Differential Expression of Circular RNAs in Rat Brain Regions with Various Degrees of Damage After Ischemia–Reperfusion
Abstract
1. Introduction
2. Results
2.1. Analysis of Differential Expression of circRNAs in Rat Striatum 24 h After tMCAO
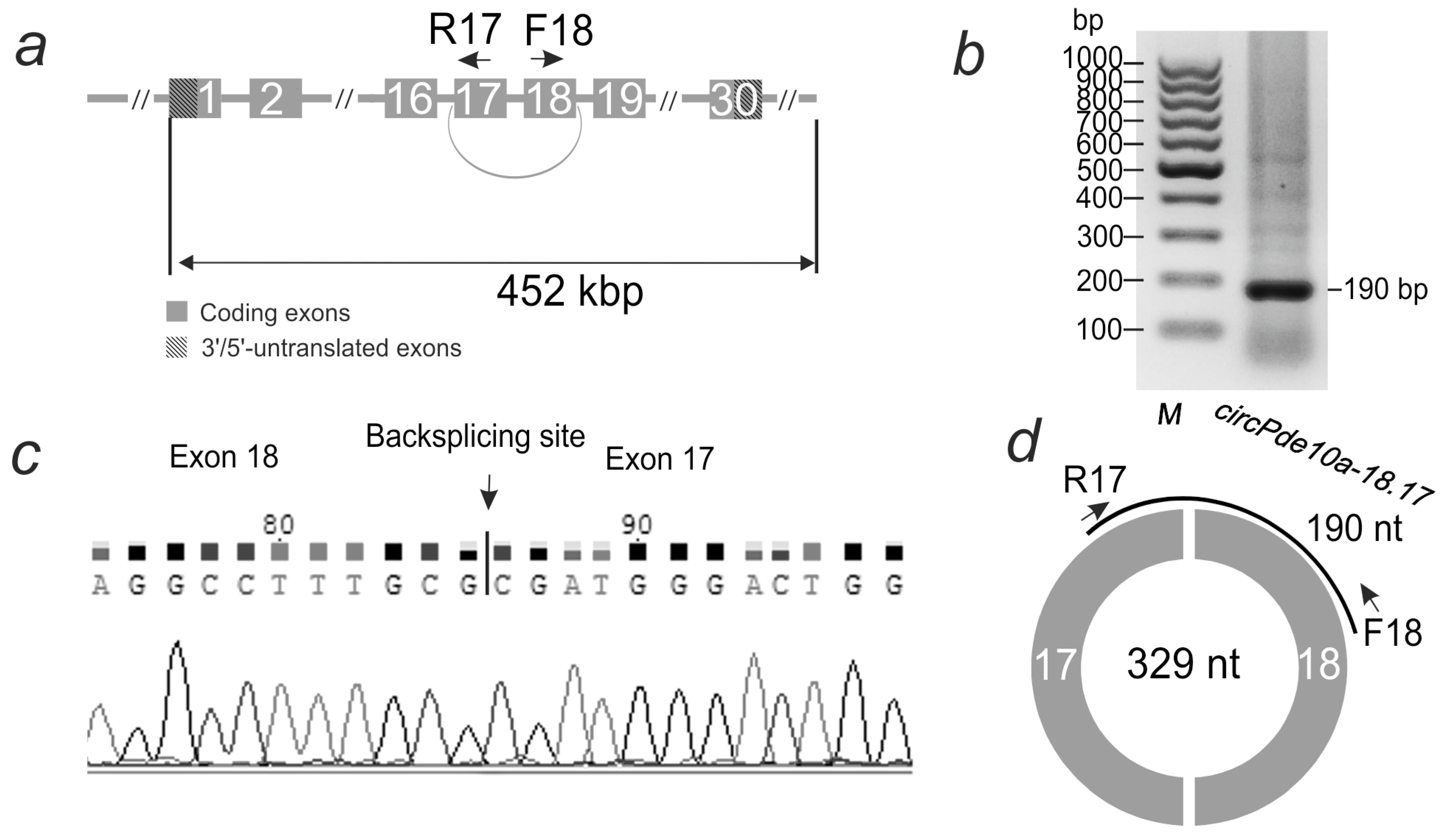
2.2. Verification of the Circular Structure of the Studied Transcripts
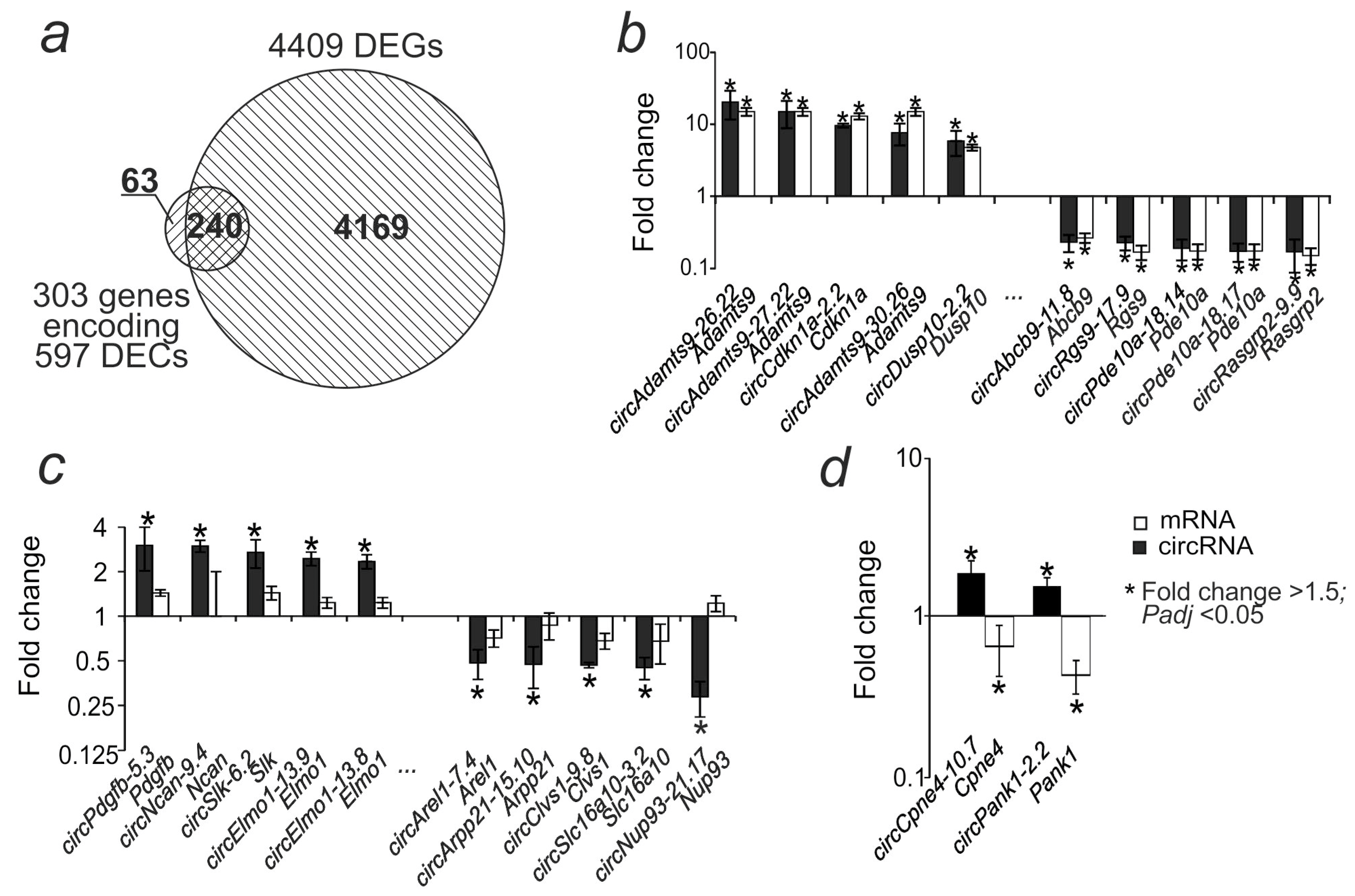
2.3. Comparative Analysis of Differential Expression of circRNA and mRNA in the Striatum at 24 h After tMCAO
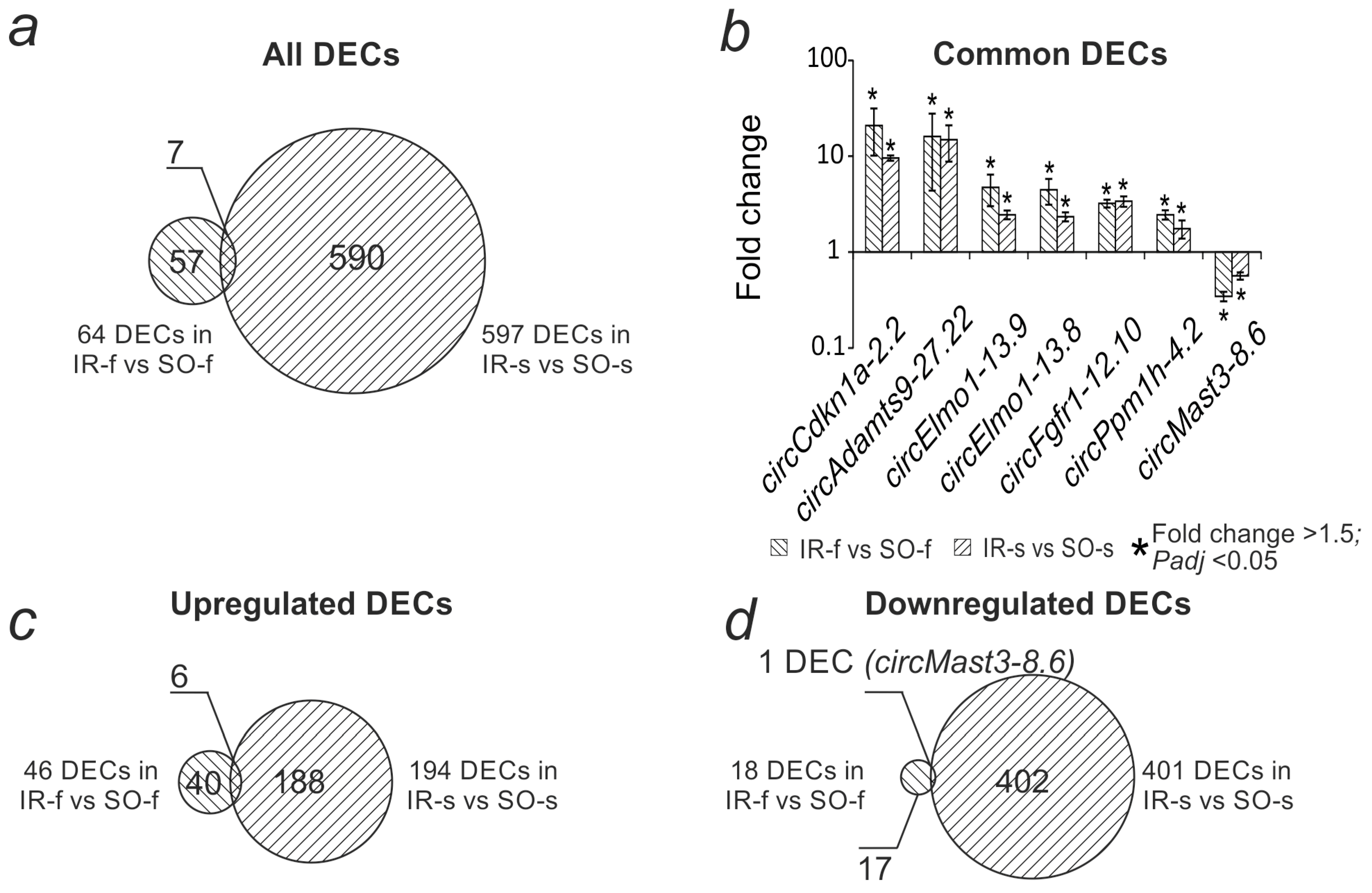
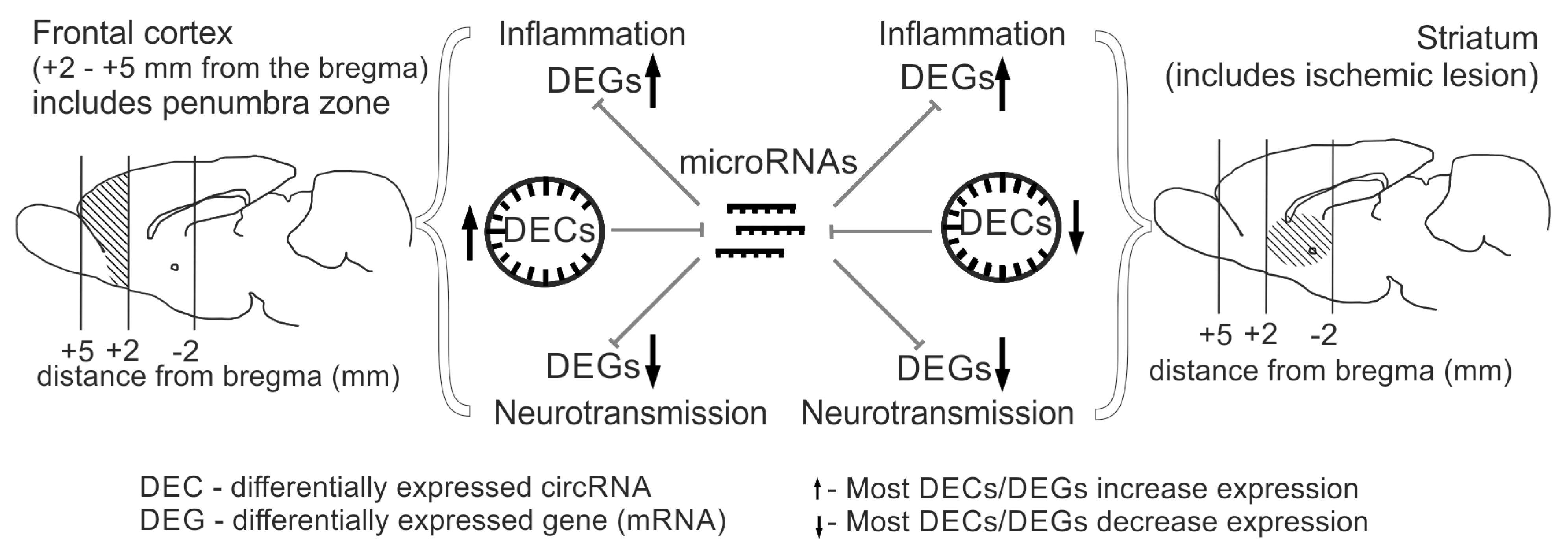
2.4. Comparative Analysis of Differential Expression of circRNAs in the Striatum and Frontal Cortex of Rats at 24 h After tMCAO
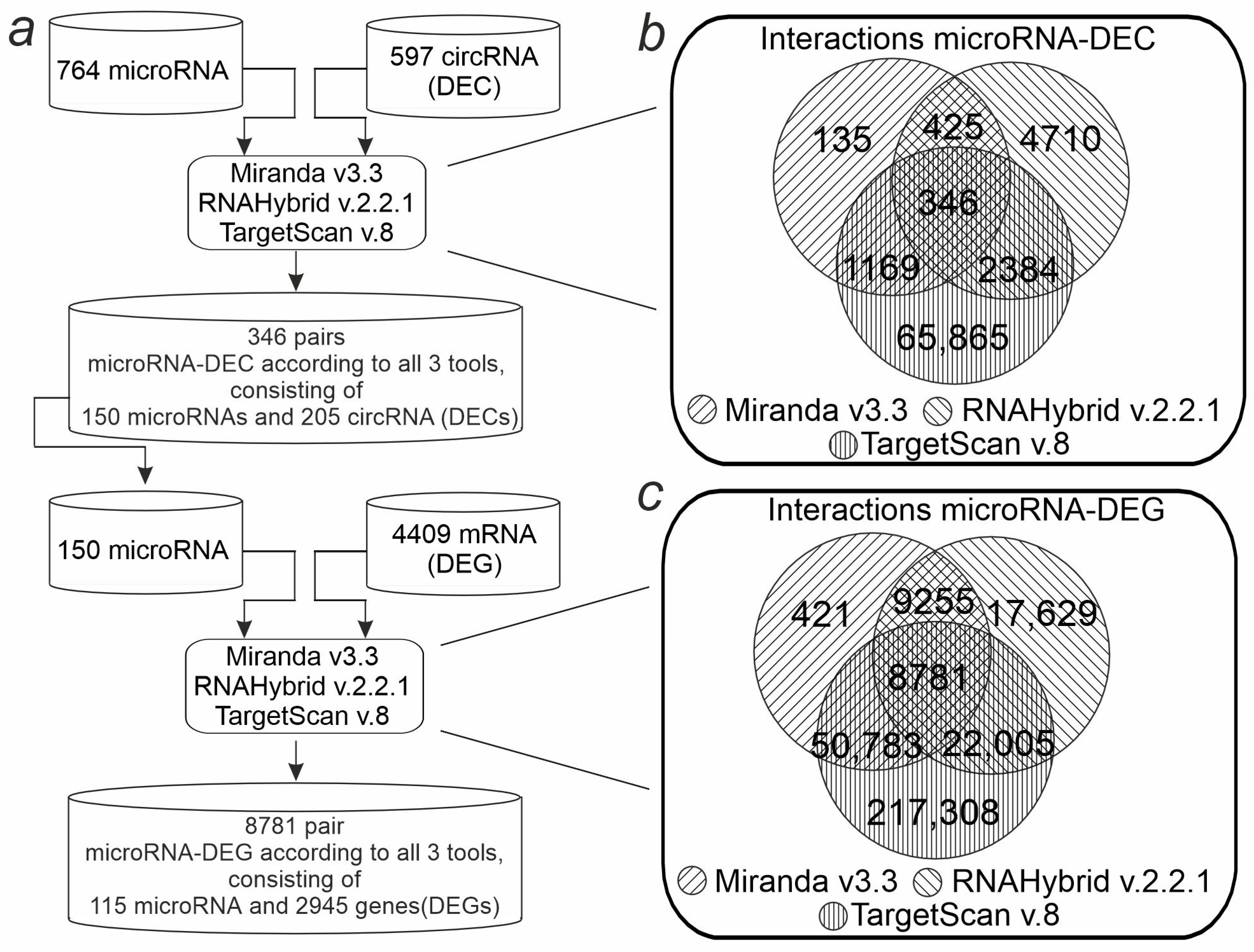
2.5. Analysis of the DEC-microRNA-DEG Interaction Network in the Rat Brain Striatum 24 h After tMCAO
2.6. Functional Annotation of DEGs Within the DEC-microRNA-DEG Network in the Rat Brain Striatum 24 h After tMCAO
2.7. Analysis of the Involvement of circRNAs in the Regulation of Gene Expression Related to the Most Significant Signaling Pathways, Both Common and Specific for the Striatum and Frontal Cortex of Rats 24 h After tMCAO
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. tMCAO Model
4.3. RNA Isolation
4.4. RNA-Seq
4.5. RNA-Seq Data Analysis
4.6. cDNA Synthesis
4.7. Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.8. Electrophoresis of PCR Products
4.9. Sanger Sequencing of PCR Products
4.10. Bioinformatic Identification of circRNA-microRNA-mRNA Networks
4.11. Functional Analysis
4.12. Hierarchical Cluster Analysis
4.13. Availability of Data and Material
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, Functions and Interactions with Proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Panda, A.C. Circular RNAs Act as MiRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Chinnaiyan, A.M.; Conn, S.J. Circular RNA in Cancer. Nat. Rev. Cancer 2024, 24, 597–613. [Google Scholar] [CrossRef]
