Gene Expression Profiling of Transcription Factors and Acclimation-Related Genes in Ribes spp.
Abstract
1. Introduction
2. Results
2.1. Protein-Protein Interaction (PPI) Network
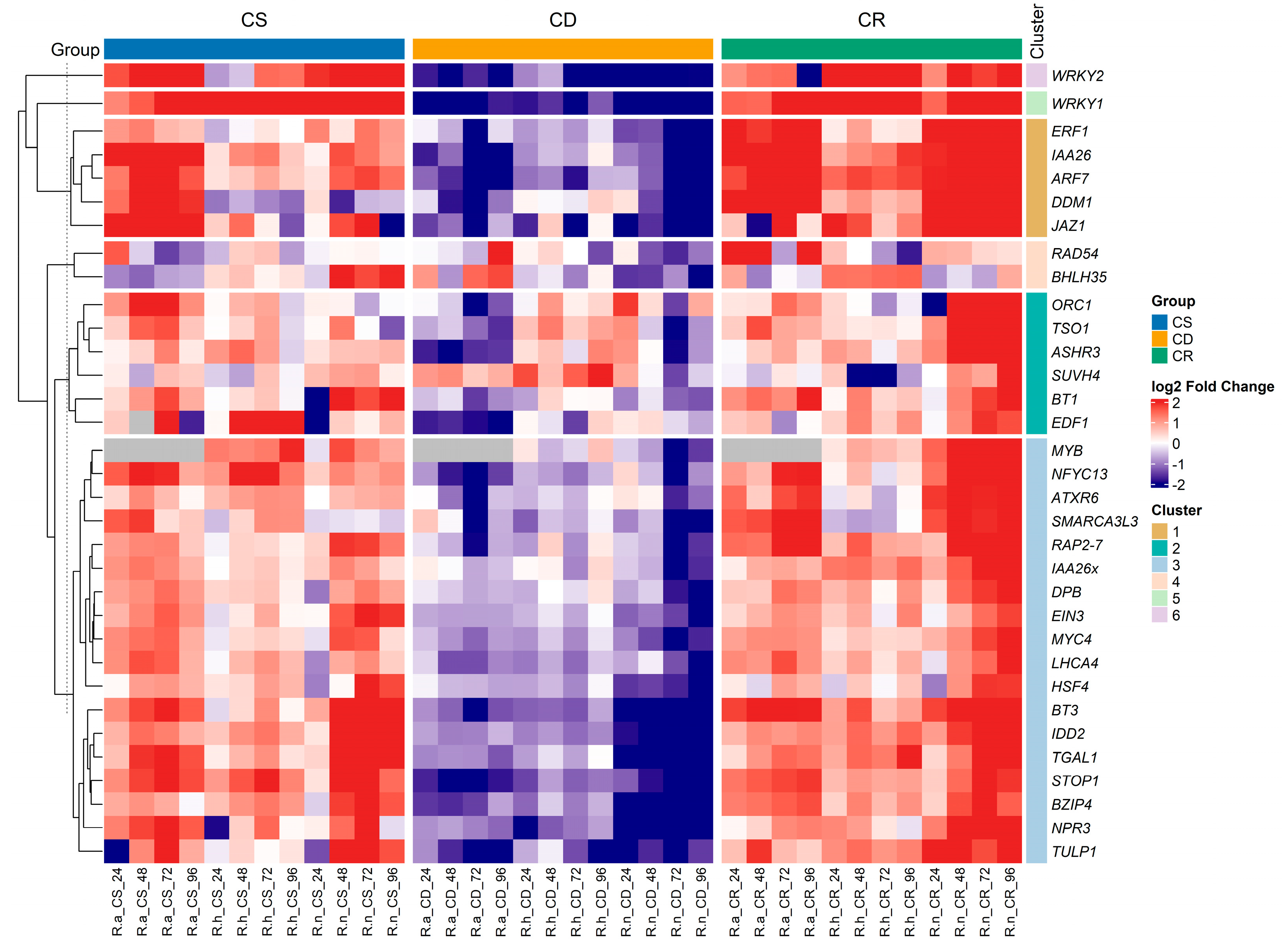
2.2. Hierarchical Clustering of Transcription Factors in Response to Hardening
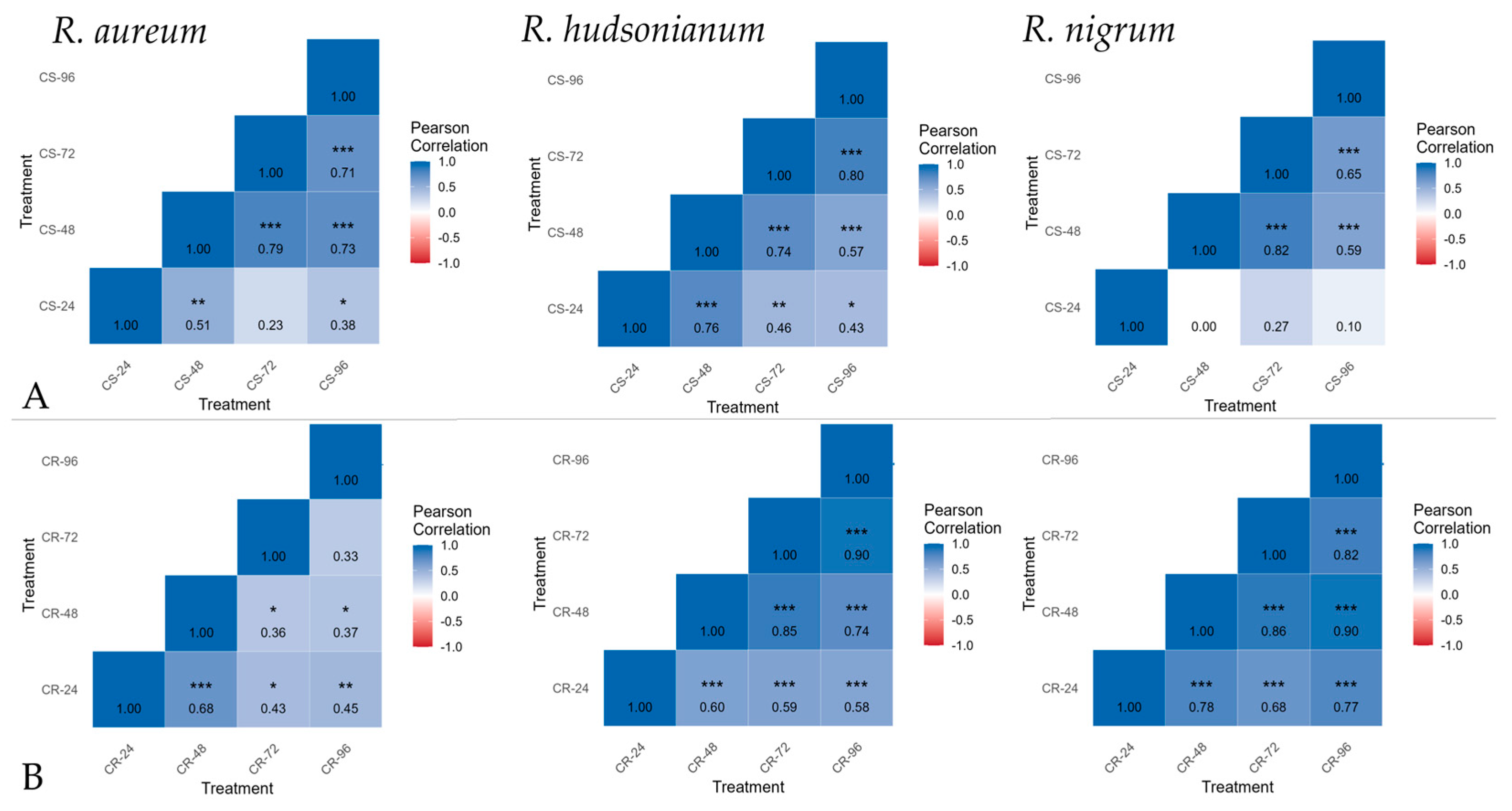
2.3. Deacclimation Impact on Genes Expression for Hardening in Ribes spp.
3. Discussion
4. Materials and Methods
4.1. Plant Material and Stress Conditions
4.2. RNA Extraction and qRT-PCR Analysis
4.3. Statistical Analysis and Data Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Brennan, R.M. Currants and gooseberries. In Fruit Breeding, Volume II: Vine and Small Fruit Crops; Janick, J., Moore, J.N., Eds.; Wiley: New York, 1996; pp. 191–295. [Google Scholar]
- Preedy, K.; Brennan, R.M.; Jones, H.; Gordon, S. Improved models of the effects of winter chilling on blackcurrant (Ribes nigrum L.) show cultivar-specific sensitivity to warm winters. Agric. For. Meteorol. 2020, 280, 107777. [Google Scholar] [CrossRef]
- Barney, D.; Hummer, K. Currants, Gooseberries and Jostaberries: A Guide for Growers, Marketers and Researchers in North America; Haworth Press: Binghampton, NY, 2005; 266p. [Google Scholar]
- Jones, H.G.; Gordon, S.L.; Brennan, R.M. Chilling requirement of Ribes cultivars. Front. Plant Sci. 2015, 5, 767. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.W.; Harding, K.; Benson, E.E. Antioxidant status and genotypic tolerance of Ribes in vitro cultures to cryopreservation. Plant Sci. 2007, 172, 524–534. [Google Scholar] [CrossRef]
- Kjær, K.H.; Winde, J.; Petersen, K.K.; Yde, C.C.; Pagter, M. Cold deacclimation mechanisms and reacclimation potential in flower buds of blackcurrant (Ribes nigrum). Physiol. Plant. 2019, 167, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Palonen, P.; Lettojärvi, I.; Luoranen, J.; Ruhanen, H.; Rantanen, M.; Haikonen, T.; Finni, S. Deacclimation and reacclimation of apple (Malus×domestica Borkh.), blackcurrant (Ribes nigrum L.) and raspberry (Rubus idaeus L.) shoots and buds under controlled conditions. Sci. Hortic. 2021, 289, 110430. [Google Scholar] [CrossRef]
- Brennan, R.M.; Millam, S.; Davidson, D.; Wilshin, A. Establishment of an in vitro Ribes germplasm collection and preliminary investigations into long-term low-temperature germplasm storage. In Proceedings of the I International Symposium on In Vitro Culture and Horticultural Breeding, Cesena, Italy, 30 May–3 June 1989; ISHS Acta Horticulturae: Leuven, Belgium, 1989; 280, pp. 109–112. [Google Scholar] [CrossRef]
