Small RNA-Seq Reveals the Effect of Formaldehyde Treatment on Chicken Embryo Liver microRNA Profiles
Abstract
1. Introduction
2. Results
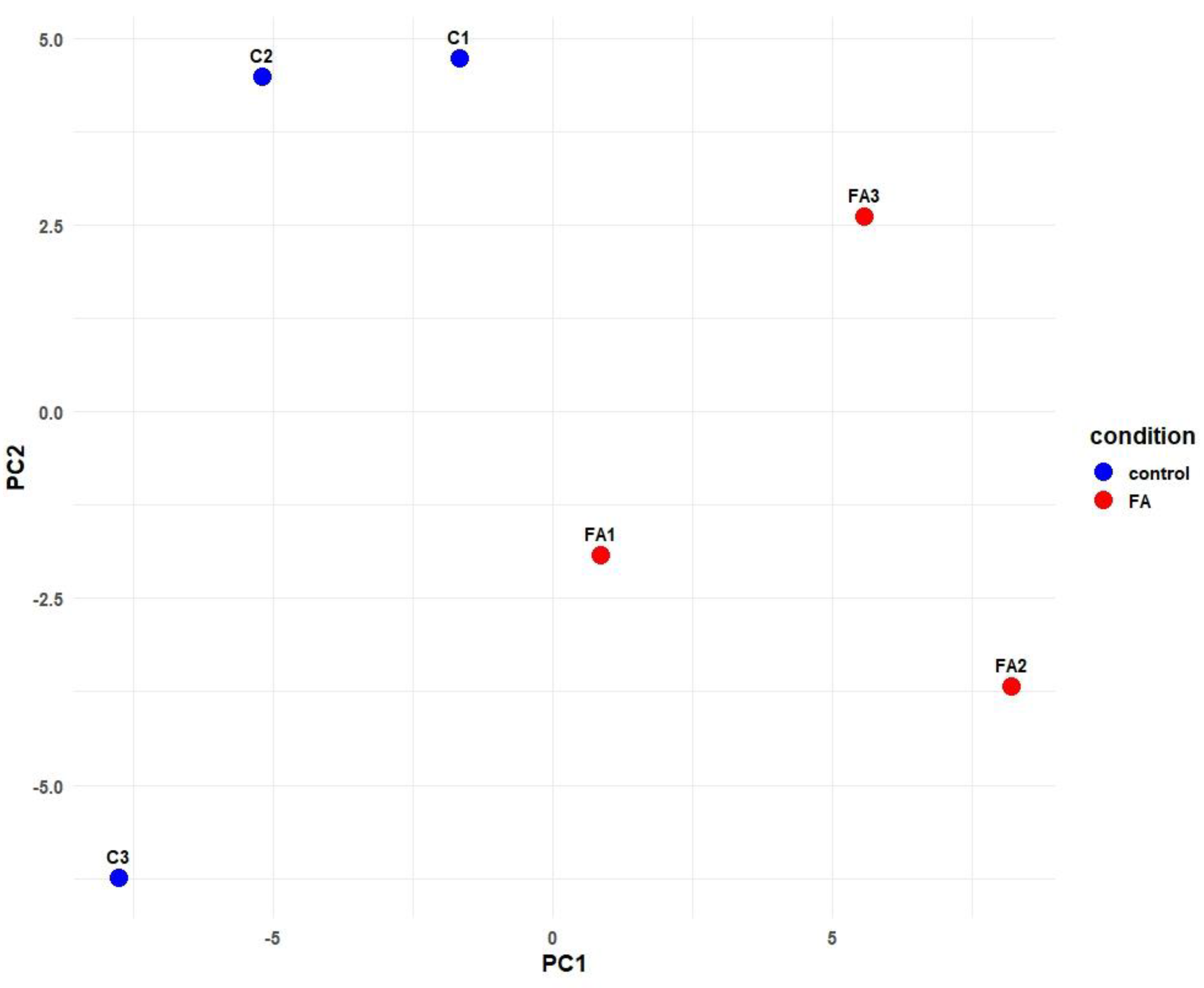
2.1. miRNAs Sequencing Data
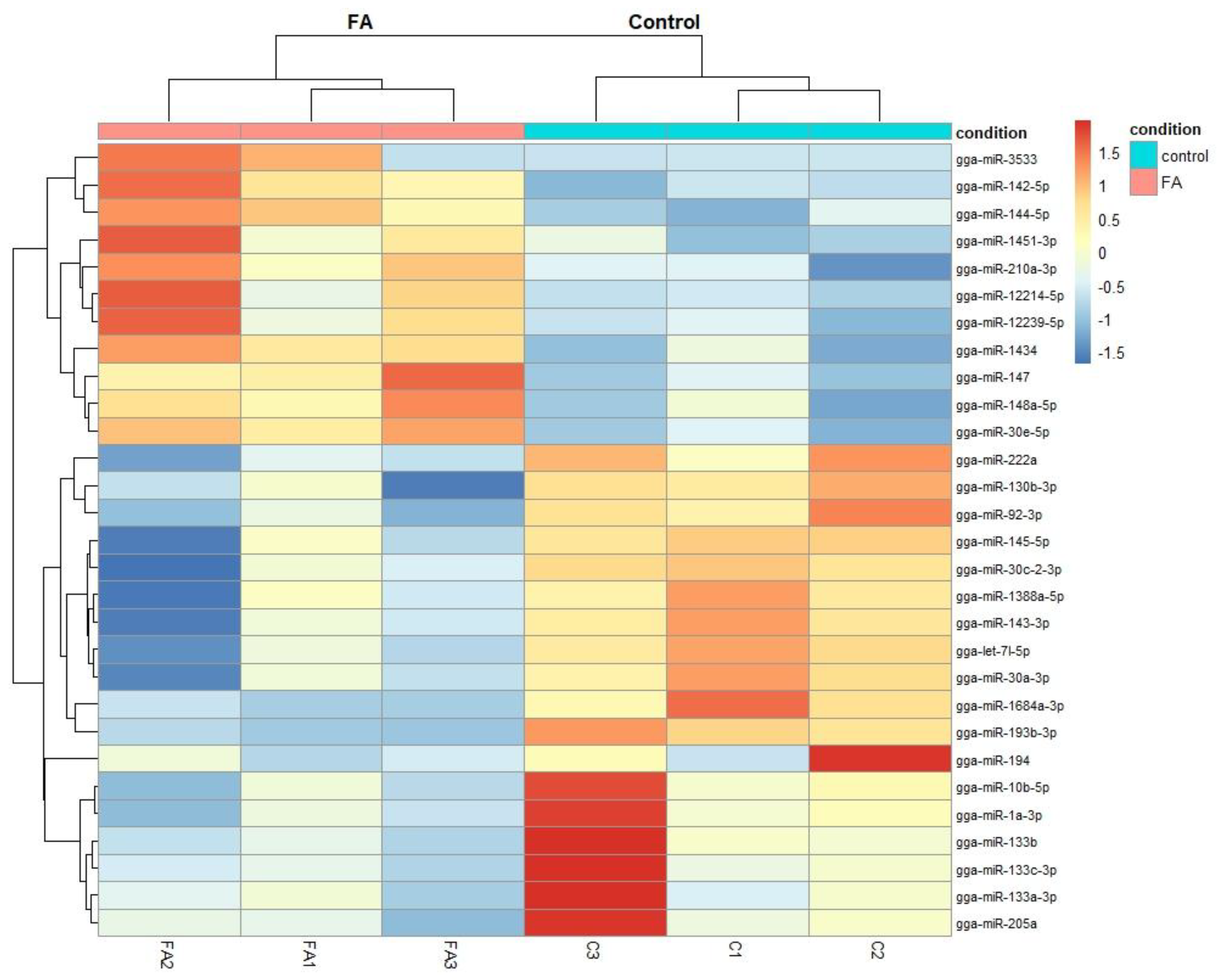
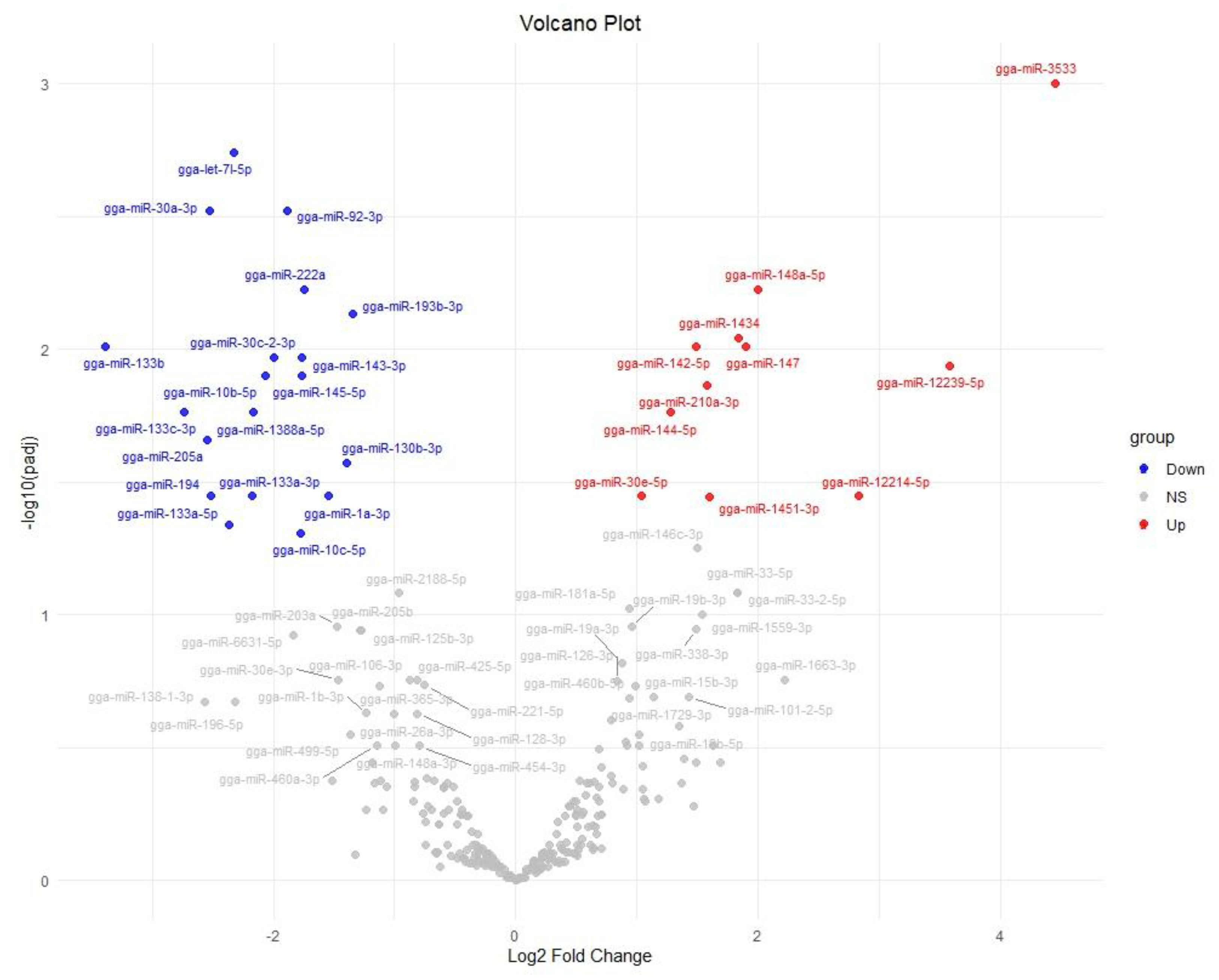
2.2. Differentially Expressed miRNA in the Chicken Between FA and Control
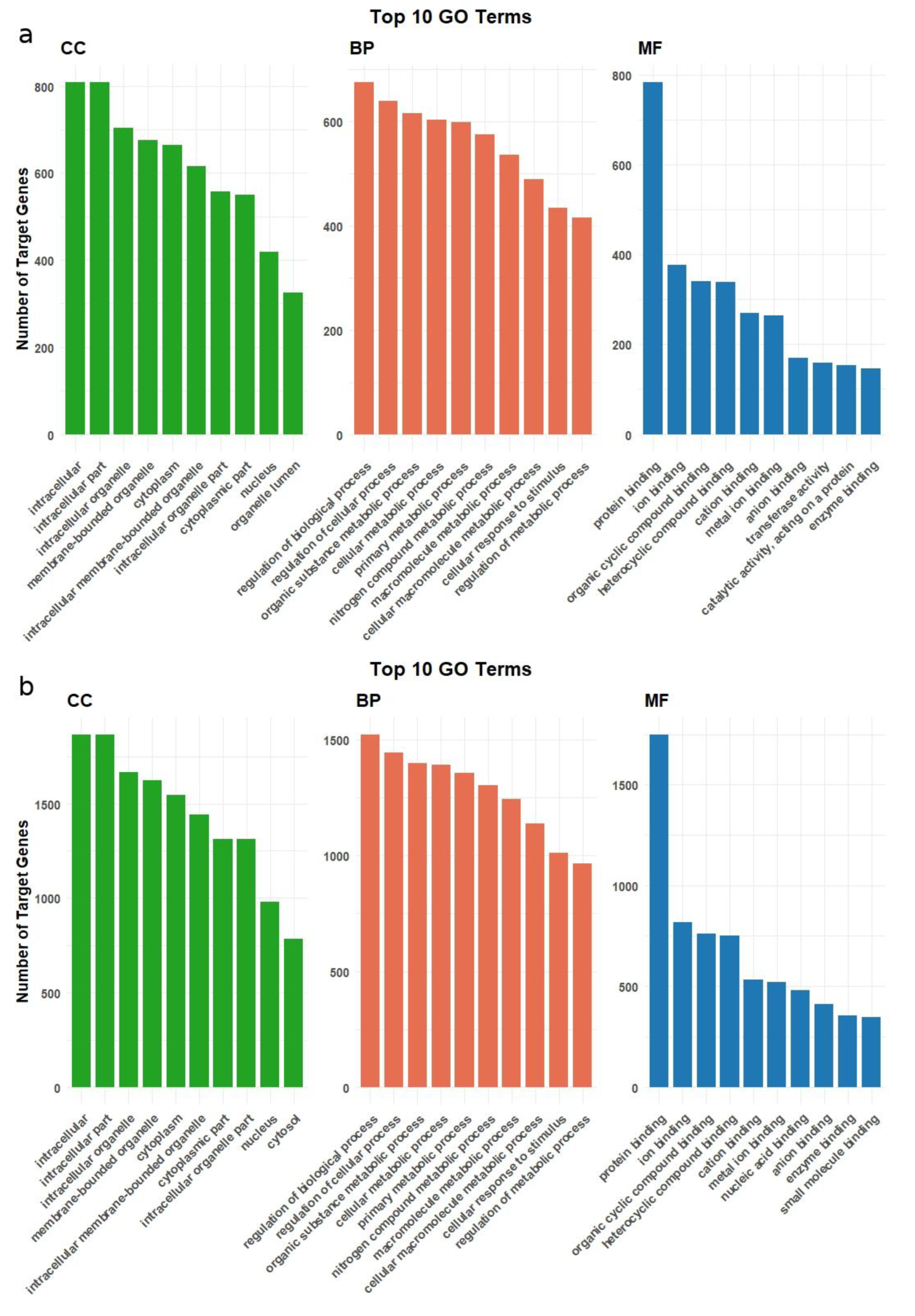
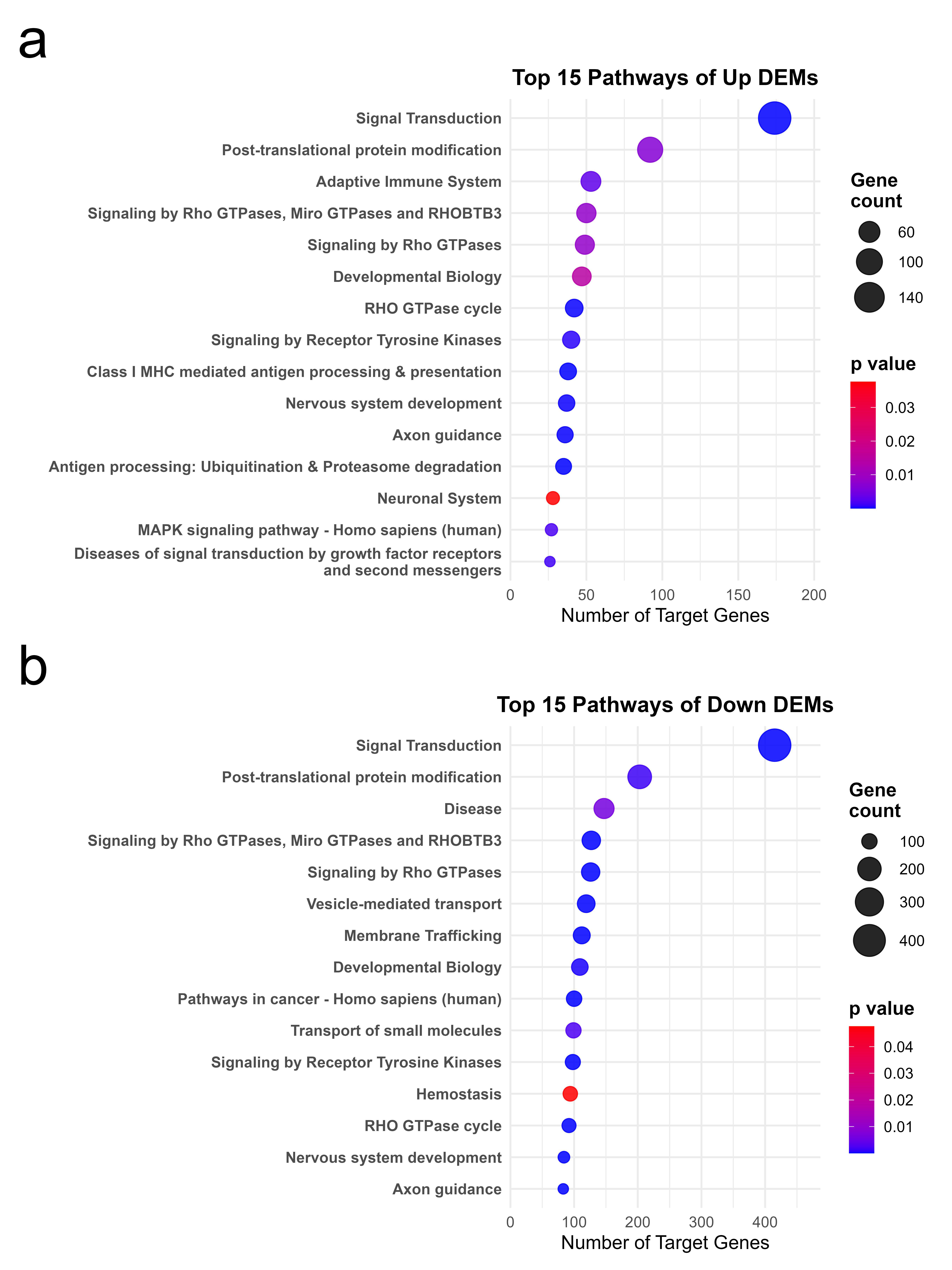
2.3. Functional Enrichment and Pathway Analysis of Target Genes of Differentially Expressed miRNAs
3. Discussion
4. Materials and Methods
4.1. Animal Ethics
4.2. Experimental Design and Sample Collection
4.3. RNA Extraction and Quality Control
4.4. Small RNA Library Preparation and Sequencing
4.5. MiRNAs Sequence Data Analysis
4.6. Differentially Expressed miRNAs Analysis
4.7. Target Gene Prediction and Functional Analysis of DEMs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FA | Formaldehyde |
| DEMs | Differentially expressed miRNAs |
| PCA | Principal component analysis |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Zhang, Y.; Yang, Y.; He, X.; Yang, P.; Zong, T.; Sun, P.; Sun, R.; Yu, T.; Jiang, Z. The Cellular Function and Molecular Mechanism of Formaldehyde in Cardiovascular Disease and Heart Development. J. Cell. Mol. Med. 2021, 25, 5358–5371. [Google Scholar] [CrossRef]
- Salthammer, T.; Mentese, S.; Marutzky, R. Formaldehyde in the Indoor Environment. Chem. Rev. 2010, 110, 2536–2572. [Google Scholar] [CrossRef]
- WHO. WHO Guidelines for Indoor Air Quality: Selected Pollutants; WHO: Geneva, Switzerland, 2010. [Google Scholar]
- Cadirci, S. Disinfection of Hatching Eggs by Formaldehyde Fumigation–a Review. Arch. Geflügelk 2009, 73, 116–123. [Google Scholar] [CrossRef]
- Avila, L.P.; Sweeney, K.M.; Schaeffer, C.; Holcombe, N.; Selby, C.; Montiel, E.; Wilson, J.L. Broiler Breeder Feed Treatment with a Formaldehyde-Based Sanitizer and Its Consequences on Reproduction, Feed and Egg Contamination, and Offspring Livability. J. Appl. Poult. Res. 2023, 32, 100330. [Google Scholar] [CrossRef]
