Central Roles of Glucosylceramide in Driving Cancer Pathogenesis
Abstract
1. Introduction
2. Glucosylceramide Architecture, Tissue Distribution, and Oncogenic Potential
3. Metabolic Rewiring of Glucosylceramide Pathways in Cancer Development
4. Glucosylceramide-Driven Crosstalk Between Energy Metabolism, Inflammatory Signaling, and Oncogenesis
5. Discussion
6. Conclusions and Outlook
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quinville, B.M.; Deschenes, N.M.; Ryckman, A.E.; Walia, J.S. A Comprehensive Review: Sphingolipid Metabolism and Implications of Disruption in Sphingolipid Homeostasis. Int. J. Mol. Sci. 2021, 22, 5793. [Google Scholar] [CrossRef]
- Futerman, A.H.; Platt, F.M. The metabolism of glucocerebrosides-From 1965 to the present. Mol. Genet. Metab. 2017, 120, 22–26. [Google Scholar] [CrossRef]
- Lanska, D. Gaucher disease. In MedLink Neurology; Lewis, S., Ed.; MedLink, LLC: San Diego, CA, USA, 2025. [Google Scholar]
- Messner, M.C.; Cabot, M.C. Glucosylceramide in humans. Adv. Exp. Med. Biol. 2010, 688, 156–164. [Google Scholar] [CrossRef]
- Yamashita, T.; Wada, R.; Sasaki, T.; Deng, C.; Bierfreund, U.; Sandhoff, K.; Proia, R.L. A vital role for glycosphingolipid synthesis during development and differentiation. Proc. Natl. Acad. Sci. USA 1999, 96, 9142–9147. [Google Scholar] [CrossRef] [PubMed]
- Steiner, R.; Saied, E.M.; Othman, A.; Arenz, C.; Maccarone, A.T.; Poad, B.L.; Blanksby, S.J.; von Eckardstein, A.; Hornemann, T. Elucidating the chemical structure of native 1-deoxysphingosine. J. Lipid Res. 2016, 57, 1194–1203. [Google Scholar] [CrossRef]
- Maula, T.; Artetxe, I.; Grandell, P.M.; Slotte, J.P. Importance of the sphingoid base length for the membrane properties of ceramides. Biophys. J. 2012, 103, 1870–1879. [Google Scholar] [CrossRef]
- Merrill, A.H., Jr. Don’t Be Surprised When These Surprise You: Some Infrequently Studied Sphingoid Bases, Metabolites, and Factors That Should Be Kept in Mind During Sphingolipidomic Studies. Int. J. Mol. Sci. 2025, 26, 650. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Arai, H.; Cocco, M.J.; White, S.H. pH dependence of sphingosine aggregation. Biophys. J. 2009, 96, 2727–2733. [Google Scholar] [CrossRef]
- Sandhoff, R. Very long chain sphingolipids: Tissue expression, function and synthesis. FEBS Lett. 2010, 584, 1907–1913. [Google Scholar] [CrossRef] [PubMed]
- Hamanaka, S.; Hara, M.; Nishio, H.; Otsuka, F.; Suzuki, A.; Uchida, Y. Human epidermal glucosylceramides are major precursors of stratum corneum ceramides. J. Investig. Dermatol. 2002, 119, 416–423. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Many ceramides. J. Biol. Chem. 2011, 286, 27855–27862. [Google Scholar] [CrossRef]
- Reza, S.; Ugorski, M.; Suchanski, J. Glucosylceramide and galactosylceramide, small glycosphingolipids with significant impact on health and disease. Glycobiology 2021, 31, 1416–1434. [Google Scholar] [CrossRef]
- Körschen, H.G.; Yildiz, Y.; Raju, D.N.; Schonauer, S.; Bönigk, W.; Jansen, V.; Kremmer, E.; Kaupp, U.B.; Wachten, D. The non-lysosomal β-glucosidase GBA2 is a non-integral membrane-associated protein at the endoplasmic reticulum (ER) and Golgi. J. Biol. Chem. 2013, 288, 3381–3393. [Google Scholar] [CrossRef] [PubMed]