- Brennan, R.M. The effects of simulated frost on black currant (Ribes nigrum L.). J. Hortic. Sci. 1991, 66, 607–612. [Google Scholar] [CrossRef]
- Woznicki, T.L.; Sønsteby, A.; Aaby, K.; Martinsen, B.K.; Heide, O.M.; Wold, A.B.; Remberg, S.F. Ascorbate pool, sugars and organic acids in black currant (Ribes nigrum L.) berries are strongly influenced by genotype and post-flowering temperature. J. Sci. Food Agric. 2017, 97, 1302–1309. [Google Scholar] [CrossRef]
- Mehrotra, S.; Verma, S.; Kumar, S.; Kumari, S.; Mishra, B.N. Transcriptional regulation and signalling of cold stress response in plants: An overview of current understanding. Environ. Exp. Bot. 2020, 180, 104243. [Google Scholar] [CrossRef]
- Qian, Z.; He, L.; Li, F. Understanding cold stress response mechanisms in plants: An overview. Front. Plant Sci. 2024, 15, 1443317. [Google Scholar] [CrossRef]
- Li, W.; Li, H.; Wei, Y.; Han, J.; Wang, Y.; Li, X.; Zhang, L.; Han, D. Overexpression of a Fragaria vesca NAM, ATAF, and CUC (NAC) transcription factor gene (FvNAC29) increases salt and cold tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 4088. [Google Scholar] [CrossRef]
- Zhang, L.; Xing, L.; Dai, J.; Li, Z.; Zhang, A.; Wang, T.; Liu, W.; Li, X.; Han, D. Overexpression of a Grape WRKY Transcription Factor VhWRKY44 Improves the Resistance to Cold and Salt of Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 7437. [Google Scholar] [CrossRef]
- Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional Factors Regulate Plant Stress Responses through Mediating Secondary Metabolism. Genes 2020, 11, 346. [Google Scholar] [CrossRef]
- Abdullah, S.N.A.; Azzeme, A.M.; Yousefi, K. Fine-Tuning Cold Stress Response Through Regulated Cellular Abundance and Mechanistic Actions of Transcription Factors. Front. Plant Sci. 2022, 13, 850216. [Google Scholar] [CrossRef] [PubMed]
- Medina, J.; Catalá, R.; Salinas, J. The CBFs: The CBFs: Three arabidopsis transcription factors to cold acclimate. Plant Sci. 2011, 180, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF transcription factor responses and tolerance to various abiotic stresses in plants: A comprehensive review. Int. J. Mol. Sci. 2024, 25, 893. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Khoso, M.A.; Xu, H.; Zhang, C.; Liu, Z.; Wagan, S.; Dinislam, K.; Liu, L. WRKY Transcription Factors (TFs) as Key Regulators of Plant Resilience to Environmental Stresses: Current Perspective. Agronomy 2024, 14, 2421. [Google Scholar] [CrossRef]