- Grafstrom, R.C.; Fornace, A.J.; Autrup, H.; Lechner, J.F.; Harris, C.C. Formaldehyde Damage to DNA and Inhibition of DNA Repair in Human Bronchial Cells. Science 1983, 220, 216–218. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Ospina, M.; Tse, C.; Toth, S.; Caudill, S.P.; Vesper, H.W. Ultraperformance Liquid Chromatography Tandem Mass Spectrometry Method to Determine Formaldehyde Hemoglobin Adducts in Humans as Biomarker for Formaldehyde Exposure. Chem. Res. Toxicol. 2017, 30, 1592–1598. [Google Scholar] [CrossRef]
- Leso, V.; Macrini, M.C.; Russo, F.; Iavicoli, I. Formaldehyde Exposure and Epigenetic Effects: A Systematic Review. Appl. Sci. 2020, 10, 2319. [Google Scholar] [CrossRef]
- Rager, J.E.; Moeller, B.C.; Miller, S.K.; Kracko, D.; Doyle-Eisele, M.; Swenberg, J.A.; Fry, R.C. Formaldehyde-Associated Changes in microRNAs: Tissue and Temporal Specificity in the Rat Nose, White Blood Cells, and Bone Marrow. Toxicol. Sci. 2014, 138, 36–46. [Google Scholar] [CrossRef]
- Ho, D.H.; Burggren, W.W. Epigenetics and Transgenerational Transfer: A Physiological Perspective. J. Exp. Biol. 2010, 213, 3–16. [Google Scholar] [CrossRef]
- Qu, X.; Neuhoff, C.; Cinar, M.U.; Pröll, M.; Tholen, E.; Tesfaye, D.; Hölker, M.; Schellander, K.; Uddin, M.J. Epigenetic Modulation of TLR4 Expression by Sulforaphane Increases Anti-Inflammatory Capacity in Porcine Monocyte-Derived Dendritic Cells. Biology 2021, 10, 490. [Google Scholar] [CrossRef]
- Akyüz, B.; Sohel, M.M.H.; Konca, Y.; Arslan, K.; Gürbulak, K.; Abay, M.; Kaliber, M.; White, S.N.; Cinar, M.U. Effects of Low and High Maternal Protein Intake on Fetal Skeletal Muscle miRNAome in Sheep. Animals 2024, 14, 1594. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA Profiling: Approaches and Considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are microRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.A.; Tembhurne, P.; Liu, H.-C. MicroRNA Expression in Chicken Embryos. Poult. Sci. 2008, 87, 2335–2343. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. microRNA Functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef]
- Bednarczyk, M.; Dunislawska, A.; Stadnicka, K.; Grochowska, E. Chicken Embryo as a Model in Epigenetic Research. Poult. Sci. 2021, 100, 101164. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Baltimore, D. MicroRNAs and Immunity: Tiny Players in a Big Field. Immunity 2007, 26, 133–137. [Google Scholar] [CrossRef]
