BTN2A1 and BTN3A1 as Novel Coeliac Disease Risk Loci: An In Silico Analysis
Abstract
1. Introduction
1.1. Background to Coeliac Disease
1.2. The Emerging Role of the Butyrophilin Family of Genes and Their Role in Maintaining γδ T Cells
1.3. A Hypothesis for the Role of Butyrophilin Variation and γδ T Cells in CeD Risk
2. Results
2.1. BTN2A1 SNPs Were Significantly Associated with CeD Risk in a Study of 94 Samples
2.1.1. Risk-Associated HLA Genotypes Were Significantly More Frequent in CeD Patients
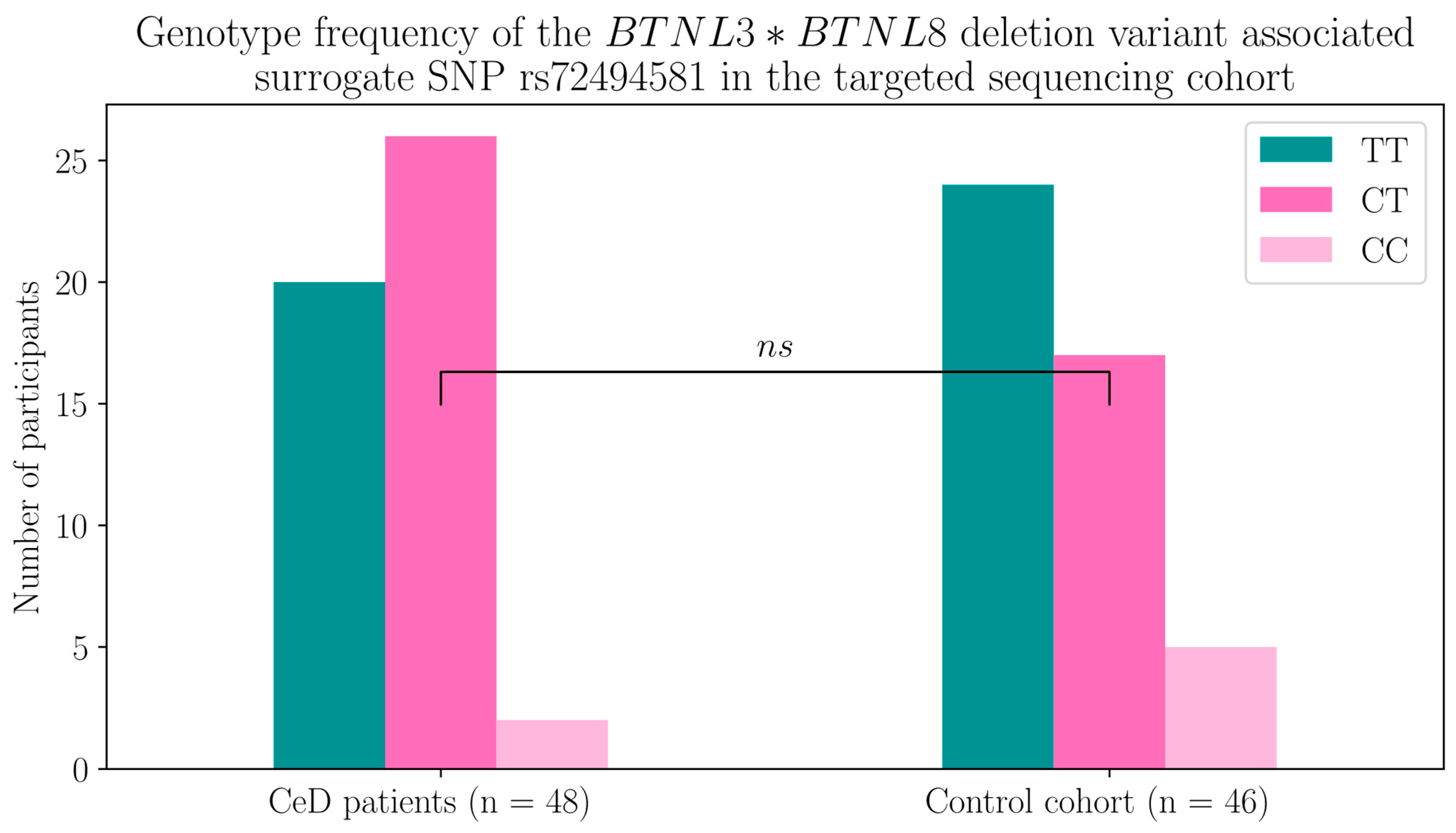
2.1.2. The BTNL8*BTNL3 Copy Number Variant Was Not Associated with CeD
2.1.3. BTN2A1 Gene Burden Was Significantly Higher in CeD Patients
2.2. BTN3A1, BTN3A2, and BTN2A1 Genes Were Significantly Associated with CeD in HLA-DQ2.5-Matched Participants of the UK Biobank Database
2.2.1. Risk-Associated HLA Genotypes Were Significantly More Frequent in CeD Patients of the UK Biobank
2.2.2. BTN2A1, BTN3A1, and BTN3A2 SNPs Were Significantly Associated with CeD Status in the UK Biobank
2.2.3. Twenty Butyrophilin SNPs from the UK Biobank Remained Significantly Associated with CeD Status When the Participants’ Risk HLA Genotypes Were Taken into Account
2.2.4. Twenty-One Butyrophilin SNPs Were Significantly Associated with CeD Status in HLA-DQ2.5-Matched Case-Control Groups of UK Biobank Participants
2.3. HV4 Variation Was Not Significantly Associated with CeD Risk in a Study of 379 Samples
2.3.1. TRGV Usage Was Not Significantly Different Between CeD and Control Samples
2.3.2. HV4 Sequence Variation Was Not Significantly Associated with CeD Risk
3. Discussion
4. Materials and Methods
4.1. Participant Selection Criteria
- Has CeD diagnosis;
- Malabsorption;
- Anaemia;
- Lymphocytosis;
- On a GFD;
- Diarrhoea.
4.1.1. Participant Selection for the Butyrophilin Family Gene Sequencing
4.1.2. Validation Cohort Participant Selection from the UK Biobank for Single-Variant Analysis
- Hospital diagnosis record includes coeliac disease: ICD9 (5790), ICD10 (K90.0);
- Cause of death includes coeliac disease: ICD10 (K90.0).
4.1.3. Samples Selected for the HV4 Analysis
4.2. Analysis of Butyrophilin Family Variation in the Targeted Sequencing Cohort
4.2.1. Sequencing of HLA Loci and Selected Butyrophilin Family Genes by Hybridisation Capture
4.2.2. Germline Short-Variant Discovery and HLA Genotyping
4.2.3. Copy Number Variation (CNV) Analysis of the BTNL8-BTNL3 Loci
4.2.4. Burden Testing Analysis
4.3. Single-Variant Testing of Butyrophilin Family Variance in the UK Biobank Database
4.3.1. CeD Risk-Associated HLA Genotyping in the UK Biobank Cohort
4.3.2. Single-Variant Testing Using Binomial Regression Models
4.4. Analysis of TRGV Usage and HV4 Variation in CeD and Control Samples
4.4.1. Processing Samples and TCR Sequencing
4.4.2. TRGV and HV4 Analysis Pipeline
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BTN/BTNL | Butyrophilin/butyrophilin-like |
| CeD | Coeliac disease |
| CNV | Copy number variation |
| DC | Dendritic cell |
| GFD | Gluten-free diet |
| FFPE | Formalin-fixed, paraffin-embedded |
| HLA | Human leukocyte antigen |
| HPA | Human Protein Atlas |
| HV4 | Hypervariable region 4 |
| IEL | Intraepithelial lymphocyte |
| NK cell | Natural Killer cell |
| TRGV | T cell receptor γ variable region |
| SNP/SNV | Single-nucleotide polymorphism/variation |
| WT | Wild-type |
Appendix A. The Molecular Background of the 56 kb BTNL3*BTNL8 Deletion Variant
| Population | Hom. for Deletion | Het. for Deletion | Hom. for Full Sequences | Deletion Allele N | Deletion Allele Frequency | Group N | Carriers N | Carriers % | |
|---|---|---|---|---|---|---|---|---|---|
| HapMap | CEU | 17 | 56 | 68 | 90 | 0.319 | 141 | 73 | 51.8 |
| Toskani, Italia | 9 | 45 | 34 | 63 | 0.358 | 88 | 54 | 61.4 | |
| HGDP | France | 7 | 28 | 17 | 42 | 0.404 | 52 | 35 | 67.3 |
| Italy | 5 | 18 | 13 | 28 | 0.389 | 36 | 23 | 63.9 | |
| Italy (Bergamo) | 3 | 2 | 9 | 8 | 0.286 | 14 | 5 | 35.7 | |
| Orkney Islands | 1 | 11 | 3 | 13 | 0.433 | 15 | 12 | 80.0 | |
| Total | European ancestry | 42 | 160 | 144 | 244 | 0.353 | 346 | 202 | 58.4 |
Appendix B. Selecting and Sequencing the Butyrophilin Genes of Interest
Appendix B.1. HPA Expression Profiles of Butyrophilin Family Genes
| (a) Butyrophilin family expression in the duodenum | ||||||
| Reliability as Defined by the HPA | Protein Expression in Duodenum (IHC) *If RNA Data Only* | Comment | Included? | |||
| ERMAP | Uncertain | High | Uncertain Tissue Atlas reliability score High expression in digestive tissues Low–medium expression in immune cells | Yes | ||
| MOG | Enhanced | None | Not expressed in immune cells or digestive tissues Genomic variance control for the significance of butyrophilin variation in CeD risk | Yes | ||
| BTN1A1 | Supported | None | Not expressed in digestive tissues or immune cells | No | ||
| BTN2A1 | Approved | Medium | Included due to reliability score and expression Implicated in the stimulation of Vγ9Vδ2+ T cells [37] | Yes | ||
| BTN2A2 | Uncertain | High | Uncertain Tissue Atlas reliability score Medium–high expression in digestive tissues Expressed in immune cells | Yes | ||
| BTN3A1 | Approved | Medium | Included due to reliability score and expression Implicated in the stimulation of Vγ9Vδ2+ T cells [30] | Yes | ||
| BTN3A2 | Uncertain | Medium | Uncertain Tissue Atlas reliability score Implicated in the stimulation of Vγ9Vδ2+ T cells [27] | Yes | ||
| BTN3A3 | Enhanced | Medium | Included due to reliability score and expression | Yes | ||
| BTNL2 | Pending | *None* | Pending Tissue Atlas reliability score HLA-independent significant association with CeD [19] | Yes | ||
| BTNL3 | Pending | *High* | Pending Tissue Atlas reliability score No protein expression data available for the intestinal tissues Previously documented role in CeD [21] | Yes | ||
| BTNL8 | Enhanced | Medium | Previously documented role in CeD [21] | Yes | ||
| BTNL9 | Pending | *Low* | Pending Tissue Atlas reliability score No protein expression data available for the intestinal tissues Not expressed in immune cells | No | ||
| BTNL10 | NA | NA | No entry | No | ||
| SKINT1L | NA | NA | No entry | No | ||
| BTN2A3P | NA | NA | No entry | No | ||
| (b) Expression of selected butyrophilin family members in the small intestine, colon, and rectum | ||||||
| Tissue Expression (IHC) *If RNA Data Only* | Included in the Panel? | |||||
| Reliability as Defined by the HPA | Small Intestine (Glandular) | Duodenum (Glandular) | Rectum (Glandular) | Colo (Glandular) | ||
| ERMAP | Uncertain | High | High | High | High | Yes |
| MOG | Enhanced | none | none | none | none | Yes |
| BTN2A1 | Approved | Low | Medium | Medium | Medium | Yes |
| BTN2A2 | Uncertain | High | High | Medium | Medium | Yes |
| BTN3A1 | Approved | High | Medium | High | High | Yes |
| BTN3A2 | Uncertain | High | Medium | Medium | Medium | Yes |
| BTN3A3 | Enhanced | Medium | Medium | Medium | Medium | Yes |
| BTNL2 | Pending | *Very low* | none | none | none | Yes |
| BTNL3 | Pending | *High* | *High* | *High* | *High* | Yes |
| BTNL8 | Enhanced | Medium | Medium | none | none | Yes |
| Immune Cell Expression (RNA Sequencing) | Included in the Panel? | |||||||
|---|---|---|---|---|---|---|---|---|
| Reliability as Defined by the HPA | γδ T Cells | T Cells | T-Reg | DCs | Macrophages | NK Cells | ||
| ERMAP | Uncertain | Low | Medium | Medium | Medium | Medium | Low | Yes |
| MOG | Enhanced | Very low | None | Very low | None | None | None | Yes |
| BTN2A1 | Approved | Medium | Medium | Medium | Medium | High | Low | Yes |
| BTN2A2 | Uncertain | Low | Medium | Medium | High | High | Medium | Yes |
| BTN3A1 | Approved | High | High | High | Low | Medium | High | Yes |
| BTN3A2 | Uncertain | High | High | High | Medium | High | High | Yes |
| BTN3A3 | Enhanced | High | High | High | Medium | Medium | High | Yes |
| BTNL2 | Pending | None | None | None | None | None | None | Yes |
| BTNL3 | Pending | None | None | None | None | None | None | Yes |
| BTNL8 | Enhanced | None | None | None | None | None | None | Yes |
| Gene of Interest | Location (GRCh38.p12) |
|---|---|
| BTN2A1 | chr6:26,457,955–26,476,622 |
| BTN2A2 | chr6:26,382,893–26,394,874 |
| BTN3A1 | chr6:26,402,269–26,415,216 |
| BTN3A2 | chr6:26,365,170–26,378,320 |
| BTN3A3 | chr6:26,440,504–26,453,415 |
| BTNL2 | chr6:32,393,339–32,408,879 |
| BTNL3 | chr5:180,988,846–181,006,727 |
| BTNL8 | chr5:180,899,097–180,952,166 |
| ERMAP | chr1:42,817,122–42,844,991 |
| MOG | chr6:29,657,092–29,672,365 |
Appendix B.2. Modified Nonacus Cell3 Hybridisation Capture and Illumina Sequencing
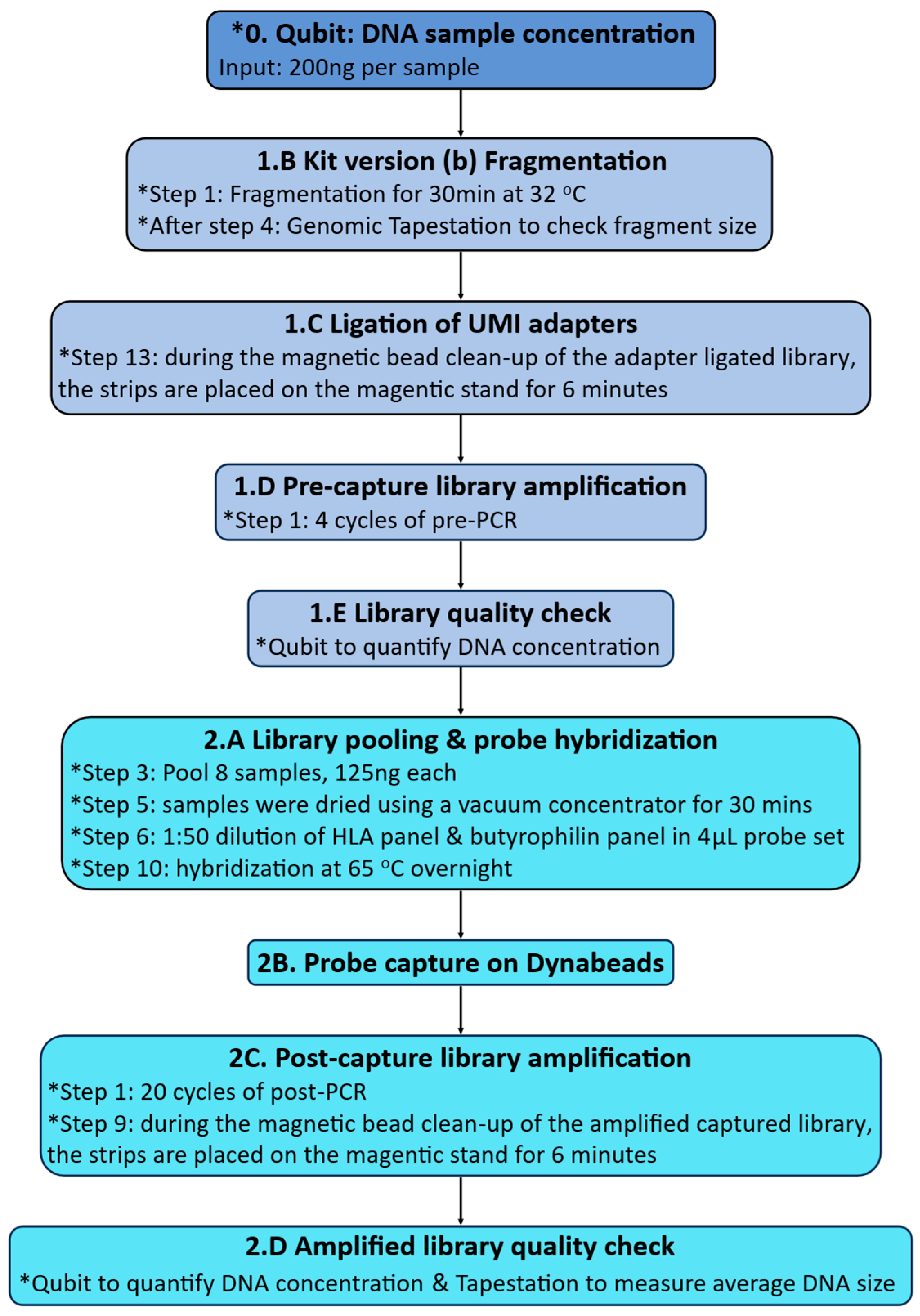
Appendix B.3. Measuring DNA Quantity and Fragment Size
Appendix C. Detailed Germline Short-Variant Discovery Protocol
Appendix C.1. Per Sample Preprocesses and Variant Call Using GATK

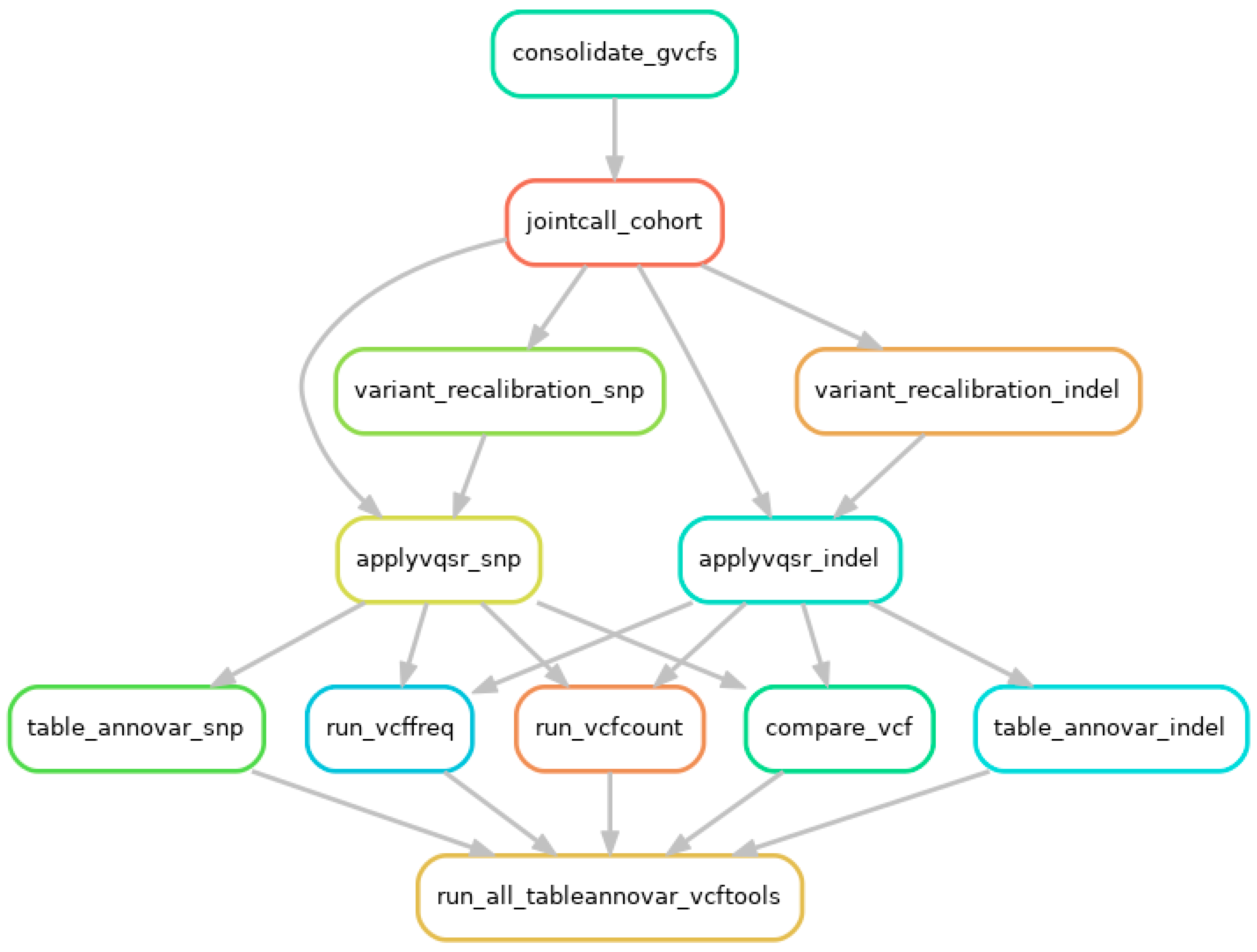
Appendix C.2. Joint Genotyping Using GATK and Variant Annotation Using VCFtools and ANNOVAR
Appendix D. Detailed CNV Analysis and Burden Testing Protocols
Appendix D.1. BTNL8*BTNL3 CNV Analysis
| rs72494581 Genotypes | Associated CNV at BTNL8-BTNL3 Region of Chromosome 5 | |
|---|---|---|
| TT | Homozygous for reference allele | Full-length BTNL8-BTNL3 region on both copies of chromosome 5 |
| CT | Heterozygous | One copy has full-length BTNL8-BTNL3 region One copy has BTNL8*BTNL3 deletion |
| CC | Homozygous for alternative allele | BTNL8*BTNL3 deletion on both copies of chromosome 5 |
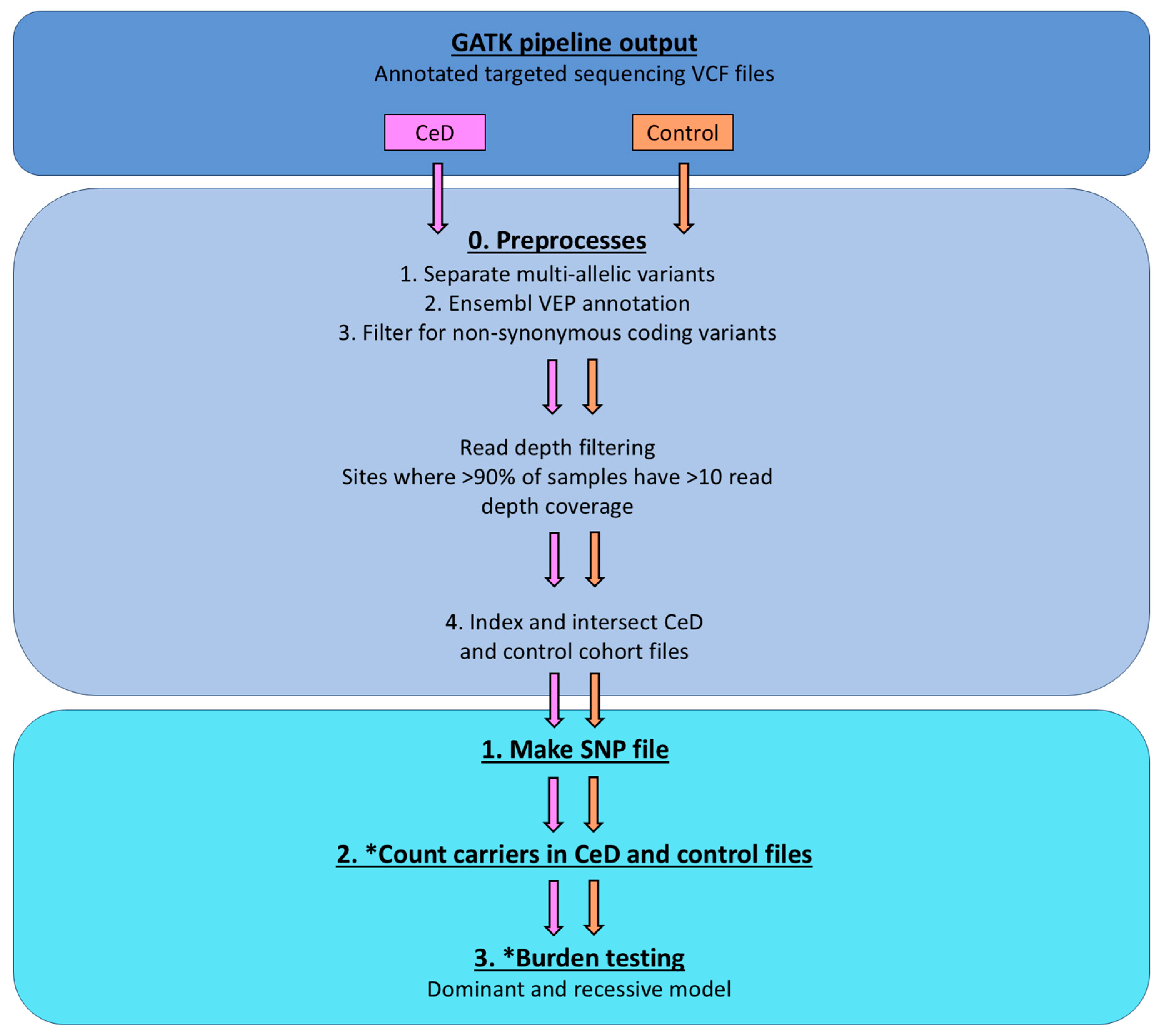
Appendix D.2. Detailed Burden Testing Protocol
Appendix E. Detailed Protocol Single-Variant Testing Analysis of Selected Butyrophilin SNPs in the UK Biobank
Appendix E.1. HLA Genotyping in the UK Biobank Using the HLA Imputation Data
Appendix E.2. Detailed Single-Variant Testing of BTN2A1, BTN3A1, and BTN3A2 SNPs in the UK Biobank
| Test/Group Number | Association Being Tested | Predictor Variable(s) | Response Variable |
|---|---|---|---|
| First test | Association between HLA risk genotypes and CeD risk | HLA risk genotype | CeD status: CeD or control |
| Second group of tests (101 models) | Association between individual butyrophilin SNPs and CeD risk | Butyrophilin SNP: 2 reference alleles 1 reference allele 0 reference allele | CeD status: CeD or control |
| Third group of tests (101 models) | Association between the combined effect of HLA genotypes and butyrophilin SNPs and CeD risk | HLA risk genotype Butyrophilin SNP: 2 reference alleles 1 reference allele 0 reference allele | CeD status: CeD or control |
| Fourth group of tests | Association between butyrophilin SNPs and CeD risk in HLA-matched groups | Butyrophilin SNP: 2 reference alleles 1 reference allele 0 reference allele | CeD status: CeD or control |
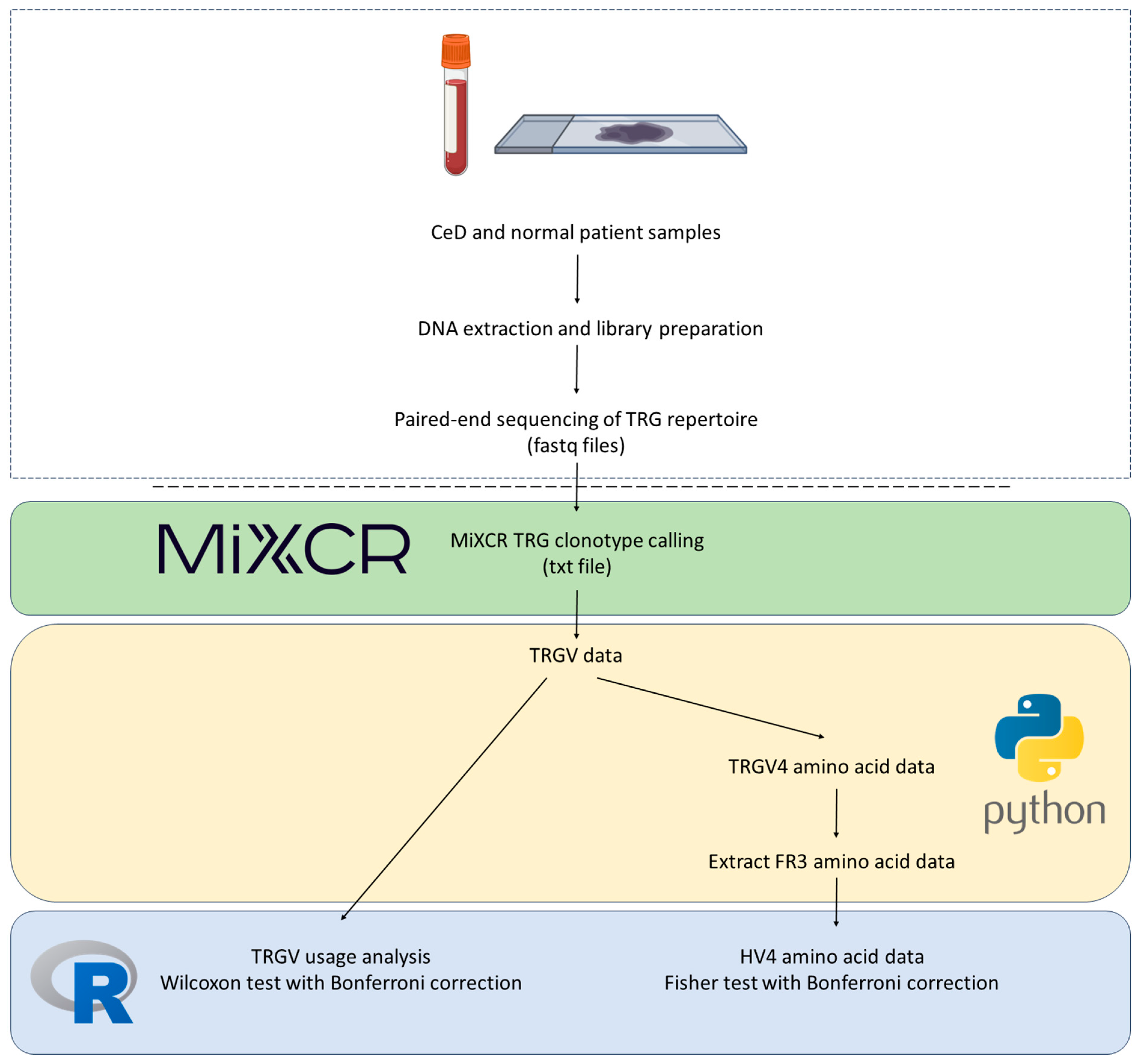
Appendix F. Detailed TRGV4 Usage and HV4 Amino Acid Sequence Analysis Pipeline
Appendix G. Supplementary Materials for Results Section 2.1

| (a) | |||||||
| HLA Genotypes | HLA-DQ2.5 | HLA-DQ8 | HLA-DQ2.2 | HLA-DQ2.5 and DQ8 | HLA-DQ2.5 and DQ2.2 | HLA-DQ8 and DQ2.2 | Other |
| CeD patients | 22 | 3 | 2 | 6 | 13 | 0 | 2 |
| Control participants | 10 | 2 | 6 | 0 | 1 | 2 | 25 |
| (b) | |||||||
| Sample | HLA-DQA1 Allele | HLA-DQB1 Allele | Potential HLA-DQ Type | ||||