- Mariyam, S.; Kumar, V.; Roychoudhury, A.; Ghodake, G.S.; Muneer, S.; Duhan, J.S.; Ahmad, F.; Sharma, R.K.; Singh, J.; Seth, C.S. Functional Diversification and Mechanistic Insights of MYB Transcription Factors in Mediating Plant Growth and Development, Secondary Metabolism, and Stress Responses. J. Plant Growth Regul. 2025, 44, 1465–1484. [Google Scholar] [CrossRef]
- Chen, C.; Li, X.; Wen, K.; Yin, T.; Tian, P.; Zhao, K.; Zhang, L.; Zhou, X.; Liu, X.; Zhang, H. Identification of the bHLH Transcription Factor Family in Orah Mandarin and the Response of CrbHLH46 to Low-Temperature Stress. Plants 2025, 14, 882. [Google Scholar] [CrossRef]
- Kavi Kishor, P.B.; Ganie, S.A.; Wani, S.H.; Guddimalli, R.; Karumanchi, A.R.; Edupuganti, S.; Naravula, J.; Kumar, V.; Polavarapu, R.; Suravajhala, P.; et al. Nuclear Factor-Y (NF-Y): Developmental and Stress-Responsive Roles in the Plant Lineage. J. Plant Growth Regul. 2023, 42, 2711–2735. [Google Scholar] [CrossRef]
- Su, Y.; Dai, S.; Li, N.; Gentile, A.; He, C.; Xu, J.; Duan, K.; Wang, X.; Wang, B.; Li, D. Unleashing the Potential of EIL Transcription Factors in Enhancing Sweet Orange Resistance to Bacterial Pathologies: Genome-Wide Identification and Expression Profiling. Int. J. Mol. Sci. 2023, 24, 12644. [Google Scholar] [CrossRef]
- Deng, S.; Ma, J.; Zhang, L.; Chen, F.; Sang, Z.; Jia, Z.; Ma, L. De novo transcriptome sequencing and gene expression profiling of Magnolia wufengensis in response to cold stress. BMC Plant Biol. 2019, 19, 321. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Soren, K.R.; Bharadwaj, C.; Sneha Priya, P.R.; Shrivastava, A.K.; Pal, M.; Roorkiwal, M.; Kumar, K.; Patil, B.S.; Soni, A.; et al. Genome-wide transcriptome analysis and physiological variation modulates gene regulatory networks acclimating salinity tolerance in chickpea. Environ. Exp. Bot. 2021, 187, 104478. [Google Scholar] [CrossRef]
- Mažeikienė, I.; Juškytė, A.D.; Bendokas, V.; Stanys, V. De novo transcriptome analysis of R. nigrum cv. Aldoniai in response to blackcurrant reversion virus infection. Int. J. Mol. Sci. 2011, 23, 9560. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, Y.; Lan, J.; Zou, Y.; Gao, J. Comparative transcriptomic analysis of the response to cold acclimation in Eucalyptus dunnii. PLoS ONE 2014, 9, e113091. [Google Scholar] [CrossRef]
- Mousavi, S.; Alisoltani, A.; Shiran, B.; Fallahi, H.; Ebrahimie, E.; Imani, A.; Houshmand, S. De novo transcriptome assembly and comparative analysis of differentially expressed genes in Prunus dulcis Mill. in response to freezing stress. PLoS ONE 2014, 9, e104541. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, T.; Rao, P.; Gao, K.; Yang, X.; Chen, Z.; An, X. Transcriptome profiling of Populus tomentosa under cold stress. Ind. Crops Prod. 2019, 135, 283–293. [Google Scholar] [CrossRef]
