DArTseq-Based, High-Throughput Identification of Novel Molecular Markers for the Detection of Fusarium Resistance in Maize
Abstract
1. Introduction
2. Results
2.1. Field Study
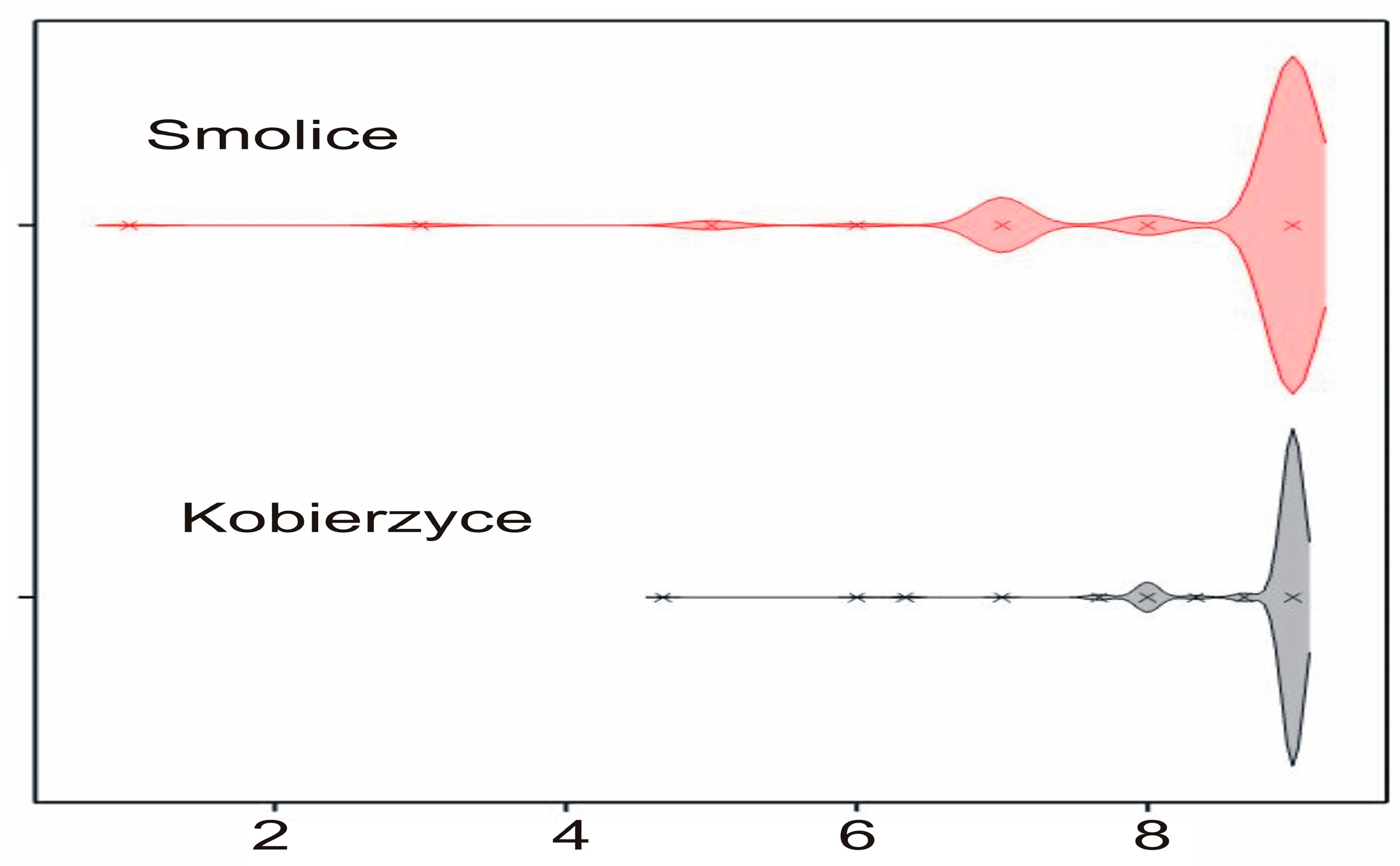
2.2. Phenotyping
2.3. Assessment of Pathogenic Fungal Species
2.4. The Impact of Ostrinia nubilalis Hbn. on Maize Infection by Fungi of the Genus Fusarium
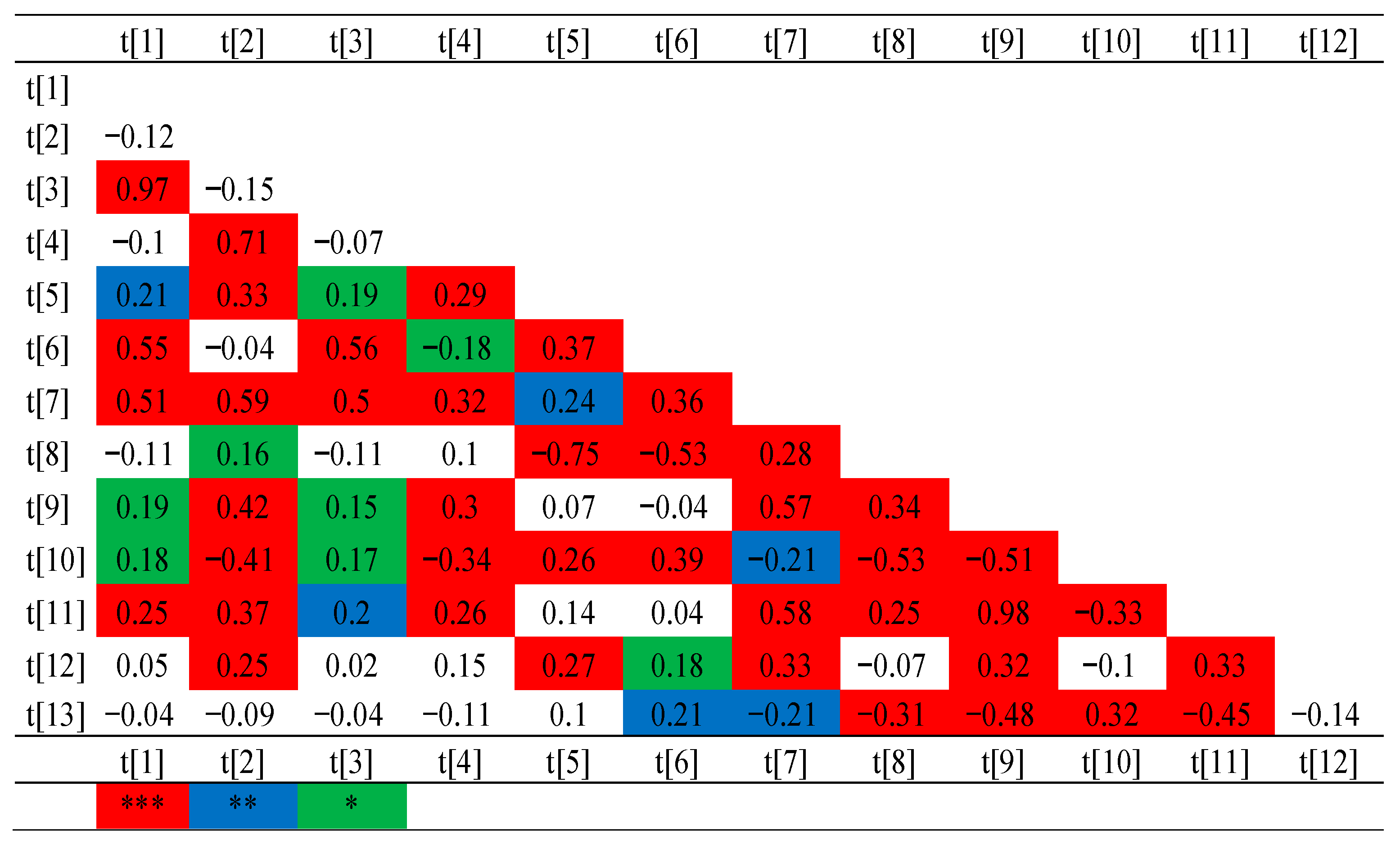
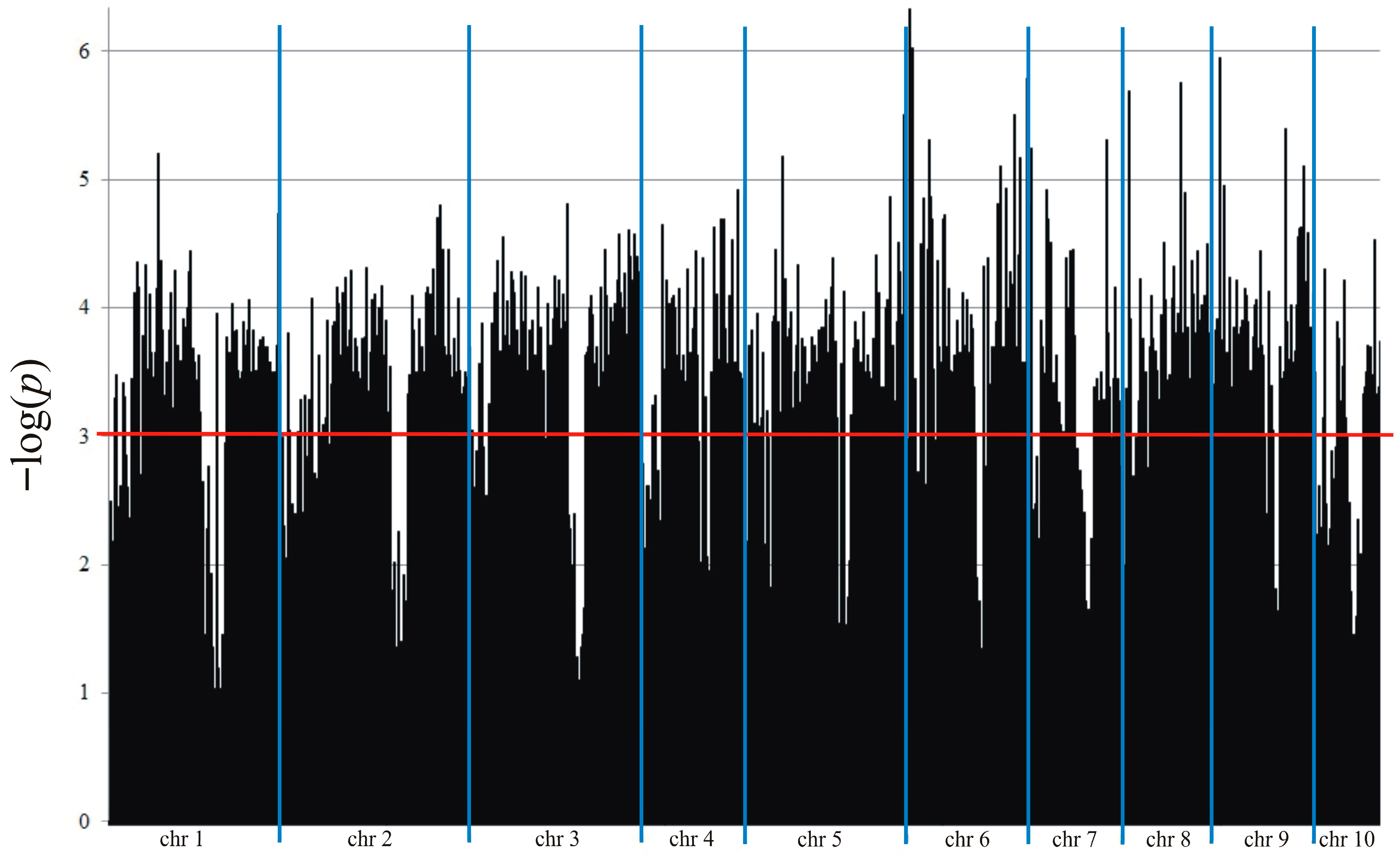
2.5. Genotyping and Association Mapping
2.6. Physical Mapping, Functional Gene Analysis
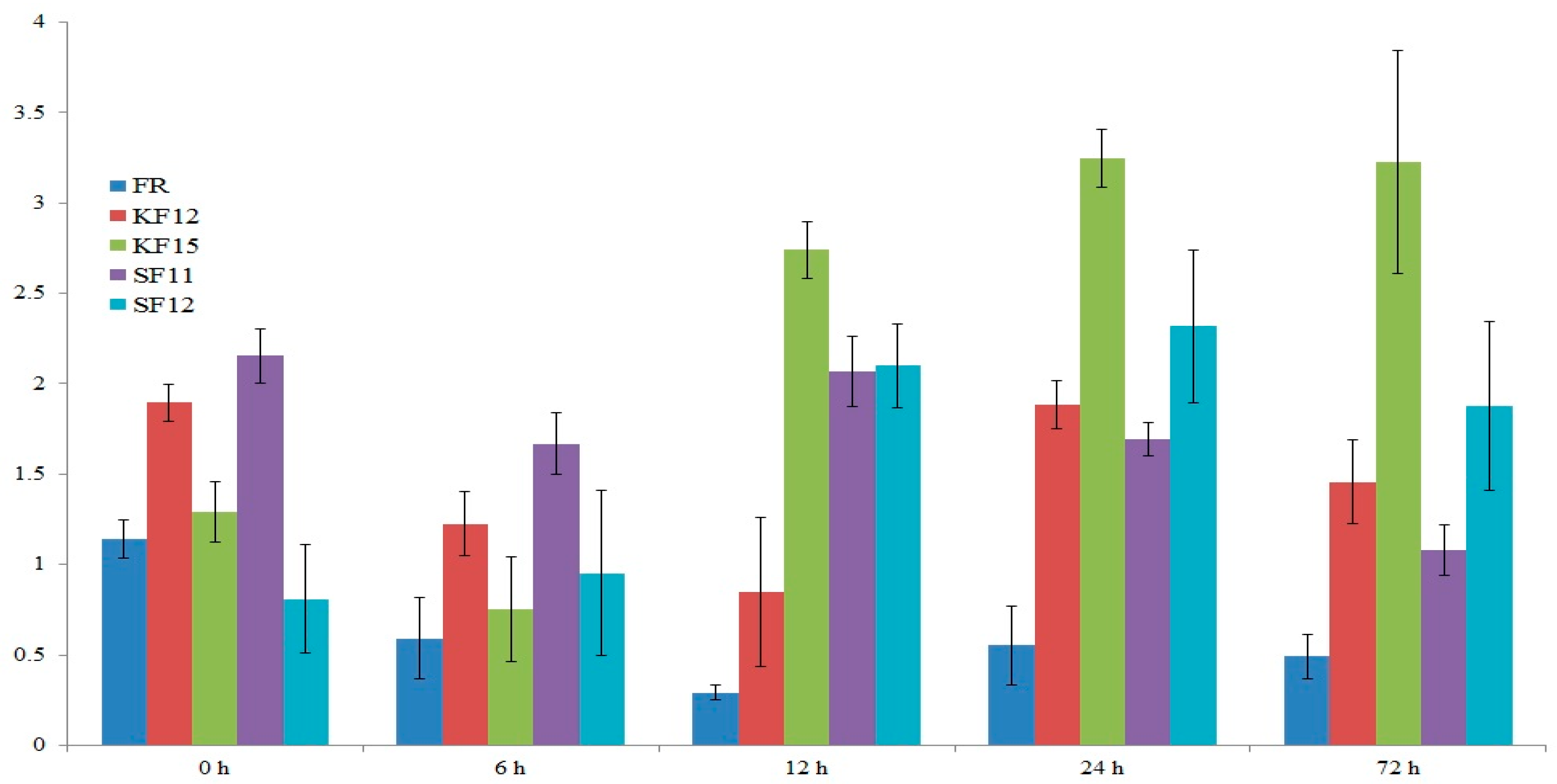
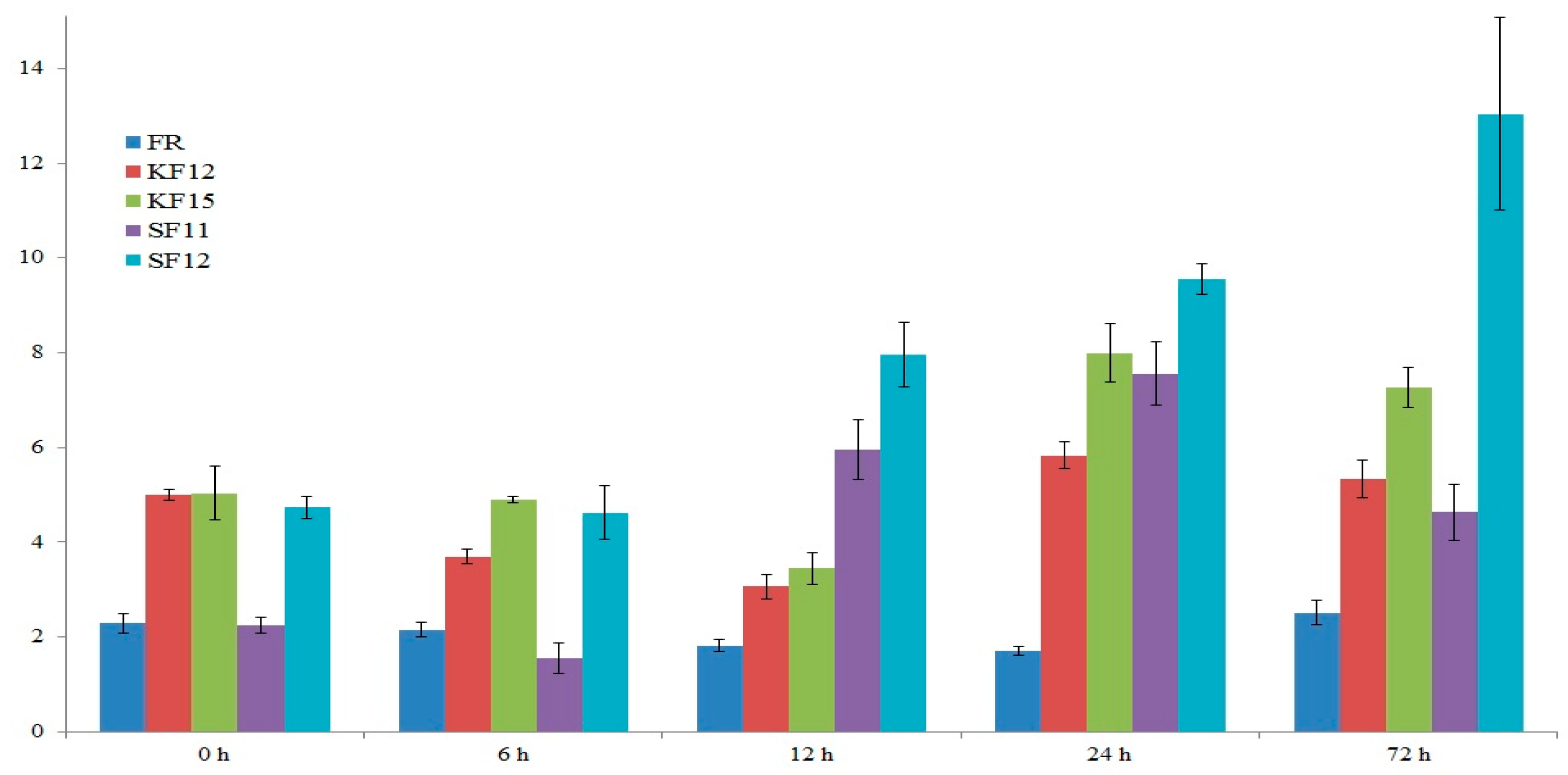
2.7. Gene Expression Analysis
2.8. Transcriptomic Data Analysis
3. Discussion
4. Materials and Methods
4.1. The Plant Material