| CB26 | HLA-DQA1*01:01:01, HLA-DQA1*02:01 | HLA-DQB1*02:01:01, HLA-DQB1*05:01:01 | HLA-DQ5.1 | ||||
| CD1 | Not typed | HLA-DQB1*02:01:01, HLA-DQB1*06:02:01 | Unknown | ||||
| rs72494581 Genotype | TT | CT | CC |
|---|---|---|---|
| BTNL8-BTNL3 genes | Homozygous for full-length sequence | Heterozygous for BTNL8*BTNL3 deletion | Homozygous for BTNL8*BTNL3 deletion |
| Coeliac disease patients | 20 | 26 | 2 |
| Control participants | 24 | 17 | 5 |
| (a) | ||||
| Number of Butyrophilin Family Sites | Sites Unique to Group | Shared Sites | Non-Matching Overlapping Sites | |
| Coeliac | 1168 | 701 | 435 | 32 |
| Control | 769 | 302 | ||
| (b) | ||||
| Number of Butyrophilin Family non-Synonymous Coding Sites | Sites Unique to Group | Shared Sites | ||
| Coeliac | 108 | 79 | 21 | |
| Control | 58 | 29 | ||
| (c) | ||||
| Number of Butyrophilin Family Non-Synonymous Coding Sites (>90% Samples in Group dp > 10) | Sites Unique to Group | Shared Sites | ||
| Coeliac | 60 | 54 | 6 | |
| Control | 21 | 15 | ||
| Gene | Qual. SNPs | CeD %(≥1 HET) | CeD %(≥2 HET) | CeD %(HOM ALT) | CeD Total Qual. allele Freq | Control %(≥1 HET) | Control %(≥2 HET) | Control %(HOM ALT) | Control Total Qual. Allele Freq | Dominant Model p-Value | Recessive Model p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BTN2A1 | 3 | 45.8 | 43.8 | 6.3 | 0.281 | 10.9 | 8.7 | 0.0 | 0.047 | 1.46 × 10−5 | 3.70 × 10−8 |
| BTN3A2 | 1 | 10.4 | 0.0 | 2.1 | 0.073 | 19.6 | 0.0 | 2.2 | 0.120 | 0.929 | 0.946 |
| ERMAP | 1 | 43.8 | 0.0 | 16.7 | 0.385 | 43.5 | 0.0 | 15.2 | 0.370 | 0.516 | 0.988 |
| (a) CeD participants | |||
| Position | 6:26463432 | 6:26468098 | 6:26468317 |
| rsID | rs13195509 | rs3734542 | rs3734543 |
| CB22 | hom alt | hom alt | hom alt |
| CB24 | hom alt | hom alt | hom alt |
| CB26 | het | het | het |
| CB27 | het | het | het |
| CB29 | het | het | het |
| CB31 | het | het | het |
| CB32 | hom ref | hom ref | hom ref |
| CB33 | hom ref | hom ref | hom ref |
| CB34 | het | het | het |
| CB35 | het | het | het |
| CB36 | het | het | het |
| CB37 | het | het | het |
| CB38 | het | het | het |
| CB39 | hom ref | hom ref | hom ref |
| CB40 | hom ref | hom ref | hom ref |
| CB41 | hom ref | hom ref | hom ref |
| CB43 | hom ref | hom ref | hom ref |
| CB44 | hom ref | hom ref | hom ref |
| CB45 | het | het | het |
| CB46 | hom ref | hom ref | hom ref |
| CB48 | hom ref | hom ref | hom ref |
| CB49 | hom ref | hom ref | hom ref |
| CB50 | hom ref | hom ref | hom ref |
| CB51 | het | het | het |
| CB52 | hom ref | hom ref | hom ref |
| CB53 | het | het | het |
| CB54 | het | het | het |
| CB55 | het | het | het |
| CB56 | hom ref | hom ref | hom ref |
| CB57 | hom ref | hom ref | hom ref |
| CB58 | het | het | het |
| CB59 | hom ref | hom ref | hom ref |
| CB60 | hom ref | hom ref | hom ref |
| CB61 | het | het | het |
| CB62 | het | het | het |
| CB63 | hom alt | hom alt | hom alt |
| CB65 | hom ref | hom ref | hom ref |
| CB67 | het | het | het |
| CB69 | het | het | het |
| CB70 | hom ref | hom ref | hom ref |
| CD1 | hom ref | het | hom ref |
| CD2 | hom ref | hom ref | hom ref |
| CD3 | hom ref | hom ref | hom ref |
| CD4 | het | het | het |
| CD5 | het | het | het |
| CD6 | hom ref | hom ref | hom ref |
| CD7 | hom ref | hom ref | hom ref |
| CD8 | hom ref | hom ref | hom ref |
| (b) Control participants | |||
| Position | 6:26463432 | 6:26468098 | 6:26468317 |
| rsID | rs13195509 | rs3734542 | rs3734543 |
| NB11 | hom ref | hom ref | hom ref |
| NB13 | hom ref | hom ref | hom ref |
| NB14 | hom ref | hom ref | hom ref |
| NB16 | hom ref | hom ref | hom ref |
| NB19 | hom ref | hom ref | hom ref |
| NB2 | hom ref | hom ref | hom ref |
| NB20 | hom ref | hom ref | hom ref |
| NB21 | het | het | het |
| NB22 | hom ref | hom ref | hom ref |
| NB25 | hom ref | hom ref | hom ref |
| NB28 | het | het | het |
| NB29 | hom ref | hom ref | hom ref |
| NB3 | hom ref | hom ref | hom ref |
| NB30 | hom ref | hom ref | hom ref |
| NB31 | hom ref | hom ref | hom ref |
| NB32 | hom ref | hom ref | hom ref |
| NB34 | hom ref | hom ref | hom ref |
| NB35 | hom ref | hom ref | hom ref |
| NB37 | hom ref | hom ref | hom ref |
| NB38 | hom ref | hom ref | hom ref |
| NB39 | hom ref | hom ref | hom ref |
| NB41 | hom ref | hom ref | hom ref |
| NB42 | hom ref | hom ref | hom ref |
| NB44 | hom ref | hom ref | hom ref |
| NB46 | hom ref | hom ref | hom ref |
| NB47 | hom ref | hom ref | hom ref |
| NB48 | hom ref | hom ref | hom ref |
| NB49 | hom ref | hom ref | hom ref |
| NB5 | hom ref | hom ref | hom ref |
| NB50 | het | het | het |
| NB52 | hom ref | hom ref | hom ref |
| NB56 | hom ref | hom ref | hom ref |
| NB58 | hom ref | hom ref | hom ref |
| NB68 | hom ref | hom ref | hom ref |
| NB69 | hom ref | hom ref | hom ref |
| NB7 | hom ref | hom ref | hom ref |
| NB70 | hom ref | hom ref | hom ref |
| NB71 | hom ref | hom ref | hom ref |
| ND1 | hom ref | hom ref | hom ref |
| ND10 | hom ref | hom ref | hom ref |
| ND2 | hom ref | hom ref | hom ref |
| ND5 | het | het | het |
| ND6 | hom ref | hom ref | hom ref |
| ND7 | hom ref | hom ref | hom ref |
| ND8 | hom ref | hom ref | hom ref |
| ND9 | hom ref | het | hom ref |
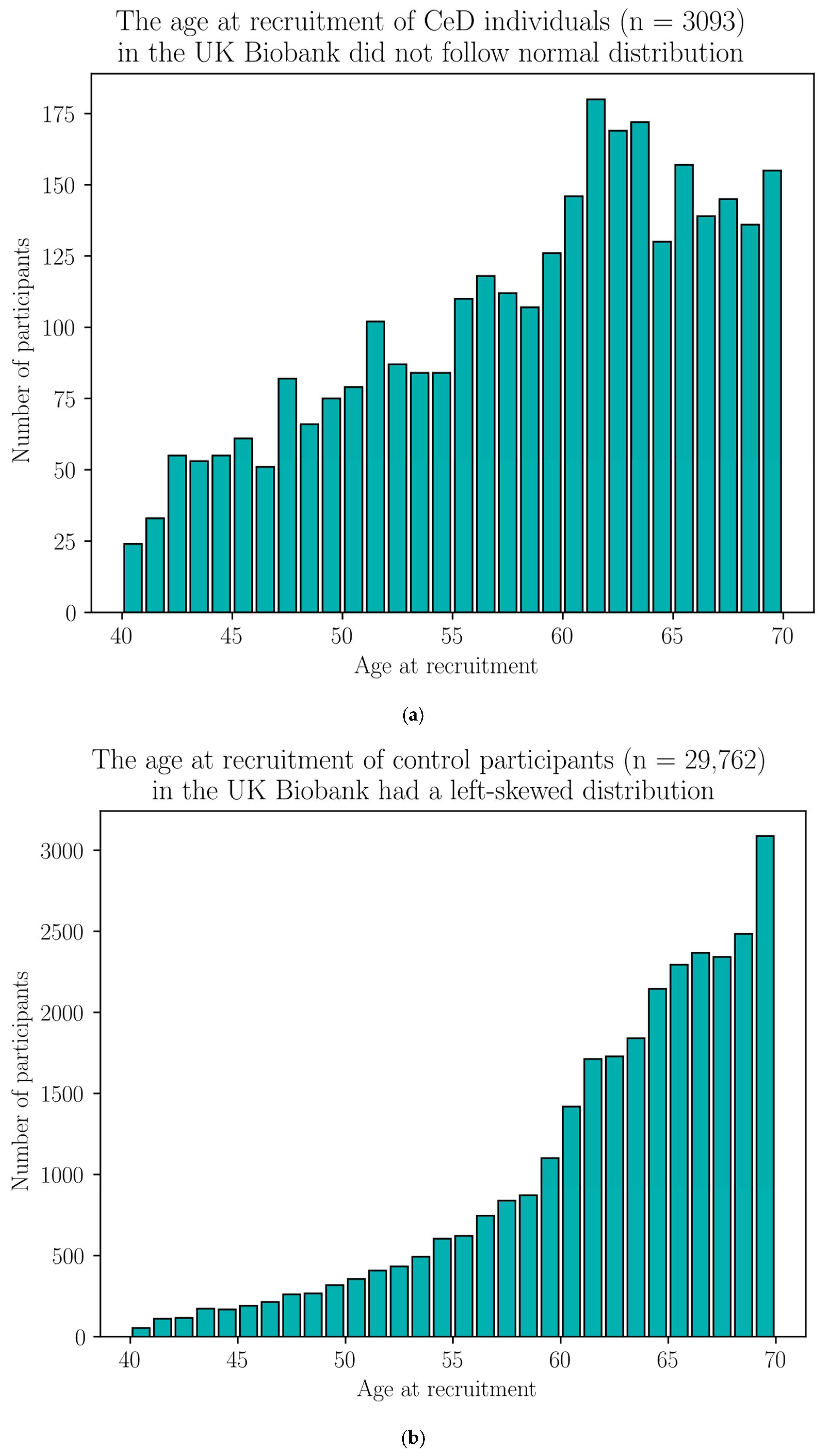
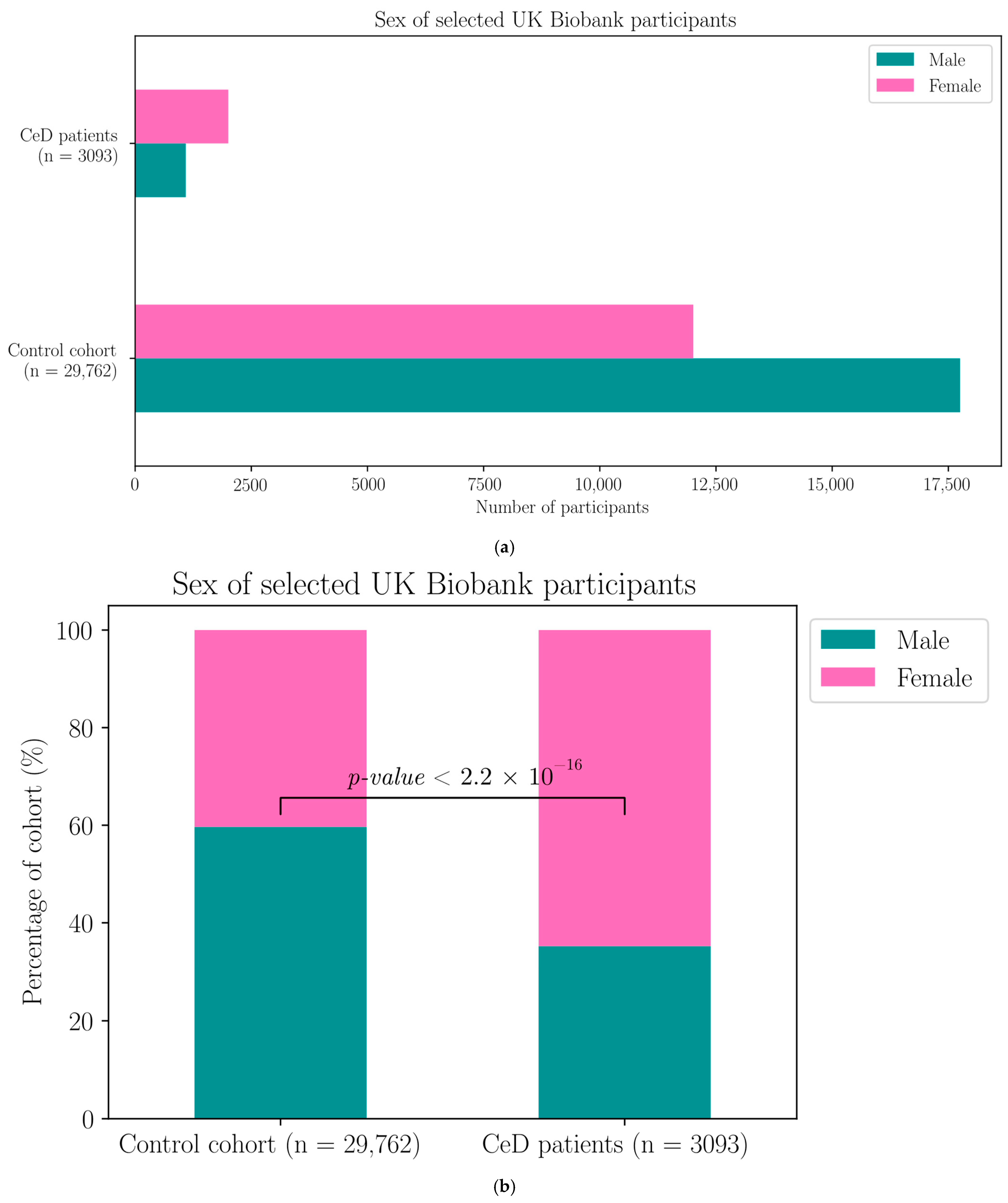
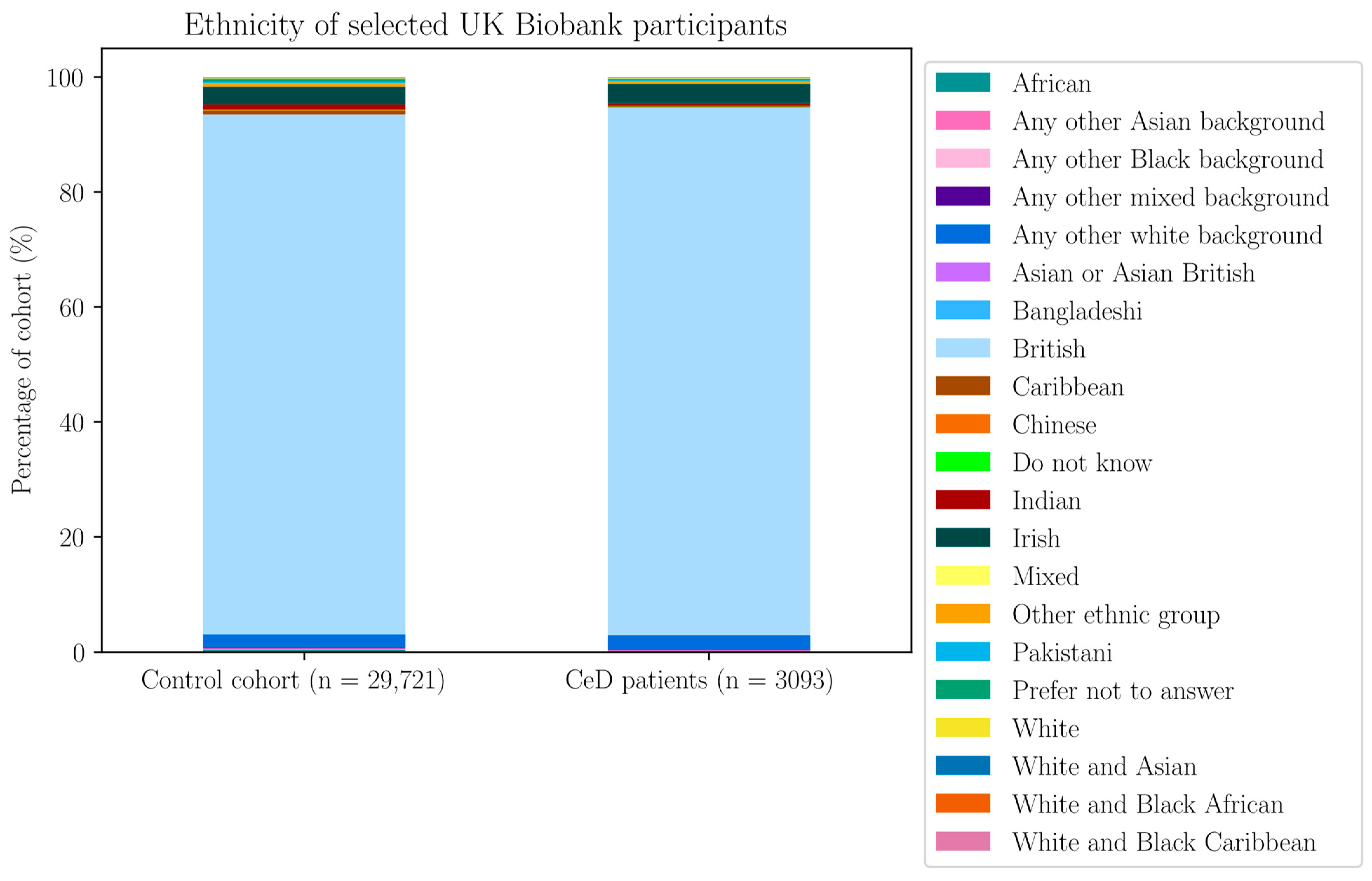
Appendix H. Demographic Data of the Selected UK Biobank Participants

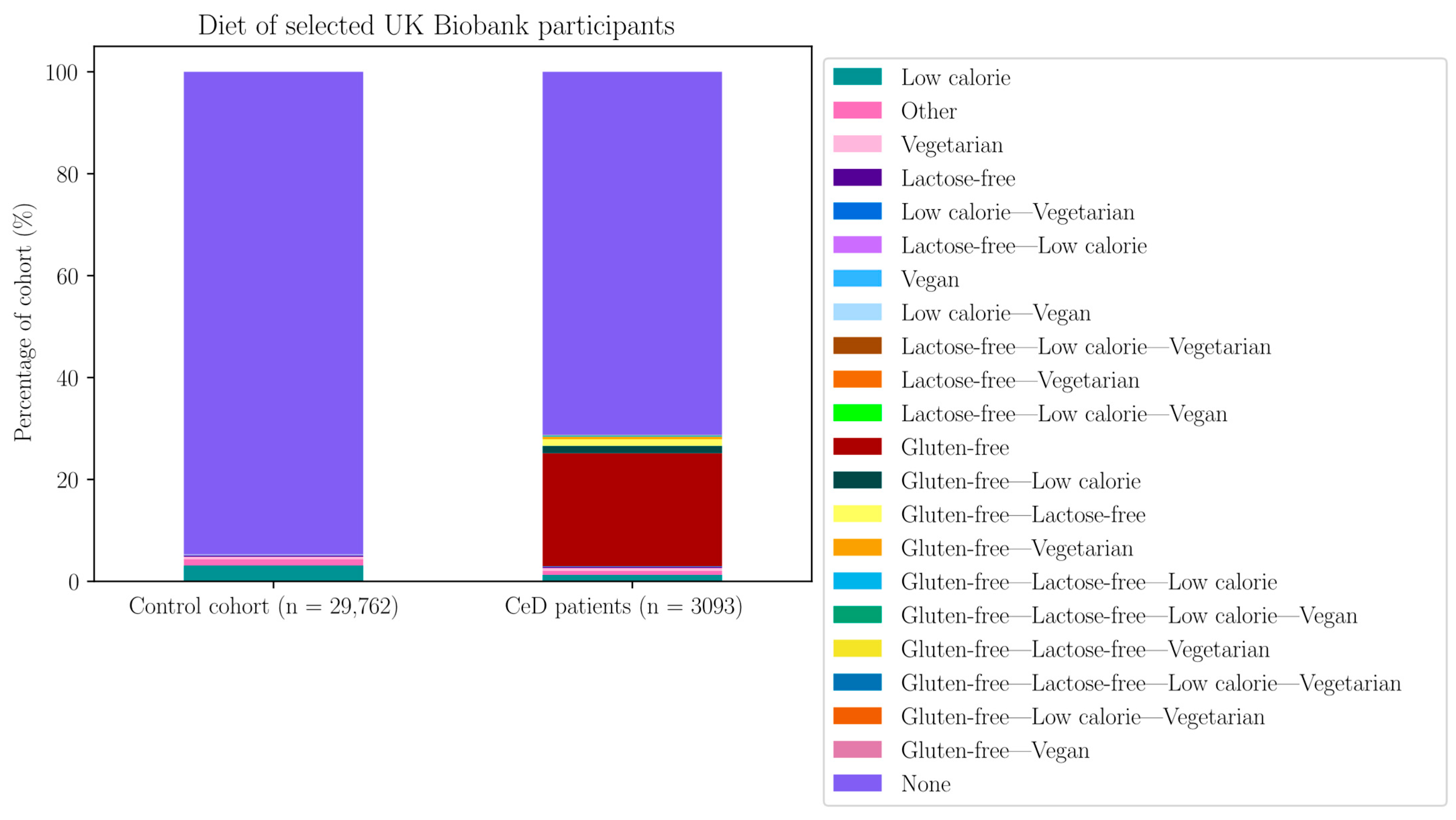
| Without GFD | With GFD | |||||
|---|---|---|---|---|---|---|
| Diet | N Controls on Diet | % Controls on Diet | N CeD on Diet | % CeD on Diet (Out of 2296) | N CeD on Diet | % CeD on Diet (Out of 3094) |
| Lactose-free–low calorie | 20 | 0.067 | 0 | 0 | 5 | 0.162 |
| Vegan | 15 | 0.050 | 0 | 0 | 1 | 0.032 |
| Lactose-free–low calorie–vegetarian | 2 | 0.007 | 0 | 0 | 1 | 0.032 |
| Lactose-free–vegetarian | 2 | 0.007 | 0 | 0 | 1 | 0.032 |
| Lactose-free–low calorie–vegan | 1 | 0.003 | 0 | 0 | 2 | 0.065 |
Appendix I. Supplementary Materials for Results Section 2.2
| HLA Genotypes | HLA-DQ2.5 | HLA-DQ8 | HLA-DQ2.2 | HLA-DQ2.5 and DQ8 | HLA-DQ2.5 and DQ2.2 | HLA-DQ8 and DQ2.2 | Other |
|---|---|---|---|---|---|---|---|
| CeD participants | 1652 | 171 | 199 | 606 | 182 | 50 | 234 |
| Control participants | 6416 | 4203 | 4154 | 895 | 886 | 590 | 12,618 |
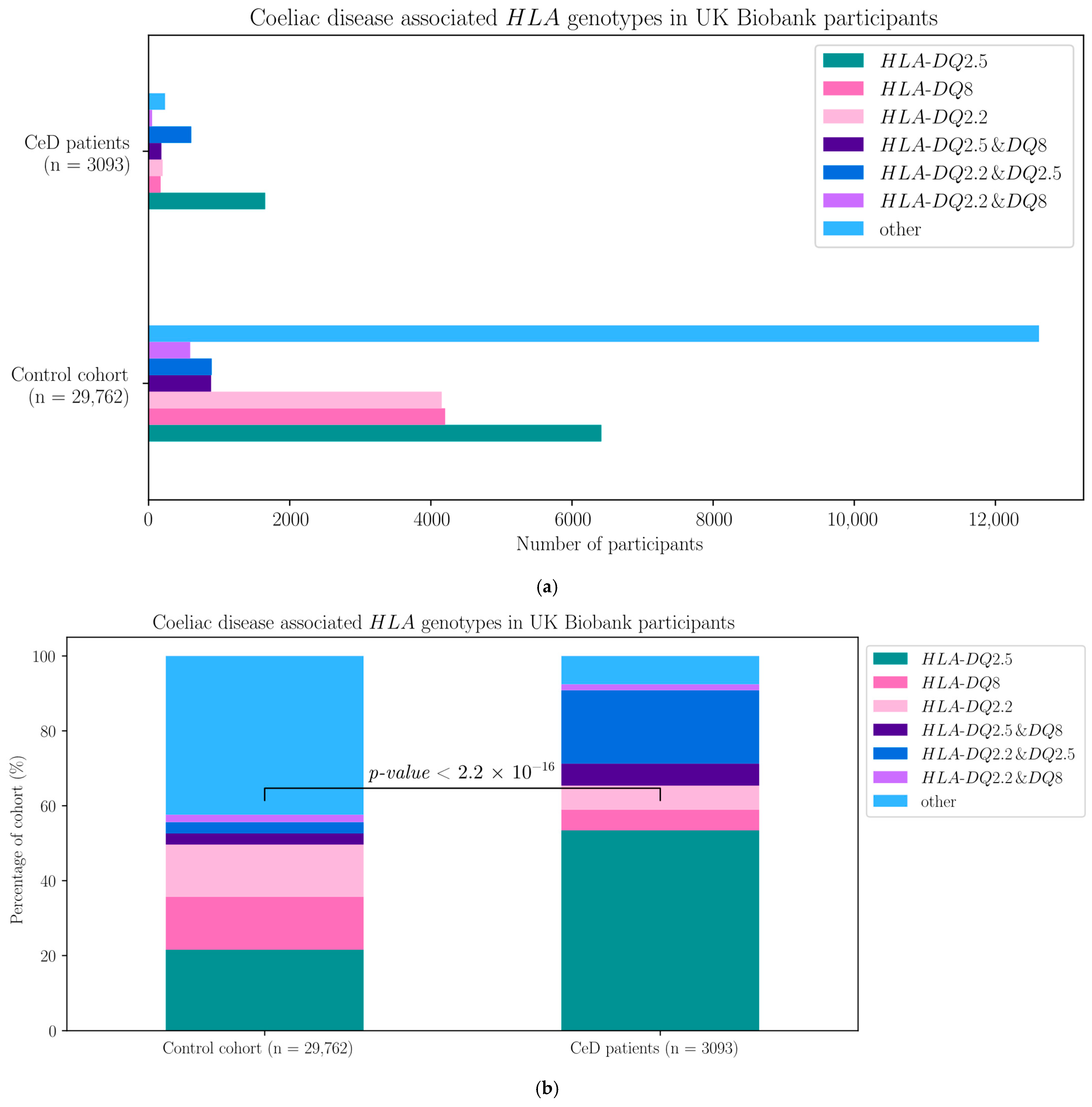
| HLA Genotype | Coefficient Estimate | Standard Error | z Value | p-Value | CeD Risk |
|---|---|---|---|---|---|
| HLA-DQ2.2, HLA-DQ2.5 | 2.649 | 0.090 | 29.550 | <2 × 10−16 | Increase |
| HLA-DQ2.2, HLA-DQ8 | 0.570 | 0.164 | 3.474 | 5.13 × 10−4 | Increase |
| HLA-DQ2.5 | 1.682 | 0.078 | 21.662 | <2 × 10−16 | Increase |
| HLA-DQ2.5, HLA-DQ8 | 1.456 | 0.109 | 13.352 | <2 × 10−16 | Increase |
| HLA-DQ8 | −0.163 | 0.107 | −1.533 | 0.125 | ns |
| Other HLA genotype | −0.949 | 0.098 | −9.677 | <2 × 10−16 | Decrease |
| Constant | −3.039 | 0.073 | −41.872 | <2 × 10−16 | NA |
| SNP Name | Gene | OR | Upper | Lower | Adjusted p-Value | ln(OR) |
|---|---|---|---|---|---|---|
| rs10484441_G | BTN2A1 | 1.138386 | 1.249955 | 1.038835 | 0.60789 | 0.129612 |
| rs12660069_C | BTN2A1 | 1.106915 | 1.299592 | 0.948994 | 1 | 0.101577 |
| rs13195402_G | BTN2A1 | 0.397002 | 0.424644 | 0.371261 | 4.67 × 10−158 | −0.92381 |
| rs13195509_G | BTN2A1 | 0.424358 | 0.45231 | 0.398239 | 1.61 × 10−151 | −0.85718 |
| rs13437351_G | BTN2A1 | 1.485538 | 1.91326 | 1.176198 | 0.14151 | 0.395777 |
| rs1407045_A | BTN2A1 | 1.314112 | 1.385827 | 1.246309 | 6.07 × 10−22 | 0.273161 |
| rs142951857_A | BTN2A1 | 1.391065 | 3.107641 | 0.723169 | 1 | 0.33007 |
| rs143104579_G | BTN2A1 | 1.14068 | 1.405753 | 0.935552 | 1 | 0.131625 |
| rs146399224_T | BTN2A1 | 10963.24 | NA | 0.003938 | 1 | 9.302303 |
| rs148111655_G | BTN2A1 | 1.130613 | 2.537072 | 0.583563 | 1 | 0.12276 |
| rs2273558_A | BTN2A1 | 0.673125 | 0.711994 | 0.636428 | 1.69 × 10−41 | −0.39582 |
| rs2893856_T | BTN2A1 | 0.839708 | 0.911101 | 0.772766 | 0.0032259 | −0.1747 |
| rs2893857_C | BTN2A1 | 1.142895 | 1.255236 | 1.042695 | 0.48083 | 0.133565 |
| rs3734539_C | BTN2A1 | 4032.285 | NA | 0.007 | 1 | 8.302088 |
| rs3734542_G | BTN2A1 | 0.425283 | 0.453292 | 0.39911 | 8.59 × 10−151 | −0.855 |
| rs3734543_G | BTN2A1 | 0.430012 | 0.458993 | 0.402974 | 1.59 × 10−140 | −0.84394 |
| rs3799380_T | BTN2A1 | 0.577632 | 0.611726 | 0.545555 | 8.59 × 10−77 | −0.54882 |
| rs56296968_C | BTN2A1 | 0.54644 | 0.579504 | 0.515375 | 9.70 × 10−89 | −0.60433 |
| rs6456724_T | BTN2A1 | 0.838697 | 0.91004 | 0.771803 | 0.00287 | −0.17591 |
| rs6907857_T | BTN2A1 | 1.433106 | 1.834039 | 1.140956 | 0.29414 | 0.359844 |
| rs6911470_C | BTN2A1 | 1.472809 | 1.963726 | 1.132407 | 0.57829 | 0.387171 |
| rs6929846_T | BTN2A1 | 0.813898 | 0.875261 | 0.756001 | 3.60 × 10−6 | −0.20592 |
| rs7773913_C | BTN2A1 | 1.433061 | 1.833981 | 1.14092 | 0.29439 | 0.359813 |
| rs7773938_C | BTN2A1 | 0.548927 | 0.582092 | 0.517765 | 1.15 × 10−87 | −0.59979 |
| rs77870445_T | BTN2A1 | 1.219723 | 1.609943 | 0.942473 | 1 | 0.198624 |
| rs9348718_A | BTN2A1 | 1.267016 | 1.454243 | 1.109411 | 0.06114 | 0.236664 |
| rs9358943_C | BTN2A1 | 1.768069 | 31.86429 | 0.363137 | 1 | 0.569888 |
| rs9358944_A | BTN2A1 | 0.546443 | 0.579355 | 0.515512 | 1.55 × 10−89 | −0.60432 |
| rs9358945_A | BTN2A1 | 0.54552 | 0.578367 | 0.514649 | 4.37 × 10−90 | −0.60602 |
| rs9461254_G | BTN2A1 | 1.464419 | 1.953756 | 1.125151 | 0.66799 | 0.381458 |
| rs10456045_G | BTN3A1 | 0.638826 | 0.674371 | 0.605202 | 2.97 × 10−57 | −0.44812 |
| rs10807008_G | BTN3A1 | 1.091584 | 1.192289 | 1.001092 | 1 | 0.087629 |
| rs12200782_C | BTN3A1 | 1.138999 | 1.250121 | 1.039809 | 0.56581 | 0.13015 |
| rs12207930_C | BTN3A1 | 1.147602 | 1.241418 | 1.062201 | 0.05418 | 0.137674 |
| rs12208447_C | BTN3A1 | 1.284779 | 1.624554 | 1.030908 | 1 | 0.250586 |
| rs12214924_T | BTN3A1 | 1.147514 | 1.241076 | 1.062324 | 0.05283 | 0.137597 |
| rs143476765_A | BTN3A1 | 1.108868 | 4.616795 | 0.396888 | 1 | 0.10334 |
| rs144114619_T | BTN3A1 | 1.212854 | 2.147941 | 0.740604 | 1 | 0.192976 |
| rs145059723_A | BTN3A1 | 1.412767 | 4.034311 | 0.629895 | 1 | 0.34555 |
| rs1741738_A | BTN3A1 | 1.144367 | 1.242176 | 1.05573 | 0.11623 | 0.134851 |
| rs17610161_G | BTN3A1 | 1.097034 | 1.199223 | 1.005306 | 1 | 0.09261 |
| rs1796520_C | BTN3A1 | 0.758646 | 0.800004 | 0.719297 | 2.40 × 10−22 | −0.27622 |
| rs3799378_A | BTN3A1 | 0.585559 | 0.61957 | 0.553499 | 2.92 × 10−75 | −0.53519 |
| rs3857549_C | BTN3A1 | 1.247408 | 1.401099 | 1.114594 | 0.01526 | 0.221068 |
| rs3902051_A | BTN3A1 | 1.090365 | 1.187959 | 1.002366 | 1 | 0.086513 |
| rs41266839_G | BTN3A1 | 0.396746 | 0.423498 | 0.37178 | 2.12 × 10−168 | −0.92446 |
| rs4609015_T | BTN3A1 | 1.151594 | 1.245578 | 1.066033 | 0.03817 | 0.141147 |
| rs4712990_C | BTN3A1 | 1.101764 | 1.203269 | 1.010539 | 1 | 0.096912 |
| rs55676749_T | BTN3A1 | 1.13993 | 1.38368 | 0.948273 | 1 | 0.130967 |
| rs56161420_G | BTN3A1 | 1.138909 | 1.230815 | 1.055142 | 0.09381 | 0.130071 |
| rs6900725_T | BTN3A1 | 1.149401 | 1.242804 | 1.064341 | 0.04333 | 0.139241 |
| rs6912853_C | BTN3A1 | 1.16488 | 1.257236 | 1.080577 | 0.00785 | 0.152618 |
| rs6920986_C | BTN3A1 | 1.148238 | 1.241884 | 1.062975 | 0.04995 | 0.138228 |
| rs6921148_T | BTN3A1 | 1.165176 | 1.411589 | 0.971033 | 1 | 0.152872 |
| rs742090_A | BTN3A1 | 0.759078 | 0.800536 | 0.719639 | 3.58 × 10−22 | −0.27565 |