- Boer, D.E.C.; van Smeden, J.; Bouwstra, J.A.; Aerts, J. Glucocerebrosidase: Functions in and Beyond the Lysosome. J. Clin. Med. 2020, 9, 736. [Google Scholar] [CrossRef] [PubMed]
- Marano, M.; Zizzo, C.; Malaguti, M.C.; Bacchin, R.; Cavallieri, F.; De Micco, R.; Spagnolo, F.; Bentivoglio, A.R.; Schirinzi, T.; Bovenzi, R.; et al. Increased glucosylsphingosine levels and Gaucher disease in GBA1-associated Parkinson’s disease. Park. Relat. Disord. 2024, 124, 107023. [Google Scholar] [CrossRef]
- Ichikawa, S.; Sakiyama, H.; Suzuki, G.; Hidari, K.I.; Hirabayashi, Y. Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc. Natl. Acad. Sci. USA 1996, 93, 4638–4643. [Google Scholar] [CrossRef]
- Mao, C.; Obeid, L.M. Ceramidases: Regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim. Biophys. Acta 2008, 1781, 424–434. [Google Scholar] [CrossRef]
- Brady, R.O.; Kanfer, J.N.; Shapiro, D. Metabolism of glucocerebrosides II. Evidence of an enzymatic deficiency in Gaucher’s disease. Biochem. Biophys. Res. Commun. 1965, 18, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Schapira, A.H. Glucocerebrosidase and Parkinson disease: Recent advances. Mol. Cell Neurosci. 2015, 66 Pt A, 37–42. [Google Scholar] [CrossRef]
- Vielhaber, G.; Pfeiffer, S.; Brade, L.; Lindner, B.; Goldmann, T.; Vollmer, E.; Hintze, U.; Wittern, K.-P.; Wepf, R. Localization of Ceramide and Glucosylceramide in Human Epidermis by Immunogold Electron Microscopy. J. Investig. Dermatol. 2001, 117, 1126–1136. [Google Scholar] [CrossRef]
- Weiler, S.; Kishimoto, Y.; O’Brien, J.S.; Barranger, J.A.; Tomich, J.M. Identification of the binding and activating sites of the sphingolipid activator protein, saposin C, with glucocerebrosidase. Protein Sci. 1995, 4, 756–764. [Google Scholar] [CrossRef]
- Grabowski, G.A. Gaucher disease and other storage disorders. Hematology 2012, 2012, 13–18. [Google Scholar] [CrossRef]
- Pandey, M.K. Exploring Pro-Inflammatory Immunological Mediators: Unraveling the Mechanisms of Neuroinflammation in Lysosomal Storage Diseases. Biomedicines 2023, 11, 1067. [Google Scholar] [CrossRef] [PubMed]
- Coant, N.; Sakamoto, W.; Mao, C.; Hannun, Y.A. Ceramidases, roles in sphingolipid metabolism and in health and disease. Adv. Biol. Regul. 2017, 63, 122–131. [Google Scholar] [CrossRef]
- Park, J.H.; Schuchman, E.H. Acid ceramidase and human disease. Biochim. Biophys. Acta 2006, 1758, 2133–2138. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Airola, M.V.; Choi, Y.M.; Coant, N.; Snider, J.; Cariello, C.; Saied, E.M.; Arenz, C.; Bannister, T.; Rahaim, R., Jr.; et al. Identification of Small-Molecule Inhibitors of Neutral Ceramidase (nCDase) via Target-Based High-Throughput Screening. SLAS Discov. 2021, 26, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Lo, N.-W.; Shaper, J.H.; Pevsner, J.; Shaper, N.L. The expanding β4-galactosyltransferase gene family: Messages from the databanks. Glycobiology 1998, 8, 517–526. [Google Scholar] [CrossRef]
- Sato, T.; Furukawa, K.; Bakker, H.; Van den Eijnden, D.H.; Van Die, I. Molecular cloning of a human cDNA encoding β-1,4-galactosyltransferase with 37% identity to mammalian UDP-Gal:GlcNAc β-1,4-galactosyltransferase. Proc. Natl. Acad. Sci. USA 1998, 95, 472–477. [Google Scholar] [CrossRef]