- Wrenzycki, C.; Niemann, H. Epigenetic Reprogramming in Early Embryonic Development: Effects of in-Vitro Production and Somatic Nuclear Transfer. Reprod. Biomed. Online 2003, 7, 649–656. [Google Scholar] [CrossRef]
- Surugihalli, C.; Farley, L.S.; Beckford, R.C.; Kamkrathok, B.; Liu, H.-C.; Muralidaran, V.; Patel, K.; Porter, T.E.; Sunny, N.E. Remodeling of Hepatocyte Mitochondrial Metabolism and De Novo Lipogenesis During the Embryonic-to-Neonatal Transition in Chickens. Front. Physiol. 2022, 13, 870451. [Google Scholar] [CrossRef]
- Hicks, J.A.; Porter, T.E.; Liu, H.-C. Identification of microRNAs Controlling Hepatic mRNA Levels for Metabolic Genes during the Metabolic Transition from Embryonic to Posthatch Development in the Chicken. BMC Genom. 2017, 18, 687. [Google Scholar] [CrossRef]
- Cogburn, L.A.; Trakooljul, N.; Chen, C.; Huang, H.; Wu, C.H.; Carré, W.; Wang, X.; White, H.B. Transcriptional Profiling of Liver during the Critical Embryo-to-Hatchling Transition Period in the Chicken (Gallus Gallus). BMC Genom. 2018, 19, 1–37. [Google Scholar] [CrossRef]
- Yanai, M.; Tatsumi, N.; Endo, F.; Yokouchi, Y. Analysis of Gene Expression Patterns in the Developing Chick Liver. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2005, 233, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yatskievych, T.A.; Baker, R.K.; Antin, P.B. Regulation of Hex Gene Expression and Initial Stages of Avian Hepatogenesis by Bmp and Fgf Signaling. Dev. Biol. 2004, 268, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Fan, H.; Zhang, B.; Ning, C.; Xing, K.; Guo, Y. Dietary Genistein Supplementation in Laying Broiler Breeder Hens Alters the Development and Metabolism of Offspring Embryos as Revealed by Hepatic Transcriptome Analysis. FASEB J. 2018, 32, 4214–4228. [Google Scholar] [CrossRef]
- Tabish, A.M.; Poels, K.; Hoet, P.; Godderis, L. Epigenetic Factors in Cancer Risk: Effect of Chemical Carcinogens on Global DNA Methylation Pattern in Human TK6 Cells. PLoS ONE 2012, 7, e34674. [Google Scholar] [CrossRef][Green Version]