| rs7770214_G | BTN3A1 | 1.145779 | 1.239155 | 1.060758 | 0.06048 | 0.136085 |
| rs80153343_G | BTN3A1 | 1.136899 | 1.439622 | 0.910733 | 1 | 0.128305 |
| rs11758089_T | BTN3A2 | 1.192878 | 1.288514 | 1.105674 | 0.00063 | 0.176369 |
| rs12176317_A | BTN3A2 | 0.445307 | 0.474099 | 0.41837 | 6.72 × 10−140 | −0.80899 |
| rs12194095_C | BTN3A2 | 1.118798 | 1.235914 | 1.015164 | 1 | 0.112255 |
| rs12199613_C | BTN3A2 | 0.67037 | 0.706851 | 0.635751 | 1.76 × 10−47 | −0.39993 |
| rs12205731_G | BTN3A2 | 1.114755 | 1.232063 | 1.011029 | 1 | 0.108634 |
| rs144016445_G | BTN3A2 | 81101.36 | NA | 217.0251 | 1 | 11.30346 |
| rs1977_A | BTN3A2 | 0.445697 | 0.474831 | 0.418455 | 1.17 × 10−136 | −0.80811 |
| rs1979_G | BTN3A2 | 0.44537 | 0.474171 | 0.418426 | 8.23 × 10−140 | −0.80885 |
| rs1985732_A | BTN3A2 | 0.63289 | 0.668226 | 0.599467 | 2.87 × 10−59 | −0.45746 |
| rs2073526_G | BTN3A2 | 0.74435 | 0.785579 | 0.705115 | 9.15 × 10−25 | −0.29524 |
| rs35183513_G | BTN3A2 | 1.102861 | 1.203659 | 1.012184 | 1 | 0.097908 |
| rs58367598_T | BTN3A2 | 1.234256 | 1.449383 | 1.057523 | 0.89091 | 0.210469 |
| rs7765566_G | BTN3A2 | 1.269787 | 1.499163 | 1.083157 | 0.39847 | 0.23885 |
| rs9104_G | BTN3A2 | 1.092904 | 1.187567 | 1.007255 | 1 | 0.088838 |
| rs9358934_G | BTN3A2 | 0.447824 | 0.476845 | 0.420678 | 2.34 × 10−137 | −0.80335 |
| rs9379855_T | BTN3A2 | 0.447682 | 0.47666 | 0.420575 | 8.85 × 10−138 | −0.80367 |
| rs9379858_T | BTN3A2 | 0.448449 | 0.477474 | 0.421299 | 3.19 × 10−137 | −0.80196 |
| rs9379859_C | BTN3A2 | 0.447843 | 0.476903 | 0.420662 | 5.37 × 10−137 | −0.80331 |
| rs9379861_G | BTN3A2 | 1.225507 | 1.619363 | 0.945713 | 1 | 0.203355 |
| rs9393713_G | BTN3A2 | 0.442958 | 0.471602 | 0.416159 | 1.07 × 10−141 | −0.81428 |
| rs9393714_G | BTN3A2 | 0.443419 | 0.47214 | 0.416551 | 6.99 × 10−141 | −0.81324 |
| rs186813312_C | BTNL3 | 0.103966 | NA | NA | NA | −2.26369 |
| rs199970076_G | BTNL3 | 0.544013 | 10.42476 | 0.087697 | 1 | −0.60878 |
| rs201534771_G | BTNL3 | 0.108957 | NA | NA | NA | −2.2168 |
| rs201813197_C | BTNL3 | 1.141842 | 4.74926 | 0.409599 | 1 | 0.132642 |
| rs35157246_C | BTNL3 | 1.069831 | 1.242264 | 0.926052 | 1 | 0.067501 |
| rs4700774_G | BTNL3 | 0.943446 | 0.999778 | 0.89061 | 1 | −0.05822 |
| rs59220426_C | BTNL3 | 1.006054 | 1.132599 | 0.896669 | 1 | 0.006036 |
| rs73815153_G | BTNL3 | 1.009988 | 1.138564 | 0.899028 | 1 | 0.009938 |
| rs7713324_A | BTNL3 | 1.004294 | 1.130736 | 0.895001 | 1 | 0.004284 |
| rs7726604_C | BTNL3 | 1.004751 | 1.13121 | 0.895444 | 1 | 0.00474 |
| rs112469887_G | BTNL8 | 1.084234 | 1.330225 | 0.893133 | 1 | 0.080874 |
| rs113071395_G | BTNL8 | 0.867372 | 1.006007 | 0.751681 | 1 | −0.14229 |
| rs113534626_A | BTNL8 | 1.019956 | 1.206531 | 0.868252 | 1 | 0.019759 |
| rs141492316_T | BTNL8 | 0.891806 | 1.095588 | 0.733483 | 1 | −0.11451 |
| rs145199317_A | BTNL8 | 0.907765 | 1.254932 | 0.673198 | 1 | −0.09677 |
| rs151174174_C | BTNL8 | 0.770441 | 0.932722 | 0.641625 | 0.63031 | −0.26079 |
| rs17704291_C | BTNL8 | 0.940216 | 0.996283 | 0.88763 | 1 | −0.06165 |
| rs200633883_C | BTNL8 | 0.311552 | 1.114968 | 0.108457 | 1 | −1.16619 |
| rs201214790_T | BTNL8 | 4044.713 | NA | 1.08 × 10−7 | 1 | 8.305166 |
| rs201891387_G | BTNL8 | 0.62355 | 2.663098 | 0.210953 | 1 | −0.47233 |
| rs2276995_A | BTNL8 | 0.983681 | 1.038422 | 0.931987 | 1 | −0.01645 |
| rs2619739_C | BTNL8 | 1.101246 | 1.212402 | 1.002386 | 1 | 0.096442 |
| rs7724813_G | BTNL8 | 1.078621 | 1.169543 | 0.996169 | 1 | 0.075683 |
| SNP, Reference Allele | Gene | Number of SNPs in Control | Number of SNPs in CeD | Total Allele Count in Control | Total allele Count in CeD | Total Number of SNPs in the UK Biobank | Total Allele Count in UK Biobank |
|---|---|---|---|---|---|---|---|
| rs13195402_G | BTN2A1 | 52,060 | 4631 | 58,392 | 6028 | 56,691 | 64,420 |
| rs13195509_G | BTN2A1 | 52,271 | 4654 | 59,474 | 6176 | 56,925 | 65,650 |
| rs1407045_A | BTN2A1 | 30,596 | 3603 | 59,306 | 6172 | 34,199 | 65,478 |
| rs2273558_A | BTN2A1 | 34,599 | 3248 | 51,134 | 5570 | 37,847 | 56,704 |
| rs2893856_T | BTN2A1 | 7815 | 697 | 59,452 | 6182 | 8512 | 65,634 |
| rs3734542_G | BTN2A1 | 52,209 | 4656 | 59,432 | 6180 | 56,865 | 65,612 |
| rs3734543_G | BTN2A1 | 52,002 | 4644 | 59,146 | 6114 | 56,646 | 65,260 |
| rs3799380_T | BTN2A1 | 46,887 | 4213 | 59,354 | 6164 | 51,100 | 65,518 |
| rs56296968_C | BTN2A1 | 47,941 | 4295 | 59,422 | 6170 | 52,236 | 65,592 |
| rs6456724_T | BTN2A1 | 7813 | 696 | 59,418 | 6178 | 8509 | 65,596 |
| rs6929846_T | BTN2A1 | 10,355 | 903 | 59,460 | 6180 | 11,258 | 65,640 |
| rs7773938_C | BTN2A1 | 47,953 | 4289 | 59,472 | 6162 | 52,242 | 65,634 |
| rs9358944_A | BTN2A1 | 47,929 | 4294 | 59,462 | 6182 | 52,223 | 65,644 |
| rs9358945_A | BTN2A1 | 47,944 | 4292 | 59,478 | 6182 | 52,236 | 65,660 |
| rs10456045_G | BTN3A1 | 41,458 | 3680 | 59,434 | 6174 | 45,138 | 65,608 |
| rs1796520_C | BTN3A1 | 28,075 | 2504 | 59,240 | 6176 | 30,579 | 65,416 |
| rs3799378_A | BTN3A1 | 45,113 | 4017 | 59,206 | 6152 | 49,130 | 65,358 |
| rs3857549_C | BTN3A1 | 55,572 | 5848 | 59,430 | 6170 | 61,420 | 65,600 |
| rs41266839_G | BTN3A1 | 52,985 | 4720 | 59,430 | 6178 | 57,705 | 65,608 |
| rs4609015_T | BTN3A1 | 50,759 | 5371 | 59,452 | 6170 | 56,130 | 65,622 |
| rs6900725_T | BTN3A1 | 50,682 | 5378 | 59,392 | 6182 | 56,060 | 65,574 |
| rs6912853_C | BTN3A1 | 50,145 | 5329 | 59,434 | 6176 | 55,474 | 65,610 |
| rs6920986_C | BTN3A1 | 50,787 | 5381 | 59,464 | 6182 | 56,168 | 65,646 |
| rs742090_A | BTN3A1 | 28,172 | 2506 | 59,438 | 6174 | 30,678 | 65,612 |
| rs11758089_T | BTN3A2 | 50,176 | 5354 | 59,432 | 6182 | 55,530 | 65,614 |
| rs12176317_A | BTN3A2 | 51,604 | 4597 | 59,488 | 6182 | 56,201 | 65,670 |
| rs12199613_C | BTN3A2 | 36,321 | 3178 | 59,396 | 6178 | 39,499 | 65,574 |
| rs1977_A | BTN3A2 | 50,506 | 4497 | 58,436 | 6074 | 55,003 | 64,510 |
| rs1979_G | BTN3A2 | 51,551 | 4590 | 59,448 | 6176 | 56,141 | 65,624 |
| rs1985732_A | BTN3A2 | 41,492 | 3674 | 59,442 | 6174 | 45,166 | 65,616 |
| rs2073526_G | BTN3A2 | 26,272 | 2288 | 59,434 | 6176 | 28,560 | 65,610 |
| rs9358934_G | BTN3A2 | 51,477 | 4591 | 59,412 | 6174 | 56,068 | 65,586 |
| rs9379855_T | BTN3A2 | 51,458 | 4579 | 59,392 | 6162 | 56,037 | 65,554 |
| rs9379858_T | BTN3A2 | 51,474 | 4586 | 59,422 | 6170 | 56,060 | 65,592 |
| rs9379859_C | BTN3A2 | 51,530 | 4595 | 59,452 | 6174 | 56,125 | 65,626 |
| rs9393713_G | BTN3A2 | 51,601 | 4590 | 59,472 | 6178 | 56,191 | 65,650 |
| rs9393714_G | BTN3A2 | 51,581 | 4594 | 59,456 | 6180 | 56,175 | 65,636 |
| SNP, Reference Allele | Gene | Number of Controls Homozygous for the Reference Allele | Number of Controls Heterozygous for the Reference Allele | Number of Control Individuals Without the Reference Allele | Allele Freq in Controls | HWE Adjusted p-Value |
|---|---|---|---|---|---|---|
| rs13195402_G | BTN2A1 | 23,188 | 5684 | 324 | 0.892 | 1 |
| rs13195509_G | BTN2A1 | 23,018 | 6235 | 484 | 0.879 | 0.340 |
| rs1407045_A | BTN2A1 | 7951 | 14,694 | 7008 | 0.516 | 1 |
| rs2273558_A | BTN2A1 | 11,804 | 10,991 | 2772 | 0.677 | 0.179 |
| rs2893856_T | BTN2A1 | 514 | 6787 | 22,425 | 0.869 | 1 |
| rs3734542_G | BTN2A1 | 22,978 | 6253 | 485 | 0.878 | 1 |
| rs3734543_G | BTN2A1 | 22,866 | 6270 | 437 | 0.879 | 1 |
| rs3799380_T | BTN2A1 | 18,595 | 9697 | 1385 | 0.790 | 1 |
| rs56296968_C | BTN2A1 | 19,326 | 9289 | 1096 | 0.807 | 1 |
| rs6456724_T | BTN2A1 | 515 | 6783 | 22,411 | 0.869 | 1 |
| rs6929846_T | BTN2A1 | 971 | 8413 | 20,346 | 0.826 | 1 |
| rs7773938_C | BTN2A1 | 19,333 | 9287 | 1116 | 0.806 | 1 |
| rs9358944_A | BTN2A1 | 19,319 | 9291 | 1121 | 0.806 | 1 |
| rs9358945_A | BTN2A1 | 19,326 | 9292 | 1121 | 0.806 | 1 |
| rs10456045_G | BTN3A1 | 14,460 | 12,538 | 2719 | 0.698 | 1 |
| rs1796520_C | BTN3A1 | 6734 | 14,607 | 8279 | 0.526 | 1 |
| rs3799378_A | BTN3A1 | 17,152 | 10,809 | 1642 | 0.762 | 1 |
| rs3857549_C | BTN3A1 | 26,084 | 3404 | 227 | 0.935 | 1 |
| rs41266839_G | BTN3A1 | 23,662 | 5661 | 392 | 0.892 | 1 |
| rs4609015_T | BTN3A1 | 21,657 | 7445 | 624 | 0.854 | 1 |
| rs6900725_T | BTN3A1 | 21,625 | 7432 | 639 | 0.853 | 1 |
| rs6912853_C | BTN3A1 | 21,168 | 7809 | 740 | 0.844 | 1 |
| rs6920986_C | BTN3A1 | 21,672 | 7443 | 617 | 0.854 | 1 |
| rs742090_A | BTN3A1 | 6727 | 14,718 | 8274 | 0.526 | 1 |
| rs11758089_T | BTN3A2 | 21,182 | 7812 | 722 | 0.844 | 1 |
| rs12176317_A | BTN3A2 | 22,420 | 6764 | 560 | 0.867 | 1 |
| rs12199613_C | BTN3A2 | 11,047 | 14,227 | 4424 | 0.612 | 1 |
| rs1977_A | BTN3A2 | 21,828 | 6850 | 540 | 0.864 | 1 |
| rs1979_G | BTN3A2 | 22,388 | 6775 | 561 | 0.867 | 1 |
| rs1985732_A | BTN3A2 | 14,433 | 12,626 | 2662 | 0.698 | 1 |
| rs2073526_G | BTN3A2 | 5874 | 14,524 | 9319 | 0.558 | 1 |
| rs9358934_G | BTN3A2 | 22,330 | 6817 | 559 | 0.866 | 1 |
| rs9379855_T | BTN3A2 | 22,327 | 6804 | 565 | 0.866 | 1 |
| rs9379858_T | BTN3A2 | 22,329 | 6816 | 566 | 0.866 | 1 |
| rs9379859_C | BTN3A2 | 22,358 | 6814 | 554 | 0.867 | 1 |
| rs9393713_G | BTN3A2 | 22,422 | 6757 | 557 | 0.868 | 1 |
| rs9393714_G | BTN3A2 | 22,404 | 6773 | 551 | 0.868 | 1 |
| SNP Name | Gene | OR | Upper | Lower | Adjusted p-Value | ln(OR) |
|---|---|---|---|---|---|---|
| rs10484441_G | BTN2A1 | 0.979307 | 1.082186 | 0.887707 | 1 | −0.02091 |
| rs12660069_C | BTN2A1 | 0.987937 | 1.171444 | 0.837975 | 1 | −0.01214 |
| rs13195402_G | BTN2A1 | 0.812801 | 0.876887 | 0.753657 | 8.15 × 10−6 | −0.20727 |
| rs13195509_G | BTN2A1 | 0.824983 | 0.886694 | 0.767819 | 1.62 × 10−5 | −0.19239 |
| rs13437351_G | BTN2A1 | 1.359939 | 1.781839 | 1.056236 | 1 | 0.30744 |
| rs1407045_A | BTN2A1 | 1.061681 | 1.125065 | 1.001972 | 1 | 0.059854 |
| rs142951857_A | BTN2A1 | 1.189393 | 2.744421 | 0.589877 | 1 | 0.173443 |
| rs143104579_G | BTN2A1 | 1.045598 | 1.306706 | 0.844534 | 1 | 0.044589 |
| rs146399224_T | BTN2A1 | 2678.29 | NA | 0.001107 | 1 | 7.892934 |
| rs148111655_G | BTN2A1 | 1.06888 | 2.495456 | 0.522042 | 1 | 0.066611 |
| rs2273558_A | BTN2A1 | 0.924046 | 0.983637 | 0.868186 | 1 | −0.07899 |
| rs2893856_T | BTN2A1 | 0.978844 | 1.068109 | 0.895906 | 1 | −0.02138 |
| rs2893857_C | BTN2A1 | 0.983276 | 1.0868 | 0.891136 | 1 | −0.01687 |
| rs3734539_C | BTN2A1 | 12320.17 | NA | 2.26 × 10−5 | 1 | 9.418993 |
| rs3734542_G | BTN2A1 | 0.828358 | 0.890318 | 0.770964 | 2.94 × 10−5 | −0.18831 |
| rs3734543_G | BTN2A1 | 0.845824 | 0.910556 | 0.785966 | 0.000823 | −0.16744 |
| rs3799380_T | BTN2A1 | 0.906299 | 0.966317 | 0.850232 | 0.260718 | −0.09839 |
| rs56296968_C | BTN2A1 | 0.889118 | 0.949206 | 0.833053 | 0.042016 | −0.11753 |
| rs6456724_T | BTN2A1 | 0.975504 | 1.064451 | 0.892856 | 1 | −0.0248 |
| rs6907857_T | BTN2A1 | 1.296362 | 1.68837 | 1.012123 | 1 | 0.259562 |
| rs6911470_C | BTN2A1 | 1.225626 | 1.666065 | 0.92177 | 1 | 0.203452 |
| rs6929846_T | BTN2A1 | 0.956691 | 1.034347 | 0.884033 | 1 | −0.04427 |
| rs7773913_C | BTN2A1 | 1.29982 | 1.692843 | 1.014844 | 1 | 0.262226 |
| rs7773938_C | BTN2A1 | 0.892443 | 0.95269 | 0.83623 | 0.062934 | −0.11379 |
| rs77870445_T | BTN2A1 | 0.971098 | 1.300178 | 0.738108 | 1 | −0.02933 |
| rs9348718_A | BTN2A1 | 1.100624 | 1.274443 | 0.954703 | 1 | 0.095877 |
| rs9358943_C | BTN2A1 | 0.455962 | 8.266602 | 0.091903 | 1 | −0.78535 |
| rs9358944_A | BTN2A1 | 0.888825 | 0.948639 | 0.833001 | 0.038293 | −0.11786 |