- Ishikawa, K.; Kataoka, M.; Yanamoto, T.; Nakabayashi, M.; Watanabe, M.; Ishihara, S.; Yamaguchi, S. Crystal structure of β-galactosidase from Bacillus circulans ATCC 31382 (BgaD) and the construction of the thermophilic mutants. FEBS J. 2015, 282, 2540–2552. [Google Scholar] [CrossRef]
- Matthews, B.W. The structure of E. coli beta-galactosidase. Comptes Rendus Biol. 2005, 328, 549–556. [Google Scholar] [CrossRef]
- Juers, D.H.; Matthews, B.W.; Huber, R.E. LacZ β-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012, 21, 1792–1807. [Google Scholar] [CrossRef]
- Lee, B.Y.; Han, J.A.; Im, J.S.; Morrone, A.; Johung, K.; Goodwin, E.C.; Kleijer, W.J.; DiMaio, D.; Hwang, E.S. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell 2006, 5, 187–195. [Google Scholar] [CrossRef]
- Dadsena, S.; Bockelmann, S.; Mina, J.G.M.; Hassan, D.G.; Korneev, S.; Razzera, G.; Jahn, H.; Niekamp, P.; Muller, D.; Schneider, M.; et al. Ceramides bind VDAC2 to trigger mitochondrial apoptosis. Nat. Commun. 2019, 10, 1832. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, C.; Childs, S.; Ohotski, J.; McGlynn, L.; Riddick, M.; MacFarlane, S.; Tasker, D.; Pyne, S.; Pyne, N.J.; Edwards, J.; et al. Regulation of cell survival by sphingosine-1-phosphate receptor S1P1 via reciprocal ERK-dependent suppression of Bim and PI-3-kinase/protein kinase C-mediated upregulation of Mcl-1. Cell Death Dis. 2013, 4, e927. [Google Scholar] [CrossRef] [PubMed]
- Taha, T.A.; Mullen, T.D.; Obeid, L.M. A house divided: Ceramide, sphingosine, and sphingosine-1-phosphate in programmed cell death. Biochim. Biophys. Acta 2006, 1758, 2027–2036. [Google Scholar] [CrossRef]
- Rufail, M.L.; Bassi, R.; Giussani, P. Sphingosine-1-Phosphate Metabolic Pathway in Cancer: Implications for Therapeutic Targets. Int. J. Mol. Sci. 2025, 26, 1056. [Google Scholar] [CrossRef]
- Wegner, M.S.; Gruber, L.; Mattjus, P.; Geisslinger, G.; Grosch, S. The UDP-glucose ceramide glycosyltransferase (UGCG) and the link to multidrug resistance protein 1 (MDR1). BMC Cancer 2018, 18, 153. [Google Scholar] [CrossRef]
- Astudillo, L.; Therville, N.; Colacios, C.; Segui, B.; Andrieu-Abadie, N.; Levade, T. Glucosylceramidases and malignancies in mammals. Biochimie 2016, 125, 267–280. [Google Scholar] [CrossRef]
- Sandhoff, R.; Sandhoff, K. Emerging concepts of ganglioside metabolism. FEBS Lett. 2018, 592, 3835–3864. [Google Scholar] [CrossRef] [PubMed]
- Mattjus, P. Specificity of the mammalian glycolipid transfer proteins. Chem. Phys. Lipids 2016, 194, 72–78. [Google Scholar] [CrossRef]
- D’Angelo, G.; Uemura, T.; Chuang, C.C.; Polishchuk, E.; Santoro, M.; Ohvo-Rekila, H.; Sato, T.; Di Tullio, G.; Varriale, A.; D’Auria, S.; et al. Vesicular and non-vesicular transport feed distinct glycosylation pathways in the Golgi. Nature 2013, 501, 116–120. [Google Scholar] [CrossRef]
- Kita, N.; Hamamoto, A.; Gowda, S.G.B.; Takatsu, H.; Nakayama, K.; Arita, M.; Hui, S.P.; Shin, H.W. Glucosylceramide flippases contribute to cellular glucosylceramide homeostasis. J. Lipid Res. 2024, 65, 100508. [Google Scholar] [CrossRef]
- Budani, M.; Mylvaganam, M.; Binnington, B.; Lingwood, C. Synthesis of a novel photoactivatable glucosylceramide cross-linker. J. Lipid Res. 2016, 57, 1728–1736. [Google Scholar] [CrossRef]
- Budani, M.; Auray-Blais, C.; Lingwood, C. ATP-binding cassette transporters mediate differential biosynthesis of glycosphingolipid species. J. Lipid Res. 2021, 62, 100128. [Google Scholar] [CrossRef]