- Dergunova, L.V.; Vinogradina, M.A.; Filippenkov, I.B.; Limborska, S.A.; Dergunov, A.D. Circular RNAs Variously Participate in Coronary Atherogenesis. Curr. Issues Mol. Biol. 2023, 45, 6682–6700. [Google Scholar] [CrossRef]
- Greco, S.; Gaetano, C.; Mazzaccaro, D.; Martelli, F. Circular RNA Role in Atherosclerosis Development and Progression. Curr. Atheroscler. Rep. 2025, 27, 60. [Google Scholar] [CrossRef]
- Wang, L.; Wang, H.; Liu, T.; Zhou, X.; Kuai, S.; Ji, Z.; Shen, H. METTL3-Mediated M6A Modification of CircCSDE1 Promote Coxsackievirus Replication by Regulating the MiR-891b/BAG3 Axis. Int. Immunopharmacol. 2025, 159, 114905. [Google Scholar] [CrossRef]
- Salgado-Somoza, A.; Zhang, L.; Vausort, M.; Devaux, Y. The Circular RNA MICRA for Risk Stratification after Myocardial Infarction. IJC Heart Vasc. 2017, 17, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.G.; Yang, H.L.; Long, Y.; Li, W.L. Circular RNA in Blood Corpuscles Combined with Plasma Protein Factor for Early Prediction of Pre-Eclampsia. BJOG Int. J. Obstet. Gynaecol. 2016, 123, 2113–2118. [Google Scholar] [CrossRef]
- Ren, Z.; Chu, C.; Pang, Y.; Cai, H.; Jia, L. A Circular RNA Blood Panel That Differentiates Alzheimer’s Disease from Other Dementia Types. Biomark. Res. 2022, 10, 63. [Google Scholar] [CrossRef]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13. [Google Scholar] [CrossRef]
- Feigin, V.L.; Brainin, M.; Norrving, B.; Martins, S.O.; Pandian, J.; Lindsay, P.; Grupper, M.F.; Rautalin, I. World Stroke Organization: Global Stroke Fact Sheet 2025. Int. J. Stroke 2025, 20, 132–144. [Google Scholar] [CrossRef]
- Shin, T.H.; Lee, D.Y.; Basith, S.; Manavalan, B.; Paik, M.J.; Rybinnik, I.; Mouradian, M.M.; Ahn, J.H.; Lee, G. Metabolome Changes in Cerebral Ischemia. Cells 2020, 9, 1630. [Google Scholar] [CrossRef]
- Dergunova, L.V.; Filippenkov, I.B.; Stavchansky, V.V.; Denisova, A.E.; Yuzhakov, V.V.; Mozerov, S.A.; Gubsky, L.V.; Limborska, S.A. Genome-Wide Transcriptome Analysis Using RNA-Seq Reveals a Large Number of Differentially Expressed Genes in a Transient MCAO Rat Model. BMC Genom. 2018, 19, 655. