Genome-Wide Association Studies in Japanese Quails of the F2 Resource Population Elucidate Molecular Markers and Candidate Genes for Body Weight Parameters
Abstract
1. Introduction
2. Results
2.1. F2 Resource Population Phenotypic Data and Population Stratification
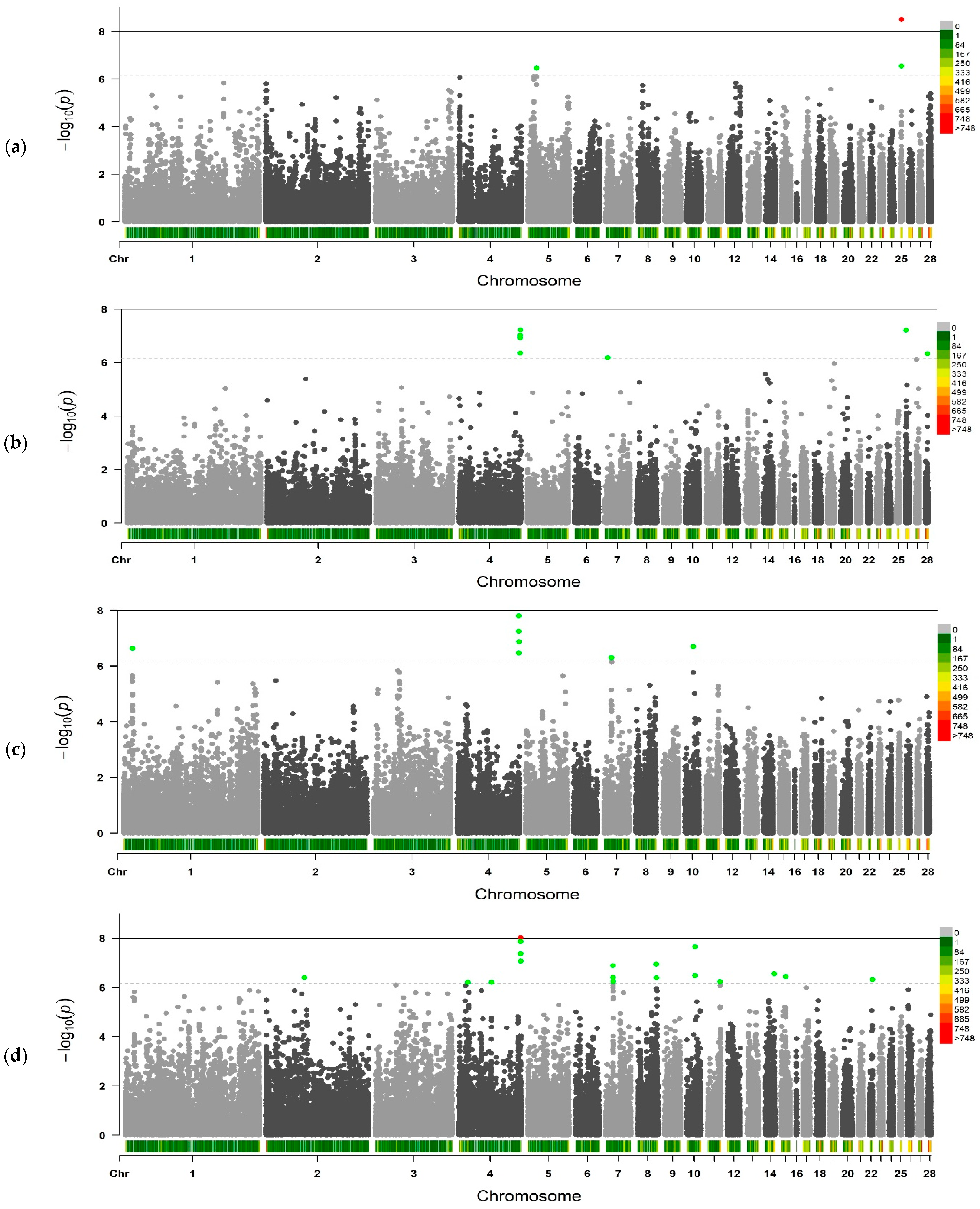
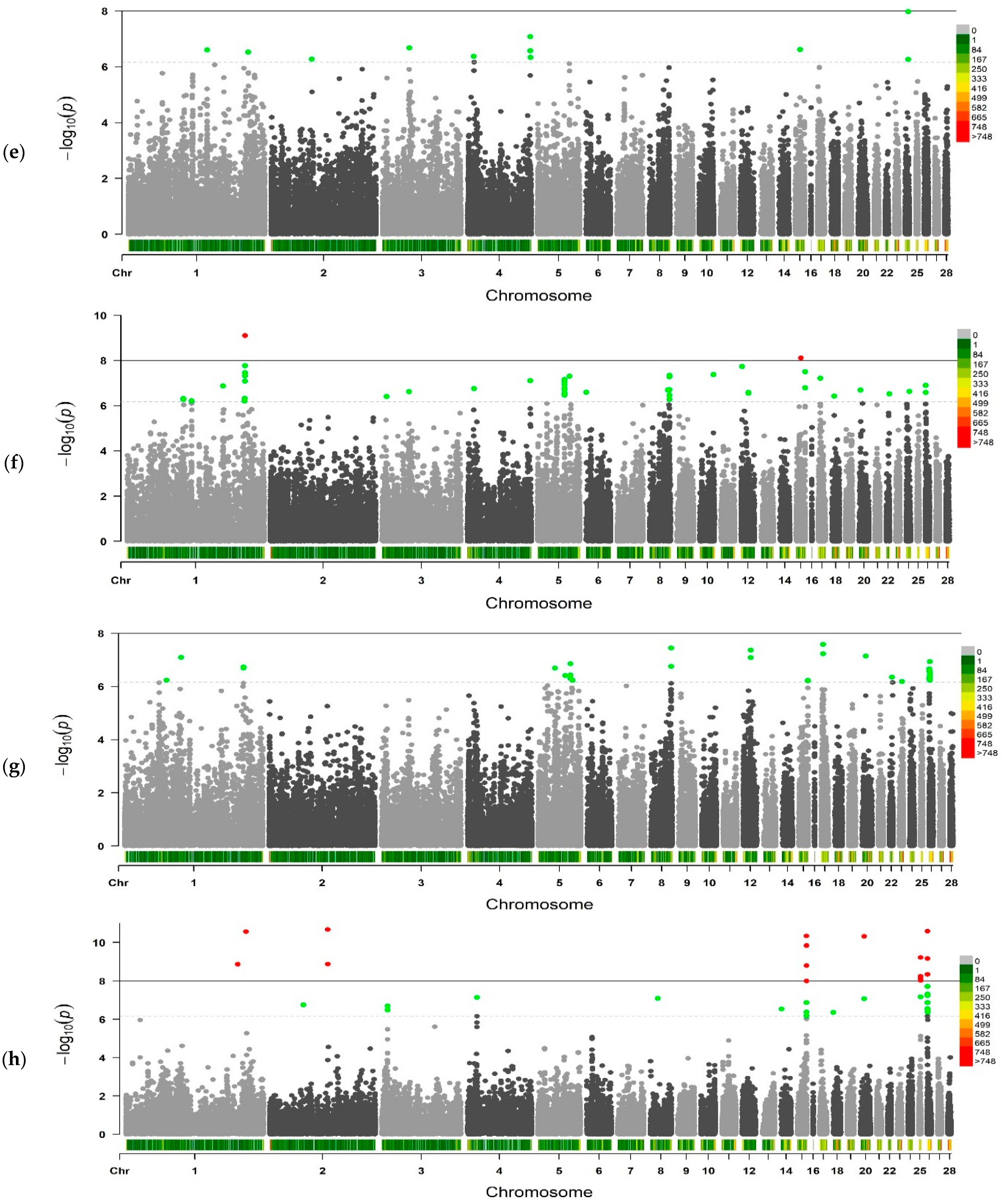
2.2. Genome-Wide Association Studies
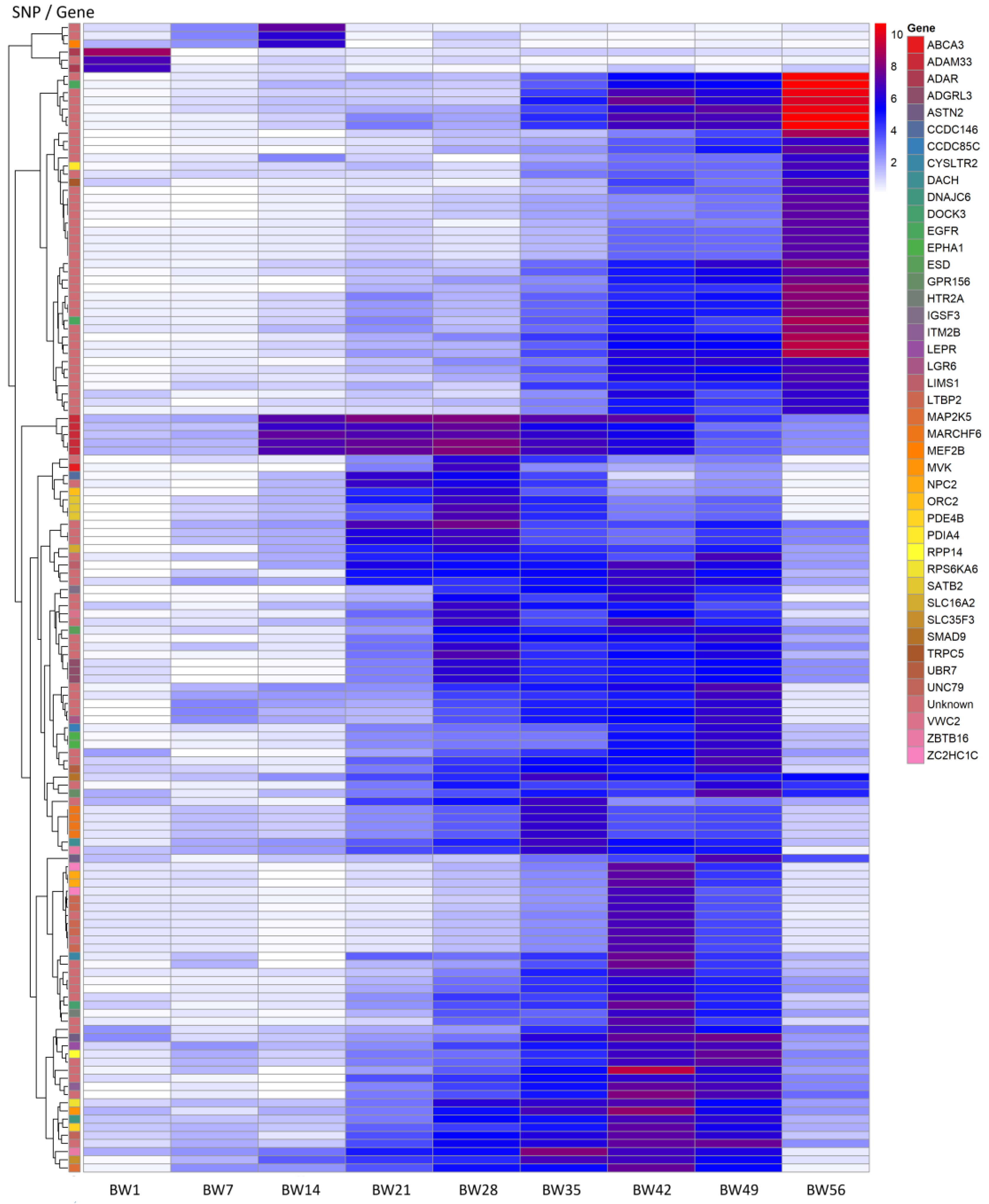
2.3. Candidate Genes Including PCGs
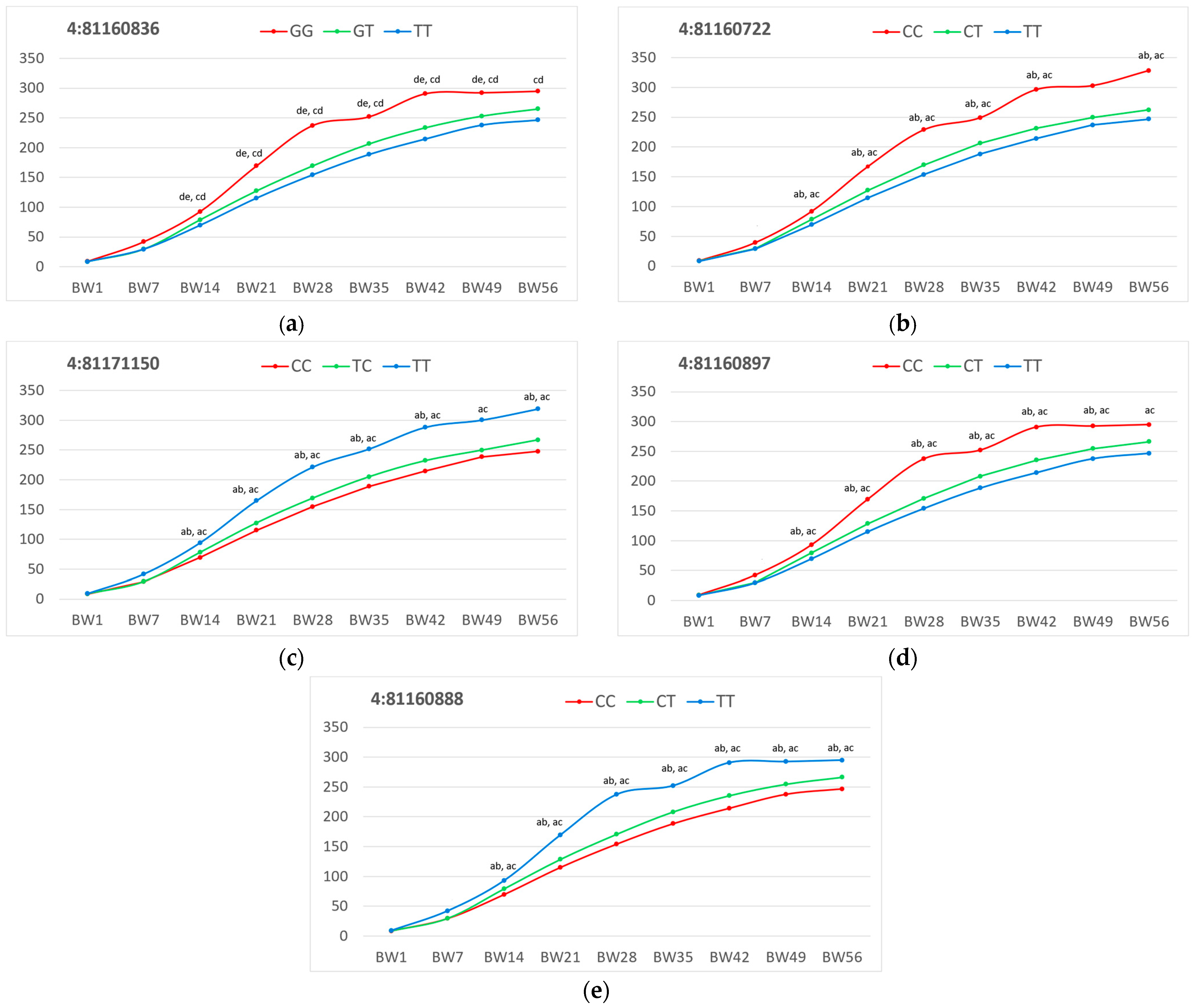
2.4. ADAM33 Association Analysis Example
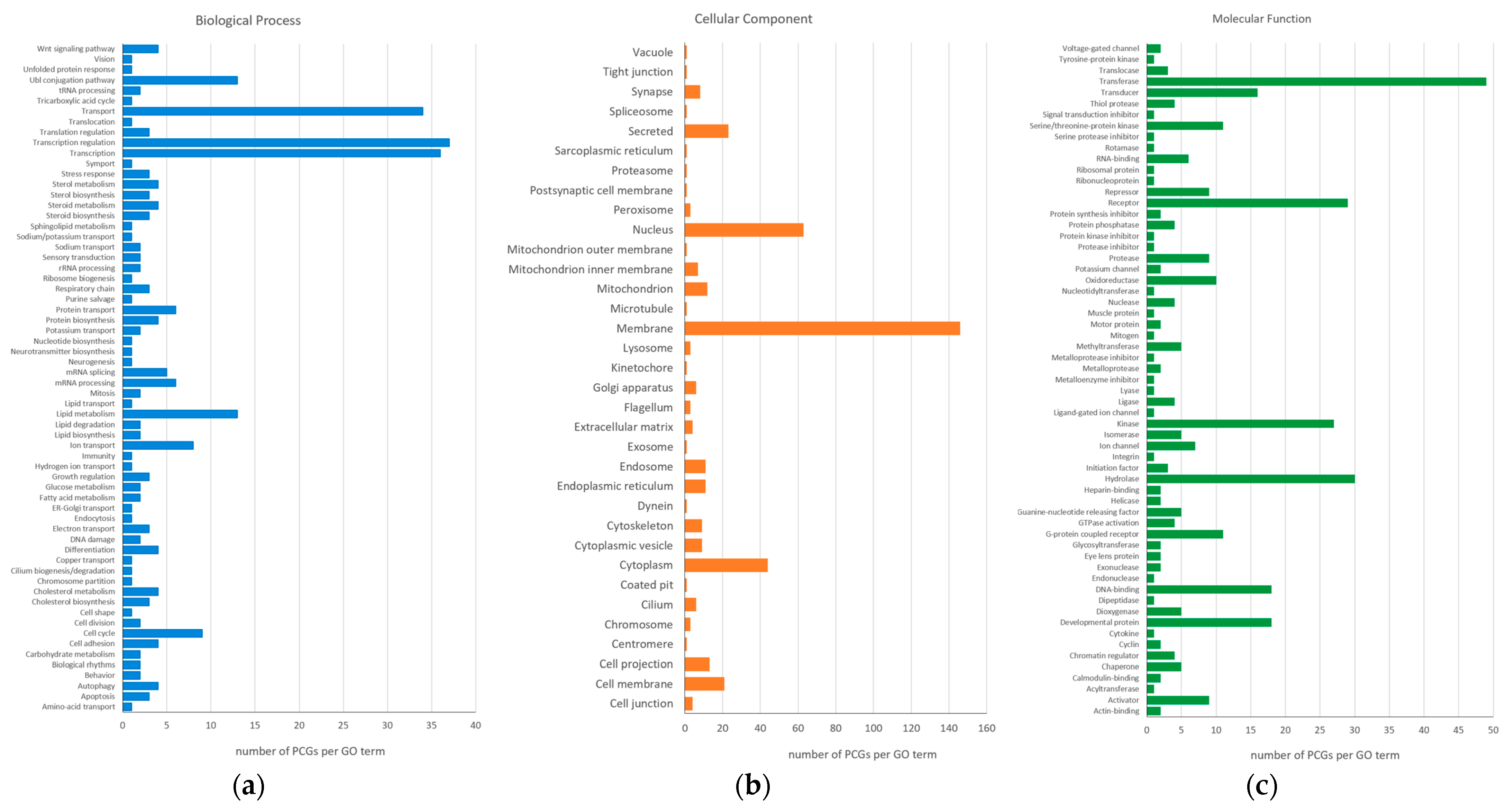
2.5. Gene Ontology Analysis
3. Discussion
3.1. Identification and Relevance of PCGs for BW Parameters
3.1.1. LEPR and ASTN2
3.1.2. EGFR and ADGRL3
3.1.3. MVK, NPC2, SATB2, ADAR, ITM2B and LTBP2
3.1.4. SLC35F3 and ADAM33
3.1.5. ADAR, UNC79 and RPP14
3.1.6. ZBTB16
3.1.7. MARCHF6, ZC2HC1C and LGR6
4. Materials and Methods
4.1. Experimental Birds
4.2. Phenotypic Traits and Their Statistical Analyses
4.3. Sampling and DNA Extraction
4.4. Sequencing, Genotyping and SNP Quality Control
4.5. PCA Procedure
4.6. Genome-Wide Association Study and GO Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BW | body weight |
| SNP(s) | single nucleotide polymorphism(s) |
| GWAS | genome-wide association study |
| PCG(s) | prioritized candidate gene(s) |
| GBS | genotyping-by-sequencing |
| CJA | Coturnix japonica |
| BW1, BW7, BW14, BW21, BW28, BW35, BW42, BW49, BW56 | 1, 7, 14, 21, 28, 35, 42, 49 and 56 days of age |
| SD | standard deviation |
| CV | coefficient of variation |
| PCA | principal component analysis |
| PC1 | first component |
| PC2 | second component |
| PC3 | third component |
| ITM2B | integral membrane protein 2B |
| SLC35F3 | solute carrier family 35 member F3 |
| ADAM33 | ADAM metallopeptidase domain 33 |
| UNC79 | unc-79 homolog, NALCN channel complex subunit |
| LEPR | leptin receptor |
| RPP14 | ribonuclease P/MRP subunit p14 |
| MVK | mevalonate kinase |
| ASTN2 | astrotactin 2 |
| ZBTB16 | zinc finger and BTB domain containing 16 |
| MARCHF6 | membrane associated ring-CH-type finger 6 |
| EGFR | epidermal growth factor receptor |
| ADGRL3 | adhesion G protein-coupled receptor L3 |
| NPC2 | NPC intracellular cholesterol transporter 2 |
| LTBP2 | latent transforming growth factor beta binding protein 2 |
| ZC2HC1C | zinc finger C2HC-type containing 1C |
| SATB2 | SATB homeobox 2 |
| ADAR | adenosine deaminase RNA specific |
| LGR6 | leucine rich repeat containing G protein-coupled receptor 6 |
| GO | gene ontology |
| ADBWG | average daily body weight gain |
| LKEFRCAH | L. K. Ernst Federal Research Centre for Animal Husbandry |
References