| rs9358945_A | BTN2A1 | 0.886765 | 0.946407 | 0.831099 | 0.02906 | −0.12018 |
| rs9461254_G | BTN2A1 | 1.206876 | 1.642976 | 0.90642 | 1 | 0.188035 |
| rs10456045_G | BTN3A1 | 0.918688 | 0.975087 | 0.865666 | 0.527112 | −0.08481 |
| rs10807008_G | BTN3A1 | 0.94089 | 1.033981 | 0.857412 | 1 | −0.06093 |
| rs12200782_C | BTN3A1 | 0.987553 | 1.090084 | 0.896183 | 1 | −0.01253 |
| rs12207930_C | BTN3A1 | 0.994811 | 1.081946 | 0.91566 | 1 | −0.0052 |
| rs12208447_C | BTN3A1 | 0.971864 | 1.246094 | 0.767755 | 1 | −0.02854 |
| rs12214924_T | BTN3A1 | 0.997777 | 1.08479 | 0.918712 | 1 | −0.00223 |
| rs143476765_A | BTN3A1 | 0.541997 | 2.391824 | 0.175983 | 1 | −0.61249 |
| rs144114619_T | BTN3A1 | 0.937876 | 1.708275 | 0.552209 | 1 | −0.06414 |
| rs145059723_A | BTN3A1 | 0.851224 | 2.534279 | 0.356178 | 1 | −0.16108 |
| rs1741738_A | BTN3A1 | 1.055187 | 1.152999 | 0.966854 | 1 | 0.053718 |
| rs17610161_G | BTN3A1 | 0.948685 | 1.04337 | 0.863865 | 1 | −0.05268 |
| rs1796520_C | BTN3A1 | 0.925332 | 0.980491 | 0.873166 | 0.877114 | −0.0776 |
| rs3799378_A | BTN3A1 | 0.866517 | 0.922476 | 0.814111 | 0.000704 | −0.14327 |
| rs3857549_C | BTN3A1 | 1.20727 | 1.366146 | 1.0703 | 0.249929 | 0.188361 |
| rs3902051_A | BTN3A1 | 0.947893 | 1.038919 | 0.865991 | 1 | −0.05351 |
| rs41266839_G | BTN3A1 | 0.806793 | 0.868508 | 0.749711 | 1.06 × 10−6 | −0.21469 |
| rs4609015_T | BTN3A1 | 0.999771 | 1.087084 | 0.920447 | 1 | −0.00023 |
| rs4712990_C | BTN3A1 | 0.953022 | 1.047148 | 0.8686 | 1 | −0.04812 |
| rs55676749_T | BTN3A1 | 1.055136 | 1.295119 | 0.867029 | 1 | 0.05367 |
| rs56161420_G | BTN3A1 | 1.003319 | 1.090026 | 0.92446 | 1 | 0.003313 |
| rs6900725_T | BTN3A1 | 0.998519 | 1.085343 | 0.919613 | 1 | −0.00148 |
| rs6912853_C | BTN3A1 | 1.060948 | 1.150125 | 0.979684 | 1 | 0.059162 |
| rs6920986_C | BTN3A1 | 0.997232 | 1.08425 | 0.918167 | 1 | −0.00277 |
| rs6921148_T | BTN3A1 | 1.075901 | 1.317605 | 0.885986 | 1 | 0.073159 |
| rs742090_A | BTN3A1 | 0.927391 | 0.982792 | 0.875004 | 1 | −0.07538 |
| rs7770214_G | BTN3A1 | 0.992737 | 1.079363 | 0.914026 | 1 | −0.00729 |
| rs80153343_G | BTN3A1 | 1.144379 | 1.465945 | 0.904679 | 1 | 0.134862 |
| rs11758089_T | BTN3A2 | 1.093163 | 1.188249 | 1.00675 | 1 | 0.089075 |
| rs12176317_A | BTN3A2 | 0.820862 | 0.880647 | 0.765377 | 3.50 × 10−6 | −0.1974 |
| rs12194095_C | BTN3A2 | 0.971352 | 1.079478 | 0.875824 | 1 | −0.02907 |
| rs12199613_C | BTN3A2 | 0.884297 | 0.93714 | 0.834446 | 0.003312 | −0.12296 |
| rs12205731_G | BTN3A2 | 0.970415 | 1.079025 | 0.874528 | 1 | −0.03003 |
| rs144016445_G | BTN3A2 | 38750.66 | NA | 151.7424 | 1 | 10.5649 |
| rs1977_A | BTN3A2 | 0.816781 | 0.87678 | 0.761121 | 2.06 × 10−6 | −0.20238 |
| rs1979_G | BTN3A2 | 0.820729 | 0.880499 | 0.765255 | 3.40 × 10−6 | −0.19756 |
| rs1985732_A | BTN3A2 | 0.896055 | 0.951463 | 0.84398 | 0.033488 | −0.10975 |
| rs2073526_G | BTN3A2 | 0.925312 | 0.980986 | 0.872648 | 0.940603 | −0.07762 |
| rs35183513_G | BTN3A2 | 0.951537 | 1.044843 | 0.867784 | 1 | −0.04968 |
| rs58367598_T | BTN3A2 | 1.089293 | 1.290835 | 0.924221 | 1 | 0.085529 |
| rs7765566_G | BTN3A2 | 1.145085 | 1.363507 | 0.967785 | 1 | 0.135479 |
| rs9104_G | BTN3A2 | 0.945089 | 1.032973 | 0.86575 | 1 | −0.05648 |
| rs9358934_G | BTN3A2 | 0.824601 | 0.884767 | 0.76877 | 7.53 × 10−6 | −0.19286 |
| rs9379855_T | BTN3A2 | 0.82361 | 0.883636 | 0.767905 | 6.04 × 10−6 | −0.19406 |
| rs9379858_T | BTN3A2 | 0.825671 | 0.885867 | 0.769811 | 8.99 × 10−6 | −0.19156 |
| rs9379859_C | BTN3A2 | 0.824802 | 0.885058 | 0.768893 | 8.10 × 10−6 | −0.19261 |
| rs9379861_G | BTN3A2 | 1.039521 | 1.399242 | 0.785634 | 1 | 0.03876 |
| rs9393713_G | BTN3A2 | 0.814157 | 0.873453 | 0.759123 | 9.27 × 10−7 | −0.2056 |
| rs9393714_G | BTN3A2 | 0.818022 | 0.877663 | 0.762671 | 2.08 × 10−6 | −0.20087 |
| rs186813312_C | BTNL3 | 0.387143 | NA | NA | NA | −0.94896 |
| rs199970076_G | BTNL3 | 0.890096 | 19.02105 | 0.105649 | 1 | −0.11643 |
| rs201534771_G | BTNL3 | 0.373628 | NA | NA | NA | −0.98449 |
| rs201813197_C | BTNL3 | 0.906025 | 3.98319 | 0.295074 | 1 | −0.09869 |
| rs35157246_C | BTNL3 | 1.061269 | 1.243749 | 0.909647 | 1 | 0.059466 |
| rs4700774_G | BTNL3 | 0.953121 | 1.014114 | 0.896074 | 1 | −0.04801 |
| rs59220426_C | BTNL3 | 0.964396 | 1.094724 | 0.852069 | 1 | −0.03625 |
| rs73815153_G | BTNL3 | 0.979858 | 1.113514 | 0.864825 | 1 | −0.02035 |
| rs7713324_A | BTNL3 | 0.962434 | 1.092563 | 0.850278 | 1 | −0.03829 |
| rs7726604_C | BTNL3 | 0.963325 | 1.093541 | 0.851096 | 1 | −0.03736 |
| rs112469887_G | BTNL8 | 1.04641 | 1.299262 | 0.850631 | 1 | 0.045365 |
| rs113071395_G | BTNL8 | 0.908391 | 1.065436 | 0.777899 | 1 | −0.09608 |
| rs113534626_A | BTNL8 | 1.008072 | 1.204841 | 0.848563 | 1 | 0.00804 |
| rs141492316_T | BTNL8 | 0.930335 | 1.160408 | 0.752616 | 1 | −0.07221 |
| rs145199317_A | BTNL8 | 0.884686 | 1.250181 | 0.639686 | 1 | −0.12252 |
| rs151174174_C | BTNL8 | 0.828069 | 1.016412 | 0.679378 | 1 | −0.18866 |
| rs17704291_C | BTNL8 | 0.952291 | 1.013025 | 0.895481 | 1 | −0.04888 |
| rs200633883_C | BTNL8 | 0.419543 | 1.675223 | 0.123618 | 1 | −0.86859 |
| rs201214790_T | BTNL8 | 740.6942 | NA | 1.97 × 10−8 | 1 | 6.607588 |
| rs201891387_G | BTNL8 | 0.495896 | 2.269513 | 0.147322 | 1 | −0.70139 |
| rs2276995_A | BTNL8 | 0.984754 | 1.043162 | 0.929755 | 1 | −0.01536 |
| rs2619739_C | BTNL8 | 1.078254 | 1.194765 | 0.974928 | 1 | 0.075343 |
| rs7724813_G | BTNL8 | 1.054983 | 1.150319 | 0.968751 | 1 | 0.053525 |
| SNP, Reference Allele | Gene | Number of Controls Homozygous for the Reference Allele | Number of Controls Heterozygous for the Reference Allele | Number of Control Individuals Without the Reference Allele | Allele Freq in Controls | HWE Adjusted p-Value |
|---|---|---|---|---|---|---|
| rs13195402_G | BTN2A1 | 23,188 | 5684 | 324 | 0.892 | 1 |
| rs13195509_G | BTN2A1 | 23,018 | 6235 | 484 | 0.879 | 0.184 |
| rs3734542_G | BTN2A1 | 22,978 | 6253 | 485 | 0.878 | 0.246 |
| rs3734543_G | BTN2A1 | 22,866 | 6270 | 437 | 0.879 | 1 |
| rs56296968_C | BTN2A1 | 19,326 | 9289 | 1096 | 0.807 | 1 |
| rs9358944_A | BTN2A1 | 19,319 | 9291 | 1121 | 0.806 | 1 |
| rs9358945_A | BTN2A1 | 19,326 | 9292 | 1121 | 0.806 | 1 |
| rs3799378_A | BTN3A1 | 17,152 | 10,809 | 1642 | 0.762 | 1 |
| rs41266839_G | BTN3A1 | 23,662 | 5661 | 392 | 0.892 | 1 |
| rs12176317_A | BTN3A2 | 22,420 | 6764 | 560 | 0.867 | 1 |
| rs12199613_C | BTN3A2 | 11,047 | 14,227 | 4424 | 0.612 | 1 |
| rs1977_A | BTN3A2 | 21,828 | 6850 | 540 | 0.864 | 1 |
| rs1979_G | BTN3A2 | 22,388 | 6775 | 561 | 0.867 | 1 |
| rs1985732_A | BTN3A2 | 14,433 | 12,626 | 2662 | 0.698 | 1 |
| rs9358934_G | BTN3A2 | 22,330 | 6817 | 559 | 0.866 | 1 |
| rs9379855_T | BTN3A2 | 22,327 | 6804 | 565 | 0.866 | 1 |
| rs9379858_T | BTN3A2 | 22,329 | 6816 | 566 | 0.866 | 1 |
| rs9379859_C | BTN3A2 | 22,358 | 6814 | 554 | 0.867 | 1 |
| rs9393713_G | BTN3A2 | 22,422 | 6757 | 557 | 0.868 | 1 |
| rs9393714_G | BTN3A2 | 22,404 | 6773 | 551 | 0.868 | 1 |
| SNP Name | Gene | OR | Upper | Lower | Adjusted p-Value | ln(OR) |
|---|---|---|---|---|---|---|
| rs10484441_G | BTN2A1 | 1.026879 | 1.182655 | 0.894513 | 1 | 0.026524 |
| rs12660069_C | BTN2A1 | 0.954382 | 1.207579 | 0.761803 | 1 | −0.04669 |
| rs13195402_G | BTN2A1 | 0.757206 | 0.831328 | 0.689864 | 5.10 × 10−7 | −0.27812 |
| rs13195509_G | BTN2A1 | 0.77459 | 0.846648 | 0.708837 | 1.75 × 10−6 | −0.25542 |
| rs13437351_G | BTN2A1 | 1.400742 | 2.090478 | 0.973921 | 1 | 0.337002 |
| rs1407045_A | BTN2A1 | 1.080193 | 1.170756 | 0.996977 | 1 | 0.07714 |
| rs142951857_A | BTN2A1 | 1.287748 | 4.431929 | 0.486536 | 1 | 0.252895 |
| rs143104579_G | BTN2A1 | 1.331616 | 1.885897 | 0.963027 | 1 | 0.286393 |
| rs146399224_T | BTN2A1 | 0.25741 | NA | NA | NA | −1.35708 |
| rs148111655_G | BTN2A1 | 0.944297 | 4.178925 | 0.294481 | 1 | −0.05731 |
| rs2273558_A | BTN2A1 | 0.892885 | 0.971388 | 0.820718 | 0.848901 | −0.1133 |
| rs2893856_T | BTN2A1 | 0.927404 | 1.048809 | 0.818043 | 1 | −0.07537 |
| rs2893857_C | BTN2A1 | 1.040565 | 1.199279 | 0.905854 | 1 | 0.039764 |
| rs3734539_C | BTN2A1 | 27166.76 | NA | 9.99 × 10−14 | 1 | 10.20975 |
| rs3734542_G | BTN2A1 | 0.77697 | 0.849181 | 0.711073 | 2.52 × 10−6 | −0.25235 |
| rs3734543_G | BTN2A1 | 0.792317 | 0.867952 | 0.723463 | 5.43 × 10−5 | −0.23279 |
| rs3799380_T | BTN2A1 | 0.869548 | 0.945573 | 0.799787 | 0.107572 | −0.13978 |
| rs56296968_C | BTN2A1 | 0.852755 | 0.928301 | 0.783513 | 0.02331 | −0.15928 |
| rs6456724_T | BTN2A1 | 0.924581 | 1.045479 | 0.815654 | 1 | −0.07841 |
| rs6907857_T | BTN2A1 | 1.401132 | 2.091061 | 0.974192 | 1 | 0.33728 |
| rs6911470_C | BTN2A1 | 1.565436 | 2.679723 | 0.978088 | 1 | 0.448164 |
| rs6929846_T | BTN2A1 | 0.870873 | 0.973832 | 0.777255 | 1 | −0.13826 |
| rs7773913_C | BTN2A1 | 1.408409 | 2.10183 | 0.979326 | 1 | 0.34246 |
| rs7773938_C | BTN2A1 | 0.854233 | 0.929778 | 0.784985 | 0.026611 | −0.15755 |
| rs77870445_T | BTN2A1 | 1.022451 | 1.530146 | 0.704151 | 1 | 0.022203 |
| rs9348718_A | BTN2A1 | 1.308813 | 1.636793 | 1.057849 | 1 | 0.26912 |
| rs9358943_C | BTN2A1 | 0.257602 | NA | NA | NA | −1.35634 |
| rs9358944_A | BTN2A1 | 0.850121 | 0.924966 | 0.781482 | 0.016059 | −0.16238 |
| rs9358945_A | BTN2A1 | 0.847905 | 0.922571 | 0.77943 | 0.012607 | −0.16499 |
| rs9461254_G | BTN2A1 | 1.565001 | 2.678973 | 0.977818 | 1 | 0.447886 |
| rs10456045_G | BTN3A1 | 0.872024 | 0.944198 | 0.805371 | 0.074344 | −0.13694 |
| rs10807008_G | BTN3A1 | 0.988264 | 1.128972 | 0.867514 | 1 | −0.01181 |
| rs12200782_C | BTN3A1 | 0.969241 | 1.112166 | 0.84719 | 1 | −0.03124 |
| rs12207930_C | BTN3A1 | 1.059407 | 1.19361 | 0.94228 | 1 | 0.05771 |
| rs12208447_C | BTN3A1 | 1.183033 | 1.71485 | 0.838562 | 1 | 0.168081 |
| rs12214924_T | BTN3A1 | 1.059617 | 1.192843 | 0.943249 | 1 | 0.057908 |
| rs143476765_A | BTN3A1 | 0.385935 | 2.931895 | 0.063895 | 1 | −0.95209 |
| rs144114619_T | BTN3A1 | 0.800306 | 1.80142 | 0.391948 | 1 | −0.22276 |
| rs145059723_A | BTN3A1 | 1.546934 | 29.22579 | 0.264012 | 1 | 0.436275 |
| rs1741738_A | BTN3A1 | 1.126398 | 1.288465 | 0.987689 | 1 | 0.119025 |
| rs17610161_G | BTN3A1 | 0.999336 | 1.142965 | 0.876283 | 1 | −0.00066 |
| rs1796520_C | BTN3A1 | 0.901974 | 0.977599 | 0.831877 | 1 | −0.10317 |
| rs3799378_A | BTN3A1 | 0.829346 | 0.900397 | 0.763968 | 0.00081 | −0.18712 |
| rs3857549_C | BTN3A1 | 1.307821 | 1.557482 | 1.105147 | 0.217446 | 0.268362 |
| rs3902051_A | BTN3A1 | 0.979611 | 1.113404 | 0.864043 | 1 | −0.0206 |
| rs41266839_G | BTN3A1 | 0.753767 | 0.824792 | 0.68903 | 7.25 × 10−8 | −0.28267 |
| rs4609015_T | BTN3A1 | 1.055122 | 1.187787 | 0.939241 | 1 | 0.053657 |
| rs4712990_C | BTN3A1 | 1.009758 | 1.153906 | 0.886126 | 1 | 0.00971 |
| rs55676749_T | BTN3A1 | 1.356015 | 1.867802 | 1.007293 | 1 | 0.30455 |
| rs56161420_G | BTN3A1 | 1.060354 | 1.193295 | 0.944184 | 1 | 0.058603 |
| rs6900725_T | BTN3A1 | 1.061349 | 1.194313 | 0.945175 | 1 | 0.059541 |
| rs6912853_C | BTN3A1 | 1.087423 | 1.216203 | 0.974151 | 1 | 0.08381 |
| rs6920986_C | BTN3A1 | 1.062734 | 1.196651 | 0.9458 | 1 | 0.060845 |
| rs6921148_T | BTN3A1 | 1.340282 | 1.834522 | 1.000858 | 1 | 0.29288 |
| rs742090_A | BTN3A1 | 0.903554 | 0.979423 | 0.833241 | 1 | −0.10142 |
| rs7770214_G | BTN3A1 | 1.053031 | 1.185369 | 0.937421 | 1 | 0.051673 |
| rs80153343_G | BTN3A1 | 0.979216 | 1.347528 | 0.724804 | 1 | −0.021 |