- Pagter, M.; Alpers, J.; Erban, A.; Kopka, J.; Zuther, E.; Hincha, D.K. Rapid transcriptional and metabolic regulation of the deacclimation process in cold-acclimated Arabidopsis thaliana. BMC Genomics 2017, 18, 731. [Google Scholar] [CrossRef]
- Edrisi Maryan, K.; Farrokhi, N.; Samizadeh Lahiji, H. Cold-responsive transcription factors in Arabidopsis and rice: A regulatory network analysis using array data and gene co-expression network. PLoS ONE 2023, 18, e0286324. [Google Scholar] [CrossRef]
- Erpen, L.; Devi, H.S.; Grosser, J.W.; Dutt, M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tiss. Organ Cult. 2018, 132, 1–25. [Google Scholar] [CrossRef]
- Diao, P.; Chen, C.; Zhang, Y.; Meng, Q.; Lv, W.; Ma, N. The role of NAC transcription factor in plant cold response. Plant Signal. Behav. 2020, 15, 1785668. [Google Scholar] [CrossRef]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Liang, Z.; Dai, X.; Zhang, Y.; Mahboub, S.; Ngu, D.W.; Roston, R.L.; Schnable, J.C. Predicting transcriptional responses to cold stress across plant species. Proc. Natl. Acad. Sci. USA 2021, 118, e2026330118. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Yang, Q.; Wang, X.; Chen, Y.; He, R.; Li, X.; Pan, H.; Zhuo, R.; Qu, T.; Qiu, W. Transcription factors involved in plant stress and growth and development: NAC. Agronomy 2025, 15, 949. [Google Scholar] [CrossRef]
- Vuliã, T.; Djordjeviã, B.; Ruml, M.; Djuroviã, D.; Fotiriã-Akðiã, M.; Radivojeviã, D.; Oparnica, C. Flowering dynamic and susceptibility of the flowers of black currant (Ribes nigrum L.) and red currant (Ribes rubrum L.) to spring frosts. Acta Hortic. 2012, 946, 373–377. [Google Scholar] [CrossRef]
- Peng, X.; Wu, Q.; Teng, L.; Tang, F.; Pi, Z.; Shen, S. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 2015, 15, 108. [Google Scholar] [CrossRef]
- Rugienius, R.; Vinskienė, J.; Andriūnaitė, E.; Morkūnaitė-Haimi, Š.; Juhani-Haimi, P.; Graham, J. Genomic design of abiotic stress-resistant berries. In Genomic Designing for Abiotic Stress Resistant Fruit Crops; Kole, C., Ed.; Springer: Cham, Switzerland, 2022; pp. 197–249. [Google Scholar] [CrossRef]
- Jun, S.H.; Yu, D.J.; Im, D.; Kim, S.J.; Hur, Y.Y.; Chung, S.W.; Lee, H.J. Searching for cold hardiness-related genes based on transcriptome profiling in grapevine canes during cold acclimation and deacclimation. J. Hortic. Sci. Biotechnol. 2024, 100, 458–472. [Google Scholar] [CrossRef]