- Sillence, D.J.; Puri, V.; Marks, D.L.; Butters, T.D.; Dwek, R.A.; Pagano, R.E.; Platt, F.M. Glucosylceramide modulates membrane traffic along the endocytic pathway. J. Lipid Res. 2002, 43, 1837–1845. [Google Scholar] [CrossRef]
- Sillence, D.J. Glucosylceramide modulates endolysosomal pH in Gaucher disease. Mol. Genet. Metab. 2013, 109, 194–200. [Google Scholar] [CrossRef]
- Wang, L.; Lin, G.; Zuo, Z.; Li, Y.; Byeon, S.K.; Pandey, A.; Bellen, H.J. Neuronal activity induces glucosylceramide that is secreted via exosomes for lysosomal degradation in glia. Sci. Adv. 2022, 8, eabn3326. [Google Scholar] [CrossRef]
- Farrer, R.G.; Quarles, R.H. Extracellular matrix upregulates synthesis of glucosylceramide-based glycosphingolipids in primary Schwann cells. J. Neurosci. Res. 1996, 45, 248–257. [Google Scholar] [CrossRef]
- Marchell, N.L.; Uchida, Y.; Brown, B.E.; Elias, P.M.; Holleran, W.M. Glucosylceramides stimulate mitogenesis in aged murine epidermis. J. Investig. Dermatol. 1998, 110, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Wertz, P. Epidermal Lamellar Granules. Ski. Pharmacol. Physiol. 2018, 31, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Sutter, C.H.; Azim, S.; Wang, A.; Bhuju, J.; Simpson, A.S.; Uberoi, A.; Grice, E.A.; Sutter, T.R. Ligand Activation of the Aryl Hydrocarbon Receptor Upregulates Epidermal Uridine Diphosphate Glucose Ceramide Glucosyltransferase and Glucosylceramides. J. Investig. Dermatol. 2023, 143, 1964–1972. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Grabowski, G.A. Immunological cells and functions in Gaucher disease. Crit. Rev. Oncog. 2013, 18, 197–220. [Google Scholar] [CrossRef]
- Pandey, M.K.; Burrow, T.A.; Rani, R.; Martin, L.J.; Witte, D.; Setchell, K.D.; McKay, M.A.; Magnusen, A.F.; Zhang, W.; Liou, B.; et al. Complement drives glucosylceramide accumulation and tissue inflammation in Gaucher disease. Nature 2017, 543, 108–112. [Google Scholar] [CrossRef]
- Pandey, M.K.; Rani, R.; Zhang, W.; Setchell, K.; Grabowski, G.A. Immunological cell type characterization and Th1-Th17 cytokine production in a mouse model of Gaucher disease. Mol. Genet. Metab. 2012, 106, 310–322. [Google Scholar] [CrossRef][Green Version]
- Magnusen, A.F.; Rani, R.; McKay, M.A.; Hatton, S.L.; Nyamajenjere, T.C.; Magnusen, D.N.A.; Köhl, J.; Grabowski, G.A.; Pandey, M.K. C-X-C Motif Chemokine Ligand 9 and Its CXCR3 Receptor Are the Salt and Pepper for T Cells Trafficking in a Mouse Model of Gaucher Disease. Int. J. Mol. Sci. 2021, 22, 12712. [Google Scholar] [CrossRef]
- Pandey, M.K.; Jabre, N.A.; Xu, Y.H.; Zhang, W.; Setchell, K.D.; Grabowski, G.A. Gaucher disease: Chemotactic factors and immunological cell invasion in a mouse model. Mol. Genet. Metab. 2014, 111, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Schutt, C.R.; Izumi, Y.; Tomiyasu, N.; Omahdi, Z.; Kano, K.; Takamatsu, H.; Aoki, J.; Bamba, T.; Kumanogoh, A.; et al. Direct activation of microglia by β-glucosylceramide causes phagocytosis of neurons that exacerbates Gaucher disease. Immunity 2023, 56, 307–319.e308. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, N.; Xu, Y.H.; Li, R.; Peng, Y.; Pandey, M.K.; Tinch, S.L.; Liou, B.; Inskeep, V.; Zhang, W.; Setchell, K.D.; et al. Neuronopathic Gaucher disease: Dysregulated mRNAs and miRNAs in brain pathogenesis and effects of pharmacologic chaperone treatment in a mouse model. Hum. Mol. Genet. 2015, 24, 7031–7048. [Google Scholar] [CrossRef]