| rs11758089_T | BTN3A2 | 1.191327 | 1.352007 | 1.052499 | 0.618146 | 0.175068 |
| rs12176317_A | BTN3A2 | 0.766628 | 0.836949 | 0.702371 | 2.82 × 10−7 | −0.26575 |
| rs12194095_C | BTN3A2 | 0.943672 | 1.092386 | 0.818034 | 1 | −0.05798 |
| rs12199613_C | BTN3A2 | 0.842231 | 0.911373 | 0.77819 | 0.002057 | −0.1717 |
| rs12205731_G | BTN3A2 | 0.933998 | 1.081909 | 0.809117 | 1 | −0.06828 |
| rs144016445_G | BTN3A2 | 73914.07 | NA | 1.75 × 10−6 | 1 | 11.21066 |
| rs1977_A | BTN3A2 | 0.764904 | 0.835801 | 0.700168 | 2.99 × 10−7 | −0.268 |
| rs1979_G | BTN3A2 | 0.76816 | 0.838582 | 0.70381 | 3.63 × 10−7 | −0.26376 |
| rs1985732_A | BTN3A2 | 0.860169 | 0.932012 | 0.793857 | 0.023502 | −0.15063 |
| rs2073526_G | BTN3A2 | 0.90252 | 0.979048 | 0.831612 | 1 | −0.10256 |
| rs35183513_G | BTN3A2 | 1.000653 | 1.140268 | 0.880486 | 1 | 0.000652 |
| rs58367598_T | BTN3A2 | 1.153984 | 1.470445 | 0.915197 | 1 | 0.143221 |
| rs7765566_G | BTN3A2 | 1.171754 | 1.497136 | 0.928192 | 1 | 0.158502 |
| rs9104_G | BTN3A2 | 1.010674 | 1.145196 | 0.894134 | 1 | 0.010617 |
| rs9358934_G | BTN3A2 | 0.772404 | 0.843549 | 0.707423 | 8.85 × 10−7 | −0.25825 |
| rs9379855_T | BTN3A2 | 0.771306 | 0.842174 | 0.706564 | 6.76 × 10−7 | −0.25967 |
| rs9379858_T | BTN3A2 | 0.774407 | 0.845625 | 0.709352 | 1.18 × 10−6 | −0.25566 |
| rs9379859_C | BTN3A2 | 0.770242 | 0.841254 | 0.705384 | 6.31 × 10−7 | −0.26105 |
| rs9379861_G | BTN3A2 | 0.987622 | 1.586757 | 0.639198 | 1 | −0.01246 |
| rs9393713_G | BTN3A2 | 0.760996 | 0.830868 | 0.697149 | 1.06 × 10−7 | −0.27313 |
| rs9393714_G | BTN3A2 | 0.765074 | 0.835356 | 0.700859 | 2.25 × 10−7 | −0.26778 |
| rs186813312_C | BTNL3 | 0.257602 | NA | NA | NA | −1.35634 |
| rs199970076_G | BTNL3 | 0.272926 | 6.904492 | 0.010788 | 1 | −1.29856 |
| rs201534771_G | BTNL3 | 0.273671 | NA | NA | NA | −1.29583 |
| rs201813197_C | BTNL3 | 0.514697 | 3.715301 | 0.100372 | 1 | −0.66418 |
| rs35157246_C | BTNL3 | 1.03751 | 1.287524 | 0.842551 | 1 | 0.036824 |
| rs4700774_G | BTNL3 | 0.978837 | 1.065525 | 0.899706 | 1 | −0.02139 |
| rs59220426_C | BTNL3 | 1.017762 | 1.216405 | 0.856252 | 1 | 0.017606 |
| rs73815153_G | BTNL3 | 1.02474 | 1.225825 | 0.861442 | 1 | 0.024438 |
| rs7713324_A | BTNL3 | 1.018119 | 1.216841 | 0.856546 | 1 | 0.017957 |
| rs7726604_C | BTNL3 | 1.020763 | 1.219953 | 0.85881 | 1 | 0.02055 |
| rs112469887_G | BTNL8 | 1.002918 | 1.333287 | 0.765187 | 1 | 0.002914 |
| rs113071395_G | BTNL8 | 0.93155 | 1.162634 | 0.752338 | 1 | −0.07091 |
| rs113534626_A | BTNL8 | 0.9371 | 1.185234 | 0.747845 | 1 | −0.06496 |
| rs141492316_T | BTNL8 | 0.963798 | 1.314391 | 0.718601 | 1 | −0.03687 |
| rs145199317_A | BTNL8 | 1.12544 | 1.912393 | 0.697448 | 1 | 0.118174 |
| rs151174174_C | BTNL8 | 0.729764 | 0.953457 | 0.564223 | 1 | −0.31503 |
| rs17704291_C | BTNL8 | 0.969451 | 1.055155 | 0.891212 | 1 | −0.03103 |
| rs200633883_C | BTNL8 | 0.171372 | 1.035119 | 0.022558 | 1 | −1.76392 |
| rs201214790_T | BTNL8 | 0.256248 | NA | NA | NA | −1.36161 |
| rs201891387_G | BTNL8 | 0.171476 | 1.035747 | 0.022572 | 1 | −1.76331 |
| rs2276995_A | BTNL8 | 1.002057 | 1.084332 | 0.926283 | 1 | 0.002055 |
| rs2619739_C | BTNL8 | 0.966968 | 1.107812 | 0.846435 | 1 | −0.03359 |
| rs7724813_G | BTNL8 | 1.041603 | 1.171988 | 0.927725 | 1 | 0.040761 |
| Participants with the HLA-DQ2.5 Genotype | |||||||
|---|---|---|---|---|---|---|---|
| SNP, Reference Allele | Gene | Number of SNPs in Controls with HLA-DQ2.5 | Number of SNPs in CeD with HLA-DQ2.5 | Total Allele count in Control with HLA-DQ2.5 | Total Allele Count in CeD with HLA-DQ2.5 | Total Number of SNPs in the UK Biobank with HLA-DQ2.5 | Total Allele Count in UK Biobank with HLA-DQ2.5 |
| rs13195402_G | BTN2A1 | 9398 | 2256 | 12,514 | 3206 | 11,654 | 15,720 |
| rs13195509_G | BTN2A1 | 9408 | 2265 | 12,820 | 3296 | 11,673 | 16,116 |
| rs3734542_G | BTN2A1 | 9386 | 2266 | 12,808 | 3300 | 11,652 | 16,108 |
| rs3734543_G | BTN2A1 | 9366 | 2265 | 12,722 | 3258 | 11,631 | 15,980 |
| rs56296968_C | BTN2A1 | 8609 | 2107 | 12,798 | 3290 | 10,716 | 16,088 |
| rs7773938_C | BTN2A1 | 8606 | 2106 | 12,804 | 3290 | 10,712 | 16,094 |
| rs9358944_A | BTN2A1 | 8603 | 2108 | 12,818 | 3304 | 10,711 | 16,122 |
| rs9358945_A | BTN2A1 | 8607 | 2106 | 12,818 | 3302 | 10,713 | 16,120 |
| rs3799378_A | BTN3A1 | 8129 | 1959 | 12,768 | 3286 | 10,088 | 16,054 |
| rs41266839_G | BTN3A1 | 9577 | 2298 | 12,820 | 3298 | 11,875 | 16,118 |
| rs12176317_A | BTN3A2 | 9347 | 2242 | 12,826 | 3302 | 11,589 | 16,128 |
| rs12199613_C | BTN3A2 | 6396 | 1515 | 12,802 | 3300 | 7911 | 16,102 |
| rs1977_A | BTN3A2 | 9135 | 2197 | 12,574 | 3248 | 11,332 | 15,822 |
| rs1979_G | BTN3A2 | 9330 | 2242 | 12,808 | 3302 | 11,572 | 16,110 |
| rs1985732_A | BTN3A2 | 7353 | 1777 | 12,816 | 3294 | 9130 | 16,110 |
| rs9358934_G | BTN3A2 | 9333 | 2240 | 12,810 | 3292 | 11,573 | 16,102 |
| rs9379855_T | BTN3A2 | 9324 | 2239 | 12,802 | 3294 | 11,563 | 16,096 |
| rs9379858_T | BTN3A2 | 9321 | 2242 | 12,804 | 3296 | 11,563 | 16,100 |
| rs9379859_C | BTN3A2 | 9340 | 2244 | 12,806 | 3296 | 11,584 | 16,102 |
| rs9393713_G | BTN3A2 | 9345 | 2237 | 12,812 | 3298 | 11,582 | 16,110 |
| rs9393714_G | BTN3A2 | 9346 | 2241 | 12,818 | 3300 | 11,587 | 16,118 |
| SNP, Reference Allele | Gene | Number of Controls Homozygous for the Reference Allele | Number of Controls Heterozygous for the Reference Allele | Number of Control Individuals Without the Reference Allele | Allele Freq in Controls | HWE Adjusted p-Value |
|---|---|---|---|---|---|---|
| rs13195402_G | BTN2A1 | 3353 | 2692 | 212 | 0.751 | 2.65 × 10−31 |
| rs13195509_G | BTN2A1 | 3303 | 2802 | 305 | 0.734 | 3.26 × 10−20 |
| rs3734542_G | BTN2A1 | 3290 | 2806 | 308 | 0.733 | 3.69 × 10−20 |
| rs3734543_G | BTN2A1 | 3278 | 2810 | 273 | 0.736 | 1.35 × 10−26 |
| rs56296968_C | BTN2A1 | 2737 | 3135 | 527 | 0.673 | 2.65 × 10−31 |
| rs7773938_C | BTN2A1 | 2737 | 3132 | 533 | 0.672 | 2.65 × 10−31 |
| rs9358944_A | BTN2A1 | 2734 | 3135 | 540 | 0.671 | 2.65 × 10−31 |
| rs9358945_A | BTN2A1 | 2737 | 3133 | 539 | 0.671 | 2.65 × 10−31 |
| rs3799378_A | BTN3A1 | 2446 | 3237 | 701 | 0.637 | 2.65 × 10−31 |
| rs41266839_G | BTN3A1 | 3429 | 2719 | 262 | 0.747 | 2.65 × 10−31 |
| rs12176317_A | BTN3A2 | 3267 | 2813 | 333 | 0.729 | 2.65 × 10−31 |
| rs12199613_C | BTN3A2 | 1505 | 3386 | 1510 | 0.500 | 2.65 × 10−31 |
| rs1977_A | BTN3A2 | 3172 | 2791 | 324 | 0.726 | 2.65 × 10−31 |
| rs1979_G | BTN3A2 | 3260 | 2810 | 334 | 0.728 | 2.65 × 10−31 |
| rs1985732_A | BTN3A2 | 1974 | 3405 | 1029 | 0.574 | 2.65 × 10−31 |
| rs9358934_G | BTN3A2 | 3259 | 2815 | 331 | 0.729 | 2.65 × 10−31 |
| rs9379855_T | BTN3A2 | 3257 | 2810 | 334 | 0.728 | 2.65 × 10−31 |
| rs9379858_T | BTN3A2 | 3253 | 2815 | 334 | 0.728 | 2.65 × 10−31 |
| rs9379859_C | BTN3A2 | 3263 | 2814 | 326 | 0.729 | 2.65 × 10−31 |
| rs9393713_G | BTN3A2 | 3269 | 2807 | 330 | 0.729 | 2.65 × 10−31 |
| rs9393714_G | BTN3A2 | 3267 | 2812 | 330 | 0.729 | 2.65 × 10−31 |
Appendix J. Supplementary Materials for Results Section 2.3
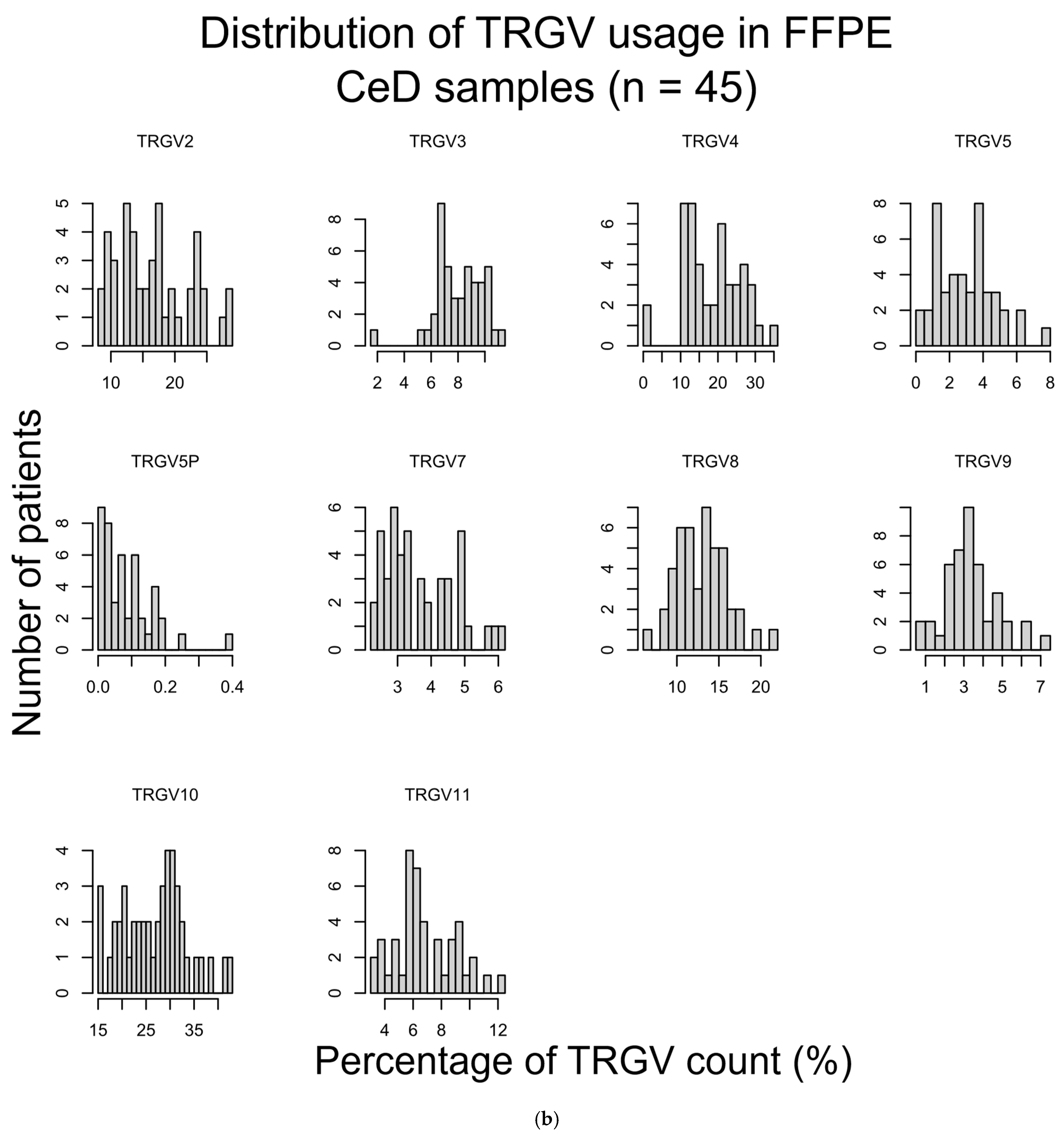
| FFPE CeD (n = 45) vs. FFPE Healthy Control (n = 108) | ||
|---|---|---|
| Raw p-Value (MWU) | Adjusted p-Value | |
| TRGV2 | 0.775 | 1 |
| TRGV3 | 0.411 | 1 |
| TRGV4 | 0.812 | 1 |
| TRGV5 | 0.906 | 1 |
| TRGV5P | 0.684 | 1 |
| TRGV7 | 0.566 | 1 |
| TRGV8 | 0.382 | 1 |
| TRGV9 | 0.070 | 0.70 |
| TRGV10 | 0.248 | 1 |
| TRGV11 | 0.025 | 0.25 |

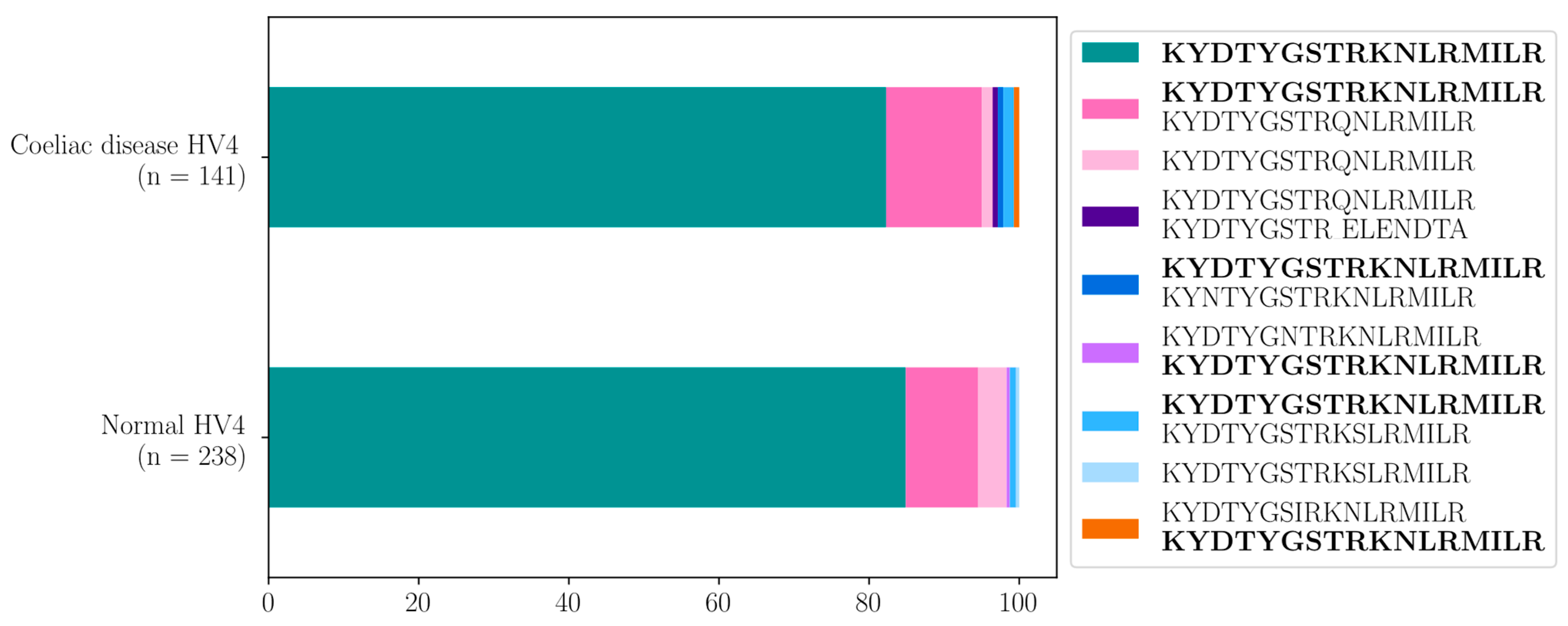
| 141 CeD vs. 238 Healthy Control Samples | ||
|---|---|---|
| Raw p-Values (Fisher) | Adjusted p-Values | |
| WT vs. WT, KYDTYGSTRQNLRMIL | 0.3925 | 1 |
| WT vs. KYDTYGSTRQNLRMILR | 0.3392 | 1 |
| WT vs. KYDTYGSTRQNLRMILR, KYDTYGSTR_ELENDTA | 0.3668 | 1 |
| WT vs. WT, KYNTYGSTRKNLRMILR | 0.3668 | 1 |
| WT vs. KYDTYGNTRKNLRMILR, WT | 1 | 1 |
| WT vs. WT, KYDTYGSTRKSLRMILR | 0.6258 | 1 |
| WT vs. KYDTYGSTRKSLRMILR | 1 | 1 |
| WT vs. WT, KYDTYGSIRKNLRMILR | 0.3668 | 1 |
| WT, KYDTYGSTRQNLRMILR vs. KYDTYGSTRQNLRMILR | 0.1697 | 1 |
| WT, KYDTYGSTRQNLRMILR vs. KYDTYGSTRQNLRMILR, KYDTYGSTR_ELENDTA | 0.4524 | 1 |
| WT, KYDTYGSTRQNLRMILR vs. WT, KYNTYGSTRKNLRMILR | 0.4524 | 1 |
| WT, KYDTYGSTRQNLRMILR vs. WT, KYDTYGNTRKNLRMILR | 1 | 1 |
| WT, KYDTYGSTRQNLRMILR vs. WT, KYDTYGSTRKSLRMILR | 1 | 1 |
| WT, KYDTYGSTRQNLRMILR vs. KYDTYGSTRKSLRMILR | 1 | 1 |
| WT, KYDTYGSTRQNLRMILR vs. WT, KYDTYGSIRKNLRMILR | 0.4524 | 1 |
| KYDTYGSTRQNLRMILR vs. KYDTYGSTRQNLRMILR, KYDTYGSTR_ELENDTA | 0.25 | 1 |
| KYDTYGSTRQNLRMILR vs. WT, KYNTYGSTRKNLRMILR | 0.25 | 1 |
| KYDTYGSTRQNLRMILR vs. WT, KYDTYGNTRKNLRMILR | 1 | 1 |
| KYDTYGSTRQNLRMILR vs. WT, KYDTYGSTRKSLRMILR | 0.5165 | 1 |
| KYDTYGSTRQNLRMILR vs. KYDTYGSTRKSLRMILR | 1 | 1 |
| KYDTYGSTRQNLRMILR vs. WT, KYDTYGSIRKNLRMILR | 0.25 | 1 |
| KYDTYGSTRQNLRMILR, KYDTYGSTR_ELENDTA vs. WT, KYNTYGSTRKNLRMILR | 1 | 1 |
| KYDTYGSTRQNLRMILR, KYDTYGSTR_ELENDTA vs. WT, KYDTYGNTRKNLRMILR | 1 | 1 |
| KYDTYGSTRQNLRMILR, KYDTYGSTR_ELENDTA vs. WT, KYDTYGSTRKSLRMILR | 1 | 1 |
| KYDTYGSTRQNLRMILR, KYDTYGSTR_ELENDTA vs. KYDTYGSTRKSLRMILR | 1 | 1 |
| KYDTYGSTRQNLRMILR, KYDTYGSTR_ELENDTA vs. WT, KYDTYGSIRKNLRMILR | 1 | 1 |
| WT, KYNTYGSTRKNLRMILR vs. WT, KYDTYGNTRKNLRMILR | 1 | 1 |
| WT, KYNTYGSTRKNLRMILR vs. WT, KYDTYGSTRKSLRMILR | 1 | 1 |
| WT, KYNTYGSTRKNLRMILR vs. KYDTYGSTRKSLRMILR | 1 | 1 |
| WT, KYNTYGSTRKNLRMILR vs. WT, KYDTYGSIRKNLRMILR | 1 | 1 |
| WT, KYDTYGNTRKNLRMILR vs. WT, KYDTYGSTRKSLRMILR | 1 | 1 |
| WT, KYDTYGNTRKNLRMILR vs. KYDTYGSTRKSLRMILR | 1 | 1 |
| WT, KYDTYGNTRKNLRMILR vs. WT, KYDTYGSIRKNLRMILR | 1 | 1 |
| WT, KYDTYGSTRKSLRMILR vs. KYDTYGSTRKSLRMILR | 1 | 1 |
| WT, KYDTYGSTRKSLRMILR vs. WT, KYDTYGSIRKNLRMILR | 1 | 1 |
| KYDTYGSTRKSLRMILR vs. WT, KYDTYGSIRKNLRMILR | 1 | 1 |
References
- Abadie, V.; Sollid, L.M.; Barreiro, L.B.; Jabri, B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu. Rev. Immunol. 2011, 29, 493–525. [Google Scholar] [CrossRef]
- Jabri, B.; Sollid, L.M. Tissue-mediated control of immunopathology in coeliac disease. Nat. Rev. Immunol. 2009, 9, 858–870. [Google Scholar] [CrossRef]
- Trier, J.S. Diagnosis of celiac sprue. Gastroenterology 1998, 115, 211–216. [Google Scholar] [CrossRef] [PubMed]
- NICE. Coeliac Disease: Recognition, Assessment and Management. Available online: https://www.nice.org.uk/guidance/ng20/chapter/Recommendations (accessed on 27 November 2020).
- Al-Toma, A.; Goerres, M.S.; Meijer, J.W.; Pena, A.S.; Crusius, J.B.; Mulder, C.J. Human leukocyte antigen-DQ2 homozygosity and the development of refractory celiac disease and enteropathy-associated T-cell lymphoma. Clin. Gastroenterol. Hepatol. 2006, 4, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Ayesh, B.M.; Zaqout, E.K.; Yassin, M.M. HLA-DQ2 and -DQ8 haplotypes frequency and diagnostic utility in celiac disease patients of Gaza strip, Palestine. Autoimmun Highlights 2017, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Björck, S.; Brundin, C.; Lörinc, E.; Lynch, K.F.; Agardh, D. Screening detects a high proportion of celiac disease in young HLA-genotyped children. J. Pediatr. Gastroenterol. Nutr. 2010, 50, 49–53. [Google Scholar] [CrossRef]
- Karell, K.; Louka, A.S.; Moodie, S.J.; Ascher, H.; Clot, F.; Greco, L.; Ciclitira, P.J.; Sollid, L.M.; Partanen, J.; European Genetics Cluster on Celiac, D. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: Results from the European Genetics Cluster on Celiac Disease. Hum. Immunol. 2003, 64, 469–477. [Google Scholar] [CrossRef]