- Niu, R.; Zhao, X.; Wang, C.; Wang, F. Transcriptome profiling of Prunus persica branches reveals candidate genes potentially involved in freezing tolerance. Sci. Hortic. 2020, 259, 108775. [Google Scholar] [CrossRef]
- Kim, J.M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef]
- Song, Z.T.; Liu, J.X.; Han, J.J. Chromatin remodeling factors regulate environmental stress responses in plants. J. Integr. Plant Biol. 2021, 63, 438–450. [Google Scholar] [CrossRef]
- Kim, J.H. Chromatin remodeling and epigenetic regulation in plant DNA damage repair. Int. J. Mol. Sci. 2019, 20, 4093. [Google Scholar] [CrossRef]
- Olatunji, D.; Geelen, D.; Verstraeten, I. Control of endogenous auxin levels in plant root development. Int. J. Mol. Sci. 2017, 18, 2587. [Google Scholar] [CrossRef]
- Eremina, M.; Rozhon, W.; Poppenberger, B. Hormonal control of cold stress responses in plants. Cell. Mol. Life Sci. 2016, 73, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Hamza, M.; ul Ain, N.; Mohamed, H.I.; Muhammad, M.; Sultan, Y.; Alam Khan, M.; Basit, A. Jasmonic acid: A key elicitor of cold stress tolerance in horticultural crops. J. Plant Growth Regul. 2025; in press. [Google Scholar] [CrossRef]
- Jones, H.G.; Hillis, R.M.; Gordon, S.L.; Brennan, R.M. An approach to the determination of winter chill requirements for different Ribes cultivars. Plant Biol. 2013, 15, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Andersen, U.B.; Kjær, K.H.; Erban, A.; Alpers, J.; Hincha, D.K.; Kopka, J.; Zuther, E.; Pagter, M. Impact of seasonal warming on overwintering and spring phenology of blackcurrant. Environ. Exp. Bot. 2017, 140, 96–109. [Google Scholar] [CrossRef]
- Barah, P.; Jayavelu, N.D.; Rasmussen, S.; Nielsen, H.B.; Mundy, J.; Bones, A.M. Genome-scale cold stress response regulatory networks in ten Arabidopsis thaliana ecotypes. BMC Genomics. 2013, 14, 722. [Google Scholar] [CrossRef]
- Baker, C.R.; Stewart, J.J.; Amstutz, C.L.; Ching, L.G.; Johnson, J.D.; Niyogi, K.K.; Adams, W.W., 3rd; Demmig-Adams, B. Genotype-dependent contribution of CBF transcription factors to long-term acclimation to high light and cool temperature. Plant Cell Environ. 2022, 45, 392–411. [Google Scholar] [CrossRef]
- Li, P.; Zheng, T.; Li, L.; Liu, W.; Qiu, L.; Ahmad, S.; Wang, J.; Cheng, T.; Zhang, Q. Integration of chromatin accessibility and gene expression reveals new regulators of cold hardening to enhance freezing tolerance in Prunus mume. J Exp Bot. 2023, 74, 2173–2187. [Google Scholar] [CrossRef]