- Farfel-Becker, T.; Vitner, E.B.; Kelly, S.L.; Bame, J.R.; Duan, J.; Shinder, V.; Merrill, A.H., Jr.; Dobrenis, K.; Futerman, A.H. Neuronal accumulation of glucosylceramide in a mouse model of neuronopathic Gaucher disease leads to neurodegeneration. Hum. Mol. Genet. 2014, 23, 843–854. [Google Scholar] [CrossRef]
- Finnie, J.; Hemsley, K.; Manavis, J.; Beard, H.; Brealey, J.; Robertson, T.; Blumbergs, P. Striking and widespread microglial activation in the brains of Southdown lambs with type II glucocerebrosidosis (neuronopathic Gaucher disease). J. Comp. Pathol. 2024, 215, 10–13. [Google Scholar] [CrossRef]
- Wang, R.; Sun, H.; Cao, Y.; Zhang, Z.; Chen, Y.; Wang, X.; Liu, L.; Wu, J.; Xu, H.; Wu, D.; et al. Glucosylceramide accumulation in microglia triggers STING-dependent neuroinflammation and neurodegeneration in mice. Sci. Signal. 2024, 17, eadk8249. [Google Scholar] [CrossRef] [PubMed]
- Duffy, H.B.D.; Byrnes, C.; Zhu, H.; Tuymetova, G.; Lee, Y.T.; Platt, F.M.; Proia, R.L. Deletion of Gba in neurons, but not microglia, causes neurodegeneration in a Gaucher mouse model. JCI Insight 2024, 9, e179126. [Google Scholar] [CrossRef]
- Boddupalli, C.S.; Nair, S.; Belinsky, G.; Gans, J.; Teeple, E.; Nguyen, T.H.; Mehta, S.; Guo, L.; Kramer, M.L.; Ruan, J.; et al. Neuroinflammation in neuronopathic Gaucher disease: Role of microglia and NK cells, biomarkers, and response to substrate reduction therapy. Elife 2022, 11, e79830. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, W.; Xu, Y.H.; Quinn, B.; Dasgupta, N.; Liou, B.; Setchell, K.D.; Grabowski, G.A. Substrate compositional variation with tissue/region and Gba1 mutations in mouse models--implications for Gaucher disease. PLoS ONE 2013, 8, e57560. [Google Scholar] [CrossRef] [PubMed]
- Burrow, T.A.; Sun, Y.; Prada, C.E.; Bailey, L.; Zhang, W.; Brewer, A.; Wu, S.W.; Setchell, K.D.R.; Witte, D.; Cohen, M.B.; et al. CNS, lung, and lymph node involvement in Gaucher disease type 3 after 11 years of therapy: Clinical, histopathologic, and biochemical findings. Mol. Genet. Metab. 2015, 114, 233–241. [Google Scholar] [CrossRef]
- Uchiyama, M.; Oguri, M.; Mojumdar, E.H.; Gooris, G.S.; Bouwstra, J.A. Free fatty acids chain length distribution affects the permeability of skin lipid model membranes. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2016, 1858, 2050–2059. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Kohyama-Koganeya, A.; Hirabayashi, Y. New insights on glucosylated lipids: Metabolism and functions. Biochim. Biophys. Acta 2013, 1831, 1475–1485. [Google Scholar] [CrossRef]
- Don, A.S.; Lim, X.Y.; Couttas, T.A. Re-configuration of sphingolipid metabolism by oncogenic transformation. Biomolecules 2014, 4, 315–353. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Merrill, A.H., Jr.; Luberto, C. The bioactive sphingolipid playbook. A primer for the uninitiated as well as sphingolipidologists. J. Lipid Res. 2025, 66, 100813. [Google Scholar] [CrossRef]