4.2. Phenotyping
4.2.1. Field and Phytotron Experiments
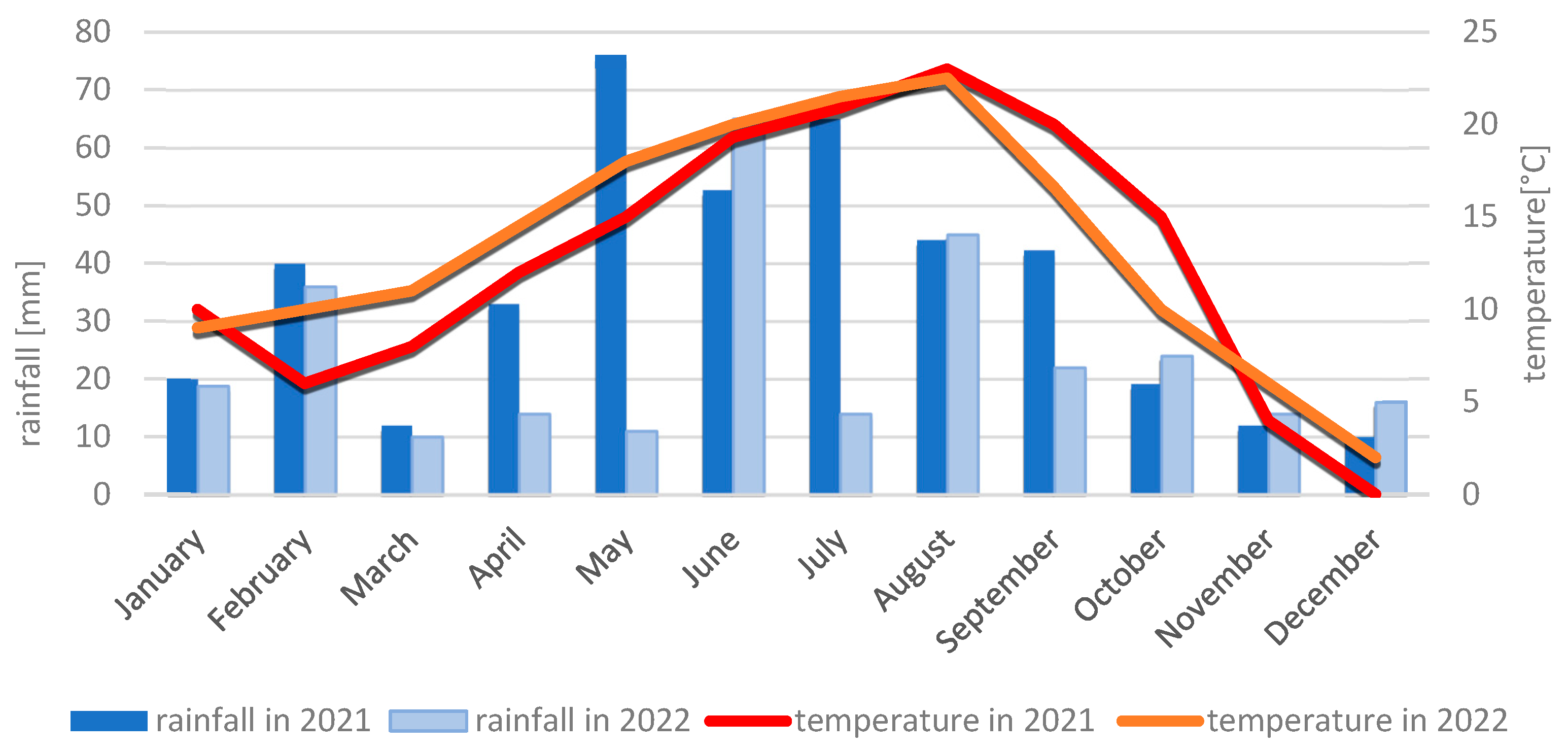
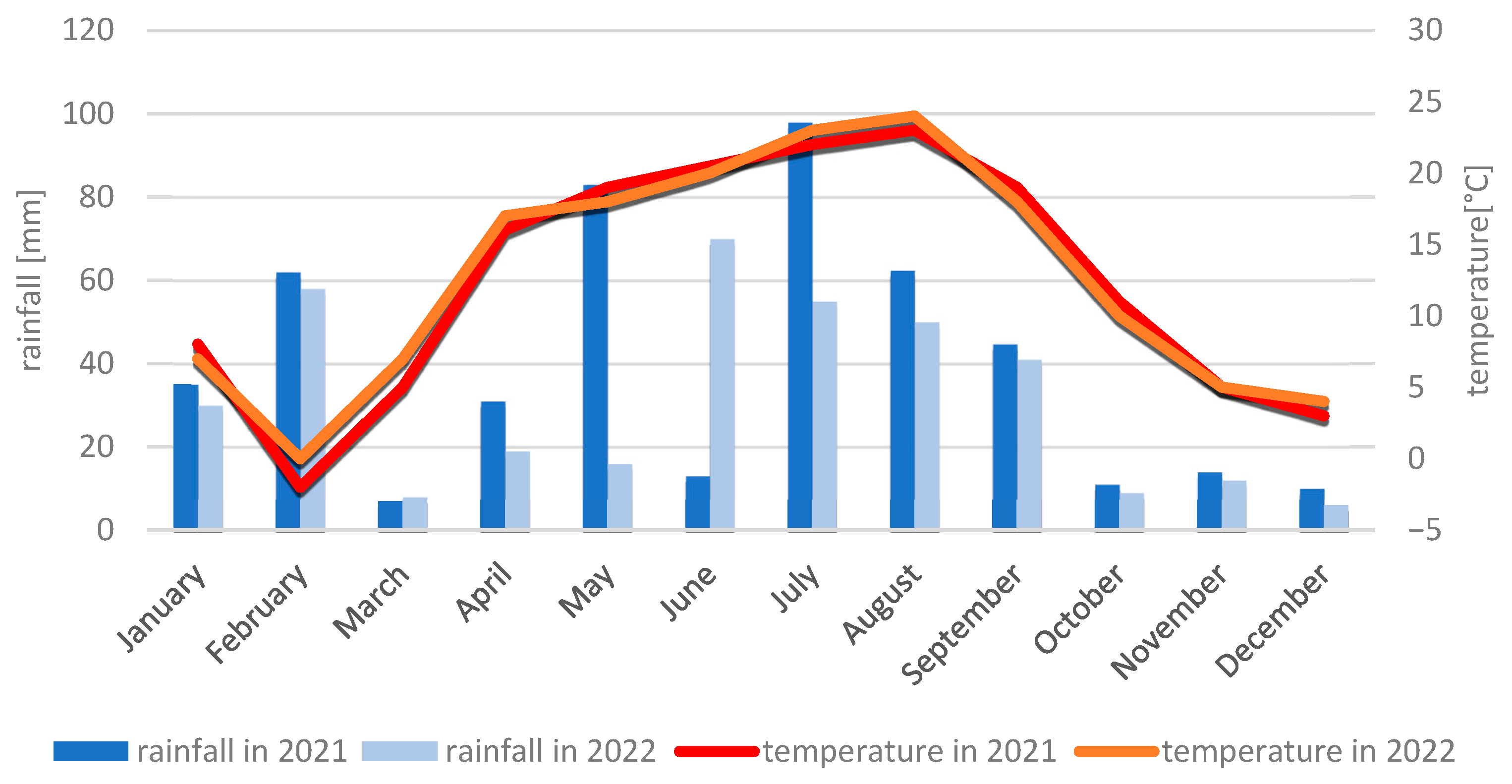
4.2.2. Meteorological Conditions During the 2021 and 2022 Growing Seasons
4.2.3. Influence of Ostrinia nubilalis Hbn. on Maize Infection by Fungi of the Genus Fusarium
4.3. Genotyping
4.3.1. DNA Isolation
4.3.2. Next-Generation Sequencing
- DNA digestion with restriction enzymes—DNA is digested with two restriction enzymes: PstI (a “frequent” cutter recognizing G/C-rich sequences) and MseI (a “rare” cutter recognizing A/T-rich sequences). Using both enzymes yields DNA fragments of different lengths.