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Chen, X.; Li, H.; Wu, Y.; Wang, S.; Shi, W.; Chen, J.; Ni, Y. Neuron-Autonomous Transcriptome Changes upon Ischemia/Reperfusion Injury. Sci. Rep. 2017, 7, 5800. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, C.; Yang, J.; Geng, X.; Du, H.; Ji, X.; Zhao, H. Screening Circular RNA Expression Patterns Following Focal Cerebral Ischemia in Mice. Oncotarget 2017, 8, 86535–86547. [Google Scholar] [CrossRef]
- Lin, S.P.; Ye, S.; Long, Y.; Fan, Y.; Mao, H.F.; Chen, M.T.; Ma, Q.J. Circular RNA Expression Alterations Are Involved in OGD/R-Induced Neuron Injury. Biochem. Biophys. Res. Commun. 2016, 471, 52–56. [Google Scholar] [CrossRef]
- Duan, X.; Li, L.; Gan, J.; Peng, C.; Wang, X.; Chen, W.; Peng, D. Identification and Functional Analysis of Circular RNAs Induced in Rats by Middle Cerebral Artery Occlusion. Gene 2019, 701, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Xie, Q.; Li, Y.; Chen, Z.; Ren, M.; Chen, H.; Li, H.; Li, J.; Wang, J. Animal Models of Cerebral Ischemia: A Review. Biomed. Pharmacother. 2020, 131, 110686. [Google Scholar] [CrossRef]
- Trotman-Lucas, M.; Gibson, C.L. A Review of Experimental Models of Focal Cerebral Ischemia Focusing on the Middle Cerebral Artery Occlusion Model. F1000Research 2021, 10, 242. [Google Scholar] [CrossRef] [PubMed]
- Velly, L.; Boumaza, D.; Simeone, P. Cerebral Ischemia: Pathophysiology, Diagnosis, and Management. In Metabolic Disorders and Critically Ill Patients: From Pathophysiology to Treatment; Springer International Publishing: Cham, Switzerland, 2018; pp. 301–325. [Google Scholar] [CrossRef]
- Paciaroni, M.; Caso, V.; Agnelli, G. The Concept of Ischemic Penumbra in Acute Stroke and Therapeutic Opportunities. Eur. Neurol. 2009, 61, 321–330. [Google Scholar] [CrossRef]
- Kalogeris, T.; Baines, C.P.; Krenz, M.; Korthuis, R.J. Ischemia/Reperfusion. Compr. Physiol. 2017, 7, 113–170. [Google Scholar] [CrossRef]