- Plemyashov, K.V.; Smaragdov, M.G.; Romanov, M.N. Molekulyarno-geneticheskiy polimorfizm v populyatsiyakh zhivotnykh i yego primeneniye v intensivnoy selektsii molochnogo skota: Obzor [Molecular Genetic Polymorphism in Animal Populations and Its Application in Intensive Breeding of Dairy Cattle—A Review]. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, 30 September 2021; Pozyabin, S.V., Kochish, I.I., Romanov, M.N., Eds.; Sel’skokhozyaistvennye tekhnologii: Moscow, Russia, 2021; pp. 368–378, (In Russian with English Summary). [Google Scholar] [CrossRef]
- Dotsev, A.V.; Moradi, M.H.; Deniskova, T.E.; Esmailizadeh, A.; Bakoev, N.F.; Koshkina, O.A.; Griffin, D.K.; Romanov, M.N.; Zinovieva, N.A. Molecular genetic assessment aids in clarifying phylogenetic status of Iranian Kerman wild sheep. Animals 2025, 15, 238. [Google Scholar] [CrossRef]
- Laptev, G.Y.; Tiurina, D.G.; Yildirim, E.A.; Gorfunkel, E.P.; Ilina, L.A.; Filippova, V.A.; Dubrovin, A.V.; Dubrovina, A.S.; Brazhnik, E.A.; Novikova, N.I.; et al. Effects of glyphosate, antibiotics and an anticoccidial drug on pancreatic gene expression and blood physiology in broilers. J. Zhejiang Univ. Sci. B 2025, 26, 185–199. [Google Scholar] [CrossRef]
- Kulibaba, R.O.; Sakhatskyi, M.I.; Griffin, D.K.; Romanov, M.N. Molecular diversity of Ukrainian native chicken breeds: A review. Worlds Poult. Sci. J. 2024, 80, 1265–1292. [Google Scholar] [CrossRef]
- Weigend, S.; Romanov, M.N. Molekulare Charakterisierung genetischer Vielfalt beim Geflügel—Molecular characterization of genetic diversity in chicken. In Jahresbericht 1998; Bundesforschungsanstalt für Landwirtschaft (FAL): Braunschweig, Germany, 1999; p. 66. Available online: https://www.researchgate.net/publication/371721279 (accessed on 30 June 2025).
- Moiseyeva, I.G.; Kovalenko, A.T.; Mosyakina, T.V.; Romanov, M.N.; Bondarenko, Y.V.; Kutnyuk, P.I.; Podstreshny, A.P.; Nikiforov, A.A.; Tkachik, T.E. Origin, History, Genetics and Economic Traits of the Poltava Chicken Breed. Elektronnyi zhurnal [Electron. J.]; Laboratory of Animal Comparative Genetics, N.I. Vavilov Institute of General Genetics: Moscow, Russia, 2006, 4. Available online: https://web.archive.org/web/20120205195904/http://www.lab-cga.ru/articles/Jornal04/Statia2.htm (accessed on 5 February 2012). (In Russian).
- Moiseyeva, I.G.; Romanov, M.N.; Alexandrov, A.V.; Nikiforov, A.A.; Sevastyanova, A.A. Evolyutsiya, geneticheskaya izmenchivost’ yurlovskoy golosistoy porody kur. Sistemnyy analiz form izmenchivosti [Evolution and genetic diversity of old domestic hen’s breed—Yurlovskaya golosistaya, System analysis of variability forms]. Izv. Timiryazev S-Kh. Akad. [Izv. Timiryazev Agric. Acad.] 2009, 3, 132–147. Available online: https://kar.kent.ac.uk/46633 (accessed on 30 June 2025). (In Russian with English Summary).