- DNA library preparation—after digestion, adaptors are ligated to the DNA fragments (the PstI adaptor contains motifs enabling amplification and identification of PstI restriction fragments, while the MseI adaptor contains motifs enabling amplification and identification of MseI restriction fragments). Adaptors carry unique sequences (so-called barcodes) that allow multiple samples to be analyzed simultaneously on a single sequencer.
- Amplification of fragment libraries—PCR is performed using primers complementary to the adaptors to create the fragment library to be sequenced. At this stage, selective PCR can be used with additional primers that include short sequences of chosen motifs to reduce the number of amplified fragments and increase specificity.
- Selection and purification of fragments—PCR products are cleaned of excess primers and enzymes. These fragments can undergo further size selection (e.g., agarose gel or bead-based size selection), typically 250–500 bp, to ensure reproducibility.
- High-throughput sequencing—using NGS platforms (Illumina), the fragments are sequenced en masse. The sequencing preparation kit includes adaptors and barcodes, enabling simultaneous sequencing of thousands to millions of fragments from many samples.
- Bioinformatic analysis—sequences are processed and quality-filtered. Segments are compared to reference sequences or to each other to identify polymorphisms: SNPs (single nucleotide polymorphisms) and indels (insertions/deletions). A database of polymorphisms is generated, which can be used for various analyses, including genetic mapping.