- Adams, K.L.; Gallo, V. The Diversity and Disparity of the Glial Scar. Nat. Neurosci. 2018, 21, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Kaya, T.; Mattugini, N.; Liu, L.; Ji, H.; Cantuti-Castelvetri, L.; Wu, J.; Schifferer, M.; Groh, J.; Martini, R.; Besson-Girard, S.; et al. CD8+ T Cells Induce Interferon-Responsive Oligodendrocytes and Microglia in White Matter Aging. Nat. Neurosci. 2022, 25, 1446–1457. [Google Scholar] [CrossRef]
- Filippenkov, I.B.; Shpetko, Y.Y.; Stavchansky, V.V.; Denisova, A.E.; Yuzhakov, V.V.; Fomina, N.K.; Gubsky, L.V.; Limborska, S.A.; Dergunova, L.V. Differentially Expressed Genes in Rat Brain Regions with Different Degrees of Ischemic Damage. Int. J. Mol. Sci. 2025, 26, 2347. [Google Scholar] [CrossRef] [PubMed]
- Mozgovoy, I.V.; Shpetko, Y.Y.; Denisova, A.E.; Stavchansky, V.V.; Vinogradina, M.A.; Gubsky, L.V.; Dergunova, L.V.; Limborska, S.A.; Filippenkov, I.B. Differential Expression of Circular RNAs in the Frontal Cortex of the Rat Brain in Ischemia–Reperfusion. Biochem. Mosc. 2025, 90, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Filippenkov, I.B.; Stavchansky, V.V.; Denisova, A.E.; Valieva, L.V.; Remizova, J.A.; Mozgovoy, I.V.; Zaytceva, E.I.; Gubsky, L.V.; Limborska, S.A.; Dergunova, L.V. Genome-Wide RNA-Sequencing Reveals Massive Circular RNA Expression Changes of the Neurotransmission Genes in the Rat Brain after Ischemia–Reperfusion. Genes 2021, 12, 1870. [Google Scholar] [CrossRef]
- He, G.; He, Y.; Ni, H.; Wang, K.; Zhu, Y.; Bao, Y. Dexmedetomidine Attenuates Neuroinflammation and Microglia Activation in LPs-Stimulated BV2 Microglia Cells through Targeting Circ-Shank3/Mir-140-3p/TLr4 Axis. Eur. J. Histochem. 2023, 67, 3766. [Google Scholar] [CrossRef]
- Li, T.; Ding, L.; Wang, Y.; Yang, O.; Wang, S.; Kong, J. Genetic Deficiency of Phactr1 Promotes Atherosclerosis Development via Facilitating M1 Macrophage Polarization and Foam Cell Formation. Clin. Sci. 2020, 134, 2353–2368. [Google Scholar] [CrossRef]
- Zou, Y.; Hu, J.; Huang, W.; Ye, S.; Han, F.; Du, J.; Shao, M.; Guo, R.; Lin, J.; Zhao, Y.; et al. Non-Mitogenic Fibroblast Growth Factor 1 Enhanced Angiogenesis Following Ischemic Stroke by Regulating the Sphingosine-1-Phosphate 1 Pathway. Front. Pharmacol. 2020, 11, 59. [Google Scholar] [CrossRef]