- Vakhrameev, A.B.; Narushin, V.G.; Larkina, T.A.; Barkova, O.Y.; Peglivanyan, G.K.; Dysin, A.P.; Dementieva, N.V.; Makarova, A.V.; Shcherbakov, Y.S.; Pozovnikova, M.V.; et al. Selection-driven chicken phenome and phenomenon of pectoral angle variation across different chicken phenotypes. Livest. Sci. 2022, 264, 105067. [Google Scholar] [CrossRef]
- Vakhrameev, A.B.; Narushin, V.G.; Larkina, T.A.; Barkova, O.Y.; Peglivanyan, G.K.; Dysin, A.P.; Dementieva, N.V.; Shcherbakov, Y.S.; Pozovnikova, M.V.; Griffin, D.K.; et al. Pectoral angle: A glance at a traditional phenotypic trait in chickens from a new perspective. J. Agric. Sci. 2023, 161, 606–615. [Google Scholar] [CrossRef]
- Romanov, M.N.; Abdelmanova, A.S.; Fisinin, V.I.; Gladyr, E.A.; Volkova, N.A.; Koshkina, O.A.; Rodionov, A.N.; Vetokh, A.N.; Gusev, I.V.; Anshakov, D.V.; et al. Selective footprints and genes relevant to cold adaptation and other phenotypic traits are unscrambled in the genomes of divergently selected chicken breeds. J. Anim. Sci. Biotechnol. 2023, 14, 35. [Google Scholar] [CrossRef]
- Moiseeva, I.G. Fenotipicheskaya i Geneticheskaya Izmenchivost’ Pokazateley Kachestva u Kur [Phenotypic and Genetic Variability of Quality Indicators in Chickens]. Abstract of Cand. Sci. (Biol.) Dissertation, Moscow, USSR, 1965. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=tFr9MGIAAAAJ:tOudhMTPpwUC (accessed on 30 June 2025). (In Russian with English Summary).
- Krutikova, A.A.; Dementieva, N.V.; Shcherbakov, Y.S.; Goncharov, V.V.; Griffin, D.K.; Romanov, M.N. Polymorphism of genes potentially affecting growth and body size suggests genetic divergence in wild and domestic reindeer (Rangifer tarandus) populations. Genes 2024, 15, 1629. [Google Scholar] [CrossRef] [PubMed]
- Deniskova, T.E.; Dotsev, A.V.; Koshkina, O.A.; Solovieva, A.D.; Churbakova, N.A.; Petrov, S.N.; Frolov, A.N.; Platonov, S.A.; Abdelmanova, A.S.; Vladimirov, M.A.; et al. Examination of runs of homozygosity distribution patterns and relevant candidate genes of potential economic interest in Russian goat breeds using whole-genome sequencing. Genes 2025, 16, 631. [Google Scholar] [CrossRef]
- Sakhatsky, N.I.; Tereshchenko, A.V.; Artemenko, A.B. [An express-method of estimation of fertilizing ability of freezing-thawing of poultry spermatozoa]. Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 1987, 22, 77–80. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19880165689 (accessed on 30 June 2025).
- Plemyashov, K.V.; Smaragdov, M.G.; Romanov, M.N. Genomnaya otsenka plemennykh bykov [Genomic Assessment of Breeding Bulls]. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, 30 September 2021; Pozyabin, S.V., Kochish, I.I., Romanov, M.N., Eds.; Sel’skokhozyaistvennye tekhnologii: Moscow, Russia, 2021; pp. 363–367. Available online: https://elibrary.ru/item.asp?id=46668864 (accessed on 30 June 2025)(In Russian with English Summary).
- Kutnyuk, P.I.; Bondarenko, Y.V.; Ivanova, T.V. Kariometrychnyy metod vyznachennya henetychnoho potentsialu nesuchosti kurey [Karyometric method of determining the genetic potential of laying hens]. Ptakhivnytstvo [Poult. Farming] 2001, 50, 75–81. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:dQ2og3OwTAUC (accessed on 30 June 2025). (In Ukrainian).
- Aslam, M.L.; Bastiaansen, J.W.; Crooijmans, R.P.; Vereijken, A.; Groenen, M. Whole genome QTL mapping for growth, meat quality and breast meat yield traits in turkey. BMC Genet. 2011, 12, 61. [Google Scholar] [CrossRef]
- Moiseyeva, I.G.; Romanov, M.N.; Kovalenko, A.T.; Mosyakina, T.V.; Bondarenko, Y.V.; Kutnyuk, P.I.; Podstreshny, A.P.; Nikiforov, A.A. The Poltava chicken breed of Ukraine: Its history, characterization and conservation. Anim. Genet. Resour. Inf. 2007, 40, 71–78. [Google Scholar] [CrossRef]
- Moiseyeva, I.G.; Sevastyanova, A.A.; Aleksandrov, A.V.; Vakhrameev, A.B.; Romanov, M.N.; Dmitriev, Y.I.; Semenova, S.K.; Sulimova, G.E. Orloff chicken breed: History, current status and studies. Izv. Timiryazev S-Kh. Akad. [Izv. Timiryazev Agric. Acad.] 2016, 1, 78–96. Available online: https://www.elibrary.ru/item.asp?id=25664565 (accessed on 30 June 2025). (In Russian with English Summary).
- Baydevlyatova, O.N.; Ogurtsova, N.S.; Shomina, N.V.; Tereshchenko, A.V. Morfologicheskiye pokazateli kachestva yaits novoy subpopulyatsii kur myaso-yaichnogo napravleniya produktivnosti [Morphological indicators of egg quality in a new chicken subpopulation of the meat-egg type of productivity]. Ptakhivnytstvo [Poult. Farming] 2009, 64, 109–115. Available online: http://avianua.com/archiv/ptahivnictvo/64/15.pdf (accessed on 30 June 2025). (In Russian with English Summary).
- Schreiweis, M.A.; Hester, P.Y.; Settar, P.; Moody, D.E. Identification of quantitative trait loci associated with egg quality, egg production, and body weight in an F2 resource population of chickens. Anim. Genet. 2006, 37, 106–112. [Google Scholar] [CrossRef]
- Moiseeva, I.G. Vliyaniye inbridinga na kachestvo kurinykh yaits [The effect of inbreeding on the quality of fowl eggs]. Genetika 1970, 6, 99–107. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19700104311 (accessed on 30 June 2025). (In Russian).
- Weigend, S.; Romanov, M.N.; Ben-Ari, G.; Hillel, J. Overview on the Use of Molecular Markers to Characterize Genetic Diversity in Chickens. In XXII World’s Poultry Congress & Exhibition, Participant List & Full Text CD + Book of Abstracts, Istanbul, Turkey, 8–13 June 2004; WPSA—Turkish Branch: Istanbul, Turkey, 2004; p. 192. Available online: https://www.researchgate.net/publication/372751440 (accessed on 30 June 2025).
- Romanov, M.N.; Ameen, Q.A.; Shaker, A.S.; Al-Obaidi, R.M.; Griffin, D.K. Conservation genetics and breeding using molecular genetic markers in Japanese quail (Coturnix japonica). Front. Biosci. (Schol. Ed.) 2024, 16, 23. [Google Scholar] [CrossRef] [PubMed]
- Weigend, S.; Romanov, M.N. Genetische Diversitätsanalysen bei Hühnern mit Hilfe molekularer Marker—Assessment of genetic diversity in chickens using molecular markers. In Jahresbericht 2001; Bundesforschungsanstalt für Landwirtschaft (FAL): Braunschweig, Germany, 2002; p. 67. Available online: https://www.researchgate.net/publication/371722377 (accessed on 30 June 2025).
- Kulibaba, R.O.; Srikulnath, K.; Singchat, W.; Liashenko, Y.V.; Griffin, D.K.; Romanov, M.N. The application of microsatellite markers as molecular tools for studying genomic variability in vertebrate populations. Curr. Issues Mol. Biol. 2025, 47, 447. [Google Scholar] [CrossRef] [PubMed]
- Trufanov, O.V.; Tereshchenko, A.V. Primeneniye molekulyarnykh i biokhimicheskikh metodov dlya diagnostiki zabolevaniy sel’skokhozyaystvennoy ptitsy [Application of molecular and biochemical methods for diagnostics of poultry diseases]. Eksklyuzivnyye tekhnologii [Excl. Technol.] 2010, 3, 42–43. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=deB5xCwAAAAJ:HtEfBTGE9r8C (accessed on 30 June 2025).
- Jiang, J.; Ma, L.; Prakapenka, D.; VanRaden, P.M.; Cole, J.B.; Da, Y. A large-scale genome-wide association study in US Holstein cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef]
- Niu, N.; Wang, H.; Shi, G.; Liu, X.; Liu, H.; Liu, Q.; Yang, M.; Wang, L.; Zhang, L. Genome scanning reveals novel candidate genes for vertebral and teat number in the Beijing Black Pig. Anim. Genet. 2021, 52, 734–738. [Google Scholar] [CrossRef]
- Niu, N.; Liu, Q.; Hou, X.; Liu, X.; Wang, L.; Zhao, F.; Gao, H.; Shi, L.; Wang, L.; Zhang, L. Genome-wide association study revealed ABCD4 on SSC7 and GREB1L and MIB1 on SSC6 as crucial candidate genes for rib number in Beijing Black pigs. Anim. Genet. 2022, 53, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Dementeva, N.V.; Romanov, M.N.; Kudinov, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Terletsky, V.P.; Fedorova, E.S.; Nikitkina, E.V.; Plemyashov, K.V. Studying the structure of a gene pool population of the Russian White chicken breed by genome-wide SNP scan. Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2017, 52, 1166–1174. [Google Scholar] [CrossRef]
- Dementieva, N.V.; Shcherbakov, Y.S.; Tyshchenko, V.I.; Terletsky, V.P.; Vakhrameev, A.B.; Nikolaeva, O.A.; Ryabova, A.E.; Azovtseva, A.I.; Mitrofanova, O.V.; Peglivanyan, G.K.; et al. Comparative analysis of molecular RFLP and SNP markers in assessing and understanding the genetic diversity of various chicken breeds. Genes 2022, 13, 1876. [Google Scholar] [CrossRef]
- Volyanskaya, A.R.; Akberdin, I.R.; Kulyashov, M.A.; Yevshin, I.S.; Romanov, M.N.; Shagimardanova, E.I.; Gusev, O.A.; Kolpakov, F.A. A bird’s-eye overview of molecular mechanisms regulating feed intake in chickens—With mammalian comparisons. Anim. Nutr. 2024, 17, 61–74. [Google Scholar] [CrossRef]
- Dementieva, N.V.; Nikitkina, E.V.; Shcherbakov, Y.S.; Pleshanov, N.V.; Ryabova, A.E.; Azovtseva, A.I.; Silyukova, Y.L.; Musidray, A.A.; Griffin, D.K.; Romanov, M.N. Genome-wide analysis of genetic predispositions linked to damaged membranes and impaired fertility as indicators of compromised sperm–egg interaction mechanisms in frozen–thawed rooster semen. Front. Biosci. (Schol. Ed.) 2025, 17, 26022. [Google Scholar] [CrossRef]
- Anthony, N.B.; Nestor, K.E.; Marks, H.L. Short-term selection for four-week body weight in Japanese quail. Poult. Sci. 1996, 75, 1192–1197. [Google Scholar] [CrossRef] [PubMed]
- Mebratie, W.; Madsen, P.; Hawken, R.; Romé, H.; Marois, D.; Henshall, J.; Bovenhuis, H.; Jensen, J. Genetic parameters for body weight and different definitions of residual feed intake in broiler chickens. Genet. Sel. Evol. 2019, 51, 53. [Google Scholar] [CrossRef]
- Bondarenko, Y.V.; Khvostik, V.P. Pokrashhennya produktyvnosti m’yaso-yayechnyh kurej vitchyznyanoyi selekciyi [Improving the productivity of meat and egg chickens of domestic selection]. Vìsn. Sumsʹkogo nac. agrar. unìv., Ser. Tvarynnytstvo [Bull. Sumy Natl. Agrar. Univ. Ser. Livest.] 2020, 2, 29–32, (In Ukrainian with English Summary). [Google Scholar] [CrossRef]
- Larkina, T.A.; Barkova, O.Y.; Peglivanyan, G.K.; Mitrofanova, O.V.; Dementieva, N.V.; Stanishevskaya, O.I.; Vakhrameev, A.B.; Makarova, A.V.; Shcherbakov, Y.S.; Pozovnikova, M.V.; et al. Evolutionary subdivision of domestic chickens: Implications for local breeds as assessed by phenotype and genotype in comparison to commercial and fancy breeds. Agriculture 2021, 11, 914. [Google Scholar] [CrossRef]
- Victoria, O.; Fayeye, T.R.; Ayorinde, K.L.; Olojede, H. Relationship between body weight and linear body measurements in Japanese quail (Coturnix coturnix japonica). J. Sci. Res. 2013, 6, 175–183. [Google Scholar] [CrossRef]
- Bekele, B.; Melesse, A.; Esatu, W.; Dessie, T. Statistical modeling of live body weight and linear body measurements of local chicken at different agro-ecologies of Ethiopia. Int. J. Poult. Sci. 2021, 20, 146–151. [Google Scholar] [CrossRef]
- Malomane, D.K.; Norris, D.; Banga, C.B.; Ngambi, J.W. Use of factor scores for predicting body weight from linear body measurements in three South African indigenous chicken breeds. Trop. Anim. Health Prod. 2014, 46, 331–335. [Google Scholar] [CrossRef]
- Mekonnen, K.T.; Lee, D.-H.; Cho, Y.-G.; Son, A.-Y.; Seo, K.-S. Estimation of carcass trait characteristics, proportions, and their correlation with preslaughter body weight in indigenous chickens in Southeastern Ethiopia. Agriculture 2024, 14, 50. [Google Scholar] [CrossRef]
- Adil, M.; Tayyab, M.; Hussain, J.; Firyal, S.; Ali, S.; Azam, M.; Wasim, M.; Awan, A.R. Relationship between body weight and linear body measurements in Pakistani quail (Coturnix japonica PK). Pak. J. Zool. 2024, 56, 1423–1431. [Google Scholar] [CrossRef]