- Karhus, L.L.; Thuesen, B.H.; Skaaby, T.; Rumessen, J.J.; Linneberg, A. The distribution of HLA DQ2 and DQ8 haplotypes and their association with health indicators in a general Danish population. United Eur. Gastroenterol. 2018, 6, 866–878. [Google Scholar] [CrossRef]
- Murad, H.; Jazairi, B.; Khansaa, I.; Olabi, D.; Khouri, L. HLA-DQ2 and -DQ8 genotype frequency in Syrian celiac disease children: HLA-DQ relative risks evaluation. BMC Gastroenterol. 2018, 18, 70. [Google Scholar] [CrossRef]
- Sollid, L.M.; Thorsby, E. HLA susceptibility genes in celiac disease: Genetic mapping and role in pathogenesis. Gastroenterology 1993, 105, 910–922. [Google Scholar] [CrossRef]
- Sollid, L.M. Molecular basis of celiac disease. Annu. Rev. Immunol. 2000, 18, 53–81. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Markussen, G.; Ek, J.; Gjerde, H.; Vartdal, F.; Thorsby, E. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J. Exp. Med. 1989, 169, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Thorsby, E. The primary association of celiac disease to a given HLA-DQ alpha/beta heterodimer explains the divergent HLA-DR associations observed in various Caucasian populations. Tissue Antigens 1990, 36, 136–137. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Tapia, A.; Hill, I.D.; Kelly, C.P.; Calderwood, A.H.; Murray, J.A.; American College of Gastroenterology. ACG clinical guidelines: Diagnosis and management of celiac disease. Am. J. Gastroenterol. 2013, 108, 656–676. [Google Scholar] [CrossRef]
- Djilali-Saiah, I.; Caillat-Zucman, S.; Schmitz, J.; Chaves-Vieira, M.L.; Bach, J.F. Polymorphism of antigen processing (TAP, LMP) and HLA class II genes in celiac disease. Hum. Immunol. 1994, 40, 8–16. [Google Scholar] [CrossRef]
- Hunt, K.A.; Zhernakova, A.; Turner, G.; Heap, G.A.; Franke, L.; Bruinenberg, M.; Romanos, J.; Dinesen, L.C.; Ryan, A.W.; Panesar, D.; et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008, 40, 395–402. [Google Scholar] [CrossRef]
- Dubois, P.C.; van Heel, D.A. Translational mini-review series on the immunogenetics of gut disease: Immunogenetics of coeliac disease. Clin. Exp. Immunol. 2008, 153, 162–173. [Google Scholar] [CrossRef]
- Goudey, B.; Abraham, G.; Kikianty, E.; Wang, Q.; Rawlinson, D.; Shi, F.; Haviv, I.; Stern, L.; Kowalczyk, A.; Inouye, M. Interactions within the MHC contribute to the genetic architecture of celiac disease. PLoS ONE 2017, 12, e0172826. [Google Scholar] [CrossRef]
- Pietz, G.; De, R.; Hedberg, M.; Sjoberg, V.; Sandstrom, O.; Hernell, O.; Hammarstrom, S.; Hammarstrom, M.L. Immunopathology of childhood celiac disease-Key role of intestinal epithelial cells. PLoS ONE 2017, 12, e0185025. [Google Scholar] [CrossRef]
- Mayassi, T.; Ladell, K.; Gudjonson, H.; McLaren, J.E.; Shaw, D.G.; Tran, M.T.; Rokicka, J.J.; Lawrence, I.; Grenier, J.C.; van Unen, V.; et al. Chronic inflammation permanently reshapes tissue-resident immunity in celiac disease. Cell 2019, 176, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.A.; Stammers, M.; Malcherek, G.; Beck, S.; Trowsdale, J. The cluster of BTN genes in the extended major histocompatibility complex. Genomics 2001, 71, 351–362. [Google Scholar] [CrossRef]
- Arnett, H.A.; Viney, J.L. Immune modulation by butyrophilins. Nat. Rev. Immunol. 2014, 14, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.A.; Reith, W.; Trowsdale, J. Regulation of immunity by butyrophilins. Annu. Rev. Immunol. 2016, 34, 151–172. [Google Scholar] [CrossRef]
- Malcherek, G.; Mayr, L.; Roda-Navarro, P.; Rhodes, D.; Miller, N.; Trowsdale, J. The B7 homolog butyrophilin BTN2A1 is a novel ligand for DC-SIGN. J. Immunol. 2007, 179, 3804–3811. [Google Scholar] [CrossRef]
- Messal, N.; Mamessier, E.; Sylvain, A.; Celis-Gutierrez, J.; Thibult, M.L.; Chetaille, B.; Firaguay, G.; Pastor, S.; Guillaume, Y.; Wang, Q.; et al. Differential role for CD277 as a co-regulator of the immune signal in T and NK cells. Eur. J. Immunol. 2011, 41, 3443–3454. [Google Scholar] [CrossRef]
- Di Marco Barros, R.; Roberts, N.A.; Dart, R.J.; Vantourout, P.; Jandke, A.; Nussbaumer, O.; Deban, L.; Cipolat, S.; Hart, R.; Iannitto, M.L.; et al. Epithelia use butyrophilin-like molecules to shape organ-specific gamma delta T cell compartments. Cell 2016, 167, 203–218. [Google Scholar] [CrossRef]
- Jandke, A.; Melandri, D.; Monin, L.; Ushakov, D.S.; Laing, A.G.; Vantourout, P.; East, P.; Nitta, T.; Narita, T.; Takayanagi, H.; et al. Butyrophilin-like proteins display combinatorial diversity in selecting and maintaining signature intraepithelial gammadelta T cell compartments. Nat. Commun. 2020, 11, 3769. [Google Scholar] [CrossRef] [PubMed]
- Melandri, D.; Zlatareva, I.; Chaleil, R.A.G.; Dart, R.J.; Chancellor, A.; Nussbaumer, O.; Polyakova, O.; Roberts, N.A.; Wesch, D.; Kabelitz, D.; et al. The γδ TCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nat. Immunol. 2018, 19, 1352–1365. [Google Scholar] [CrossRef]
- Vantourout, P.; Laing, A.; Woodward, M.J.; Zlatareva, I.; Apolonia, L.; Jones, A.W.; Snijders, A.P.; Malim, M.H.; Hayday, A.C. Heteromeric interactions regulate butyrophilin (BTN) and BTN-like molecules governing gammadelta T cell biology. Proc. Natl. Acad. Sci. USA 2018, 115, 1039–1044. [Google Scholar] [CrossRef]
- Willcox, C.R.; Vantourout, P.; Salim, M.; Zlatareva, I.; Melandri, D.; Zanardo, L.; George, R.; Kjaer, S.; Jeeves, M.; Mohammed, F.; et al. Butyrophilin-like 3 Directly Binds a Human Vγ4(+) T Cell Receptor Using a Modality Distinct from Clonally-Restricted Antigen. Immunity 2019, 51, 813–825 e814. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.M.; Girardi, M.; Roberts, S.J.; Barbee, S.D.; Hayday, A.C.; Tigelaar, R.E. Selection of the cutaneous intraepithelial γδ+ T cell repertoire by a thymic stromal determinant. Nat. Immunol. 2006, 7, 843–850. [Google Scholar] [CrossRef]
- Cano, C.E.; Pasero, C.; De Gassart, A.; Kerneur, C.; Gabriac, M.; Fullana, M.; Granarolo, E.; Hoet, R.; Scotet, E.; Rafia, C.; et al. BTN2A1, an immune checkpoint targeting Vγ9Vδ2 T cell cytotoxicity against malignant cells. Cell Rep. 2021, 36, 109359. [Google Scholar] [CrossRef] [PubMed]
- Hayday, A.C.; Vantourout, P. The innate biologies of adaptive antigen receptors. Annu. Rev. Immunol. 2020, 38, 487–510. [Google Scholar] [CrossRef]
- Karunakaran, M.M.; Gobel, T.W.; Starick, L.; Walter, L.; Herrmann, T. Vγ9 and Vδ2 T cell antigen receptor genes and butyrophilin 3 (BTN3) emerged with placental mammals and are concomitantly preserved in selected species like alpaca (Vicugna pacos). Immunogenetics 2014, 66, 243–254. [Google Scholar] [CrossRef]
- Fichtner, A.S.; Karunakaran, M.M.; Gu, S.; Boughter, C.T.; Borowska, M.T.; Starick, L.; Nohren, A.; Gobel, T.W.; Adams, E.J.; Herrmann, T. Alpaca (Vicugna pacos), the first nonprimate species with a phosphoantigen-reactive Vγ9Vδ2 T cell subset. Proc. Natl. Acad. Sci. USA 2020, 117, 6697–6707. [Google Scholar] [CrossRef] [PubMed]
- Rigau, M.; Ostrouska, S.; Fulford, T.S.; Johnson, D.N.; Woods, K.; Ruan, Z.; McWilliam, H.E.G.; Hudson, C.; Tutuka, C.; Wheatley, A.K.; et al. Butyrophilin 2A1 is essential for phosphoantigen reactivity by gammadelta T cells. Science 2020, 367, eaay5516. [Google Scholar] [CrossRef] [PubMed]
- Sandstrom, A.; Peigne, C.M.; Leger, A.; Crooks, J.E.; Konczak, F.; Gesnel, M.C.; Breathnach, R.; Bonneville, M.; Scotet, E.; Adams, E.J. The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vγ9Vδ2 T cells. Immunity 2014, 40, 490–500. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Shang, R.; Yang, J.; Chen, C.; Liu, Z.; Liang, G.; He, W.; Luo, G. Skin γδ T tells and their function in wound healing. Front. Immunol. 2022, 13, 875076. [Google Scholar] [CrossRef]
- Han, A.; Newell, E.W.; Glanville, J.; Fernandez-Becker, N.; Khosla, C.; Chien, Y.H.; Davis, M.M. Dietary gluten triggers concomitant activation of CD4+ and CD8+ alphabeta T cells and gammadelta T cells in celiac disease. Proc. Natl. Acad. Sci. USA 2013, 110, 13073–13078. [Google Scholar] [CrossRef]
- Aigner, J.; Villatoro, S.; Rabionet, R.; Roquer, J.; Jimenez-Conde, J.; Marti, E.; Estivill, X. A common 56-kilobase deletion in a primate-specific segmental duplication creates a novel butyrophilin-like protein. BMC Genet. 2013, 14, 61. [Google Scholar] [CrossRef]
- Mitsunaga, S.; Hosomichi, K.; Okudaira, Y.; Nakaoka, H.; Kunii, N.; Suzuki, Y.; Kuwana, M.; Sato, S.; Kaneko, Y.; Homma, Y.; et al. Exome sequencing identifies novel rheumatoid arthritis-susceptible variants in the BTNL2. J. Hum. Genet. 2013, 58, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Sirota, M.; Schaub, M.A.; Batzoglou, S.; Robinson, W.H.; Butte, A.J. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 2009, 5, e1000792. [Google Scholar] [CrossRef]
- Orozco, G.; Eerligh, P.; Sanchez, E.; Zhernakova, S.; Roep, B.O.; Gonzalez-Gay, M.A.; Lopez-Nevot, M.A.; Callejas, J.L.; Hidalgo, C.; Pascual-Salcedo, D.; et al. Analysis of a functional BTNL2 polymorphism in type 1 diabetes, rheumatoid arthritis, and systemic lupus erythematosus. Hum. Immunol. 2005, 66, 1235–1241. [Google Scholar] [CrossRef]
- Traherne, J.A.; Barcellos, L.F.; Sawcer, S.J.; Compston, A.; Ramsay, P.P.; Hauser, S.L.; Oksenberg, J.R.; Trowsdale, J. Association of the truncating splice site mutation in BTNL2 with multiple sclerosis is secondary to HLA-DRB1*15. Hum. Mol. Genet. 2006, 15, 155–161. [Google Scholar] [CrossRef]
- Hippich, M.; Beyerlein, A.; Hagopian, W.A.; Krischer, J.P.; Vehik, K.; Knoop, J.; Winker, C.; Toppari, J.; Lernmark, A.; Rewers, M.J.; et al. Genetic contribution to the divergence in type 1 diabetes risk between children from the general population and children from affected families. Diabetes 2019, 68, 847–857. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Hamon, S.; Li, D.; Barral-Rodriguez, S.; Ott, J.; Diabetes Genetics Consortium. MHC fine mapping of human type 1 diabetes using the T1DGC data. Diabetes Obes. Metab. 2009, 11 (Suppl. 1), 53–59. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhao, N.; Spragins, E.; Kagda, M.S.; Li, M.; Assis, P.; Jolanki, O.; Luo, Y.; Cherry, J.M.; Boyle, A.P.; et al. Annotating and prioritizing human non-coding variants with RegulomeDB. bioRxiv 2022. [Google Scholar] [CrossRef]
- Spurkland, A.; Sollid, L.M.; Polanco, I.; Vartdal, F.; Thorsby, E. HLA-DR and -DQ genotypes of celiac disease patients serologically typed to be non-DR3 or non-DR5/7. Hum. Immunol. 1992, 35, 188–192. [Google Scholar] [CrossRef]
- Dart, R.J.; Zlatareva, I.; Vantourout, P.; Theodoridis, E.; Amar, A.; Kannambath, S.; East, P.; Recaldin, T.; Mansfield, J.C.; Lamb, C.A.; et al. Conserved gammadelta T cell selection by BTNL proteins limits progression of human inflammatory bowel disease. Science 2023, 381, eadh0301. [Google Scholar] [CrossRef]
- Guo, M.H.; Plummer, L.; Chan, Y.M.; Hirschhorn, J.N.; Lippincott, M.F. Burden Testing of Rare Variants Identified through Exome Sequencing via Publicly Available Control Data. Am. J. Hum. Genet. 2018, 103, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.H. Burden Testing Against Public Controls. Available online: https://github.com/mhguo1/TRAPD (accessed on 24 April 2023).