- Barbosa, E.; dos Santos, A.L.A.; Peteffi, G.P.; Schneider, A.; Müller, D.; Rovaris, D.; Bau, C.H.D.; Linden, R.; Antunes, M.V.; Charão, M.F. Increase of Global DNA Methylation Patterns in Beauty Salon Workers Exposed to Low Levels of Formaldehyde. Environ. Sci. Pollut. Res. 2019, 26, 1304–1314. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, L.; Gong, C.; Tao, G.; Huang, H.; Liu, J.; Zhang, H.; Wu, D.; Xia, B.; Hu, G.; et al. Effects of Long-Term Low-Dose Formaldehyde Exposure on Global Genomic Hypomethylation in 16HBE Cells. Toxicol. Lett. 2011, 205, 235–240. [Google Scholar] [CrossRef]
- Yoshida, I.; Ibuki, Y. Formaldehyde-Induced Histone H3 Phosphorylation via JNK and the Expression of Proto-Oncogenes. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2014, 770, 9–18. [Google Scholar] [CrossRef]
- Ibuki, Y.; Toyooka, T.; Zhao, X.; Yoshida, I. Cigarette Sidestream Smoke Induces Histone H3 Phosphorylation via JNK and PI3K/Akt Pathways, Leading to the Expression of Proto-Oncogenes. Carcinogenesis 2014, 35, 1228–1237. [Google Scholar] [CrossRef]
- Rager, J.E.; Moeller, B.C.; Doyle-Eisele, M.; Kracko, D.; Swenberg, J.A.; Fry, R.C. Formaldehyde and Epigenetic Alterations: MicroRNA Changes in the Nasal Epithelium of Nonhuman Primates. Environ. Health Perspect. 2013, 121, 339–344. [Google Scholar] [CrossRef][Green Version]
- Rager, J.E.; Smeester, L.; Jaspers, I.; Sexton, K.G.; Fry, R.C. Epigenetic Changes Induced by Air Toxics: Formaldehyde Exposure Alters miRNA Expression Profiles in Human Lung Cells. Environ. Health Perspect. 2011, 119, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yang, J.; Ling, S. Formaldehyde Exposure Alters miRNA Expression Profiles in the Olfactory Bulb. Inhal. Toxicol. 2015, 27, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Yang, H.-S.; Song, M.-K.; Kim, D.I.; Lee, K. Formaldehyde Exposure Induces Regulatory T Cell-Mediated Immunosuppression via Calcineurin-NFAT Signalling Pathway. Sci. Rep. 2020, 10, 17023. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Yao, Z.; Kong, J.; Zhang, X.; Li, H.; Chen, W.; Xie, Q. Exploring the Role of miRNAs in Early Chicken Embryonic Development and Their Significance. Poult. Sci. 2023, 102, 103105. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, Q.; Ning, C.; Yang, M.; Zhu, Q.; Li, D.; Wang, T.; Li, F. miRNA Profiling of Chicken Follicles during Follicular Development. Sci. Rep. 2024, 14, 2212. [Google Scholar] [CrossRef]