- Mendes, A.S.; Paixão, S.J.; Restelatto, R.; Reffatti, R.; Possenti, J.C.; Moura, D.J.I.; Morello, G.M.Z.; Carvalho, T.M.R. Effects of initial body weight and litter material on broiler production. Braz. J. Poult. Sci. 2011, 13, 197–204. [Google Scholar] [CrossRef]
- Marks, H.L. Long-term selection for body weight in Japanese quail under different environments. Poult. Sci. 1996, 75, 1198–1203. [Google Scholar] [CrossRef]
- Smaldone, G.; Capezzuto, S.; Ambrosio, R.L.; Peruzy, M.F.; Marrone, R.; Peres, G.; Anastasio, A. The influence of broilers’ body weight on the efficiency of electrical stunning and meat quality under field conditions. Animals 2021, 11, 1362. [Google Scholar] [CrossRef]
- Raziq, F.; Hussain, J.; Ahmad, S.; Hussain, M.A.; Khan, M.T.; Ullah, A.; Qumar, M.; Wadood, F.; Gull-E-Faran. Effect of body weight at photostimulation on productive performance and welfare aspects of commercial layers. Anim. Biosci. 2024, 37, 500–508. [Google Scholar] [CrossRef]
- Narinc, D.; Karaman, E.; Aksoy, T.; Firat, M.Z. Genetic parameter estimates of growth curve and reproduction traits in Japanese quail. Poult. Sci. 2014, 93, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Molee, A.; Kuadsantia, P.; Kaewnakian, P. Gene effects on body weight, carcass yield, and meat quality of Thai indigenous chicken. J. Poult. Sci. 2018, 55, 94–102. [Google Scholar] [CrossRef]
- Zhong, C.; Li, X.; Guan, D.; Zhang, B.; Wang, X.; Qu, L.; Zhou, H.; Fang, L.; Sun, C.; Yang, N. Age-dependent genetic architectures of chicken body weight explored by multidimensional GWAS and MolQTL analyses. J. Genet. Genom. 2024, 51, 1423–1434. [Google Scholar] [CrossRef]
- Haqani, M.I.; Nomura, S.; Nakano, M.; Goto, T.; Nagano, A.J.; Takenouchi, A.; Nakamura, Y.; Ishikawa, A.; Tsudzuki, M. Quantitative trait loci for growth-related traits in Japanese quail (Coturnix japonica) using restriction-site associated DNA sequencing. Mol. Genet. Genom. 2021, 296, 1147–1159. [Google Scholar] [CrossRef]
- Volkova, N.A.; Romanov, M.N.; Abdelmanova, A.S.; Larionova, P.V.; German, N.Y.; Vetokh, A.N.; Shakhin, A.V.; Volkova, L.A.; Sermyagin, A.A.; Anshakov, D.V.; et al. Genome-wide association study revealed putative SNPs and candidate genes associated with growth and meat traits in Japanese quail. Genes 2024, 15, 294. [Google Scholar] [CrossRef]
- Narushin, V.G.; Volkova, N.A.; Vetokh, A.N.; Dzhagaev, A.Y.; Volkova, L.A.; Griffin, D.K.; Romanov, M.N.; Zinovieva, N.A. Metabolic rate and egg production in Japanese quails can be predicted by assessing growth parameters of laying hens. Animals 2024, 14, 258. [Google Scholar] [CrossRef]
- Cai, W.; Hu, J.; Fan, W.; Xu, Y.; Tang, J.; Xie, M.; Zhang, Y.; Guo, Z.; Zhou, Z.; Hou, S. Strategies to improve genomic predictions for 35 duck carcass traits in an F2 population. J. Anim. Sci. Biotechnol. 2023, 14, 74. [Google Scholar] [CrossRef]
- Shkurko, M.I.; Bondarenko, Y.V. Porivnyalʹna kharakterystyka molodnyaka kachok riznykh henotypiv v umovakh prysadybnoho hospodarstva [Comparative Characteristics of Young Ducks of Different Genotypes in Homestead Farming Conditions]. In Materials of the Scientific Practical Conference of Teachers, Postgraduates and Students of the Sumy National Agrarian University, Sumy, Ukraine, 20–24 April 2015; Sumy National Agrarian University: Sumy, Ukraine, 2015; Volume 2, p. 103. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:nrtMV_XWKgEC (accessed on 30 June 2025). (In Ukrainian)
- Shkurko, M.I.; Bondarenko, U.V.; Ostapenko, V.I. Produktyvnistʹ molodnyaka kachok riznykh henotypiv v umovakh prysadybnoho hospodarstva [Productivity of young ducks of different genotypes under conditions of farming household]. Vìsn. Sumsʹkogo nac. agrar. unìv., Ser. Tvarynnytstvo [Bull. Sumy Natl. Agrar. Univ. Ser. Livest.] 2015, 6, 75–78. Available online: http://www.irbis-nbuv.gov.ua/cgi-bin/irbis_nbuv/cgiirbis_64.exe?C21COM=2&I21DBN=UJRN&P21DBN=UJRN&IMAGE_FILE_DOWNLOAD=1&Image_file_name=PDF/Vsna_tvar_2015_6_18.pdf (accessed on 30 June 2025). (In Ukrainian with English Summary).
- Yang, C.; Wang, Z.; Song, Q.; Dong, B.; Bi, Y.; Bai, H.; Jiang, Y.; Chang, G.; Chen, G. Transcriptome sequencing to identify important genes and lncRNAs regulating abdominal fat deposition in ducks. Animals 2022, 12, 1256. [Google Scholar] [CrossRef] [PubMed]
- Kutnyuk, P.I.; Grechikhin, S.N.; Bondarenko, Y.V.; Gadyuchko, O.T. Geneticheskiy polimorfizm ovobelkov utok i yego adaptivnoye znacheniye [Genetic Polymorphism of Duck Proteins and Its Adaptive Significance]. In Molecular Genetic Markers of Animals, Proceedings of the International Conference, Kyiv, Ukraine, 14 May 1999; n.p.: Kyiv, Ukraine, 1999; p. 373. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:5awf1xo2G04C (accessed on 30 June 2025). (In Russian)
- Kutnyuk, P.I.; Gadyuchko, O.T.; Bondarenko, Y.V. Locus Om as Body Weight Marker in Process of Frequent-dependent Selection of Turkeys. In The Poultry Industry Towards the 21st Century, Proceedings of the 10th European Poultry Conference, Jerusalem, Israel, 21–26 June 1998; World’s Poultry Science Association: Jerusalem, Israel, 1998; p. 67. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:mlAyqtXpCwEC (accessed on 30 June 2025).
- Kaya Başar, E.; Narinç, D. Genetic parameter estimates of growth curve and feed efficiency traits in Japanese quail. Animals 2023, 13, 1765. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Lin, H.; Jiao, H.; Zhao, J.; Ma, B.; Wang, Y.; He, S.; Wang, X. Daily feeding frequency affects feed intake and body weight management of growing layers. Poult. Sci. 2024, 103, 103748. [Google Scholar] [CrossRef]
- Abdelraheem, N.; Ahmed, M.; Hou, F. Effect of feed restriction on broiler chicks prior to slaughter. Open J. Anim. Sci. 2019, 9, 12–22. [Google Scholar] [CrossRef]
- Chaikuad, N.; Loengbudnark, W.; Chankitisakul, V.; Boonkum, W. Genetic comparisons of body weight, average daily gain, and breast circumference between slow-growing Thai native chickens (Pradu Hang dum) raised on-site farm and on-station. Vet. Sci. 2023, 10, 11. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Li, Y.; Xiao, F.; Guo, H.; Gao, H.; Wang, N.; Zhang, H.; Li, H. Genome-wide association study and selective sweep analysis reveal the genetic architecture of body weights in a chicken F2 resource population. Front. Vet. Sci. 2022, 9, 875454. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Nong, Y.; Liu, Y.; Wang, Z.; Wang, J.; Li, Z. Genome-wide association study of body size traits in Luning chickens using whole-genome sequencing. Animals 2025, 15, 972. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.; Choo, H.; Srikanth, K.; Lee, S.-H.; Son, J.-W.; Park, M.-R.; Kim, N.; Jang, G.W.; Park, J.-E. Genome-wide association study identifies 12 loci associated with body weight at age 8 weeks in Korean native chickens. Genes 2021, 12, 1170. [Google Scholar] [CrossRef]
- Kanlisi, R.A.; Amuzu-Aweh, E.N.; Naazie, A.; Otsyina, H.R.; Kelly, T.R.; Gallardo, R.A.; Lamont, S.J.; Zhou, H.; Dekkers, J.; Kayang, B.B. Genetic architecture of body weight, carcass, and internal organs traits of Ghanaian local chickens. Front. Genet. 2024, 15, 1297034. [Google Scholar] [CrossRef]
- Mebratie, W.; Reyer, H.; Wimmers, K.; Bovenhuis, H.; Jensen, J. Genome wide association study of body weight and feed efficiency traits in a commercial broiler chicken population, a re-visitation. Sci. Rep. 2019, 9, 922. [Google Scholar] [CrossRef]
- Wang, J.; Liu, J.; Lei, Q.; Li, Y.; Zhang, Y.; Wang, Y.; Zhao, X.; Li, D.; Sun, G.; Kang, X.; et al. Elucidation of the genetic determination of body weight and size in Chinese local chicken breeds by large-scale genomic analyses. BMC Genom. 2024, 25, 296. [Google Scholar] [CrossRef]
- Dementeva, N.V.; Kudinov, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Fedorova, E.S.; Romanov, M.N. Genome-wide association study of reproductive traits in a gene pool breed of the Russian White chickens. Reprod. Domest. Anim. 2018, 53 (Suppl. S2), 123–124. [Google Scholar] [CrossRef]
- Volkova, N.A.; Romanov, M.N.; Vetokh, A.N.; Larionova, P.V.; Volkova, L.A.; Abdelmanova, A.S.; Sermyagin, A.A.; Griffin, D.K.; Zinovieva, N.A. Genome-wide association study reveals the genetic architecture of growth and meat production traits in a chicken F2 resource population. Genes 2024, 15, 1246. [Google Scholar] [CrossRef]
- Volkova, N.A.; Kotova, T.O.; Vetokh, A.N.; Larionova, P.V.; Volkova, L.A.; Romanov, M.N.; Zinovieva, N.A. Genome-wide association study of testes development indicators in roosters (Gallus gallus L.). Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2024, 59, 649–657. [Google Scholar] [CrossRef]
- Volkova, N.A.; Romanov, M.N.; Dzhagaev, A.Y.; Larionova, P.V.; Volkova, L.A.; Abdelmanova, A.S.; Vetokh, A.N.; Griffin, D.K.; Zinovieva, N.A. Genome-wide association studies and candidate genes for egg production traits in layers from an F2 crossbred population produced using two divergently selected chicken breeds, Russian White and Cornish White. Genes 2025, 16, 583. [Google Scholar] [CrossRef]
- Dementieva, N.V.; Shcherbakov, Y.S.; Stanishevskaya, O.I.; Vakhrameev, A.B.; Larkina, T.A.; Dysin, A.P.; Nikolaeva, O.A.; Ryabova, A.E.; Azovtseva, A.I.; Mitrofanova, O.V.; et al. Large-scale genome-wide SNP analysis reveals the rugged (and ragged) landscape of global ancestry, phylogeny, and demographic history in chicken breeds. J. Zhejiang Univ. Sci. B 2024, 25, 324–340. [Google Scholar] [CrossRef] [PubMed]
- Volkova, N.A.; Romanov, M.N.; Abdelmanova, A.S.; Larionova, P.V.; German, N.Y.; Vetokh, A.N.; Shakhin, A.V.; Volkova, L.A.; Anshakov, D.V.; Fisinin, V.I.; et al. Genotyping-by-sequencing strategy for integrating genomic structure, diversity and performance of various Japanese quail (Coturnix japonica) breeds. Animals 2023, 13, 3439. [Google Scholar] [CrossRef]
- Volkova, N.A.; German, N.Y.; Larionova, P.V.; Vetokh, A.N.; Romanov, M.N.; Zinovieva, N.A. [Identification of SNPs and candidate genes associated with abdominal fat deposition in quails (Coturnix japonica)]. Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2023, 58, 1079–1087. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Romanov, M.N.; Kochish, I.I.; Sharafetdinov, G.R.; Myasnikova, O.V.; Nikonov, I.N.; Selina, M.V.; Surai, P.F. Napravleniya sovremennykh biotekhnologicheskikh razrabotok dlya realizatsii geneticheskogo potentsiala yaichnoy ptitsy [Towards Advanced Biotechnological Developments to Realize the Genetic Potential of Egg-type Poultry]. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, 30 September 2021; Pozyabin, S.V., Kochish, I.I., Romanov, M.N., Eds.; Sel’skokhozyaistvennye tekhnologii: Moscow, Russia, 2021; pp. 40–51, (In Russian with English Summary). [Google Scholar] [CrossRef]
- Kutnyuk, P.I.; Volohovich, V.A.; Moiseeva, I.G. Elektroforeticheskij Analiz Belkov Selskohozyajstvennoj Pticy: Metodicheskie Rekomendacii [Electrophoretic Analysis of Poultry Proteins: Technical Recommendations]; Ukrainian Poultry Research Institute: Kharkov, USSR, 1986; Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=tFr9MGIAAAAJ:dshw04ExmUIC (accessed on 30 June 2025). (In Russian)
- Gadyuchko, O.T.; Sakhatsky, N.I.; Tereshchenko, A.V.; Anisimov, V.I.; Kislaya, E.D.; Svinarev, V.F. Geneticheskiy potentsial porod i populyatsiy gusey Ukrainy [Genetic potential of breeds and populations of geese in Ukraine]. Ptakhivnytstvo [Poult. Farming] 2003, 53, 54–62. Available online: https://www.researchgate.net/publication/342746669 (accessed on 30 June 2025). (In Russian with English Summary).