4.3.3. Association Mapping Using GWAS Analysis
4.3.4. Physical Mapping
4.3.5. Functional Analysis of Gene Sequences
4.3.6. Designing Primers for Identified SilicoDArT and SNP Polymorphisms Associated with Maize Plant Resistance to Fungi of the Genus Fusarium
4.4. mRNA Isolation
4.5. cDNA Synthesis
4.6. Gene Expression Analysis Using RT-qPCR
4.7. Reference Gene Analysis
- 5′CTACCTCACGGCATCTGCTATGT3′
- 3′AACACGAATCAAGCAGAG5′
- 5′CTGAGTGGTGGTCTTAGT3′
- 3′GTCACACACACTCGACTTCACG5′
4.8. Transcriptomic Analysis
4.9. Evaluation of Pathogenic Fungal Species
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Erenstein, O.; Jaleta, M.; Sonder, K.; Mottaleb, K.; Prasanna, B.M. Global maize production, consumption and trade: Trends and R&D implications. Food Secur. 2022, 14, 1295–1319. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, Y.; Ma, Y.; Fu, J.; Wang, J. Genetic and molecular control of grain yield in maize. Mol. Breed. 2021, 41, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Tong, L.; Xu, M.; Zhong, T. Genetic dissection of maize disease resistance and its applications in molecular breeding. Mol. Breed. 2021, 41, 32, Erratum in Mol. Breed. 2021, 41, 58. [Google Scholar] [CrossRef] [PubMed]
- Perry, E.D.; Moschini, G. Neonicotinoids in US maize: Insecticide substitution effects and environmental risk. J. Environ. Econ. Manag. 2020, 102, 102320. [Google Scholar] [CrossRef]
- Ji, D.; Li, L.; Wang, Y.; Zhang, J.; Cheng, M.; Sun, Y.; Liu, Z.; Wang, L.; Tang, G.; Hu, B.; et al. The heaviest particulate air-pollution episodes occurred in northern China in January, 2013: Insights gained from observation. Atmos. Environ. 2014, 92, 546–556. [Google Scholar] [CrossRef]
- Deng, B.; Carter, R.A.; Cheng, Y.; Liu, Y.; Eddy, L.; Wyss, K.M.; Ucak-Astarlioglu, M.G.; Luong, D.X.; Gao, X.; JeBailey, K.; et al. High-temperature electrothermal remediation of multi-pollutants in soil. Nat. Commun. 2023, 14, 6371. [Google Scholar] [CrossRef]
- Ranum, P.; Peña-Rosas, J.P.; Garcia-Casal, M.N. Global maize production, utilization, and consumption. Ann. New York Acad. Sci. 2014, 1312, 105–112. [Google Scholar] [CrossRef]
- Butrón, A.; Reid, L.M.; Santiago, R.; Cao, A.; Malvar, R.A. Inheritance of maize resistance to gibberella and fusarium ear rots and kernel contamination with deoxynivalenol and fumonisins. Plant Pathol. 2015, 64, 1053–1060. [Google Scholar] [CrossRef]
- Scauflaire, J.; Mahieu, O.; Louvieaux, J.; Foucart, G.; Renard, F.; Munaut, F. Biodiversity of Fusarium species in ears and stalks of maize plants in Belgium. Eur. J. Plant Pathol. 2011, 131, 59–66. [Google Scholar] [CrossRef]
- Bode, W.M.; Calvin, D.D. Yield-loss relationships and economic injury levels for European corn borer (Lepidoptera: Pyralidae) populations infesting Pennsylvania field corn. J. Econ. Entomol. 1990, 83, 1595–1603. [Google Scholar] [CrossRef]
- Szőke, C.; Zsubori, Z.; Pók, I.; Rácz, F.; Illés, O.; Szegedi, I. Significance of the European corn borer (Ostrinia nubilalis Hübn.) in maize production. Acta Agron. Hung. 2002, 50, 447–461. [Google Scholar] [CrossRef]
- Blandino, M.; Reyneri, A.; Vanara, F.; Tamietti, G.; Pietri, A. Influence of agricultural practices on Fusarium infection, fumonisin and deoxynivalenol contamination of maize kernels. World Mycotoxin J. 2009, 2, 409–418. [Google Scholar] [CrossRef]
- Sobek, E.A.; Munkvold, G.P. European corn borer (Lepidoptera: Pyralidae) larvae as vectors of Fusarium moniliforme, causing kernel rot and symptomless infection of maize kernels. J. Econ. Entomol. 1999, 92, 503–509. [Google Scholar] [CrossRef]
- Garcia-Ceron, D.; Lowe, R.G.T.; McKenna, J.A.; Brain, L.M.; Dawson, C.S.; Clark, B.; Berkowitz, O.P.; Whelan, J.; Bleackley, M.R.; Anderson, M.A. Extracellular Vesicles from Fusarium graminearum Contain Protein Effectors Expressed during Infection of Corn. J. Fungi 2021, 7, 977. [Google Scholar] [CrossRef] [PubMed]
- Ncube, E.; Truter, M.; Flett, B.C.; Van Den Berg, J.; Erasmus, A.; Viljoen, A. Fungal mycoflora associated with Busseola fusca frass in maize plants. Afr. Entomol. 2020, 28, 394–405. [Google Scholar] [CrossRef]
- Vigier, B.; Reid, L.M.; Dwyer, L.M.; Stewart, D.W.; Sinha, R.C.; Arnason, J.T.; Butler, G. Maize resistance to Gibberella ear rot: Symptoms, deoxynivalenol, and yield. Can. J. Plant Pathol. 2001, 23, 99–105. [Google Scholar] [CrossRef]
- Mesterházy, Á.; Lemmens, M.; Reid, L.M. Breeding for resistance to ear rot caused by Fusarium spp. in maize—A review. Plant Breed. 2012, 131, 1–19. [Google Scholar] [CrossRef]
- Uwe, L.; Miedaner, T.; Bürstmayr, H.; Vögele, R.T. Breeding for Resistance to Fusarium Ear Diseases in Maize and Small-Grain Cereals Using Genomic Tools. Ph.D. Thesis, University of Hohenheim, Stuttgart, Germany, 2020. [Google Scholar]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity Arrays Technology (DArT) and next-generation sequencing combined: Genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5 (Suppl. S7), P54. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Courtois, B.; Audebert, A.; Dardou, A.; Roques, S.; Ghneim-Herrera, T.; Droc, G.; Frouin, J.; Rouan, L.; Gozé, E.; Kilian, A. Genome-Wide Association Mapping of Root Traits in a Japonica Rice Panel. PLoS ONE 2013, 8, e78037. [Google Scholar] [CrossRef]
- Cruz, V.M.V.; Kilian, A.; Dierig, D.A. Development of DArT Marker Platforms and Genetic Diversity Assessment of the U.S. Collection of the New Oilseed Crop Lesquerella and Related Species. PLoS ONE 2013, 8, e64062. [Google Scholar] [CrossRef] [PubMed]
- Sakhare, A.S.; Kota, S.; Rathod, S.; Parmar, B.; Chinnusamy, V. Genome-Wide Association Study. In Genotyping by Sequencing for Crop Improvement; Wiley: Hoboken, NJ, USA, 2022. [Google Scholar] [CrossRef]