- Engeland, K. Cell Cycle Regulation: P53-P21-RB Signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.M.; Liang, X.L.; Xiong, G.F.; Xing, X.L.; Zhang, Q.J.; Zhang, B.R.; Liu, M.W. Analysis and Identification of Oxidative Stress-Ferroptosis Related Biomarkers in Ischemic Stroke. Sci. Rep. 2024, 14, 3803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xiao, N.; Cao, Y.; Peng, Y.; Lian, A.; Chen, Y.; Wang, P.; Gu, W.; Xiao, B.; Yu, J.; et al. De Novo Variants in MAST4 Related to Neurodevelopmental Disorders with Developmental Delay and Infantile Spasms: Genotype-Phenotype Association. Front. Mol. Neurosci. 2023, 16, 1097553. [Google Scholar] [CrossRef]
- Kaplanis, J.; Samocha, K.E.; Wiel, L.; Zhang, Z.; Arvai, K.J.; Eberhardt, R.Y.; Gallone, G.; Lelieveld, S.H.; Martin, H.C.; McRae, J.F.; et al. Evidence for 28 Genetic Disorders Discovered by Combining Healthcare and Research Data. Nature 2020, 586, 757–762. [Google Scholar] [CrossRef]
- Shu, L.; Xiao, N.; Qin, J.; Tian, Q.; Zhang, Y.; Li, H.; Liu, J.; Li, Q.; Gu, W.; Wang, P.; et al. The Role of Microtubule Associated Serine/Threonine Kinase 3 Variants in Neurodevelopmental Diseases: Genotype-Phenotype Association. Front. Mol. Neurosci. 2022, 14, 775479. [Google Scholar] [CrossRef]
- Buchtele, N.; Schwameis, M.; Gilbert, J.C.; Schörgenhofer, C.; Jilma, B. Targeting von Willebrand Factor in Ischaemic Stroke: Focus on Clinical Evidence. Thromb. Haemost. 2018, 118, 959–978. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, S.F.; Stoll, G.; Wagner, D.D.; Kleinschnitz, C. Von Willebrand Factor: An Emerging Target in Stroke Therapy. Stroke 2012, 43, 599–606. [Google Scholar] [CrossRef]
- Pisignano, G.; Michael, D.C.; Visal, T.H.; Pirlog, R.; Ladomery, M.; Calin, G.A. Going Circular: History, Present, and Future of CircRNAs in Cancer. Oncogene 2023, 42, 2783–2800. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of Functional MicroRNA Targets by Integrative Modeling of MicroRNA Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Lee, D.; Shin, C. MicroRNA–Target Interactions: New Insights from Genome-Wide Approaches. Ann. N. Y. Acad. Sci. 2012, 1271, 118–128. [Google Scholar] [CrossRef]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread Changes in Protein Synthesis Induced by MicroRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Nakamura, K.; Arimura, K.; Nishimura, A.; Tachibana, M.; Yoshikawa, Y.; Makihara, N.; Wakisaka, Y.; Kuroda, J.; Kamouchi, M.; Ooboshi, H.; et al. Possible Involvement of Basic FGF in the Upregulation of PDGFRβ in Pericytes after Ischemic Stroke. Brain Res. 2016, 1630, 98–108. [Google Scholar] [CrossRef]
- Wang, X.; Chen, S.; Ni, J.; Cheng, J.; Jia, J.; Zhen, X. MiRNA-3473b Contributes to Neuroinflammation Following Cerebral Ischemia. Cell Death Dis. 2018, 9, 11. [Google Scholar] [CrossRef]
- Icli, B.; Wu, W.; Ozdemir, D.; Li, H.; Cheng, H.S.; Haemmig, S.; Liu, X.; Giatsidis, G.; Avci, S.N.; Lee, N.; et al. MicroRNA-615-5p Regulates Angiogenesis and Tissue Repair by Targeting Akt/ENOS (Protein Kinase B/Endothelial Nitric Oxide Synthase) Signaling in Endothelial Cells. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1458–1474. [Google Scholar] [CrossRef]
- Theofilatos, K.; Korfiati, A.; Mavroudi, S.; Cowperthwaite, M.C.; Shpak, M. Discovery of Stroke-Related Blood Biomarkers from Gene Expression Network Models. BMC Med. Genom. 2019, 12, 118. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, Y.; Si, Y.; Lu, C.; Wang, J.; Wang, S.; Li, L.; Xie, W.; Yue, Z.; Yong, J.; et al. Shank3 Ameliorates Neuronal Injury after Cerebral Ischemia/Reperfusion via Inhibiting Oxidative Stress and Inflammation. Redox Biol. 2024, 69, 102983, Erratum in Redox Biol. 2024, 78, 103432. [Google Scholar] [CrossRef] [PubMed]
- Ey, E.; Bourgeron, T.; Boeckers, T.M.; Kim, E.; Han, K. Editorial: Shankopathies: Shank Protein Deficiency-Induced Synaptic Diseases. Front. Mol. Neurosci. 2020, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.Y.; Hsieh, B.Y.; Lin, M.H.; Huang, T.N.; Tsai, C.Y.; Pong, W.L.; Lee, S.P.; Hsueh, Y.P. CTTNBP2 Controls Synaptic Expression of Zinc-Related Autism-Associated Proteins and Regulates Synapse Formation and Autism-like Behaviors. Cell Rep. 2020, 31, 107700. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Mabarak, C.; Loaiza-Zuluaga, M.; Hernández-Ojeda, S.L.; Camacho-Carranza, R.; Espinosa-Aguirre, J.J. Neuroinflammation Is Able to Downregulate Cytochrome P450 Epoxygenases 2J3 and 2C11 in the Rat Brain. Brain Res. Bull. 2020, 163, 57–64. [Google Scholar] [CrossRef]
- Koizumi, J.; Yoshida, Y.; Nakazawa, T.; Ooneda, G. Experimental Studies of Ischemic Brain Edema. Jpn. Sci. Technol. Inf. Aggregator Electron. 1986, 8, 1–8. [Google Scholar] [CrossRef]
- Filippenkov, I.B.; Remizova, J.A.; Denisova, A.E.; Stavchansky, V.V.; Golovina, K.D.; Gubsky, L.V.; Limborska, S.A.; Dergunova, L.V. Comparative Use of Contralateral and Sham-Operated Controls Reveals Traces of a Bilateral Genetic Response in the Rat Brain after Focal Stroke. Int. J. Mol. Sci. 2022, 23, 7308. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Wilks, C.; Antonescu, V.; Charles, R. Scaling Read Aligners to Hundreds of Threads on General-Purpose Processors. Bioinformatics 2019, 35, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Izuogu, O.G.; Alhasan, A.A.; Alafghani, H.M.; Santibanez-Koref, M.; Elliot, D.J.; Jackson, M.S. PTESFinder: A Computational Method to Identify Post-Transcriptional Exon Shuffling (PTES) Events. BMC Bioinform. 2016, 17, 31, Erratum in BMC Bioinform. 2016, 17, 92. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative Expression Software Tool (REST©) for Group-Wise Comparison and Statistical Analysis of Relative Expression Results in Real-Time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA Targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA Target Prediction Easy, Fast and Flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Marín, R.M.; Vaníek, J. Efficient Use of Accessibility in MicroRNA Target Prediction. Nucleic Acids Res. 2011, 39, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-Enabled Heat Mapping for All. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. Available online: https://Dataview.Ncbi.Nlm.Nih.Gov/Object/PRJNA1151256?Reviewer=i8c1hucc02bth1d8f83uckd1ka (accessed on 25 June 2024).