- Kutnyuk, P.I.; Moiseeva, I.G. Informatsionno-statisticheskiye aspekty modelirovaniya populyatsionnykh protsessov s pomoshch’yu molekulyarno-geneticheskikh markerov [Information and Statistical Aspects of Modeling Population Processes Using Molecular Genetic Markers]. In Proceedings of the 1st International Conference “Molecular-Genetic Markers of Animals”, Kyiv, Ukraine, 27–29 January 1994; Zubets, M.V., Ed.; UAAS, Agrarna nauka: Kyiv, Ukraine, 1994; pp. 132–133. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=tFr9MGIAAAAJ:7T2F9Uy0os0C (accessed on 30 June 2025). (In Russian).
- Baumgartner, J.; Bondarenko, Y.V. Search for Autosexing Strains and Crosses in Japanese Quail. In Proceedings of the 8th International Symposium on Actual Problems of Avian Genetics, Smolenice, Czechoslovakia, 3–6 April 1989; Slovak Society for Agriculture, Forestry, Food and Veterinary Sciences of Slovak Academy of Sciences: Bratislava, Czechoslovakia; Poultry Research and Production Institute: Ivanka pri Dunaji, Bratislava, Czechoslovakia; Czechoslovak Branch of WPSA: Smolenice, Czechoslovakia, 1989; pp. 262–265. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19890171986 (accessed on 30 June 2025).
- Romanov, M.N.; Shakhin, A.V.; Abdelmanova, A.S.; Volkova, N.A.; Efimov, D.N.; Fisinin, V.I.; Korshunova, L.G.; Anshakov, D.V.; Dotsev, A.V.; Griffin, D.K.; et al. Dissecting selective signatures and candidate genes in grandparent lines subject to high selection pressure for broiler production and in a local Russian chicken breed of Ushanka. Genes 2024, 15, 524. [Google Scholar] [CrossRef] [PubMed]
- Bondarenko, Y.V. Polymorphism of proteins of eggs of chickens of different breeds and lines. Ptakhivnytstvo [Poult. Farming] 1974, 17, 32–37. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:P7Ujq4OLJYoC (accessed on 30 June 2025).
- Zhang, H.; Hunt, H.D.; Cheng, H.H.; Dodgson, J.B.; Romanov, M.N.; Bacon, L.D. Identification and Evaluation of SNPs at the 3′ End of the Tva Gene Segregating Among ALSV Resistance and Susceptible Lines of Chickens. In Proceedings of the International Plant and Animal Genome XIII Conference, San Diego, CA, USA, 15–19 January 2005; Scherago International: San Diego, CA, USA, 2005; p. 123. Available online: https://www.ars.usda.gov/research/publications/publication/?seqNo115=170524 (accessed on 30 June 2025).
- Volkovoy, S.; Bondarenko, Y. Raduga opereniya yaponskogo perepela [Japanese quail plumage rainbow]. Priusadebnoye Khoz. [Allot. Husb.] 1989, 5, 14–15. Available online: https://yablonka.net/world/zh/686-raduga-opereniya-yaponskogo-perepela.html (accessed on 30 June 2025). (In Russian).
- Podstreshnyi, O.P.; Tereshchenko, O.V.; Tkachyk, T.E.; Podstreshna, I.O.; Ishchenko, Y.B. Henetychna Identyfikatsiya i Pasportyzatsiya Porid ta Liniy Ptytsi: Metodychni Rekomendatsiyi [Genetic Identification and Passportization of Poultry Breeds and Lines: Methodical Recommendations]; Poultry Research Institute of the Ukrainian Academy of Agrarian Sciences: Birky, Ukraine, 2009; Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=deB5xCwAAAAJ:j8SEvjWlNXcC (accessed on 30 June 2025). (In Ukrainian)
- Raetskii, A.; Mamontova, I.; Moiseeva, I. Rezul’taty ispol’zovaniya moskovskikh kur linii M5 v skreshchivanii s belymi leggornami krossa “Khayseks” [Results of using Moscow chickens of M5 line for crossing with White Leghorns of Highsex cross]. Izv. Timiryazev S-Kh. Akad. [Izv. Timiryazev Agric. Acad.] 1981, 6, 136–142. Available online: https://eurekamag.com/research/016/935/016935374.php (accessed on 30 June 2025).
- Moiseyeva, I.G.; Semyenova, S.K.; Gorbachyova, N.S. Chicken breed differentiation estimated by body measurements. In Proceedings of the Scientific Poultry Conference; Pigarev, N., Ed.; n.p.: Sergiev Posad, Russia, 1995; pp. 45–46. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=tFr9MGIAAAAJ:SdhP9T11ey4C (accessed on 30 June 2025). (In Russian).
- Narushin, V.G.; Romanov, M.N. Modelling growth of chick embryo. In Proceedings of the 9th European Poultry Conference: Plenary Papers and Contributed Papers; Glasgow, Scotland, UK, 7–12 August 1994; Great Britain, Ministry of Agriculture, Fisheries and Food; World’s Poultry Science Association, UK Branch: Andover, UK, 1994; Volume 1, pp. 411–412. Available online: https://kar.kent.ac.uk/46310/ (accessed on 30 June 2025).
- Narushin, V.G.; Romanov, M.N.; Sakhatsky, N.I. Modelling growth of chick embryo: Correction for egg weight [Modelowanie wzrostu zarodka kurzego z poprawką na masę jaja]. Zeszyty Naukowe. Przegląd Hodowlany [Anim. Prod. Rev., Appl. Sci. Rep.] 1997, 31, 55–57. Available online: https://www.researchgate.net/publication/355808171 (accessed on 30 June 2025).
- Bondarenko, Y.V.; Romanov, M.N. Osoblyvosti statevoyi minlyvosti masy tila u dobovoho molodnyaka sviysʹkykh ptakhiv [Peculiarities of sexual variation of body weight in day-old poults of domestic birds]. In Genetics of Animal Performance, Proceedings of the All-Ukrainian Anniversary Scientific and Practical Conference, Kyiv, Ukraine, 20 December 1994; n.p.: Kyiv, Ukraine, 1994; p. 36. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:HDshCWvjkbEC (accessed on 30 June 2025).
- Kochish, I.I.; Titov, V.Y.; Nikonov, I.N.; Brazhnik, E.A.; Vorobyov, N.I.; Korenyuga, M.V.; Myasnikova, O.V.; Dolgorukova, A.M.; Griffin, D.K.; Romanov, M.N. Unraveling signatures of chicken genetic diversity and divergent selection in breed-specific patterns of early myogenesis, nitric oxide metabolism and post-hatch growth. Front. Genet. 2023, 13, 1092242. [Google Scholar] [CrossRef]
- Khvostik, V.P.; Bondarenko, Y.V. Ekoloho-henetychni parametry zhyvoyi masy m’yaso-yayechnykh kurey riznoho henetychnoho pokhodzhennya [Ecological and genetic parameters of live weight meat and eggs hens of different genetic origin]. Vìsn. Sumsʹkogo nac. agrar. unìv., Ser. Tvarynnytstvo [Bull. Sumy Natl. Agrar. Univ. Ser. Livest.] 2024, 3, 106–109, (In Ukrainian with English Summary). [Google Scholar] [CrossRef]
- Lei, M.; Luo, C.; Peng, X.; Fang, M.; Nie, Q.; Zhang, D.; Yang, G.; Zhang, X. Polymorphism of growth-correlated genes associated with fatness and muscle fiber traits in chickens. Poult. Sci. 2007, 86, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Haqani, M.I.; Nomura, S.; Nakano, M.; Goto, T.; Nagano, A.J.; Takenouchi, A.; Nakamura, Y.; Ishikawa, A.; Tsudzuki, M. Mapping of quantitative trait loci controlling egg-quality and -production traits in Japanese quail (Coturnix japonica) using restriction-site associated DNA sequencing. Genes 2021, 12, 735. [Google Scholar] [CrossRef]
- Vollmar, S.; Haas, V.; Schmid, M.; Preuß, S.; Joshi, R.; Rodehutscord, M.; Bennewitz, J. Mapping genes for phosphorus utilization and correlated traits using a 4k SNP linkage map in Japanese quail (Coturnix japonica). Anim. Genet. 2020, 52, 90–98. [Google Scholar] [CrossRef]
- Recoquillay, J.; Pitel, F.; Arnould, C.; Leroux, S.; Dehais, P.; Moréno, C.; Calandreau, L.; Bertin, A.; Gourichon, D.; Bouchez, O.; et al. A medium density genetic map and QTL for behavioral and production traits in Japanese quail. BMC Genom. 2015, 16, 10. [Google Scholar] [CrossRef]
- Chen, S.; Jin, S.; Lu, Y.; Zhang, D.; Ji, C.; Yang, N. Association of leptin receptor gene polymorphisms with growth and feed efficiency in meat-type chickens. Poult. Sci. 2014, 93, 1910–1915. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, Y.; Bai, J. Research Note: Association of LEPR gene polymorphism with growth and carcass traits in Savimalt and French Giant meat-type quails. Poult. Sci. 2023, 102, 103047. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.D.; Wang, W.J.; Li, Z.W.; Ning, W.; Fan, X.I.A.O.; Gao, H.H.; Guo, H.S.; Hui, L.I.; Wang, S.Z. Integration of association and computational methods reveals functional variants of LEPR gene for abdominal fat content in chickens. J. Integr. Agric. 2021, 20, 2734–2748. [Google Scholar] [CrossRef]
- da Silva, R.C.; Ferraz, J.B.; Meirelles, F.V.; Eler, J.P.; Balieiro, J.C.; Cucco, D.C.; Mattos, E.C.; Rezende, F.M.; Silva, S.L. Association of single nucleotide polymorphisms in the bovine leptin and leptin receptor genes with growth and ultrasound carcass traits in Nellore cattle. Genet. Mol. Res. 2012, 11, 3721–3728. [Google Scholar] [CrossRef]
- Ferraz, J.B.S.; Pinto, L.F.B.; Meirelles, F.V.; Eler, J.P.; De Rezende, F.M.; Oliveira, E.C.M.; Almeida, H.B.; Woodward, B.; Nkrumah, D. Association of single nucleotide polymorphisms with carcass traits in Nellore cattle. Genet. Mol. Res. 2009, 8, 1360–1366. [Google Scholar] [CrossRef]
- Ovilo, C.; Fernández, A.; Noguera, J.L.; Barragán, C.; Letón, R.; Rodríguez, C.; Mercadé, A.; Alves, E.; Folch, J.M.; Varona, L. Fine mapping of porcine chromosome 6 QTL and LEPR effects on body composition in multiple generations of an Iberian by Landrace intercross. Genet. Res. 2005, 85, 57–67. [Google Scholar] [CrossRef]
- Pérez-Montarelo, D.; Fernández, A.; Folch, J.M.; Pena, R.N.; Ovilo, C.; Rodríguez, C.; Silió, L.; Fernández, A.I. Joint effects of porcine leptin and leptin receptor polymorphisms on productivity and quality traits. Anim. Genet. 2012, 43, 805–809. [Google Scholar] [CrossRef]
- Suárez-Mesa, R.; Ros-Freixedes, R.; Pena, R.N.; Reixach, J.; Estany, J. Impact of the leptin receptor gene on pig performance and quality traits. Sci. Rep. 2024, 14, 10652. [Google Scholar] [CrossRef]