- Abdurakhmonov, I.Y.; Abdukarimov, A. Application of Association Mapping to Understanding the Genetic Diversity of Plant Germplasm Resources. Int. J. Plant Genom. 2008, 2008, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Rafalski, J.A. Association genetics in crop improvement. Curr. Opin. Plant Biol. 2010, 13, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and Prospects of Association Mapping in Plants. Plant Genome J. 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Rakoczy-Trojanowska, M.; Krajewski, P.; Bocianowski, J.; Schollenberger, M.; Wakuliński, W.; Milczarski, P.; Masojć, P.; Targońska-Karasek, M.; Banaszak, Z.; Banaszak, K. Identification of Single Nucleotide Polymorphisms Associated with Brown Rust Resistance, α-Amylase Activity and Pre-harvest Sprouting in Rye (Secale cereale L.). Plant Mol. Biol. Rep. 2017, 35, 366–378. [Google Scholar] [CrossRef]
- Edwards, D.; Batley, J.; Snowdon, R.J. Accessing complex crop genomes with next-generation sequencing. Theor. Appl. Genet. 2013, 126, 1–11. [Google Scholar] [CrossRef]
- Bar-Hen, A.; Charcosset, A.; Bourgoin, M.; Guiard, J. Relationship between genetic markers and morphological traits in a maize inbred lines collection. Euphytica 1995, 84, 145–154. [Google Scholar] [CrossRef]
- Pritchard, J.K. Deconstructing Maize Population Structure. Nat. Genet. 2001, 28, 203–204. [Google Scholar] [CrossRef]
- Lanubile, A.; Pasini, L.; Lo Pinto, M.; Battilani, P.; Prandini, A.; Marocco, A. Evaluation of broad spectrum, sources of resistance to Fusarium verticillioides and advanced maize breeding lines. World Mycotoxin J. 2011, 4, 43–51. [Google Scholar] [CrossRef]
- Clements, M.J.; Kleinschmidt, C.E.; Maragos, C.M.; Pataky, J.K.; White, D.G. Evaluation of inoculation techniques for Fusarium ear rot and fumonisin contamination of corn. Plant Dis. 2003, 87, 147–153. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, F.; Wan, X.Q.; Pan, G.T. Molecular mapping of QTL for resistance to maize ear rot caused by Fusarium moniliforme. Agric. Sci. China 2007, 6, 491–496. [Google Scholar]
- Zhang, X.; Zheng, S.; Yu, M.; Xu, C.; Li, Y.; Sun, L.; Qiu, X. Evaluation of Resistance Resources and Analysis of Resistance Mechanisms of Maize to Stalk Rot Caused by Fusarium graminearum. Plant Dis. 2024, 108, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Zila, C.T.; Ogut, F.; Romay, M.C.; Gardner, C.A.; Buckler, E.S. Genome wide association study of Fusarium ear rot disease in the U.S.A. maize inbred line collection. BMC Plant Biol. 2014, 14, 372. [Google Scholar] [CrossRef] [PubMed]
- Zila, C.T.; Samayoa, L.F.; Santiago, R.; Butrón, A.; Holland, J.B. A genome-wide association study reveals genes associated with fusarium ear rot resistance in a maize core diversity panel. G3 Genes Genomes Genet. 2013, 3, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- De Jong, G.; Pamplona, A.K.A.; Von Pinho, R.G.; Balestre, M. Genome-wide association analysis of ear rot resistance caused by Fusarium verticillioides in maize. Genomics 2018, 110, 291–303. [Google Scholar] [CrossRef]
- Chen, J.; Ding, J.; Li, H.; Li, Z.; Sun, X.; Li, J.; Dai, X.; Dong, H.; Song, W.; Chen, W. Detection and verification of quantitative trait loci for resistance to Fusarium ear rot in maize. Mol. Breed. 2012, 30, 1649–1656. [Google Scholar] [CrossRef]
- Li, Z.M.; Ding, J.Q.; Wang, R.X.; Chen, J.; Sun, X.; Chen, W.; Song, W.; Dong, H.; Dai, X.; Xia, Z.; et al. A new QTL for resistance to Fusarium ear rot in maize. J. Appl. Genet. 2011, 52, 403–406. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, Z.; Dong, C.; Chen, J.; Ding, J.; Zhang, X.; Mu, C.; Chen, Y.; Li, X.; Li, H.; et al. Linkage mapping and genome-wide association study reveals conservative QTL and candidate genes for Fusarium rot resistance in maize. BMC Genom. 2020, 21, 357. [Google Scholar] [CrossRef]
- Baran, M.; Roik, K.; Bocianowski, J. Analysis of monitoring of maize damage caused by the European corn borer (Ostrinia nubilalis Hbn.) in Poland in 2006–2016. Prog. Plant Prot. 2022, 62, 199–207. [Google Scholar] [CrossRef]
- Lanubile, A.; Ferrarini, A.; Maschietto, V.; Delledonne, M.; Marocco, A.; Bellin, D. Functional genomic analysis of constitutive and inducible defense responses to Fusarium verticillioides infection in maize genotypes with contrasting ear rot resistance. BMC Genom. 2014, 15, 710. [Google Scholar] [CrossRef]
- Escrivá, L.; Font, G.; Manyes, L. In vivo toxicity studies of fusarium mycotoxins in the last decade: A review. Food Chem. Toxicol. 2015, 78, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Shi, J.; Zeng, C.; Shi, H.; Yang, Y.; Zhang, C.; Shen, Y. Integrated analysis of transcriptomics and defense-related phytohormones to discover hub genes conferring maize Gibberella ear rot caused by Fusarium Graminearum. BMC Genom. 2024, 25, 733. [Google Scholar] [CrossRef] [PubMed]
- Mesterhazy, A. Food Safety Aspects of Breeding Maize to Multi-Resistance against the Major (Fusarium graminearum, F. verticillioides, Aspergillus flavus) and Minor Toxigenic Fungi (Fusarium spp.) as Well as to Toxin Accumulation, Trends, and Solutions—A Review. J. Fungi 2024, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Dhillon, B.S.; Miedaner, T.; Melchinger, A.E. Inheritance of resistance to gibberella ear rot and deoxynivalenol contamination in five flint maize crosses. Plant Breed. 2012, 131, 28–32. [Google Scholar] [CrossRef]
- Hoenisch, R.W.; Davis, R.M. Relationship between kernel pericarp thickness and susceptibility to Fusarium ear rot in field corn. Plant Dis. 1994, 78, 578–580. [Google Scholar] [CrossRef]
- Sampietro, D.A.; Vattuone, M.A.; Presello, D.A.; Fauguel, C.M.; Catalán, C.A.N. The pericarp and its surface wax layer in maize kernels as resistance factors to fumonisin accumulation by Fusarium verticillioides. Crop Prot. 2009, 28, 196–200. [Google Scholar] [CrossRef]