- Chen, B.; Yu, J.; Guo, L.; Byers, M.S.; Wang, Z.; Chen, X.; Xu, H.; Nie, Q. Circular RNA circHIPK3 Promotes the Proliferation and Differentiation of Chicken Myoblast Cells by Sponging miR-30a-3p. Cells 2019, 8, 177. [Google Scholar] [CrossRef]
- Chen, J.-F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.-Z. The Role of microRNA-1 and microRNA-133 in Skeletal Muscle Proliferation and Differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Shi, J.; Li, W.; Liu, A.; Ren, L.; Zhang, P.; Jiang, T.; Han, Y.; Liu, L. MiRNA Sequencing of Embryonic Myogenesis in Chengkou Mountain Chicken. BMC Genom. 2022, 23, 571. [Google Scholar] [CrossRef]
- Zhou, Y.; Lu, Y.; Xu, H.; Ji, X.; Deng, Q.; Wang, X.; Zhang, Y.; Li, Q.; Lu, Y.; Rustempasic, A.; et al. The Effect of miR-205a with RUNX2 towards Proliferation and Differentiation of Chicken Chondrocytes in Thiram-Induced Tibial Dyschondroplasia. Poult. Sci. 2024, 103, 104535. [Google Scholar] [CrossRef]
- Wang, Z.; Ouyang, H.; Chen, X.; Yu, J.; Abdalla, B.A.; Chen, B.; Nie, Q. Gga-miR-205a Affecting Myoblast Proliferation and Differentiation by Targeting CDH11. Front. Genet. 2018, 9, 414. [Google Scholar] [CrossRef]
- Ouyang, H.; He, X.; Li, G.; Xu, H.; Jia, X.; Nie, Q.; Zhang, X. Deep Sequencing Analysis of miRNA Expression in Breast Muscle of Fast-Growing and Slow-Growing Broilers. Int. J. Mol. Sci. 2015, 16, 16242–16262. [Google Scholar] [CrossRef]
- Jia, X.; Nie, Q.; Zhang, X.; Nolan, L.K.; Lamont, S.J. Novel MicroRNA Involved in Host Response to Avian Pathogenic Escherichia Coli Identified by Deep Sequencing and Integration Analysis. Infect. Immun. 2016, 85, 10–1128. [Google Scholar] [CrossRef]
