. Nature 2020, 579, 123–129. [Google Scholar] [CrossRef]
- Schoefer, L.; Braune, A.; Blaut, M. Cloning and Expression of a Phloretin Hydrolase Gene from Eubacterium Ramulus and Characterization of the Recombinant Enzyme. Appl. Environ. Microbiol. 2004, 70, 6131–6137. [Google Scholar] [CrossRef]
- Zhuang, X.; Li, T.; Li, M.; Huang, S.; Qiu, Y.; Feng, R.; Zhang, S.; Chen, M.; Xiong, L.; Zeng, Z. Systematic Review and Meta-Analysis: Short-Chain Fatty Acid Characterization in Patients With Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2019, 25, 1751–1763. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.-K.; Lee, J.-Y.; Lim, S.-J.; Kim, M.-J.; Kim, G.-B.; Kim, J.-H.; Hong, S.-K.; Kang, D.-K. Molecular Cloning and Characterization of a Bile Salt Hydrolase from Lactobacillus Acidophilus PF01. J. Microbiol. Biotechnol. 2008, 18, 449–456. [Google Scholar] [PubMed]
- Chae, J.P.; Valeriano, V.D.; Kim, G.-B.; Kang, D.-K. Molecular Cloning, Characterization and Comparison of Bile Salt Hydrolases from Lactobacillus Johnsonii PF01. J. Appl. Microbiol. 2013, 114, 121–133. [Google Scholar] [CrossRef]
- Fiorucci, S.; Carino, A.; Baldoni, M.; Santucci, L.; Costanzi, E.; Graziosi, L.; Distrutti, E.; Biagioli, M. Bile Acid Signaling in Inflammatory Bowel Diseases. Dig. Dis. Sci. 2021, 66, 674–693. [Google Scholar] [CrossRef] [PubMed]
- Jiao, N.; Baker, S.S.; Chapa-Rodriguez, A.; Liu, W.; Nugent, C.A.; Tsompana, M.; Mastrandrea, L.; Buck, M.J.; Baker, R.D.; Genco, R.J.; et al. Suppressed Hepatic Bile Acid Signalling despite Elevated Production of Primary and Secondary Bile Acids in NAFLD. Gut 2018, 67, 1881–1891. [Google Scholar] [CrossRef]
- Jena, P.K.; Sheng, L.; Di Lucente, J.; Jin, L.-W.; Maezawa, I.; Wan, Y.-J.Y. Dysregulated Bile Acid Synthesis and Dysbiosis Are Implicated in Western Diet–Induced Systemic Inflammation, Microglial Activation, and Reduced Neuroplasticity. FASEB J. 2018, 32, 2866–2877. [Google Scholar] [CrossRef]
- Gadaleta, R.M.; Oldenburg, B.; Willemsen, E.C.L.; Spit, M.; Murzilli, S.; Salvatore, L.; Klomp, L.W.J.; Siersema, P.D.; van Erpecum, K.J.; van Mil, S.W.C. Activation of Bile Salt Nuclear Receptor FXR Is Repressed by Pro-Inflammatory Cytokines Activating NF-κB Signaling in the Intestine. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2011, 1812, 851–858. [Google Scholar] [CrossRef]
- Gadaleta, R.M.; van Erpecum, K.J.; Oldenburg, B.; Willemsen, E.C.L.; Renooij, W.; Murzilli, S.; Klomp, L.W.J.; Siersema, P.D.; Schipper, M.E.I.; Danese, S.; et al. Farnesoid X Receptor Activation Inhibits Inflammation and Preserves the Intestinal Barrier in Inflammatory Bowel Disease. Gut 2011, 60, 463–472. [Google Scholar] [CrossRef]
- D’Aldebert, E.; Biyeyeme Bi Mve, M.; Mergey, M.; Wendum, D.; Firrincieli, D.; Coilly, A.; Fouassier, L.; Corpechot, C.; Poupon, R.; Housset, C.; et al. Bile Salts Control the Antimicrobial Peptide Cathelicidin Through Nuclear Receptors in the Human Biliary Epithelium. Gastroenterology 2009, 136, 1435–1443. [Google Scholar] [CrossRef]
- Yersin, S.; Vonaesch, P. Small Intestinal Microbiota: From Taxonomic Composition to Metabolism. Trends Microbiol. 2024, 32, 970–983. [Google Scholar] [CrossRef] [PubMed]
- Masclee, A.; Tangerman, A.; Schaik, A.V.; Van Der Hoek, E.W.; Van Tongeren, J.H.M. Unconjugated Serum Bile Acids as a Marker of Small Intestinal Bacterial Overgrowth. Eur. J. Clin. Investig. 1989, 19, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Gong, Z.; Zhou, J.; Tian, C.; Gao, Y.; Xu, C.; Chen, Y.; Cai, W.; Wu, J. Deoxycholic Acid Triggers NLRP3 Inflammasome Activation and Aggravates DSS-Induced Colitis in Mice. Front. Immunol. 2016, 7, 536. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhou, J.; Wang, L.; Gong, Z.; Le, H.; Huang, Y.; Xu, C.; Tian, C.; Cai, W.; Wu, J. Gut Microbial Metabolite Deoxycholic Acid Facilitates Th17 Differentiation through Modulating Cholesterol Biosynthesis and Participates in High-Fat Diet-Associated Colonic Inflammation. Cell Biosci. 2023, 13, 186. [Google Scholar] [CrossRef]