- Haldar, A.; French, M.C.; Brauning, R.; Edwards, S.J.; O’Connell, A.R.; Farquhar, P.A.; Davis, G.H.; Johnstone, P.D.; Juengel, J.L. Single-nucleotide polymorphisms in the LEPR gene are associated with divergent phenotypes for age at onset of puberty in Davisdale ewes. Biol. Reprod. 2014, 90, 33. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, G.; Ovilo, C.; Silió, L.; Tomás, A.; Noguera, J.L.; Rodríguez, M.C. Single- and joint-population analyses of two experimental pig crosses to confirm quantitative trait loci on Sus scrofa chromosome 6 and leptin receptor effects on fatness and growth traits. J. Anim. Sci. 2009, 87, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Lakhssassi, K.; Serrano, M.; Lahoz, B.; Sarto, M.P.; Iguácel, L.P.; Folch, J.; Alabart, J.L.; Calvo, J.H. The LEPR gene is associated with reproductive seasonality traits in Rasa Aragonesa sheep. Animals 2020, 10, 2448. [Google Scholar] [CrossRef]
- Deng, S.; Qiu, Y.; Zhuang, Z.; Wu, J.; Li, X.; Ruan, D.; Xu, C.; Zheng, E.; Yang, M.; Cai, G.; et al. Genome-wide association study of body conformation traits in a three-way crossbred commercial pig population. Animals 2023, 13, 2414. [Google Scholar] [CrossRef]
- Meng, G.; Bao, Q.; Ma, X.; Chu, M.; Huang, C.; Guo, X.; Liang, C.; Yan, P. Analysis of copy number variation in the whole genome of normal-haired and long-haired Tianzhu white yaks. Genes 2022, 13, 2405. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Sánchez, F.A.; Sifuentes-Rincón, A.M.; Segura Cabrera, A.; García Pérez, C.A.; Parra Bracamonte, G.M.; Ambriz Morales, P. Associations of SNPs located at candidate genes to bovine growth traits, prioritized with an interaction networks construction approach. BMC Genet. 2015, 16, 91. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lv, G.; Liu, Z.; Cheng, Y.; Ding, R.; Yang, G.; Yu, T. Whole genome and transcriptome analyses identify genetic markers associated with growth traits in Qinchuan black pig. BMC Genom. 2025, 26, 469. [Google Scholar] [CrossRef]
- Ladeira, G.C.; Pilonetto, F.; Fernandes, A.C.; Bóscollo, P.P.; Dauria, B.D.; Titto, C.G.; Coutinho, L.L.; E Silva, F.F.; Pinto, L.F.B.; Mourao, G.B. CNV detection and their association with growth, efficiency and carcass traits in Santa Inês sheep. J. Anim. Breed Genet. 2022, 139, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, X.; Wang, Y.; Liu, L.; Wang, Y.; Zhao, G.; Wen, J.; Cui, H. Genome-wide association study revealed the effect of rs312715211 in ZNF652 gene on abdominal fat percentage of chickens. Biology 2022, 11, 1849. [Google Scholar] [CrossRef]
- Wu, Y.; Xiao, H.; Pi, J.; Zhang, H.; Pan, A.; Pu, Y.; Liang, Z.; Shen, J.; Du, J. EGFR promotes the proliferation of quail follicular granulosa cells through the MAPK/extracellular signal-regulated kinase (ERK) signaling pathway. Cell Cycle 2019, 18, 2742–2756. [Google Scholar] [CrossRef]
- Ji, G.; Zhang, M.; Tu, Y.; Liu, Y.; Shan, Y.; Ju, X.; Zou, J.; Shu, J.; Sheng, Z.; Li, H. Molecular regulatory mechanisms in chicken feather follicle morphogenesis. Genes 2023, 14, 1646. [Google Scholar] [CrossRef]
- Reis, H.B.D.; Carvalho, M.E.; Espigolan, R.; Poleti, M.D.; Ambrizi, D.R.; Berton, M.P.; Ferraz, J.B.S.; de Mattos Oliveira, E.C.; Eler, J.P. Genome-wide association (GWAS) applied to carcass and meat traits of Nellore cattle. Metabolites 2024, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Moreira, G.C.M.; Boschiero, C.; Cesar, A.S.M.; Reecy, J.M.; Godoy, T.F.; Trevisoli, P.A.; Cantão, M.E.; Ledur, M.C.; Ibelli, A.M.G.; Peixoto, J.D.O.; et al. A genome-wide association study reveals novel genomic regions and positional candidate genes for fat deposition in broiler chickens. BMC Genom. 2018, 19, 374. [Google Scholar] [CrossRef]
- Kogelman, L.J.; Pant, S.D.; Fredholm, M.; Kadarmideen, H.N. Systems genetics of obesity in an F2 pig model by genome-wide association, genetic network, and pathway analyses. Front. Genet. 2014, 5, 214. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, J.; Fan, X.; Chen, J.; Wang, Z.; Liu, X.; Yi, G.; Liu, Y.; Niu, Y.; Zhang, L.; et al. The genome variation and developmental transcriptome maps reveal genetic differentiation of skeletal muscle in pigs. PLoS Genet. 2021, 17, e1009910. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Tang, Q.; Qi, J.; Han, X.; Tao, Q.; Lu, Y.; Bai, Y.; Hu, S.; Li, L.; Bai, L.; et al. Integration of GWAS and transcriptomic analyses reveal candidate genes for duck gonadal development during puberty onset. BMC Genom. 2024, 25, 1151. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.W.; Liang, Y.C.; Wu, P.; Chen, C.K.; Lai, Y.C.; Jiang, T.X.; Haung, Y.H.; Chuong, C.M. Regional specific differentiation of integumentary organs: SATB2 is involved in α- and β-keratin gene cluster switching in the chicken. Dev. Dyn. 2022, 251, 1490–1508. [Google Scholar] [CrossRef] [PubMed]
- Lü, S.J.; Chen, F.Y.; Jin, L.; Zhang, Z.J.; Zhu, X.T.; Shi, Q.T.; Xin, X.L.; Chu, Q.X.; Bai, Z.L.; Wang, E.Y.; et al. Identification of growth-related genes under selection in Angus cattle using SLAF-seq. Acta Vet. Zootech. Sin. 2020, 51, 713–721. [Google Scholar] [CrossRef]
- Park, H.B.; Han, S.H.; Lee, J.B.; Cho, I.C. Rapid Communication: High-resolution quantitative trait loci analysis identifies LTBP2 encoding latent transforming growth factor beta binding protein 2 associated with thoracic vertebrae number in a large F2 intercross between Landrace and Korean native pigs. J. Anim. Sci. 2017, 95, 1957–1962. [Google Scholar] [CrossRef]
- Park, J.; Do, K.T.; Park, K.D.; Lee, H.K. Genome-wide association study using a single-step approach for teat number in Duroc, Landrace and Yorkshire pigs in Korea. Anim. Genet. 2023, 54, 743–751. [Google Scholar] [CrossRef]
- Lee, Y.; Tjeerdema, E.; Kling, S.; Chang, N.; Hamdoun, A. Solute carrier (SLC) expression reveals skeletogenic cell diversity. Dev. Biol. 2023, 503, 68–82. [Google Scholar] [CrossRef]
- Lin, J.; Redies, C.; Luo, J. Regionalized expression of ADAM13 during chicken embryonic development. Dev. Dyn. 2007, 236, 862–870. [Google Scholar] [CrossRef]
- Lin, J.; Yan, X.; Markus, A.; Redies, C.; Rolfs, A.; Luo, J. Expression of seven members of the ADAM family in developing chicken spinal cord. Dev. Dyn. 2010, 239, 1246–1254. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Luo, J.; Redies, C. Differential regional expression of multiple ADAMs during feather bud formation. Dev. Dyn. 2011, 240, 2142–2152. [Google Scholar] [CrossRef] [PubMed]
- Lukic, B.; Curik, I.; Drzaic, I.; Galić, V.; Shihabi, M.; Vostry, L.; Cubric-Curik, V. Genomic signatures of selection, local adaptation and production type characterisation of East Adriatic sheep breeds. J. Anim. Sci. Biotechnol. 2023, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Holgate, S.T.; Davies, D.E.; Powell, R.M.; Holloway, J.W. ADAM33: A newly identified protease involved in airway remodelling. Pulm. Pharmacol. Ther. 2006, 19, 3–11. [Google Scholar] [CrossRef]
- Davis, R.V.N. Comparative Transcriptional Profiling and Differential Expression Analysis of Pectoralis Major Muscle in Modern and Legacy Gallus Broiler Lines. Ph.D. Thesis, University of Delaware, Newark, DE, USA, 2017. Available online: https://udspace.udel.edu/server/api/core/bitstreams/509dbb29-d100-4f7f-ae41-bc6ae75466c5/content (accessed on 30 June 2025).
- Eydivandi, S.; Roudbar, M.A.; Karimi, M.O.; Sahana, G. Genomic scans for selective sweeps through haplotype homozygosity and allelic fixation in 14 indigenous sheep breeds from Middle East and South Asia. Sci. Rep. 2021, 11, 2834. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Y.; Chen, Q.; Wang, D.; Zhang, X.; Huang, X.; Xu, L. Genome-wide association study on body conformation traits in Xinjiang Brown cattle. Int. J. Mol. Sci. 2024, 25, 10557. [Google Scholar] [CrossRef]
- Kschonsak, M.; Chua, H.C.; Weidling, C.; Chakouri, N.; Noland, C.L.; Schott, K.; Chang, T.; Tam, C.; Patel, N.; Arthur, C.P.; et al. Structural architecture of the human NALCN channelosome. Nature 2022, 603, 180–186. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, H.; Zhao, Q.; Wu, J.; Yan, Z. Architecture of the human NALCN channelosome. Cell Discov. 2022, 8, 33. [Google Scholar] [CrossRef]
- Bayat, A.; Liu, Z.; Luo, S.; Fenger, C.D.; Højte, A.F.; Isidor, B.; Cogne, B.; Larson, A.; Zanus, C.; Faletra, F.; et al. A new neurodevelopmental disorder linked to heterozygous variants in UNC79. Genet. Med. 2023, 25, 100894. [Google Scholar] [CrossRef]
- Zhou, X.; Ouyang, M.; Zhang, Y.; Ikhwanuddin, M.; Ma, H.; Ye, S. Whole-genome resequencing provides novel insights into the genetic diversity, population structure, and patterns of runs of homozygosity in mud crab (Scylla paramamosain). SSRN 2025, 5128826. [Google Scholar] [CrossRef]
- Murakami, K.; Palermo, J.; Stanhope, B.A.; Gibbs, A.G.; Keene, A.C. A screen for sleep and starvation resistance identifies a wake-promoting role for the auxiliary channel unc79. G3 2021, 11, jkab199. [Google Scholar] [CrossRef]
- Albrecht, E.; Komolka, K.; Ponsuksili, S.; Gotoh, T.; Wimmers, K.; Maak, S. Transcriptome profiling of Musculus longissimus dorsi in two cattle breeds with different intramuscular fat deposition. Genom. Data 2016, 7, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Yao, Z.; Kong, J.; Zhang, X.; Li, H.; Chen, W.; Xie, Q. Transcriptomic analysis reveals the dynamic changes of transcription factors during early development of chicken embryo. BMC Genom. 2022, 23, 825. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.Y.; He, T.T.; Gao, X.M.; Zhao, Y.; Wang, J. ZBTB transcription factors: Key regulators of the development, differentiation and effector function of T cells. Front. Immunol. 2021, 12, 713294. [Google Scholar] [CrossRef] [PubMed]