- Viken, M.K.; Blomhoff, A.; Olsson, M.; Akselsen, H.E.; Pociot, F.; Nerup, J.; Kockum, I.; Cambon-Thomsen, A.; Thorsby, E.; Undlien, D.E.; et al. Reproducible association with type 1 diabetes in the extended class I region of the major histocompatibility complex. Genes Immun. 2009, 10, 323–333. [Google Scholar] [CrossRef]
- Horton, R.; Wilming, L.; Rand, V.; Lovering, R.C.; Bruford, E.A.; Khodiyar, V.K.; Lush, M.J.; Povey, S.; Talbot, C.C., Jr.; Wright, M.W.; et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004, 5, 889–899. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Foers, A.D.; Shoukat, M.S.; Welsh, O.E.; Donovan, K.; Petry, R.; Evans, S.C.; FitzPatrick, M.E.; Collins, N.; Klenerman, P.; Fowler, A.; et al. Classification of intestinal T-cell receptor repertoires using machine learning methods can identify patients with coeliac disease regardless of dietary gluten status. J. Pathol. 2021, 253, 279–291. [Google Scholar] [CrossRef]
- Falchuk, Z.M.; Rogentine, G.N.; Strober, W. Predominance of histocompatibility antigen HL-A8 in patients with gluten-sensitive enteropathy. J. Clin. Investig. 1972, 51, 1602–1605. [Google Scholar] [CrossRef] [PubMed]
- Stokes, P.L.; Asquith, P.; Holmes, G.K.; Mackintosh, P.; Cooke, W.T. Histocompatibility antigens associated with adult coeliac disease. Lancet 1972, 2, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Lindfors, K.; Ciacci, C.; Kurppa, K.; Lundin, K.E.A.; Makharia, G.K.; Mearin, M.L.; Murray, J.A.; Verdu, E.F.; Kaukinen, K. Coeliac disease. Nat. Rev. Dis. Primers 2019, 5, 3. [Google Scholar] [CrossRef]
- Karunakaran, M.M.; Willcox, C.R.; Salim, M.; Paletta, D.; Fichtner, A.S.; Noll, A.; Starick, L.; Nohren, A.; Begley, C.R.; Berwick, K.A.; et al. Butyrophilin-2A1 directly binds germline-encoded regions of the Vγ9Vδ2 TCR and is essential for phosphoantigen sensing. Immunity 2020, 52, 487–498 e486. [Google Scholar] [CrossRef]
- Rhodes, D.A.; Chen, H.C.; Price, A.J.; Keeble, A.H.; Davey, M.S.; James, L.C.; Eberl, M.; Trowsdale, J. Activation of human gammadelta T cells by cytosolic interactions of BTN3A1 with soluble phosphoantigens and the cytoskeletal adaptor periplakin. J. Immunol. 2015, 194, 2390–2398. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Human Protein Atlas, P. Human Protein Atlas. Available online: http://www.proteinatlas.org (accessed on 20 April 2021).
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Nonacus. Nonacus Probe Design Tool. Available online: https://mynonacus.nonacus.com/view-panel-designs (accessed on 8 February 2021).
- Simms, V. 5 Tips for Using the Nonacus Panel Design Tool. Available online: https://nonacus.com/blog-get-great-coverage-for-the-genes-you-care-about/ (accessed on 27 September 2024).
- Nonacus. Custom NGS Panel Design Tool. Available online: https://nonacus.com/panel-design/ (accessed on 2 October 2024).
- Andrews, S. FastQC: A Quality Control Analysis Tool for High Throughput Sequencing Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 2 October 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra, 1st ed.; O’Reilly Media: Sebastopol, CA, USA, 2020. [Google Scholar]
- Zhao, S.; Agafonov, O.; Azab, A.; Stokowy, T.; Hovig, E. Accuracy and efficiency of germline variant calling pipelines for human genome data. Sci. Rep. 2020, 10, 20222. [Google Scholar] [CrossRef]
- Cucco, F.; Barrans, S.; Sha, C.; Clipson, A.; Crouch, S.; Dobson, R.; Chen, Z.; Thompson, J.S.; Care, M.A.; Cummin, T.; et al. Distinct genetic changes reveal evolutionary history and heterogeneous molecular grade of DLBCL with MYC/BCL2 double-hit. Leukemia 2020, 34, 1329–1341. [Google Scholar] [CrossRef]
- Cucco, F.; Clipson, A.; Kennedy, H.; Sneath Thompson, J.; Wang, M.; Barrans, S.; van Hoppe, M.; Ochoa Ruiz, E.; Caddy, J.; Hamid, D.; et al. Mutation screening using formalin-fixed paraffin-embedded tissues: A stratified approach according to DNA quality. Lab Investig. 2018, 98, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Matthews, J. A Snakemake Pipeline for Analysing (Cancer) DNA Sequencing Data. Available online: https://gitlab.com/jdm204/dnaseq_snakemake (accessed on 21 October 2022).
- Kawaguchi, S. HLA-HD. Available online: https://w3.genome.med.kyoto-u.ac.jp/HLA-HD/ (accessed on 17 March 2023).
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Fedele, G.; Celardo, I.; Loh, S.H.Y.; Martins, L.M. Parp mutations protect from mitochondrial toxicity in Alzheimer’s disease. Cell Death Dis. 2021, 12, 651, Erratum in Cell Death Dis. 2021, 12, 720. [Google Scholar] [CrossRef] [PubMed]
- Grueneberg, A.; de Los Campos, G. BGData—A Suite of R Packages for Genomic Analysis with Big Data. G3 2019, 9, 1377–1383. [Google Scholar] [CrossRef]
- NCBI. SNP. Available online: https://www.ncbi.nlm.nih.gov/snp (accessed on 25 June 2024).
- Lefranc, M.P.; Giudicelli, V.; Duroux, P.; Jabado-Michaloud, J.; Folch, G.; Aouinti, S.; Carillon, E.; Duvergey, H.; Houles, A.; Paysan-Lafosse, T.; et al. IMGT(R), the international ImMunoGeneTics information system(R) 25 years on. Nucleic Acids Res. 2015, 43, D413–D422. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Davydov, A.N.; Frenkel, F.E.; Fanchi, L.; Zolotareva, O.I.; Hemmers, S.; Putintseva, E.V.; Obraztsova, A.S.; Shugay, M.; et al. Antigen receptor repertoire profiling from RNA-seq data. Nat. Biotechnol. 2017, 35, 908–911. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.H. Handbook of Biological Statistics, 3rd ed.; Sparky House Publishing: Baltimore, MD, USA, 2014. [Google Scholar]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Mölder, F.; Jablonski, K.; Letcher, B.; Hall, M.; Tomkins-Tinch, C.; Sochat, V.; Forster, J.; Lee, S.; Twardziok, S.; Kanitz, A.; et al. Sustainable data analysis with Snakemake [version 1; peer review: 1 approved, 1 approved with reservations]. F1000Research 2021, 10, 33. [Google Scholar] [CrossRef]
- Schneider, V.A.; Graves-Lindsay, T.; Howe, K.; Bouk, N.; Chen, H.C.; Kitts, P.A.; Murphy, T.D.; Pruitt, K.D.; Thibaud-Nissen, F.; Albracht, D.; et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017, 27, 849–864. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Dilthey, A.; Leslie, S.; Moutsianas, L.; Shen, J.; Cox, C.; Nelson, M.R.; McVean, G. Multi-population classical HLA type imputation. PLoS Comput. Biol. 2013, 9, e1002877. [Google Scholar] [CrossRef]
- Gustavsen, J.; Rüeger, S.; Chamberlain, S.; Ushey, K.; Zhu, H. rsnps: Get ‘SNP’ (‘Single-Nucleotide’ ‘Polymorphism’) Data on the Web. 2024. Available online: https://github.com/ropensci/rsnps/ (accessed on 21 July 2024).
- Graffelman, J. Exploring Diallelic Genetic Markers: The HardyWeinberg Package. J. Stat. Softw. 2015, 64, 1–23. [Google Scholar] [CrossRef]
- Kawaguchi, S.; Higasa, K.; Shimizu, M.; Yamada, R.; Matsuda, F. HLA-HD: An accurate HLA typing algorithm for next-generation sequencing data. Hum. Mutat. 2017, 38, 788–797. [Google Scholar] [CrossRef] [PubMed]
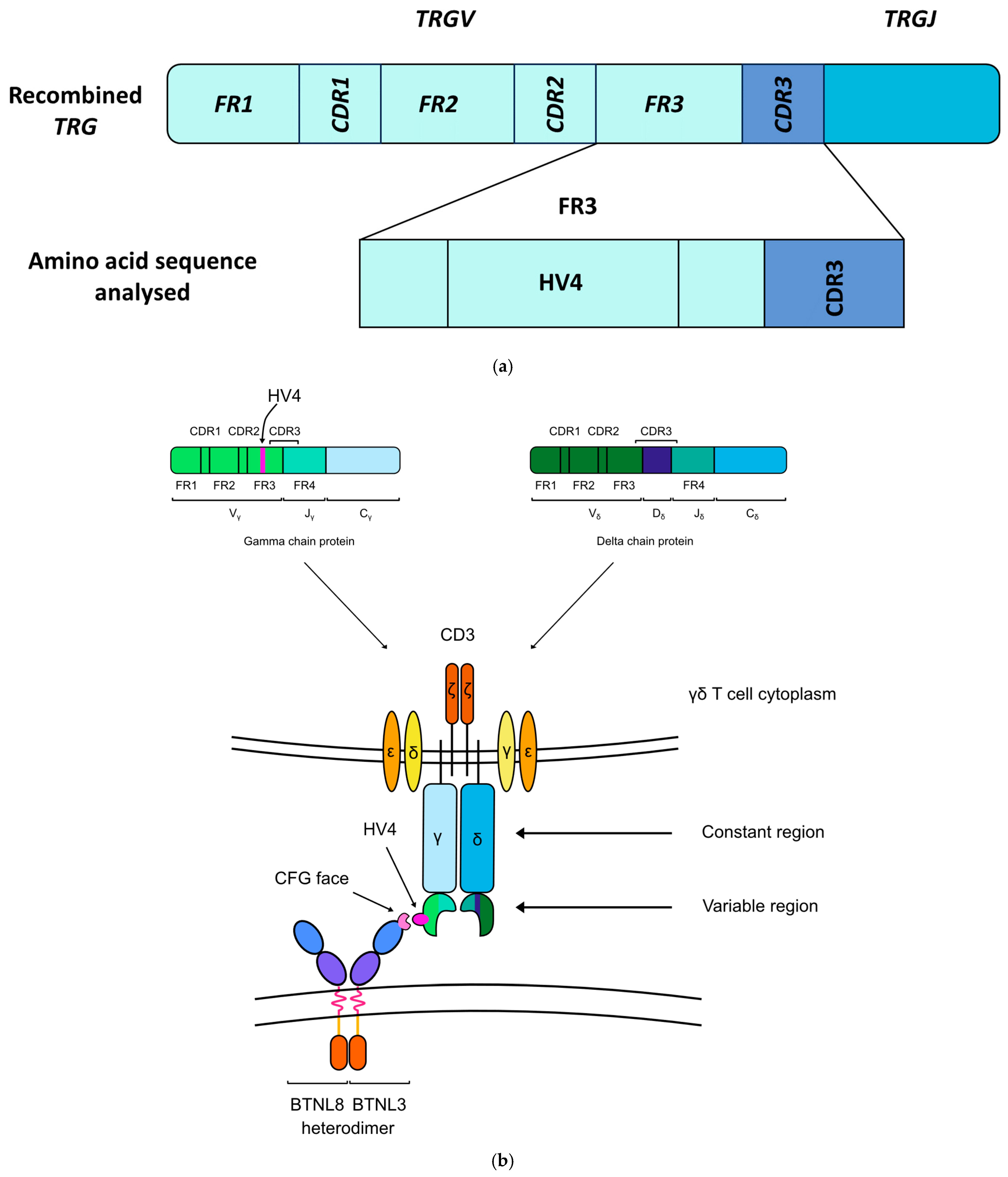
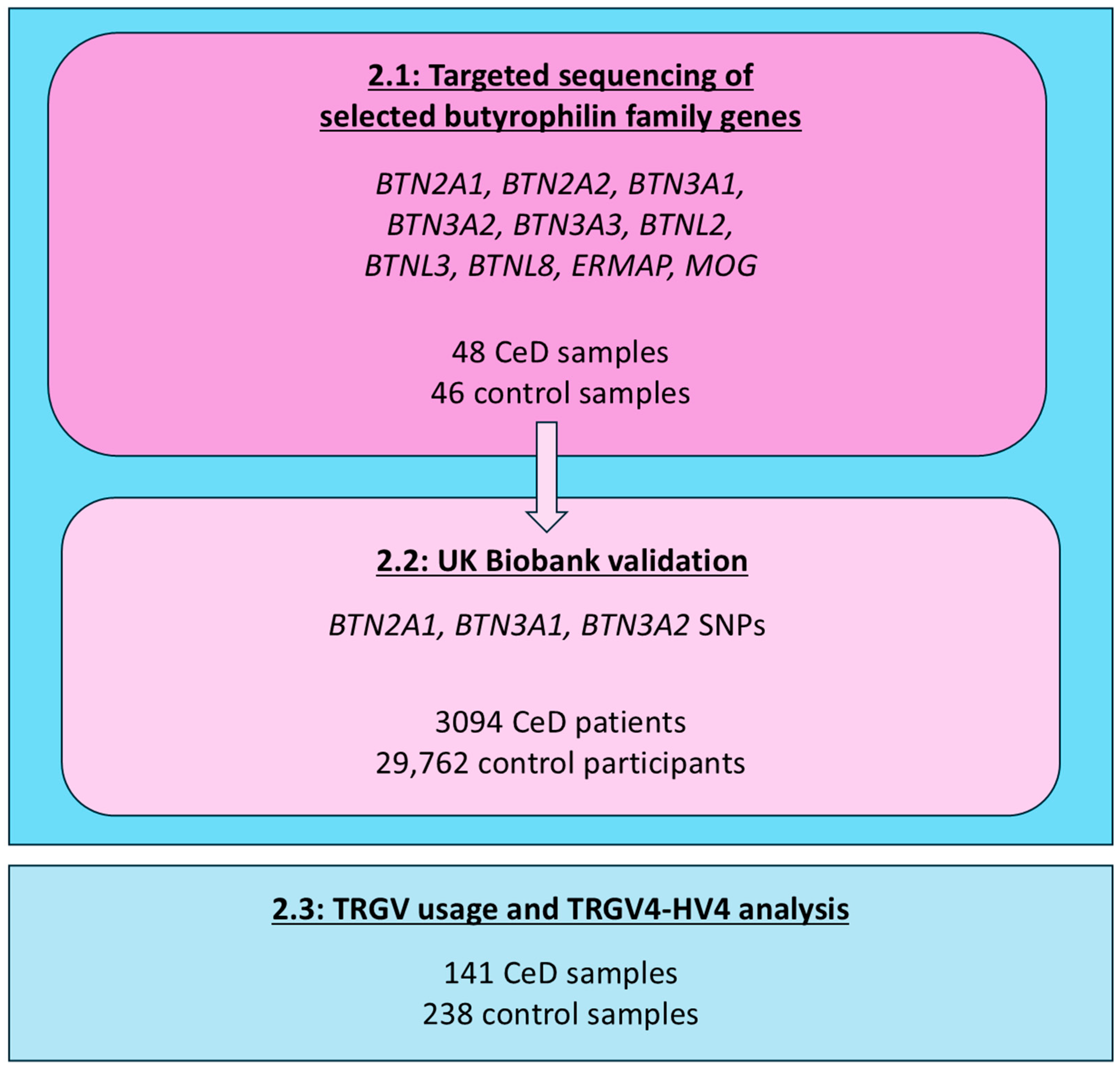
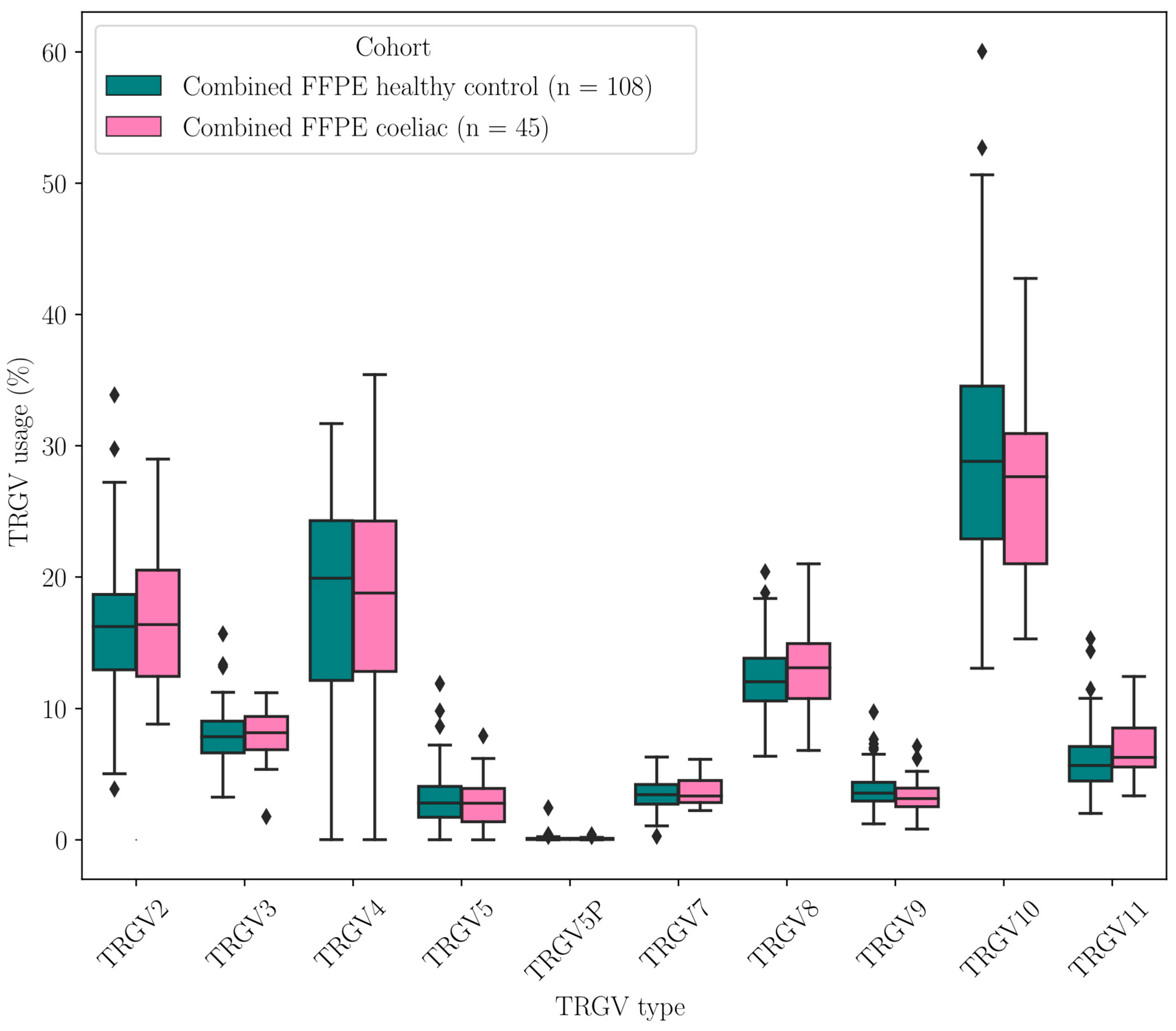

| Butyrophilins | γδ T Cell Subset | Role of Butyrophilins | References | |
|---|---|---|---|---|
| Peripheral blood | Mouse unidentified | Unidentified | Unidentified | NA |
| Alpaca BTN3 | Vγ9Vδ2+ T cells | No interaction has been identified | [35,36] | |
| Human BTN3A homodimers/ heterodimers and BTN2A1 homodimer | Vγ9Vδ2+ T cells | Phosphoantigen-mediated, CDR3-independent γδ T cell activation | [30,33,37,38] | |
| Skin | Mouse Skint1 and Skint2 | Vγ5Vδ1+ DETC | Thymic selection, tissue homing of dendritic epidermal T cells to the skin | [27,28,32] |
| Human? | Vδ1+ T cells | Unidentified, unknown if there is butyrophilin involvement | [39] | |
| Intestinal epithelium | Mouse Btnl1 and Btnl6 | Vγ7+ IEL | Phenotypic maintenance of the intestinal IEL compartment | [27,28] |
| Human BTNL3 and BTNL8 | Vγ4Vδ1+ IEL | Phenotypic maintenance of the intestinal IEL compartment | [21,27,29] |
| (a) | |||||||||||
| Gene | Qual. SNPs | CeD N(≥1 HET) | CeD N(≥2 HET) | CeD N(HOM ALT) | CeD Total Allele Count | Control N(≥1 HET) | Control N(≥2 HET) | Control N(HOM ALT) | Control Total Allele Count | Dominant Model p-Value | Recessive Model p-Value |
| BTN2A1 | 3 | 22 | 21 | 3 | 81 | 5 | 4 | 0 | 13 | 1.46 × 10−5 | 3.70 × 10−8 |
| BTN3A2 | 1 | 5 | 0 | 1 | 7 | 9 | 0 | 1 | 11 | 0.929 | 0.946 |
| ERMAP | 1 | 21 | 0 | 8 | 37 | 20 | 0 | 7 | 34 | 0.516 | 0.988 |
| (b) BTN2A1 variants significantly associated with CeD risk | |||||||||||
| Position (GRCh38) | rsID | Variation | Impact | HET CeD | HOM ALT CeD | HET Control | HOM ALT Control | Alt Allele in CeD | Alt Allele in Controls | ||
| 6:26463432 | rs13195509 | G > A | Missense variant, Val > Met | 43.8% (21) | 6.3% (3) | 8.7% (4) | 0.0% (0) | 27.1% (26) | 4.3% (4) | ||
| 6:26468098 | rs3734542 | G > A | Missense variant, Arg > Gln | 45.8% (22) | 6.3% (3) | 10.9% (5) | 0.0% (0) | 29.2% (28) | 5.4% (5) | ||
| 6:26468317 | rs3734543 | G > C | Missense variant, Gly > Ala | 43.8% (21) | 6.3% (3) | 8.7% (4) | 0.0% (0) | 28.1% (27) | 4.3% (4) | ||
| Gene | SNPs in NCBI | Unique SNPs in NCBI | SNPs in UK Biobank |
|---|---|---|---|
| BTN2A1 | 7912 | 7605 | 30 |
| BTN3A1 | 5348 | 5164 | 27 |
| BTN3A2 | 5905 | 5611 | 21 |
| BTNL3 | 6164 | 5929 | 10 |
| BTNL8 | 18,889 | 18,197 | 13 |
| Position (GRCh38) | SNP, Reference Allele | Gene | SNP Consequence | CeD Allele Freq | Control Allele Freq | Total Allele Freq | ln(OR) | CeD Risk | Adjusted p-Value |
|---|---|---|---|---|---|---|---|---|---|
| 6:26463347 | rs13195402 | BTN2A1 | STOP gained | 0.768 | 0.892 | 0.880 | −0.924 | decrease | 4.67 × 10−158 |
| 6:26463432 | rs13195509 | BTN2A1 | missense | 0.754 | 0.879 | 0.867 | −0.857 | decrease | 1.61 × 10−151 |
| 6:26475927 | rs1407045 | BTN2A1 | intronic | 0.584 | 0.516 | 0.522 | 0.273 | increase | 6.07 × 10−22 |
| 6:26465807 | rs2273558 | BTN2A1 | intronic | 0.583 | 0.677 | 0.667 | −0.396 | decrease | 1.69 × 10−41 |
| 6:26460493 | rs2893856 | BTN2A1 | intronic | 0.113 | 0.131 | 0.130 | −0.175 | decrease | 3.23 × 10−3 |
| 6:26468098 | rs3734542 | BTN2A1 | missense | 0.753 | 0.878 | 0.867 | −0.855 | decrease | 8.59 × 10−151 |
| 6:26468317 | rs3734543 | BTN2A1 | missense | 0.760 | 0.879 | 0.868 | −0.844 | decrease | 1.59 × 10−140 |
| 6:26466954 | rs3799380 | BTN2A1 | intronic | 0.683 | 0.790 | 0.780 | −0.549 | decrease | 8.59 × 10−77 |