- Medani, M.; Collins, D.; Docherty, N.G.; Baird, A.W.; O’Connell, P.R.; Winter, D.C. Emerging Role of Hydrogen Sulfide in Colonic Physiology and Pathophysiology. Inflamm. Bowel Dis. 2011, 17, 1620–1625. [Google Scholar] [CrossRef]
- Possemiers, S.; Verthé, K.; Uyttendaele, S.; Verstraete, W. PCR-DGGE-Based Quantification of Stability of the Microbial Community in a Simulator of the Human Intestinal Microbial Ecosystem. FEMS Microbiol. Ecol. 2004, 49, 495–507. [Google Scholar] [CrossRef]
- Han, J.; Lin, K.; Sequeira, C.; Borchers, C.H. An Isotope-Labeled Chemical Derivatization Method for the Quantitation of Short-Chain Fatty Acids in Human Feces by Liquid Chromatography–Tandem Mass Spectrometry. Anal. Chim. Acta 2015, 854, 86–94. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Amplicon, P.; Clean-Up, P.; Index, P. 16s Metagenomic Sequencing Library Preparation; Illumina: San Diego, CA, USA, 2013; p. 21. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- García-Jaramillo, M.; Beaver, L.M.; Truong, L.; Axton, E.R.; Keller, R.M.; Prater, M.C.; Magnusson, K.R.; Tanguay, R.L.; Stevens, J.F.; Hord, N.G. Nitrate and Nitrite Exposure Leads to Mild Anxiogenic-like Behavior and Alters Brain Metabolomic Profile in Zebrafish. PLoS ONE 2020, 15, e0240070. [Google Scholar] [CrossRef]
- Kirkwood, J.S.; Lebold, K.M.; Miranda, C.L.; Wright, C.L.; Miller, G.W.; Tanguay, R.L.; Barton, C.L.; Traber, M.G.; Stevens, J.F. Vitamin C Deficiency Activates the Purine Nucleotide Cycle in Zebrafish. J. Biol. Chem. 2012, 287, 3833–3841. [Google Scholar] [CrossRef] [PubMed]
- Bouranis, J.A.; Beaver, L.M.; Jiang, D.; Choi, J.; Wong, C.P.; Davis, E.W.; Williams, D.E.; Sharpton, T.J.; Stevens, J.F.; Ho, E. Interplay between Cruciferous Vegetables and the Gut Microbiome: A Multi-Omic Approach. Nutrients 2023, 15, 42. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Shi, J.; Gail, M.H.; Ravel, J.; Goedert, J.J. Assessment of the Human Faecal Microbiota: I. Measurement and Reproducibility of Selected Enzymatic Activities. Eur. J. Clin. Investig. 2018, 48, 848–854, Erratum in Eur. J. Clin. Investig. 2018, 48, e12881. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamieson, P.E.; Reichart, N.J.; Maier, C.S.; Sharpton, T.J.; Bradley, R.; Metz, T.O.; Stevens, J.F. Xanthohumol Alters Gut Microbiota Metabolism and Bile Acid Dynamics in Gastrointestinal Simulation Models of Eubiotic and Dysbiotic States. Int. J. Mol. Sci. 2025, 26, 10698. https://doi.org/10.3390/ijms262110698
Jamieson PE, Reichart NJ, Maier CS, Sharpton TJ, Bradley R, Metz TO, Stevens JF. Xanthohumol Alters Gut Microbiota Metabolism and Bile Acid Dynamics in Gastrointestinal Simulation Models of Eubiotic and Dysbiotic States. International Journal of Molecular Sciences. 2025; 26(21):10698. https://doi.org/10.3390/ijms262110698
Chicago/Turabian StyleJamieson, Paige E., Nicholas J. Reichart, Claudia S. Maier, Thomas J. Sharpton, Ryan Bradley, Thomas O. Metz, and Jan F. Stevens. 2025. "Xanthohumol Alters Gut Microbiota Metabolism and Bile Acid Dynamics in Gastrointestinal Simulation Models of Eubiotic and Dysbiotic States" International Journal of Molecular Sciences 26, no. 21: 10698. https://doi.org/10.3390/ijms262110698
APA StyleJamieson, P. E., Reichart, N. J., Maier, C. S., Sharpton, T. J., Bradley, R., Metz, T. O., & Stevens, J. F. (2025). Xanthohumol Alters Gut Microbiota Metabolism and Bile Acid Dynamics in Gastrointestinal Simulation Models of Eubiotic and Dysbiotic States. International Journal of Molecular Sciences, 26(21), 10698. https://doi.org/10.3390/ijms262110698