- Mao, A.P.; Ishizuka, I.E.; Kasal, D.N.; Mandal, M.; Bendelac, A. A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development. Nat. Commun. 2017, 8, 863. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Kitamura, Y.; Maezawa, S.; Namekawa, S.H.; Cairns, B.R. ZBTB16/PLZF regulates juvenile spermatogonial stem cell development through an extensive transcription factor poising network. Nat. Struct. Mol. Biol. 2025, 32, 1213–1226. [Google Scholar] [CrossRef]
- Plaisier, C.L.; Bennett, B.J.; He, A.; Guan, B.; Lusis, A.J.; Reue, K.; Vergnes, L. Zbtb16 has a role in brown adipocyte bioenergetics. Nutr. Diabetes 2012, 2, e46. [Google Scholar] [CrossRef]
- Ying, F.; Gu, H.; Xiong, Y.; Zuo, B. Analysis of differentially expressed genes in gastrocnemius muscle between DGAT1 transgenic mice and wild-type mice. Biomed. Res. Int. 2017, 2017, 5404682. [Google Scholar] [CrossRef]
- Wei, S.; Zhang, M.; Zheng, Y.; Yan, P. ZBTB16 overexpression enhances white adipogenesis and induces brown-like adipocyte formation of bovine white intramuscular preadipocytes. Cell Physiol. Biochem. 2018, 48, 2528–2538. [Google Scholar] [CrossRef]
- Chen, J.F.; Wang, J.; Chai, J.; Jin, W.; Ren, Q.L.; Ma, Q.; Lu, Q.X.; Sun, J.J.; Mo, D.L.; Zhang, J.Q.; et al. Transcriptome profiling of longissimus dorsi during different prenatal stages to identify genes involved in intramuscular fat deposition in lean and obese pig breeds. Mol. Biol. Rep. 2024, 51, 386. [Google Scholar] [CrossRef]
- Liška, F.; Landa, V.; Zídek, V.; Mlejnek, P.; Šilhavý, J.; Šimáková, M.; Strnad, H.; Trnovská, J.; Škop, V.; Kazdová, L.; et al. Downregulation of Plzf gene ameliorates metabolic and cardiac traits in the spontaneously hypertensive rat. Hypertension 2017, 69, 1084–1091. [Google Scholar] [CrossRef]
- Bendlová, B.; Vaňková, M.; Hill, M.; Vacínová, G.; Lukášová, P.; Vejražková, D.; Šedová, L.; Šeda, O.; Včelák, J. ZBTB16 gene variability influences obesity-related parameters and serum lipid levels in Czech adults. Physiol. Res. 2017, 66 (Suppl. S3), S425–S431. [Google Scholar] [CrossRef]
- Topno, N.A.; Kesarwani, V.; Kushwaha, S.K.; Azam, S.; Kadivella, M.; Gandham, R.K.; Majumdar, S.S. Non-synonymous variants in fat QTL genes among high- and low-milk-yielding indigenous breeds. Animals 2023, 13, 884. [Google Scholar] [CrossRef]
- Xu, S.; Donnelly, L.; Kober, D.L.; Mak, M.; Radhakrishnan, A. Development of a monoclonal antibody to study MARCH6, an E3 ligase that regulates proteins that control lipid homeostasis. J. Lipid Res. 2024, 65, 100650. [Google Scholar] [CrossRef]
- Scott, N.A.; Sharpe, L.J.; Brown, A.J. The E3 ubiquitin ligase MARCHF6 as a metabolic integrator in cholesterol synthesis and beyond. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158837. [Google Scholar] [CrossRef]
- Liu, Q.; Yue, J.; Niu, N.; Liu, X.; Yan, H.; Zhao, F.; Hou, X.; Gao, H.; Shi, L.; Wang, L.; et al. Genome-wide association analysis identified BMPR1A as a novel candidate gene affecting the number of thoracic vertebrae in a Large White × Minzhu intercross pig population. Animals 2020, 10, 2186. [Google Scholar] [CrossRef]
- Zhang, L.C.; Yue, J.W.; Pu, L.; Wang, L.G.; Liu, X.; Liang, J.; Yan, H.; Zhao, K.B.; Li, N.; Shi, H.B.; et al. Genome-wide study refines the quantitative trait locus for number of ribs in a Large White × Minzhu intercross pig population and reveals a new candidate gene. Mol. Genet. Genom. 2016, 291, 1885–1890. [Google Scholar] [CrossRef] [PubMed]
- Ogunbawo, A.R.; Hidalgo, J.; Mulim, H.A.; Carrara, E.R.; Ventura, H.T.; Souza, N.O.; Lourenco, D.; Oliveira, H.R. Applying the Algorithm for Proven and Eoung in GWAS reveals high polygenicity for key traits in Nellore cattle. Front. Genet. 2025, 16, 1549284. [Google Scholar] [CrossRef] [PubMed]
- Derežanin, L.; Blažytė, A.; Dobrynin, P.; Duchêne, D.A.; Grau, J.H.; Jeon, S.; Kliver, S.; Koepfli, K.P.; Meneghini, D.; Preick, M.; et al. Multiple types of genomic variation contribute to adaptive traits in the mustelid subfamily Guloninae. Mol. Ecol. 2022, 31, 2898–2919. [Google Scholar] [CrossRef]
- Kitakaze, T.; Tatsumi, R.; Yamaguchi, M.; Kubota, M.; Nakatsuji, A.; Harada, N.; Yamaji, R. All-trans retinoic acid-responsive LGR6 is transiently expressed during myogenic differentiation and is required for myoblast differentiation and fusion. Int. J. Mol. Sci. 2023, 24, 9035. [Google Scholar] [CrossRef] [PubMed]
- King, J.S.; Wan, M.; Wagley, Y.; Stestiv, M.; Kalajzic, I.; Hankenson, K.D.; Sanjay, A. Signaling pathways associated with Lgr6 to regulate osteogenesis. Bone 2024, 187, 117207. [Google Scholar] [CrossRef] [PubMed]
- Podstreshnyi, O.P.; Tereshchenko, O.V.; Katerynych, O.O.; Tkachyk, T.E.; Podstreshna, I.O. Vyrobnytstvo Perepelynykh Yayets’ ta M″yasa: Metodychni Rekomendatsiyi [Production of Quail Eggs and Meat: Methodical Recommendations], 2nd ed.; Tereshchenko, O.V., Ed.; Poultry Research Institute, NAAS of Ukraine: Birky, Ukraine, 2010; Available online: https://www.researchgate.net/publication/342802513 (accessed on 30 June 2025). (In Ukrainian)
- Podstreshnyi, O.; Tereshchenko, O. Hodivlya molodnyaka perepeliv [Feeding young quails]. Ahrar. Krayina [Agrar. Country] 2012, 7, 6. Available online: https://www.researchgate.net/publication/342832583 (accessed on 30 June 2025). (In Ukrainian).
- Podstreshnyi, O.; Tereshchenko, O. Utrymannya doroslykh perepeliv [Maintenance of adult quails]. Ahrar. Krayina [Agrar. Country] 2012, 6, 8–9. Available online: https://www.researchgate.net/publication/342832587 (accessed on 30 June 2025). (In Ukrainian).
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPF90 Family of Programs; University of Georgia: Athens, GA, USA, 2022; Available online: https://nce.ads.uga.edu/html/projects/programs/docs/blupf90_all8.pdf (accessed on 30 June 2025).
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Dodds, K.G.; McEwan, J.C.; Brauning, R.; Anderson, R.M.; van Stijn, T.C.; Kristjánsson, T.; Clarke, S.M. Construction of relatedness matrices using genotyping-by-sequencing data. BMC Genom. 2015, 16, 1047. [Google Scholar] [CrossRef]
- AgResearch. DECONVQC; GitHub, Inc.: San Francisco, CA, USA, 2016; Available online: https://github.com/AgResearch/DECONVQC (accessed on 30 June 2025).
- Jacobs, J.; Clarke, S.; Faville, M.; Griffiths, A.; Cao, M.; Tan, R.; Van Stijn, T.; Anderson, R.; Ashby, R.; Rowe, S.; et al. Genotyping-by-sequencing applications in biology. In Proceedings of the Plant and Animal Genome XXV Conference, San Diego, CA, USA, 13–18 January 2017; Scherago International: Surfside, FL, USA, 2017. Abstract P0128. Available online: https://pag.confex.com/pag/xxv/meetingapp.cgi/Paper/24487 (accessed on 30 June 2025).
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data, Version 0.10.1; Bioinformatics Group, Babraham Institute: Cambridge, UK, 2012; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 30 June 2025).
- Morris, K.M.; Hindle, M.M.; Boitard, S.; Burt, D.W.; Danner, A.F.; Eory, L.; Forrest, H.L.; Gourichon, D.; Gros, J.; Hillier, L.W.; et al. The quail genome: Insights into social behaviour, seasonal biology and infectious disease response. BMC Biol. 2020, 18, 14. [Google Scholar] [CrossRef]
- Szpak, M. Ensembl 104 Has Been Released. Ensembl Blog. 2021. Available online: https://www.ensembl.info/2021/05/05/ensembl-104-has-been-released/ (accessed on 30 June 2025).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt, Version 3.4; GitHub, Inc.: San Francisco, CA, USA, 2021. Available online: https://github.com/marcelm/cutadapt (accessed on 30 June 2025).
- Langmead, B. bowtie2: A Fast and Sensitive Gapped Read Aligner, Version 2.4.4; GitHub, Inc.: San Francisco, CA, USA, 2021. Available online: https://github.com/BenLangmead/bowtie2 (accessed on 30 June 2025).
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- AgResearch. snpGBS; GitHub, Inc.: San Francisco, CA, USA, 2021; Available online: https://github.com/AgResearch/snpGBS (accessed on 30 June 2025).
- R Core Team. R-4; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://cran.r-project.org/bin/windows/base/old/4.4.0/NEWS.R-4.4.0.html (accessed on 30 June 2025).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.r-project.org/ (accessed on 30 June 2025).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S. PLINK 1.9; Center for Human Genetic Research, Massachusetts General Hospital, and the Broad Institute of Harvard & MIT: Boston, MA, USA, 2017; Available online: https://zzz.bwh.harvard.edu/plink/index.shtml (accessed on 30 June 2025).
- DataCamp. Principal Component Analysis in R Tutorial; DataCamp, Inc.: New York, NY, USA, 2023; Available online: https://www.datacamp.com/tutorial/pca-analysis-r (accessed on 30 June 2025).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98141-3. [Google Scholar] [CrossRef]
- GitHub. ggplot2. tidyverse; GitHub, Inc.: San Francisco, CA, USA, 2023; Available online: https://github.com/tidyverse/ggplot2 (accessed on 30 June 2025).
- GitHub. qqman; GitHub, Inc.: San Francisco, CA, USA, 2025; Available online: https://github.com/qqman (accessed on 30 June 2025).
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and Manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]

| Traits 1 | F2 (n = 240) | Japanese Breed (n = 30) | Texas White Breed (n = 20) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD 2 | CV 3, % | Mean | SD | CV, % | Mean | SD | CV, % | |
| BW1 | 8.8 | 0.9 | 10.5 | 8.5 | 0.4 | 4.2 | 9.3 | 0.8 | 8.6 |
| BW7 | 31.7 | 6.3 | 21.7 | 28.9 | 3.2 | 10.1 | 40.0 | 3.7 | 9.4 |
| BW14 | 77.4 | 12.1 | 17.4 | 69.3 | 7.4 | 9.6 | 93.2 | 4.2 | 4.5 |
| BW21 | 116.4 | 19.6 | 16.8 | 103.8 | 18.4 | 17.7 | 157.8 | 25.7 | 16.3 |
| BW28 | 158.5 | 22.1 | 13.9 | 125.5 | 24.8 | 19.8 | 204.9 | 32.1 | 15.6 |
| BW35 | 191.7 | 25.2 | 13.2 | 164.5 | 22.4 | 13.6 | 255.7 | 29.3 | 11.5 |