- Okino, N.; Li, M.; Qu, Q.; Nakagawa, T.; Hayashi, Y.; Matsumoto, M.; Ishibashi, Y.; Ito, M. Two bacterial glycosphingolipid synthases responsible for the synthesis of glucuronosylceramide and alpha-galactosylceramide. J. Biol. Chem. 2020, 295, 10709–10725. [Google Scholar] [CrossRef]
- Sugawara, T. Sphingolipids as Functional Food Components: Benefits in Skin Improvement and Disease Prevention. J. Agric. Food Chem. 2022, 70, 9597–9609. [Google Scholar] [CrossRef]
- Dai, H.; Otsuka, A.; Tanabe, K.; Yanagita, T.; Nakayama, J.; Kitagaki, H. Glucosylceramide Changes Bacterial Metabolism and Increases Gram-Positive Bacteria through Tolerance to Secondary Bile Acids In Vitro. Int. J. Mol. Sci. 2022, 23, 5300. [Google Scholar] [CrossRef]
- Brown, E.M.; Clardy, J.; Xavier, R.J. Gut microbiome lipid metabolism and its impact on host physiology. Cell Host Microbe 2023, 31, 173–186. [Google Scholar] [CrossRef]
- Long, L.; Wang, L.; Liang, Y.; Ye, F.; Jin, Y.; Luo, D.; Li, X.; Wang, Y.; Li, Y.; Han, D.; et al. UGCG promotes chemoresistance and breast cancer progression via NF-kappaB and Wnt/beta-catenin pathway activation. Transl. Oncol. 2025, 52, 102241. [Google Scholar] [CrossRef] [PubMed]
- Schomel, N.; Gruber, L.; Alexopoulos, S.J.; Trautmann, S.; Olzomer, E.M.; Byrne, F.L.; Hoehn, K.L.; Gurke, R.; Thomas, D.; Ferreiros, N.; et al. UGCG overexpression leads to increased glycolysis and increased oxidative phosphorylation of breast cancer cells. Sci. Rep. 2020, 10, 8182. [Google Scholar] [CrossRef]
- Nguyen Van Long, F.; Le, T.; Caron, P.; Valcourt-Gendron, D.; Sergerie, R.; Laverdiere, I.; Vanura, K.; Guillemette, C. Targeting sphingolipid metabolism in chronic lymphocytic leukemia. Clin. Exp. Med. 2024, 24, 174. [Google Scholar] [CrossRef]
- Nguyen Van Long, F.; Valcourt-Gendron, D.; Caron, P.; Rouleau, M.; Villeneuve, L.; Simonyan, D.; Le, T.; Sergerie, R.; Laverdiere, I.; Vanura, K.; et al. Untargeted metabolomics identifies metabolic dysregulation of sphingolipids associated with aggressive chronic lymphocytic leukaemia and poor survival. Clin. Transl. Med. 2023, 13, e1442. [Google Scholar] [CrossRef]
- Faedo, R.R.; da Silva, G.; da Silva, R.M.; Ushida, T.R.; da Silva, R.R.; Lacchini, R.; Matos, L.L.; Kowalski, L.P.; Lopes, N.P.; Leopoldino, A.M. Sphingolipids signature in plasma and tissue as diagnostic and prognostic tools in oral squamous cell carcinoma. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2022, 1867, 159057. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Wang, X.; Yang, Z.; Liao, S.; Dong, W.; Sun, T.; Wu, H.; Zhang, Q.; Pan, Z.; Lam, S.M.; et al. GBA1-dependent membrane glucosylceramide reprogramming promotes liver cancer metastasis via activation of the Wnt/beta-catenin signalling pathway. Cell Death Dis. 2022, 13, 508. [Google Scholar] [CrossRef]
- Merz, N.; Hartel, J.C.; Grosch, S. How ceramides affect the development of colon cancer: From normal colon to carcinoma. Pflug. Arch. 2024, 476, 1803–1816. [Google Scholar] [CrossRef] [PubMed]
- Patterson, L.; Allen, J.; Posey, I.; Shaw, J.J.P.; Costa-Pinheiro, P.; Walker, S.J.; Gademsey, A.; Wu, X.; Wu, S.; Zachos, N.C.; et al. Glucosylceramide production maintains colon integrity in response to Bacteroides fragilis toxin-induced colon epithelial cell signaling. FASEB J. 2020, 34, 15922–15945. [Google Scholar] [CrossRef]