- National Center for Biotechnology Information. Available online: https://Dataview.Ncbi.Nlm.Nih.Gov/Object/PRJNA1119923?Reviewer=plp0lftpig94lmq8vmup84at3d (accessed on 23 September 2025).
- National Center for Biotechnology Information. Available online: https://Dataview.Ncbi.Nlm.Nih.Gov/Object/PRJNA1151230?Reviewer=aci8msf8dea3o2plha0dc2bno2 (accessed on 23 September 2025).
- National Center for Biotechnology Information. Available online: https://Dataview.Ncbi.Nlm.Nih.Gov/Object/PRJNA1128447 (accessed on 23 September 2025).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mozgovoy, I.V.; Tsareva, E.V.; Denisova, A.E.; Stavchansky, V.V.; Gubsky, L.V.; Dergunova, L.V.; Limborska, S.A.; Filippenkov, I.B. Differential Expression of Circular RNAs in Rat Brain Regions with Various Degrees of Damage After Ischemia–Reperfusion. Int. J. Mol. Sci. 2025, 26, 10555. https://doi.org/10.3390/ijms262110555
Mozgovoy IV, Tsareva EV, Denisova AE, Stavchansky VV, Gubsky LV, Dergunova LV, Limborska SA, Filippenkov IB. Differential Expression of Circular RNAs in Rat Brain Regions with Various Degrees of Damage After Ischemia–Reperfusion. International Journal of Molecular Sciences. 2025; 26(21):10555. https://doi.org/10.3390/ijms262110555
Chicago/Turabian StyleMozgovoy, Ivan V., Ekaterina V. Tsareva, Alina E. Denisova, Vasily V. Stavchansky, Leonid V. Gubsky, Lyudmila V. Dergunova, Svetlana A. Limborska, and Ivan B. Filippenkov. 2025. "Differential Expression of Circular RNAs in Rat Brain Regions with Various Degrees of Damage After Ischemia–Reperfusion" International Journal of Molecular Sciences 26, no. 21: 10555. https://doi.org/10.3390/ijms262110555
APA StyleMozgovoy, I. V., Tsareva, E. V., Denisova, A. E., Stavchansky, V. V., Gubsky, L. V., Dergunova, L. V., Limborska, S. A., & Filippenkov, I. B. (2025). Differential Expression of Circular RNAs in Rat Brain Regions with Various Degrees of Damage After Ischemia–Reperfusion. International Journal of Molecular Sciences, 26(21), 10555. https://doi.org/10.3390/ijms262110555