- Netshifhefhe, N.E.I.; Flett, B.C.; Viljoen, A.; Rose, L.J. Inheritance and genotype by environment analyses of resistance to Fusarium verticillioides and fumonisin contamination in maize F1 hybrids. Euphytica 2018, 214, 235. [Google Scholar] [CrossRef]
- Fritsch, L.; Fischer, R.; Wambach, C.; Dudek, M.; Schillberg, S.; Schröper, F. Next-generation sequencing is a robust strategy for the high-throughput detection of zygosity in transgenic maize. Transgenic Res. 2015, 24, 615–623. [Google Scholar] [CrossRef]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef]
- Huttner, S.; Veit, C.; Vavra, U.; Schoberer, J.; Liebminger, E.; Maresch, D.; Grass, J.; Altmann, F.; Mach, L.; Strasser, R. Arabidopsis Class I α-mannosidases MNS4 and MNS5 are involved in endoplasmic reticulum-associated degradation of misfolded glycoproteins. Plant Cell 2014, 26, 1712–1728. [Google Scholar] [CrossRef]
- Gidalevitz, T.; Stevens, F.; Argon, Y. Orchestration of secretory protein folding by ER chaperones. Biochim. Biophys. Acta 2013, 1833, 2410–2424. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R. Protein quality control in the endoplasmic reticulum of plants. Annu. Rev. Plant Biol. 2018, 69, 147–172. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Guo, C.; Ali, K.; Zheng, Q.; Wei, Q.; Zhu, Y.; Wang, L.; Li, G.; Li, W.; Zheng, B.; et al. A Non-redundant Function of MNS5: A Class I α-1, 2 Mannosidase, in the Regulation of Endoplasmic Reticulum-Associated Degradation of Misfolded Glycoproteins. Front. Plant Sci. 2022, 13, 873688. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Baker, A.; Bartel, B.; Linka, N.; Mullen, R.T.; Reumann, S.; Zolman, B.K. Plant Peroxisomes: Biogenesis and Function. The Plant Cell 2012, 24, 2279–2303. [Google Scholar] [CrossRef]
- Terrón-Camero, L.; Peláez-Vico, M.A.; Rodriguez-Gonzalez, A.; del Val, C.; Sandalio, L.M.; Romero-Puertas, M.C. Gene network downstream plant stress response modulated by peroxisomal H2O2. Front. Plant Sci. 2022, 13, 930721. [Google Scholar] [CrossRef]
- Beaugelin, I.; Chevalier, A.; D’Alessandro, S.; Ksas, B.; Havaux, M. Endoplasmic reticulum-mediated unfolded protein response is an integral part of singlet oxygen signalling in plants. Plant J. 2020, 102, 1266–1280. [Google Scholar] [CrossRef]
- Darino, M.; Jaiswal, N.; Darma, R.; Kroll, E.; Urban, M.; Xiang, Y.; Srivastava, M.; Kim, H.S.; Myers, A.; Scofield, S.R.; et al. The Fusarium graminearum Effector Protease FgTPP1 Suppresses Immune Responses and Facilitates Fusarium Head Blight Disease. Mol. Plant-Microbe Interact. 2025, 38, 297–314. [Google Scholar] [CrossRef]
- Jarzyniak, K.; Narożna, D. Rapid Identification of Rhizobia Nodulating Soybean by a High-Resolution Melting Analysis. Agronomy 2024, 14, 1305. [Google Scholar] [CrossRef]
- Tang, J.; Yan, J.; Ma, X.; Teng, W.; Wu, W.; Dai, J.; Dhillon, B.; Melchinger, D.; Li, J. Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theor. Appl. Genet. 2010, 120, 333–340. [Google Scholar] [CrossRef]
- Sobiech, A.; Tomkowiak, A.; Jamruszka, T.; Kosiada, T.; Spychała, J.; Lenort, M.; Bocianowski, J. Transcriptomic Characterization of Candidate Genes for Fusarium Resistance in Maize (Zea mays L.). Pathogens 2025, 14, 779. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- van Eeuwijk, F.A.; Bink, M.C.A.M.; Chenu, K.; Chapman, S.C.; Malosetti, M.; Ribaut, J.-M.; van Eeuwijk, F.A. The statistical analysis of multi-environment data: Modeling genotype-by-environment interaction and its genetic basis. Curr. Opin. Plant Biol. 2010, 13, 193–205. [Google Scholar] [CrossRef]
- VSN International. VSN International Genstat for Windows, 23rd ed.; VSN International: Hemel Hempstead, UK, 2023. [Google Scholar]
- Bocianowski, J. Using NGS Technology and Association Mapping to Identify Candidate Genes Associated with Fusarium Stalk Rot Resistance. Genes 2024, 15, 106. [Google Scholar] [CrossRef]
- Bocianowski, J.; Leśniewska-Bocianowska, A. Towards the Identification of Candidate Genes for Pollen Morphological Traits in Rubus L. Using Association Mapping. Forests 2025, 16, 1395. [Google Scholar] [CrossRef]

| Infection Degree | Description |
|---|---|
| 9 | clean ears, without mycelium |
| 7 | isolated fungal colonies on the cobs |
| 5 | 50% of the cobs colonized by mycelium |
| 3 | 75% of the cobs colonized by mycelium |
| 1 | entire cobs colonized by mycelium |
| Source of Variability | Hybrid | Residual |
|---|---|---|
| Number of degrees of freedom | 172 | 346 |
| Cob fusariosis [1,2,3,4,5,6,7,8,9] | 0.393 *** | 0.2004 |
| % Colonization of Kernels by Fungi | % Colonization of Kernels by Fungi | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | F. poae | F. culmorum | F. graminearum | F. pseudograminerarum | F. boothii | F. subglutinans | Sample | F. poae | F. culmorum | F. graminearum | F. pseudograminerarum | F. boothii | F. subglutinans |
| G21.08 | 45 | 0 | 0 | 0 | 0 | 8 | G23.03 | 24 | 8 | 0 | 21 | 0 | 0 |
| G21.09 | 4 | 11 | 0 | 7 | 0 | 0 | G23.04 | 19 | 12 | 0 | 0 | 0 | 0 |
| G21.10 | 64 | 9 | 5 | 0 | 0 | 0 | G23.05 | 61 | 0 | 0 | 0 | 0 | 0 |
| G21.11 | 23 | 21 | 0 | 8 | 0 | 1 | G23.06 | 38 | 0 | 0 | 0 | 0 | 0 |
| G21.12 | 16 | 6 | 0 | 0 | 12 | 6 | G23.07 | 14 | 0 | 0 | 0 | 0 | 1 |
| G21.13 | 7 | 8 | 0 | 0 | 0 | 0 | G23.08 | 45 | 0 | 0 | 0 | 0 | 1 |
| G21.14 | 27 | 14 | 0 | 0 | 0 | 0 | G23.09 | 12 | 0 | 0 | 0 | 0 | 9 |
| G21.15 | 78 | 9 | 0 | 5 | 0 | 0 | G23.10 | 54 | 1 | 0 | 16 | 0 | 9 |
| G21.16 | 41 | 15 | 0 | 0 | 0 | 0 | G23.11 | 6 | 0 | 0 | 0 | 0 | 1 |
| G21.17 | 9 | 10 | 0 | 0 | 0 | 12 | G23.12 | 1 | 0 | 0 | 0 | 0 | 1 |
| G21.18 | 12 | 6 | 0 | 0 | 0 | 11 | G23.13 | 1 | 0 | 0 | 0 | 0 | 3 |