- Dinh, H.; Hong, Y.H.; Lillehoj, H.S. Modulation of microRNAs in Two Genetically Disparate Chicken Lines Showing Different Necrotic Enteritis Disease Susceptibility. Vet. Immunol. Immunopathol. 2014, 159, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Jebessa, E.; Bello, S.F.; Guo, L.; Tuli, M.D.; Hanotte, O.; Nie, Q. MicroRNA Expression Profile of Chicken Jejunum in Different Time Points Eimeria Maxima Infection. Front. Immunol. 2024, 14, 1532. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Hu, X.; Han, H.; Wang, J.; Liu, W.; Zhou, Y.; Cao, D.; Li, F.; Liu, J. Integrative Analysis of circRNA, miRNA, and mRNA Profiles to Reveal ceRNA Regulation in Chicken Muscle Development from the Embryonic to Post-Hatching Periods. BMC Genom. 2022, 23, 342. [Google Scholar] [CrossRef] [PubMed]
- Sajewicz-Krukowska, J.; Mirosław, P.; Jastrzębski, J.P.; Domańska-Blicharz, K.; Tarasiuk, K.; Marzec-Kotarska, B. miRNA Expression Signatures Induced by Chicken Astrovirus Infection in Chickens. Int. J. Mol. Sci. 2023, 24, 15128. [Google Scholar] [CrossRef]
- Al-Shaer, A.E.; Flentke, G.R.; Berres, M.E.; Garic, A.; Smith, S.M. Exon Level Machine Learning Analyses Elucidate Novel Candidate miRNA Targets in an Avian Model of Fetal Alcohol Spectrum Disorder. PLoS Comput. Biol. 2019, 15, e1006937. [Google Scholar] [CrossRef]
- Do, D.N.; Dudemaine, P.-L.; Mathur, M.; Suravajhala, P.; Zhao, X.; Ibeagha-Awemu, E.M. miRNA Regulatory Functions in Farm Animal Diseases, and Biomarker Potentials for Effective Therapies. Int. J. Mol. Sci. 2021, 22, 3080. [Google Scholar] [CrossRef]
- Coulibaly, F.; Onbaşılar, E.E.; Bakır, B.; Sarıçam İnce, S. The Effects of Using UV Light Instead of Formaldehyde in Disinfection of Hatching Eggs on Shell Microbial Load, Embryo Development, Hatchability, and Chick Characteristics. Int. J. Environ. Health Res. 2024, 34, 2852–2862. [Google Scholar] [CrossRef]
- Taşdemir, A.N.; Onbaşılar, E.E.; Yalçın, S.; Boyalı, B.; Aygören, H.; Tülek, E.; Sarıçam, S.; Akan, M. Effects of Oregano Juice on Eggshell Microbial Load, Layer Embryo Development, Hatching Results, and Growth at the First 2 Weeks after Hatch. Trop Anim Health Prod 2021, 53, 404. [Google Scholar] [CrossRef]
- Hamburger, V.; Hamilton, H.L. A Series of Normal Stages in the Development of the Chick Embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 Accurately Identifies Known and Hundreds of Novel microRNA Genes in Seven Animal Clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Ferreira, J.A.; Zwinderman, A.H. On the Benjamini–Hochberg Method. Ann. Stat. 2006, 34, 1827–1849. [Google Scholar] [CrossRef]