| BW42 | 218.9 | 29.6 | 13.5 | 174.5 | 18.1 | 10.4 | 275.6 | 37.3 | 13.5 |
| BW49 | 241.4 | 30.1 | 12.5 | 205.8 | 32.0 | 15.5 | 301.0 | 41.2 | 13.7 |
| BW56 | 249.3 | 33.2 | 13.3 | – | – | – | – | – | – |
| Traits 1 | Genetic Variance (VarA) | Residual Variance (VarE) | Heritability (h2) |
|---|---|---|---|
| BW1 | 0.395 | 0.304 | 0.565 |
| BW7 | 13.49 | 24.69 | 0.353 |
| BW14 | 48.22 | 106.2 | 0.312 |
| BW21 | 87.44 | 226.4 | 0.279 |
| BW28 | 122.9 | 310.4 | 0.284 |
| BW35 | 207.8 | 500.4 | 0.293 |
| BW42 | 234.9 | 577.3 | 0.289 |
| BW49 | 246.2 | 627.3 | 0.282 |
| BW56 | 281.0 | 732.0 | 0.277 |
| Traits 1 | BW1 | BW7 | BW14 | BW21 | BW28 | BW35 | BW42 | BW49 | BW56 |
|---|---|---|---|---|---|---|---|---|---|
| BW1 | – | ||||||||
| BW7 | 0.039 | – | |||||||
| BW14 | 0.074 | 0.518 | – | ||||||
| BW21 | 0.049 | 0.427 | 0.680 | – | |||||
| BW28 | 0.026 | 0.386 | 0.659 | 0.665 | – | ||||
| BW35 | 0.026 | 0.336 | 0.570 | 0.628 | 0.702 | – | |||
| BW42 | 0.008 | 0.314 | 0.542 | 0.543 | 0.668 | 0.711 | – | ||
| BW49 | −0.002 | 0.314 | 0.487 | 0.494 | 0.580 | 0.629 | 0.744 | – | |
| BW56 | 0.009 | 0.308 | 0.428 | 0.436 | 0.492 | 0.508 | 0.601 | 0.751 | – |
| Traits 1 | No. of SNPs | Chromosomes |
|---|---|---|
| BW1 | 3 | CJA5, CJA25 |
| BW14 | 8 | CJA4, CJA7, CJA26, CJA28 |
| BW21 | 8 | CJA4, CJA7, CJA10 |
| BW28 | 23 | CJA2, CJA4, CJA7, CJA8, CJA10, CJA11, CJA14, CJA15, CJA22 |
| BW35 | 15 | CJA1, CJA2, CJA3, CJA4, CJA15, CJA24 |
| BW42 | 50 | CJA1, CJA3-CJA6, CJA8, CJA10, CJA12, CJA15, CJA17, CJA18, CJA20, CJA22, CJA24, CJA26 |
| BW49 | 31 | CJA1, CJA5, CJA8, CJA12, CJA17, CJA20, CJA22, CJA23, CJA26 |
| BW56 | 42 | CJA1-CJA4, CJA8, CJA14, CJA15, CJA18, CJA20, CJA25, CJA26 |
| Chromosome | SNP Position (in bp) | Traits 1 | PCG (at SNP Position) |
|---|---|---|---|
| CJA1 | 151,007,527 | BW42, BW49 | ITM2B |
| CJA1 | 151,084,995 | BW42, BW49 | – |
| CJA3 | 34,595,032 | BW35, BW42 | SLC35F3 |
| CJA4 | 81,160,722 | BW14, BW21, BW28, BW35, BW42 | ADAM33 |
| CJA4 | 81,160,836 | BW14, BW21, BW28 | ADAM33 |
| CJA4 | 81,160,888 | BW14, BW21, BW28, BW35 | ADAM33 |
| CJA4 | 81,160,897 | BW14, BW21, BW28, BW35 | ADAM33 |
| CJA4 | 81,171,150 | BW14, BW21, BW28, BW35 | ADAM33 |
| CJA5 | 40,981,270 | BW42, BW49 | UNC79 |
| CJA8 | 24,869,812 | BW28, BW42 | – |
| CJA8 | 25,524,051 | BW42, BW49 | LEPR |
| CJA8 | 25,548,415 | BW42, BW49 | – |
| CJA10 | 10,505,840 | BW21, BW28 | – |
| CJA12 | 9,300,025 | BW42, BW49 | RPP14 |
| CJA12 | 9,302,400 | BW42, BW49 | – |
| CJA15 | 5,904,595 | BW35, BW42 | MVK |
| CJA15 | 11,552,880 | BW42, BW56 | – |
| CJA15 | 11,552,881 | BW42, BW49, BW56 | – |
| CJA15 | 11,622,693 | BW49, BW56 | – |
| CJA17 | 2,261,513 | BW42, BW49 | ASTN2 |
| CJA20 | 4,345,582 | BW49, BW56 | – |
| CJA24 | 3,829,762 | BW35, BW42 | ZBTB16 |
| CJA26 | 68,898 | BW49, BW56 | – |
| CJA26 | 243,899 | BW42, BW49, BW56 | – |
| CJA26 | 243,912 | BW42, BW49, BW56 | – |
| Chromosome | PCG | SNP Position (in bp) | Traits 1 | p-Value |
|---|---|---|---|---|
| CJA2 | MARCHF6 | 52,281,470 | BW35 | 5.35 × 10−7 |
| 52,281,495 | BW35 | 5.35 × 10−7 | ||
| 52,281,496 | BW35 | 5.35 × 10−7 | ||
| 52,281,497 | BW35 | 5.35 × 10−7 | ||
| EGFR | 73,896,881 | BW56 | 2.12 × 10−11 | |
| 73,896,952 | BW56 | 1.32 × 10−9 | ||
| CJA4 | ADGRL3 | 43,060,269 | BW28 | 6.18 × 10−7 |
| 43,060,333 | BW28 | 6.18 × 10−7 | ||
| 43,060,345 | BW28 | 6.18 × 10−7 | ||
| ADAM33 | 81,160,722 | BW14 | 9.46 × 10−8 | |
| BW21 | 1.59 × 10−8 | |||
| BW28 | 1.36 × 10−8 | |||
| BW35 | 8.27 × 10−8 | |||
| BW42 | 7.96 × 10−8 | |||
| 81,160,836 | BW14 | 4.46 × 10−8 | ||
| BW21 | 3.39 × 10−7 | |||
| BW28 | 4.27 × 10−8 | |||
| 81,160,888 | BW14 | 1.20 × 10−7 | ||
| BW21 | 5.73 × 10−8 | |||
| BW28 | 9.55 × 10−9 | |||
| BW35 | 2.64 × 10−7 | |||
| 81,160,897 | BW14 | 1.20 × 10−7 | ||
| BW21 | 5.73 × 10−8 | |||
| BW28 | 9.55 × 10−9 | |||
| BW35 | 2.64 × 10−7 | |||
| 81,171,150 | BW14 | 6.12 × 10−8 | ||
| BW21 | 1.35 × 10−7 | |||
| BW28 | 8.41 × 10−8 | |||
| BW35 | 4.58 × 10−7 | |||
| CJA5 | NPC2 | 34,403,827 | BW42 | 8.89 × 10−8 |
| 34,403,875 | BW42 | 8.89 × 10−8 | ||
| LTBP2 | 34,409,135 | BW42 | 1.70 × 10−7 | |
| 34,409,184 | BW42 | 3.45 × 10−7 | ||
| 34,411,616 | BW42 | 1.70 × 10−7 | ||
| 34,412,981 | BW42 | 3.07 × 10−7 | ||
| 34,417,379 | BW42 | 2.75 × 10−7 | ||
| ZC2HC1C | 34,609,626 | BW42 | 2.72 × 10−7 | |
| 34,610,313 | BW42 | 6.96 × 10−8 | ||
| CJA7 | SATB2 | 9,470,112 | BW28 | 3.91 × 10−7 |
| 9,470,188 | BW28 | 1.30 × 10−7 | ||
| 9,470,245 | BW28 | 1.30 × 10−7 | ||
| CJA17 | ASTN2 | 2,218,419 | BW49 | 5.83 × 10−8 |
| 2,261,513 | BW42 | 6.15 × 10−8 | ||
| BW49 | 2.61 × 10−8 | |||
| CJA24 | ZBTB16 | 3,829,762 | BW35 | 1.05 × 10−8 |
| BW42 | 2.36 × 10−7 | |||
| 3,847,375 | BW35 | 5.43 × 10−7 | ||
| CJA25 | ADAR | 1,503,153 | BW1 | 2.82 × 10−7 |
| 1,503,237 | BW1 | 3.14 × 10−9 | ||
| CJA26 | LGR6 | 798,927 | BW49 | 3.76 × 10−7 |
| 850,193 | BW49 | 2.94 × 10−7 |
| SNP Geno-type | n 1 | Traits 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BW1 | BW7 | BW14 | BW21 | BW28 | BW35 | BW42 | BW49 | BW56 | ||
| 4:81160722 | ||||||||||
| CC | 13 | 9.2 ± 0.2 | 39.7 ± 1.2 | 91.9 ± 1.0 ab, ac | 167.0 ± 3.8 ab, ac | 228.8 ± 5.5 ab, ac | 249.0 ± 5.2 ab, ac | 296.2 ± 9.7 ab, ac | 302.8 ± 9.7 | 328.0 ± 8.2 ab, ac |
| CT | 47 | 8.8 ± 0.1 | 29.4 ± 1.0 | 78.5 ± 1.8 ab, bc | 127.3 ± 3.7 bc | 169.7 ± 4.1 ab, bc | 206.4 ± 4.4 ab, bc | 231.5 ± 4.9 ab, bc | 249.4 ± 5.4 | 262.4 ± 5.7 ab |
| TT | 173 | 8.5 ± 0.1 | 29.1 ± 0.5 | 69.5 ± 0.9 ac | 114.5 ± 1.5 ac | 153.6 ± 1.9 ac | 188.0 ± 2.1 ac | 214.1 ± 2.4 ac | 236.8 ± 2.2 | 246.8 ± 2.4 ac |
| 4:81160836 | ||||||||||
| GG | 11 | 9.0 ± 0.2 | 42.0 ± 1.4 | 92.7 ± 1.4 de, cd | 169.3 ± 5.5 de, cd | 237.3 ± 5.9 de, cd | 252.0 ± 5.8 de, cd | 290.7 ± 10.4 de, cd | 292.3 ± 10.9 de, cd | 294.8 ± 8.9 cd |
| GT | 50 | 8.7 ± 0.1 | 29.4 ± 1.0 | 78.7 ± 1.8 ce, de | 127.3 ± 3.8 ce, de | 169.5 ± 4.0 ce, de | 206.4 ± 4.3 ce, de | 233.6 ± 5.0 ce, de | 253.1 ± 5.5 ce | 265.1 ± 5.8 ce, de |
| TT | 172 | 8.5 ± 0.1 | 29.3 ± 0.5 | 69.5 ± 0.9 cd, ce | 115.1 ± 1.5 cd, ce | 154.4 ± 1.9 cd, ce | 188.6 ± 2.2 cd, ce | 214.6 ± 2.4 cd, ce | 237.9 ± 2.5 cd | 246.4 ± 2.3 cd, ce |
| 4:81160888 | ||||||||||
| CC | 171 | 8.5 ± 0.1 | 29.2 ± 1.4 | 69.4 ± 0.9 ab, ac | 114.9 ± 1.5 ab, ac | 154.1 ± 1.9 ab, ac | 188.3 ± 2.1 ab, ac | 214.2 ± 2.4 ab, ac | 237.6 ± 2.5 ab, ac | 246.7 ± 2.5 ab, ac |
| TC | 51 | 8.8 ± 0.1 | 29.7 ± 1.0 | 79.1 ± 1.8 ab, bc | 128.5 ± 3.8 ab, bc | 170.7 ± 4.0 ab, bc | 208.1 ± 4.4 ab, bc | 235.3 ± 5.1 ab, bc | 254.4 ± 5.5 ab, bc | 266.1 ± 5.4 ab |
| TT | 11 | 9.0 ± 0.2 | 42.0 ± 0.2 | 92.7 ± 1.4 ac, bc | 169.3 ± 5.5 ac, bc | 237.3 ± 5.9 ac, bc | 252.0 ± 5.8 ac, bc | 290.7 ± 10.4 ac, bc | 292.3 ± 10.9 ac, bc | 294.8 ± 8.9 ac |
| 4:81160897 | ||||||||||
| CC | 11 | 9.0 ± 0.2 | 42.0 ± 1.4 | 92.7 ± 1.4 ab, ac | 169.3 ± 5.5 ab, ac | 237.3 ± 5.9 ab, ac | 252.0 ± 5.8 ab, ac | 290.7 ± 10.4 ab, ac | 292.3 ± 10.9 ab, ac | 294.8 ± 8.9 ac |
| CT | 51 | 8.8 ± 0.1 | 29.7 ± 1.0 | 79.1 ± 1.8 ab, bc | 128.5 ± 3.8 ab, bc | 170.7 ± 4.0 ab, bc | 208.1 ± 4.4 ab, bc | 235.3 ± 5.1 ab, bc | 254.4 ± 5.5 ab, bc | 266.1 ± 5.4 bc |
| TT | 171 | 8.5 ± 0.1 | 29.2 ± 0.5 | 69.4 ± 0.9 ac, bc | 114.9 ± 1.5 ac, bc | 154.1 ± 1.9 ac, bc | 188.3 ± 2.1 ac, bc | 214.2 ± 2.4 ac, bc | 237.6 ± 2.5 ac, bc | 246.7 ± 2.5 ac, bc |
| 4:81171150 | ||||||||||
| CC | 174 | 8.5 ± 0.1 ac | 29.2 ± 0.5 | 69.5 ± 0.9 ab, ac | 115.2 ± 1.5 ab, ac | 154.8 ± 1.9 ab, ac | 189.0 ± 2.1 ab, ac | 214.8 ± 2.4 ab, ac | 238.4 ± 2.5 ac | 247.6 ± 2.5 ab, ac |
| TC | 45 | 8.7 ± 0.1 | 29.0 ± 1.0 | 77.9 ± 1.8 ab, bc | 127.4 ± 4.0 ab, bc | 169.1 ± 4.4 ab, bc | 205.0 ± ±4.4 ab, bc | 232.1 ± 5.3 ab, bc | 249.7 ± 5.5 bc | 267.0 ± 5.9 ab, bc |
| TT | 14 | 9.3 ± 0.2 ac | 41.8 ± 1.2 | 94.3 ± 1.4 ac, bc | 164.8 ± 4.6 ac, bc | 221.5 ± 6.2 ac, bc | 251.8 ± 5.5 ac, bc | 288.3 ± 10.8 ac, bc | 300.3 ± 10.0 ac, bc | 319.1 ± 8.7 ac, bc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volkova, N.A.; Romanov, M.N.; German, N.Y.; Larionova, P.V.; Vetokh, A.N.; Volkova, L.A.; Sermyagin, A.A.; Shakhin, A.V.; Griffin, D.K.; Sölkner, J.; et al. Genome-Wide Association Studies in Japanese Quails of the F2 Resource Population Elucidate Molecular Markers and Candidate Genes for Body Weight Parameters. Int. J. Mol. Sci. 2025, 26, 8243. https://doi.org/10.3390/ijms26178243
Volkova NA, Romanov MN, German NY, Larionova PV, Vetokh AN, Volkova LA, Sermyagin AA, Shakhin AV, Griffin DK, Sölkner J, et al. Genome-Wide Association Studies in Japanese Quails of the F2 Resource Population Elucidate Molecular Markers and Candidate Genes for Body Weight Parameters. International Journal of Molecular Sciences. 2025; 26(17):8243. https://doi.org/10.3390/ijms26178243
Chicago/Turabian StyleVolkova, Natalia A., Michael N. Romanov, Nadezhda Yu. German, Polina V. Larionova, Anastasia N. Vetokh, Ludmila A. Volkova, Alexander A. Sermyagin, Alexey V. Shakhin, Darren K. Griffin, Johann Sölkner, and et al. 2025. "Genome-Wide Association Studies in Japanese Quails of the F2 Resource Population Elucidate Molecular Markers and Candidate Genes for Body Weight Parameters" International Journal of Molecular Sciences 26, no. 17: 8243. https://doi.org/10.3390/ijms26178243
APA StyleVolkova, N. A., Romanov, M. N., German, N. Y., Larionova, P. V., Vetokh, A. N., Volkova, L. A., Sermyagin, A. A., Shakhin, A. V., Griffin, D. K., Sölkner, J., McEwan, J., Brauning, R., & Zinovieva, N. A. (2025). Genome-Wide Association Studies in Japanese Quails of the F2 Resource Population Elucidate Molecular Markers and Candidate Genes for Body Weight Parameters. International Journal of Molecular Sciences, 26(17), 8243. https://doi.org/10.3390/ijms26178243