- Yamaji, T.; Horie, A.; Tachida, Y.; Sakuma, C.; Suzuki, Y.; Kushi, Y.; Hanada, K. Role of Intracellular Lipid Logistics in the Preferential Usage of Very Long Chain-Ceramides in Glucosylceramide. Int. J. Mol. Sci. 2016, 17, 1761. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, H. UDP-Glucose Ceramide Glycosyltransferase Contributes to the Proliferation and Glycolysis of Cervical Cancer Cells by Regulating the PI3K/AKT Pathway. Ann. Clin. Lab Sci. 2021, 51, 663–669. [Google Scholar] [PubMed]
- Li, J.; Chen, Q.; Guo, L.; Li, J.; Jin, B.; Wu, X.; Shi, Y.; Xu, H.; Zheng, Y.; Wang, Y.; et al. In situ Detecting Lipids as Potential Biomarkers for the Diagnosis and Prognosis of Intrahepatic Cholangiocarcinoma. Cancer Manag. Res. 2022, 14, 2903–2912. [Google Scholar] [CrossRef] [PubMed]
- Guri, Y.; Colombi, M.; Dazert, E.; Hindupur, S.K.; Roszik, J.; Moes, S.; Jenoe, P.; Heim, M.H.; Riezman, I.; Riezman, H.; et al. mTORC2 Promotes Tumorigenesis via Lipid Synthesis. Cancer Cell 2017, 32, 807–823. [Google Scholar] [CrossRef]
- Dubot, P.; Astudillo, L.; Therville, N.; Sabourdy, F.; Stirnemann, J.; Levade, T.; Andrieu-Abadie, N. Are Glucosylceramide-Related Sphingolipids Involved in the Increased Risk for Cancer in Gaucher Disease Patients? Review and Hypotheses. Cancers 2020, 12, 475. [Google Scholar] [CrossRef]
- Rosenbloom, B.E.; Cappellini, M.D.; Weinreb, N.J.; Dragosky, M.; Revel-Vilk, S.; Batista, J.L.; Sekulic, D.; Mistry, P.K. Cancer risk and gammopathies in 2123 adults with Gaucher disease type 1 in the International Gaucher Group Gaucher Registry. Am. J. Hematol. 2022, 97, 1337–1347. [Google Scholar] [CrossRef]
- Chu, Q.; Liu, P.; Song, Y.; Yang, R.; An, J.; Zhai, X.; Niu, J.; Yang, C.; Li, B. Stearate-derived very long-chain fatty acids are indispensable to tumor growth. EMBO J. 2023, 42, e114802. [Google Scholar] [CrossRef]
- Jennemann, R.; Volz, M.; Bestvater, F.; Schmidt, C.; Richter, K.; Kaden, S.; Müthing, J.; Gröne, H.-J.; Sandhoff, R. Blockade of Glycosphingolipid Synthesis Inhibits Cell Cycle and Spheroid Growth of Colon Cancer Cells In Vitro and Experimental Colon Cancer Incidence In Vivo. Int. J. Mol. Sci. 2021, 22, 10539. [Google Scholar] [CrossRef]
- Roh, J.-L.; Kim, E.H.; Park, J.Y.; Kim, J.W. Inhibition of Glucosylceramide Synthase Sensitizes Head and Neck Cancer to Cisplatin. Mol. Cancer Ther. 2015, 14, 1907–1915. [Google Scholar] [CrossRef]
- Chatterjee, S.; Alsaeedi, N.; Hou, J.; Bandaru, V.V.; Wu, L.; Halushka, M.K.; Pili, R.; Ndikuyeze, G.; Haughey, N.J. Use of a glycolipid inhibitor to ameliorate renal cancer in a mouse model. PLoS ONE 2013, 8, e63726. [Google Scholar] [CrossRef]
- Wannemacher, R.; Jubran-Rudolf, L.; Zdora, I.; Leitzen, E.; Rohn, K.; Sippel, V.; Paschen, C.; Blattmann, P.; Baumgartner, W.; Gerhauser, I.; et al. Sinbaglustat ameliorates disease pathology in a murine model of G(M1) gangliosidosis without affecting CNS ganglioside levels. Neurobiol. Dis. 2025, 210, 106917. [Google Scholar] [CrossRef]
- Roecker, A.J.; Schirripa, K.M.; Loughran, H.M.; Tong, L.; Liang, T.; Fillgrove, K.L.; Kuo, Y.; Bleasby, K.; Collier, H.; Altman, M.D.; et al. Pyrazole Ureas as Low Dose, CNS Penetrant Glucosylceramide Synthase Inhibitors for the Treatment of Parkinson’s Disease. ACS Med. Chem. Lett. 2023, 14, 146–155. [Google Scholar] [CrossRef]