- Herwig, R.; Hardt, C.; Lienhard, M.; Kamburov, A. Analyzing and Interpreting Genome Data at the Network Level with ConsensusPathDB. Nat. Protoc. 2016, 11, 1889–1907. [Google Scholar] [CrossRef]
| Raw Reads | Trimmed Reads | |||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Raw Reads | % GC | Trimmed Reads | % GC | Q20 | Q30 | Mapped Reads | Mapped % |
| C1 | 6,256,428 | 50% | 5,377,299 | 46% | 99.24% | 97.32% | 3,865,384 | 72.379 |
| C2 | 6,974,290 | 49% | 6,257,434 | 44% | 99.24% | 97.43% | 5,181,287 | 83.227 |
| C3 | 5,489,007 | 49% | 4,993,507 | 44% | 99.27% | 97.49% | 4,357,515 | 87.961 |
| FA1 | 10,220,093 | 48% | 8,814,039 | 43% | 99.33% | 97.71% | 7,511,638 | 87.132 |
| FA2 | 5,919,854 | 48% | 3,820,899 | 42% | 99.12% | 96.97% | 3,399,960 | 93.602 |
| FA3 | 5,016,966 | 50% | 4,118,223 | 46% | 99.05% | 96.69% | 3,309,574 | 81.247 |
| miRNA | baseMean | Log2FC | p-Value | p-Adj |
|---|---|---|---|---|
| gga-miR-3533 | 38.164892 | 4.453057 | 0.000003 | 0.000997 |
| gga-miR-12239-5p | 13.882909 | 3.582699 | 0.000577 | 0.011505 |
| gga-miR-12214-5p | 10.217137 | 2.826239 | 0.003372 | 0.035794 |
| gga-miR-148a-5p | 1205.717617 | 1.994005 | 0.000121 | 0.005943 |
| gga-miR-147 | 66.211133 | 1.893574 | 0.000335 | 0.009736 |
| gga-miR-1434 | 77.881529 | 1.838995 | 0.000260 | 0.009074 |
| gga-miR-1451-3p | 73.627625 | 1.600179 | 0.003596 | 0.035836 |
| gga-miR-142-5p | 396.305198 | 1.485202 | 0.000382 | 0.009736 |
| gga-miR-30e-5p | 64,373.223801 | 1.035841 | 0.003019 | 0.035794 |
| gga-miR-210a-3p | 284.758497 | 1.580432 | 0.000832 | 0.013660 |
| gga-miR-144-5p | 725.246217 | 1.271770 | 0.001146 | 0.017249 |
| gga-miR-30c-2-3p | 190.978155 | −1.994259 | 0.000499 | 0.010709 |
| gga-miR-10b-5p | 367.784900 | −2.067845 | 0.000711 | 0.012542 |
| gga-miR-193b-3p | 447.476617 | −1.344009 | 0.000183 | 0.007331 |
| gga-miR-130b-3p | 9184.792929 | −1.393183 | 0.002106 | 0.026710 |
| gga-miR-1a-3p | 11,719.852639 | −1.542965 | 0.003463 | 0.035794 |
| gga-miR-222a | 2048.361237 | −1.740781 | 0.000127 | 0.005943 |
| gga-miR-143-3p | 121,035.324778 | −1.762389 | 0.000463 | 0.010709 |