- Juškytė, A.D.; Mažeikienė, I.; Stanys, V. An effective method of Ribes spp. inoculation with blackcurrant reversion virus under in vitro conditions. Plants 2022, 11, 1635. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wickham, H.; Bryan, J. readxl: Read Excel Files. R Package Version 1.4.5. Available online: https://readxl.tidyverse.org (accessed on 3 July 2025).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation. R Package Version 1.1.4. Available online: https://dplyr.tidyverse.org (accessed on 3 July 2025).
- Wickham, H.; Vaughan, D.; Girlich, M. tidyr: Tidy Messy Data. R Package Version 1.3.1. Available online: https://tidyr.tidyverse.org (accessed on 3 July 2025).
- Müller, K.; Wickham, H. Tibble: Simple Data Frames. R Package Version 3.3.0. Available online: https://tibble.tidyverse.org (accessed on 3 July 2025).
- Gu, Z. Complex Heatmap Visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes (Version 1.1-2) [R Package]. Available online: https://CRAN.R-project.org/package=RColorBrewer (accessed on 3 July 2025).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer Nature: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 3 July 2025).
- de Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research (Version 1.3-5) [R Package]. Available online: https://CRAN.R-project.org/package=agricolae (accessed on 3 July 2025).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zubauskienė, A.D.; Misiukevičius, E.; Bendokas, V.; Njoku, E.G.; Mažeikienė, I. Gene Expression Profiling of Transcription Factors and Acclimation-Related Genes in Ribes spp. Int. J. Mol. Sci. 2025, 26, 10367. https://doi.org/10.3390/ijms262110367
Zubauskienė AD, Misiukevičius E, Bendokas V, Njoku EG, Mažeikienė I. Gene Expression Profiling of Transcription Factors and Acclimation-Related Genes in Ribes spp. International Journal of Molecular Sciences. 2025; 26(21):10367. https://doi.org/10.3390/ijms262110367
Chicago/Turabian StyleZubauskienė, Ana Dovilė, Edvinas Misiukevičius, Vidmantas Bendokas, Emmanuel Gabriel Njoku, and Ingrida Mažeikienė. 2025. "Gene Expression Profiling of Transcription Factors and Acclimation-Related Genes in Ribes spp." International Journal of Molecular Sciences 26, no. 21: 10367. https://doi.org/10.3390/ijms262110367
APA StyleZubauskienė, A. D., Misiukevičius, E., Bendokas, V., Njoku, E. G., & Mažeikienė, I. (2025). Gene Expression Profiling of Transcription Factors and Acclimation-Related Genes in Ribes spp. International Journal of Molecular Sciences, 26(21), 10367. https://doi.org/10.3390/ijms262110367