| 6:26474343 | rs56296968 | BTN2A1 | intronic | 0.696 | 0.807 | 0.796 | −0.604 | decrease | 9.70 × 10−89 |
| 6:26456215 | rs6456724 | BTN2A1 | 2 kb upstream | 0.113 | 0.131 | 0.130 | −0.176 | decrease | 2.87 × 10−3 |
| 6:26458037 | rs6929846 | BTN2A1 | 5′ UTR | 0.146 | 0.174 | 0.172 | −0.206 | decrease | 3.60 × 10−6 |
| 6:26473816 | rs7773938 | BTN2A1 | intronic | 0.696 | 0.806 | 0.796 | −0.600 | decrease | 1.15 × 10−87 |
| 6:26469647 | rs9358944 | BTN2A1 | intronic | 0.695 | 0.806 | 0.796 | −0.604 | decrease | 1.55 × 10−89 |
| 6:26471886 | rs9358945 | BTN2A1 | intronic | 0.694 | 0.806 | 0.796 | −0.606 | decrease | 4.37 × 10−90 |
| 6:26404730 | rs10456045 | BTN3A1 | intronic | 0.596 | 0.698 | 0.688 | −0.448 | decrease | 2.97 × 10−57 |
| 6:26410572 | rs1796520 | BTN3A1 | intronic | 0.405 | 0.474 | 0.467 | −0.276 | decrease | 2.40 × 10−22 |
| 6:26404146 | rs3799378 | BTN3A1 | intronic | 0.653 | 0.762 | 0.752 | −0.535 | decrease | 2.92 × 10−75 |
| 6:26405825 | rs3857549 | BTN3A1 | intronic | 0.948 | 0.935 | 0.936 | 0.221 | increase | 1.53 × 10−2 |
| 6:26409662 | rs41266839 | BTN3A1 | missense | 0.764 | 0.892 | 0.880 | −0.924 | decrease | 2.12 × 10−168 |
| 6:26407180 | rs4609015 | BTN3A1 | intronic | 0.871 | 0.854 | 0.855 | 0.141 | increase | 3.82 × 10−2 |
| 6:26412860 | rs6900725 | BTN3A1 | intronic | 0.870 | 0.853 | 0.855 | 0.139 | increase | 4.33 × 10−2 |
| 6:26401210 | rs6912853 | BTN3A1 | 2 kb upstream | 0.863 | 0.844 | 0.846 | 0.153 | increase | 7.85 × 10−3 |
| 6:26413007 | rs6920986 | BTN3A1 | intronic | 0.870 | 0.854 | 0.856 | 0.138 | increase | 4.99 × 10−2 |
| 6:26415409 | rs742090 | BTN3A1 | 500 b downstream | 0.406 | 0.474 | 0.468 | −0.276 | decrease | 3.58 × 10−22 |
| 6:26374321 | rs11758089 | BTN3A2 | intronic | 0.866 | 0.844 | 0.846 | 0.176 | increase | 6.30 × 10−4 |
| 6:26372558 | rs12176317 | BTN3A2 | intronic | 0.744 | 0.867 | 0.856 | −0.809 | decrease | 6.72 × 10−140 |
| 6:26366990 | rs12199613 | BTN3A2 | intronic | 0.514 | 0.612 | 0.602 | −0.400 | decrease | 1.76 × 10−47 |
| 6:26377318 | rs1977 | BTN3A2 | 3′ UTR | 0.740 | 0.864 | 0.853 | −0.808 | decrease | 1.17 × 10−136 |
| 6:26377363 | rs1979 | BTN3A2 | 3′ UTR | 0.743 | 0.867 | 0.855 | −0.809 | decrease | 8.23 × 10−140 |
| 6:26375933 | rs1985732 | BTN3A2 | intronic | 0.595 | 0.698 | 0.688 | −0.457 | decrease | 2.87 × 10−59 |
| 6:26374430 | rs2073526 | BTN3A2 | intronic | 0.370 | 0.442 | 0.435 | −0.295 | decrease | 9.15 × 10−25 |
| 6:26363527 | rs9358934 | BTN3A2 | 2 kb upstream | 0.744 | 0.866 | 0.855 | −0.803 | decrease | 2.34 × 10−137 |
| 6:26364702 | rs9379855 | BTN3A2 | 2 kb upstream | 0.743 | 0.866 | 0.855 | −0.804 | decrease | 8.85 × 10−138 |
| 6:26367461 | rs9379858 | BTN3A2 | intronic | 0.743 | 0.866 | 0.855 | −0.802 | decrease | 3.19 × 10−137 |
| 6:26369321 | rs9379859 | BTN3A2 | intronic | 0.744 | 0.867 | 0.855 | −0.803 | decrease | 5.37 × 10−137 |
| 6:26373450 | rs9393713 | BTN3A2 | intronic | 0.743 | 0.868 | 0.856 | −0.814 | decrease | 1.07 × 10−141 |
| 6:26373512 | rs9393714 | BTN3A2 | intronic | 0.743 | 0.868 | 0.856 | −0.813 | decrease | 6.99 × 10−141 |
| SNP, Reference Allele | Gene | SNP Consequence | ln(OR) | CeD Risk | Adjusted p-Value |
|---|---|---|---|---|---|
| rs13195402 | BTN2A1 | STOP gained | −0.20727 | decrease | 8.15 × 10−6 |
| rs13195509 | BTN2A1 | missense | −0.19239 | decrease | 1.62 × 10−5 |
| rs3734542 | BTN2A1 | missense | −0.18831 | decrease | 2.94 × 10−5 |
| rs3734543 | BTN2A1 | missense | −0.16744 | decrease | 8.23 × 10−4 |
| rs56296968 | BTN2A1 | intronic | −0.11753 | decrease | 4.20 × 10−2 |
| rs9358944 | BTN2A1 | intronic | −0.11786 | decrease | 3.83 × 10−2 |
| rs9358945 | BTN2A1 | intronic | −0.12018 | decrease | 2.91 × 10−2 |
| rs3799378 | BTN3A1 | intronic | −0.14327 | decrease | 7.04 × 10−4 |
| rs41266839 | BTN3A1 | missense | −0.21469 | decrease | 1.06 × 10−6 |
| rs12176317 | BTN3A2 | intronic | −0.1974 | decrease | 3.50 × 10−6 |
| rs12199613 | BTN3A2 | intronic | −0.12296 | decrease | 3.31 × 10−3 |
| rs1977 | BTN3A2 | 3′ UTR | −0.20238 | decrease | 2.06 × 10−6 |
| rs1979 | BTN3A2 | 3′ UTR | −0.19756 | decrease | 3.40 × 10−6 |
| rs1985732 | BTN3A2 | intronic | −0.10975 | decrease | 3.35 × 10−2 |
| rs9358934 | BTN3A2 | 2 kb upstream | −0.19286 | decrease | 7.53 × 10−6 |
| rs9379855 | BTN3A2 | 2 kb upstream | −0.19406 | decrease | 6.04 × 10−6 |
| rs9379858 | BTN3A2 | intronic | −0.19156 | decrease | 8.99 × 10−6 |
| rs9379859 | BTN3A2 | intronic | −0.19261 | decrease | 8.10 × 10−6 |
| rs9393713 | BTN3A2 | intronic | −0.2056 | decrease | 9.27 × 10−7 |
| rs9393714 | BTN3A2 | intronic | −0.20087 | decrease | 2.08 × 10−6 |
| HLA Genotype of Individuals in Model | Number of CeD Participants | Number of Controls | Number of Significant SNPs |
|---|---|---|---|
| HLA-DQ2.2 | 199 | 4154 | 0 |
| HLA-DQ2.5 | 1652 | 6416 | 21 |
| HLA-DQ8 | 171 | 4203 | 0 |
| HLA-DQ2.2, HLA-DQ2.5 | 606 | 895 | 0 |
| HLA-DQ2.2, HLA-DQ8 | 50 | 590 | 0 |
| HLA-DQ2.5, HLA-DQ8 | 182 | 886 | 0 |
| Other | 234 | 12,618 | 0 |
| SNP, Reference Allele | Gene | SNP Consequence | CeD Allele Freq | Control Allele Freq | Total Allele Freq | ln(OR) | CeD Risk | Adjusted p-Value |
|---|---|---|---|---|---|---|---|---|
| rs13195402 | BTN2A1 | STOP gained | 0.704 | 0.751 | 0.741 | −0.27812 | decrease | 5.10 × 10−7 |
| rs13195509 | BTN2A1 | missense | 0.687 | 0.734 | 0.724 | −0.25542 | decrease | 1.75 × 10−6 |
| rs3734542 | BTN2A1 | missense | 0.687 | 0.733 | 0.723 | −0.25235 | decrease | 2.52 × 10−6 |
| rs3734543 | BTN2A1 | missense | 0.695 | 0.736 | 0.728 | −0.23279 | decrease | 5.43 × 10−5 |
| rs56296968 | BTN2A1 | intronic | 0.640 | 0.673 | 0.666 | −0.15928 | decrease | 2.33 × 10−2 |
| rs7773938 | BTN2A1 | intronic | 0.640 | 0.672 | 0.666 | −0.15755 | decrease | 2.66 × 10−2 |
| rs9358944 | BTN2A1 | intronic | 0.638 | 0.671 | 0.664 | −0.16238 | decrease | 1.61 × 10−2 |
| rs9358945 | BTN2A1 | intronic | 0.638 | 0.671 | 0.665 | −0.16499 | decrease | 1.26 × 10−2 |
| rs3799378 | BTN3A1 | intronic | 0.596 | 0.637 | 0.628 | −0.18712 | decrease | 8.10 × 10−4 |
| rs41266839 | BTN3A1 | missense | 0.697 | 0.747 | 0.737 | −0.28267 | decrease | 7.25 × 10−8 |
| rs12176317 | BTN3A2 | intronic | 0.679 | 0.729 | 0.719 | −0.26575 | decrease | 2.82 × 10−7 |
| rs12199613 | BTN3A2 | intronic | 0.459 | 0.500 | 0.491 | −0.1717 | decrease | 2.06 × 10−3 |
| rs1977 | BTN3A2 | 3′ UTR | 0.676 | 0.726 | 0.716 | −0.268 | decrease | 2.99 × 10−7 |
| rs1979 | BTN3A2 | 3′ UTR | 0.679 | 0.728 | 0.718 | −0.26376 | decrease | 3.63 × 10−7 |
| rs1985732 | BTN3A2 | intronic | 0.539 | 0.574 | 0.567 | −0.15063 | decrease | 2.35 × 10−2 |
| rs9358934 | BTN3A2 | 2 kb upstream | 0.680 | 0.729 | 0.719 | −0.25825 | decrease | 8.85 × 10−7 |
| rs9379855 | BTN3A2 | 2 kb upstream | 0.680 | 0.728 | 0.718 | −0.25967 | decrease | 6.76 × 10−7 |
| rs9379858 | BTN3A2 | intronic | 0.680 | 0.728 | 0.718 | −0.25566 | decrease | 1.18 × 10−6 |
| rs9379859 | BTN3A2 | intronic | 0.681 | 0.729 | 0.719 | −0.26105 | decrease | 6.31 × 10−7 |
| rs9393713 | BTN3A2 | intronic | 0.678 | 0.729 | 0.719 | −0.27313 | decrease | 1.06 × 10−7 |
| rs9393714 | BTN3A2 | intronic | 0.679 | 0.729 | 0.719 | −0.26778 | decrease | 2.25 × 10−7 |
| Coeliac Disease | Healthy Control | Sequencing Method | |
|---|---|---|---|
| Dataset 1 | 34 FFPE, 12 fresh frozen duodenal | 97 FFPE duodenal | Lymphotrack (Invivoscribe Inc., San Diego, CA, USA) and Illumina Miseq micro (San Diego, CA, USA) |
| Dataset 2 | 11 FFPE duodenal | 11 FFPE duodenal | Lymphotrack (Invivoscribe Inc.) and Illumina Miseq |
| Dataset 3 | 84 blood | 130 blood | Illumina NextSeq |
| Combined | 84 blood, 48 FFPE duodenal, 12 fresh frozen duodenal | 130 blood, 108 FFPE duodenal | NA |
| (a) | |||||
| HV4 Amino Acid Sequence | Amino Acid Change | Effect | Freq. in Healthy Control Samples (n = 238) | Freq. in CeD Samples (n = 141) | Predicted Change in Binding [31] |
| KYDTYGSTRKNLRMILR (WT) | - | - | 430/476 = 0.903 | 254/282 = 0.901 | - |
| KYDTYGSTRQNLRMILR | Lysine > Glutamine | Positive charge > polar uncharged | 41/476 = 0.086 | 23/282 = 0.082 | Marginal reduction in binding |
| KYDTYGSTRKSLRMILR | Asparagine > Serine | Polar uncharged > polar uncharged | 4/476 = 0.008 | 2/282 = 0.007 | Unknown |
| KYDTYGSTR_ELENDTA | Lysine > frameshift | Positive charge > different sequence | 0 | 1/282 = 0.003 | Unknown |
| KYNTYGSTRKNLRMILR | Aspartic acid > Asparagine | Negative charge > polar uncharged | 0 | 1/282 = 0.003 | Disrupted binding |
| KYDTYGNTRKNLRMILR | Serine > Asparagine | Polar uncharged > polar uncharged | 1/476 = 0.002 | 0 | Unknown |
| KYDTYGSIRKNLRMILR | Threonine > Isoleucine | Polar uncharged > apolar | 0 | 1/282 = 0.003 | Unknown |
| (b) | |||||
| Phenotype | Combined Healthy Control Samples (n = 238) | Combined CeD Samples (n = 141) | |||
| WT | 202 | 116 | |||
| WT, KYDTYGSTRQNLRMILR | 23 | 18 | |||
| KYDTYGSTRQNLRMILR | 9 | 2 | |||
| WT, KYDTYGSTRKSLRMILR | 2 | 2 | |||
| KYDTYGSTRQNLRMILR, KYDTYGSTR_ELENDTA | 0 | 1 | |||
| WT, KYNTYGSTRKNLRMILR | 0 | 1 | |||
| WT, KYDTYGNTRKNLRMILR | 1 | 0 | |||
| KYDTYGSTRKSLRMILR | 1 | 0 | |||
| WT, KYDTYGSIRKNLRMILR | 0 | 1 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luu Hoang, K.N.; Evans, S.; Willis, T.W.; Davies, K.; Kockelbergh, H.; Silcock, L.; Piechocki, K.; Fowler, A.; Soilleux, E.J. BTN2A1 and BTN3A1 as Novel Coeliac Disease Risk Loci: An In Silico Analysis. Int. J. Mol. Sci. 2025, 26, 10697. https://doi.org/10.3390/ijms262110697
Luu Hoang KN, Evans S, Willis TW, Davies K, Kockelbergh H, Silcock L, Piechocki K, Fowler A, Soilleux EJ. BTN2A1 and BTN3A1 as Novel Coeliac Disease Risk Loci: An In Silico Analysis. International Journal of Molecular Sciences. 2025; 26(21):10697. https://doi.org/10.3390/ijms262110697
Chicago/Turabian StyleLuu Hoang, Kim Ngan, Shelley Evans, Thomas W. Willis, Kate Davies, Hannah Kockelbergh, Lee Silcock, Kim Piechocki, Anna Fowler, and Elizabeth J. Soilleux. 2025. "BTN2A1 and BTN3A1 as Novel Coeliac Disease Risk Loci: An In Silico Analysis" International Journal of Molecular Sciences 26, no. 21: 10697. https://doi.org/10.3390/ijms262110697
APA StyleLuu Hoang, K. N., Evans, S., Willis, T. W., Davies, K., Kockelbergh, H., Silcock, L., Piechocki, K., Fowler, A., & Soilleux, E. J. (2025). BTN2A1 and BTN3A1 as Novel Coeliac Disease Risk Loci: An In Silico Analysis. International Journal of Molecular Sciences, 26(21), 10697. https://doi.org/10.3390/ijms262110697