| G21.19 | 15 | 8 | 0 | 4 | 25 | 7 | G23.14 | 8 | 0 | 0 | 0 | 0 | 0 |
| G21.20 | 32 | 2 | 0 | 0 | 0 | 9 | G23.15 | 34 | 21 | 0 | 0 | 0 | 0 |
| G21.21 | 12 | 1 | 0 | 0 | 0 | 12 | G15.21 | 38 | 12 | 0 | 0 | 23 | 0 |
| G22.01 | 6 | 6 | 0 | 0 | 28 | 3 | G16.01 | 12 | 16 | 0 | 0 | 0 | 0 |
| G22.02 | 1 | 9 | 0 | 0 | 0 | 0 | G16.02 | 17 | 9 | 0 | 0 | 0 | 4 |
| G22.03 | 37 | 22 | 0 | 12 | 0 | 0 | G16.03 | 26 | 9 | 6 | 0 | 0 | 4 |
| G22.04 | 29 | 27 | 0 | 0 | 0 | 0 | G16.04 | 3 | 14 | 0 | 0 | 0 | 4 |
| G22.05 | 8 | 19 | 0 | 0 | 0 | 0 | G16.05 | 3 | 11 | 0 | 0 | 17 | 1 |
| G22.06 | 7 | 7 | 0 | 0 | 0 | 0 | G16.06 | 45 | 6 | 0 | 0 | 0 | 1 |
| G22.07 | 12 | 21 | 0 | 1 | 8 | 2 | G16.07 | 39 | 2 | 0 | 0 | 0 | 0 |
| G22.21 | 38 | 9 | 0 | 0 | 0 | 1 | G16.08 | 22 | 15 | 0 | 0 | 0 | 0 |
| G23.01 | 45 | 14 | 0 | 0 | 0 | 11 | G16.09 | 12 | 7 | 0 | 0 | 0 | 0 |
| G23.02 | 29 | 8 | 0 | 0 | 0 | 0 | G16.10 | 16 | 8 | 1 | 0 | 0 | 0 |
| Trait | Number of SilicoDArT and SNP Markers | Effect (Min.) | Effect (Max.) | Effect (Average) | Percentage of Explained Variance (Min.) | Percentage of Explained Variance (Max.) | Percentage of Explained Variance (Average) | LOD (Min.) | LOD (Max.) | LOD (Average) |
|---|---|---|---|---|---|---|---|---|---|---|
| Cob fusariosis [scale 1–9] | 5714 | −0.759 | 0.2999 | −0.016 | 1.7 | 13.3 | 4.65 | 1.3 | 6.33 | 2.59 |
| Marker Number (Marker Type) | Marker Effect (Percentage Variance Accounted for) | Chromosomal Localization | Marker Sequence (5′-3′) | Neighboring Genes |
|---|---|---|---|---|
| 14586781 (SilicoDArT) | 0.2999 (13.3%) | Chr6, 145045072 bp | TGCAGAATAAAGGCCGTAGCTACTAGCATGAGATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGAT |
|
| 7049252 (SilicoDArT) | 0.2871 (12.6%) | Chr6, 145473013 bp | TGCAGAGCAGAAGCCTTCCGCTGAAACGAGCCGGCCAGCCGGGTCAAAGCGGCGGGCGAATGCATGAGA |
|
| 4778172 (SilicoDArT) | 0.2744 (12.5%) | Chr9, 62605002 bp | TGCAGTCTCCAGCCGGCAGTGGCTGCGAACCAGTGACGAGATGAGCACGTCATCTGAAGGTCCCTCCTG |
|
| 2414058 (SNP) | 0.2753 (12.1%) | Chr6, 151006006 bp | TGCAGCACACCTTCAAACCGTTTCCCCTCTAAACTGGCAAGATCATTGCATAGATCAGCAATACAAGAC |
|
| 4579116 (SilicoDArT) | −0.2759 (12.0%) | Chr8, 116991138 bp | TGCAGGCTGAAGCCGTTCCGGAAGGCATACCAAACTGATTCATACCAAACTTTGAGGCATGAGATCGGA |
|
| 4587705 (SilicoDArT) | 0.2660 (11.9%) | Chr8, 170156900 bp | TGCAGTAGCCTCGTCGTCACCGACATAACCTGAAAAAATCATTCAATTGACTCATGTAGTAGCGCCCCC |
|
| 25947704 (SNP) | 0.2586 (11.4%) | Chr6, 148365780 bp | TGCAGCAACGAGGCGGAGGAGGAGGCCGGGTTCAACCTCCTGGGGCTGCTGGTCGCCGCCATCATCGCG |
|
| 4583014 (SNP) | 0.2586 (11.4%) | Chr5, 214247330 bp | TGCAGCTTCATATCTAGAATCACCAGTCAAACGTGACAACACACCCATTTCAAGTATAAGGGAACCTGT |
|
| 4777510 (SNP) | 0.2668 (11.2%) | Chr9, 117791088 bp | TGCAGATGAATAAATATTAGATATATTGACAACTTAAGTATCTGAGTGGCGCAAATTGAAGTTCTGATC |
|
| 4584918 (SNP) | 0.2574 (11.0%) | Chr7, 144651551 bp | TGCAGTGCTCTAGGAACTTGGTTCTTCTCAGTTGCGGGTGCTCTTGTTGCTATTCCTGTGGGCATAAAG |
|
| Marker Number | Primer Sequences | PCR Product (bp) |
|---|---|---|
| 4586781 | Forward: ACAAAAGCTCTATAAATCTCTTAAA Reverse: CAAATATTCAGTAGTAAAGGATATC | 218 |
| 7049252 | Forward: GCGTCTCATGCATTCGCAC Reverse: CTACACTCAAGCAACTAAGGTCATC | 235 |
| 4778172 | Forward: GCGAACCAGTGACGAGATGAGCAAG Reverse: GGTCCTAGTCGGTCCCTGGTCG | 258 |
| 2414058 | Forward: ATCATTGCATAGATCAGCAATACCG Reverse: ATTCGTTGTATCAAGTGAAAACGCT | 490 |
| 4579116 | Forward: TTTGATATGGCTCCTGCAAG Reverse: GAATAAGGTGTGTATCTGGGG | 489 |
| 4587705 | Forward: CGTCACCGACATAACCTGAAAAAGT Reverse: TTTTCTTAAGGATTCTGCCACAATC | 210 |
| 25947704 | Forward: CACATGCTGAAGCTGATCCGAAACC Reverse: AGGAGGAGGCCGGGTTCAACCGT | 241 |
| 4583014 | Forward: CTATCAGCTAAAATGATAAGAATG Reverse: CCATTTCAAGTATAAGGGCG | 321 |
| 4777510 | Forward: GCAGCCAACAAATCCATC Reverse: AAGTTGTCAATATGTCTAATACG | 309 |
| 4584918 | Forward: GGCTCGGTGGAGTCAGCTTGTG Reverse: ATAGCAACAAGAGCACCCGCAACCG | 200 |
| Marker | Gene Locus | Accession Number | F. verticillioides Infection | |
|---|---|---|---|---|
| Susceptible Genotype | Resistant Genotype | |||
| 4778172 | LOC100274139 | GRMZM2G093598 | −6.4% | +7.5% |
| 4579116 | LOC100282644 | GRMZM2G149211 | +73.8% | +126.2% |
| 4587705 | LOC100272376 | GRMZM5G805585 | −11.4% | +59.4% |
| 4583014 | LOC103627708 | GRMZM2G501450 | +12.11% | −29.5% |
| 4777510 | LOC103652964 | GRMZM2G162859 | −2.4% | −14.4% |
| 4584918 | LOC103633003 | GRMZM5G854301 | +9.3% | +66.3% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lenort, M.; Tomkowiak, A.; Sobiech, A.; Bocianowski, J.; Jarzyniak, K.; Olejnik, P.; Jamruszka, T.; Gawrysiak, P. DArTseq-Based, High-Throughput Identification of Novel Molecular Markers for the Detection of Fusarium Resistance in Maize. Int. J. Mol. Sci. 2025, 26, 10534. https://doi.org/10.3390/ijms262110534
Lenort M, Tomkowiak A, Sobiech A, Bocianowski J, Jarzyniak K, Olejnik P, Jamruszka T, Gawrysiak P. DArTseq-Based, High-Throughput Identification of Novel Molecular Markers for the Detection of Fusarium Resistance in Maize. International Journal of Molecular Sciences. 2025; 26(21):10534. https://doi.org/10.3390/ijms262110534
Chicago/Turabian StyleLenort, Maciej, Agnieszka Tomkowiak, Aleksandra Sobiech, Jan Bocianowski, Karolina Jarzyniak, Przemysław Olejnik, Tomasz Jamruszka, and Przemysław Gawrysiak. 2025. "DArTseq-Based, High-Throughput Identification of Novel Molecular Markers for the Detection of Fusarium Resistance in Maize" International Journal of Molecular Sciences 26, no. 21: 10534. https://doi.org/10.3390/ijms262110534
APA StyleLenort, M., Tomkowiak, A., Sobiech, A., Bocianowski, J., Jarzyniak, K., Olejnik, P., Jamruszka, T., & Gawrysiak, P. (2025). DArTseq-Based, High-Throughput Identification of Novel Molecular Markers for the Detection of Fusarium Resistance in Maize. International Journal of Molecular Sciences, 26(21), 10534. https://doi.org/10.3390/ijms262110534