| gga-miR-145-5p | 8553.054840 | −1.764542 | 0.000719 | 0.012542 |
| gga-miR-10c-5p | 55.671853 | −1.772399 | 0.005286 | 0.049166 |
| gga-miR-92-3p | 3139.439756 | −1.886186 | 0.000043 | 0.003015 |
| gga-miR-1388a-5p | 350.525535 | −2.168952 | 0.001207 | 0.017249 |
| gga-miR-133a-3p | 236.212541 | −2.173803 | 0.003326 | 0.035794 |
| gga-let-7l-5p | 396.995234 | −2.322389 | 0.000013 | 0.001814 |
| gga-miR-133a-5p | 47.417866 | −2.362333 | 0.004741 | 0.045612 |
| gga-miR-194 | 43.556299 | −2.514918 | 0.003255 | 0.035794 |
| gga-miR-30a-3p | 482.244321 | −2.530401 | 0.000036 | 0.003015 |
| gga-miR-205a | 59.202319 | −2.547512 | 0.001648 | 0.021905 |
| gga-miR-133c-3p | 42.136850 | −2.731904 | 0.001236 | 0.017249 |
| gga-miR-133b | 28.789600 | −3.383927 | 0.000383 | 0.009736 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teber, S.; Özdemir, M.; Sajid, G.A.; Büyükkılıç Beyzi, S.; Kizilaslan, M.; Arzık, Y.; Yalçın, S.; White, S.N.; Cinar, M.U. Small RNA-Seq Reveals the Effect of Formaldehyde Treatment on Chicken Embryo Liver microRNA Profiles. Int. J. Mol. Sci. 2025, 26, 10633. https://doi.org/10.3390/ijms262110633
Teber S, Özdemir M, Sajid GA, Büyükkılıç Beyzi S, Kizilaslan M, Arzık Y, Yalçın S, White SN, Cinar MU. Small RNA-Seq Reveals the Effect of Formaldehyde Treatment on Chicken Embryo Liver microRNA Profiles. International Journal of Molecular Sciences. 2025; 26(21):10633. https://doi.org/10.3390/ijms262110633
Chicago/Turabian StyleTeber, Saffet, Mustafa Özdemir, Ghulam Asghar Sajid, Selma Büyükkılıç Beyzi, Mehmet Kizilaslan, Yunus Arzık, Servet Yalçın, Stephen N. White, and Mehmet Ulas Cinar. 2025. "Small RNA-Seq Reveals the Effect of Formaldehyde Treatment on Chicken Embryo Liver microRNA Profiles" International Journal of Molecular Sciences 26, no. 21: 10633. https://doi.org/10.3390/ijms262110633
APA StyleTeber, S., Özdemir, M., Sajid, G. A., Büyükkılıç Beyzi, S., Kizilaslan, M., Arzık, Y., Yalçın, S., White, S. N., & Cinar, M. U. (2025). Small RNA-Seq Reveals the Effect of Formaldehyde Treatment on Chicken Embryo Liver microRNA Profiles. International Journal of Molecular Sciences, 26(21), 10633. https://doi.org/10.3390/ijms262110633