- Pavlova, E.V.; Archer, J.; Wang, S.; Dekker, N.; Aerts, J.M.; Karlsson, S.; Cox, T.M. Inhibition of UDP-glucosylceramide synthase in mice prevents Gaucher disease-associated B-cell malignancy. J. Pathol. 2015, 235, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Jennemann, R.; Volz, M.; Frias-Soler, R.C.; Schulze, A.; Richter, K.; Kaden, S.; Sandhoff, R. Glucosylceramide Synthase Inhibition in Combination with Aripiprazole Sensitizes Hepatocellular Cancer Cells to Sorafenib and Doxorubicin. Int. J. Mol. Sci. 2024, 26, 304. [Google Scholar] [CrossRef] [PubMed]
- Jain, V.; Harper, S.L.; Versace, A.M.; Fingerman, D.; Brown, G.S.; Bhardwaj, M.; Crissey, M.A.S.; Goldman, A.R.; Ruthel, G.; Liu, Q.; et al. Targeting UGCG Overcomes Resistance to Lysosomal Autophagy Inhibition. Cancer Discov. 2023, 13, 454–473. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Cao, Z.; Chen, M.; Liu, Y.; Ma, X.; Lu, Y.; Zhang, Y.; Feng, K.; Zhang, Y.; Meng, Z.; et al. Inhibition of glycosphingolipid synthesis with eliglustat in combination with immune checkpoint inhibitors in advanced cancers: Preclinical evidence and phase I clinical trial. Nat. Commun. 2024, 15, 6970. [Google Scholar] [CrossRef]
- Wingerter, A.; El Malki, K.; Sandhoff, R.; Seidmann, L.; Wagner, D.C.; Lehmann, N.; Vewinger, N.; Frauenknecht, K.B.M.; Sommer, C.J.; Traub, F.; et al. Exploiting Gangliosides for the Therapy of Ewing’s Sarcoma and H3K27M-Mutant Diffuse Midline Glioma. Cancers 2021, 13, 520. [Google Scholar] [CrossRef]
- Guan, S.; Liu, Y.Y.; Yan, T.; Zhou, J. Inhibition of ceramide glucosylation sensitizes lung cancer cells to ABC294640, a first-in-class small molecule SphK2 inhibitor. Biochem. Biophys. Res. Commun. 2016, 476, 230–236. [Google Scholar] [CrossRef]
- Moreira, I.B.; Buettner, F.F.R. Glycosphingolipids as emerging attack points in bladder cancer. Discov. Oncol. 2025, 16, 569. [Google Scholar] [CrossRef]
- Ghosh, S.; Juin, S.K.; Bhattacharyya Majumdar, S.; Majumdar, S. Crucial role of glucosylceramide synthase in the regulation of stem cell-like cancer cells in B16F10 murine melanoma. Mol. Carcinog. 2021, 60, 840–858. [Google Scholar] [CrossRef]
- Lin, Z.; Hua, G.; Hu, X. Lipid metabolism associated crosstalk: The bidirectional interaction between cancer cells and immune/stromal cells within the tumor microenvironment for prognostic insight. Cancer Cell Int. 2024, 24, 295. [Google Scholar] [CrossRef]
- Ma, Q.; Kang, R.; Xu, R.; Guan, Y.; Chang, S.; Li, S. Crosstalk between stromal, immune, and ovarian cancer cells in lipid-rich tumor microenvironment exhibits proliferative features. Front. Immunol. 2025, 16, 1614815. [Google Scholar] [CrossRef] [PubMed]
- Seager, R.J.; Hajal, C.; Spill, F.; Kamm, R.D.; Zaman, M.H. Dynamic interplay between tumour, stroma and immune system can drive or prevent tumour progression. Converg. Sci. Phys. Oncol. 2017, 3, 034002. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef]
- Companioni, O.; Mir, C.; Garcia-Mayea, Y.; ME, L.L. Targeting Sphingolipids for Cancer Therapy. Front. Oncol. 2021, 11, 745092. [Google Scholar] [CrossRef]
- Lewis, A.C.; Wallington-Beddoe, C.T.; Powell, J.A.; Pitson, S.M. Targeting sphingolipid metabolism as an approach for combination therapies in haematological malignancies. Cell Death Discov. 2018, 4, 72. [Google Scholar] [CrossRef] [PubMed]
- Janneh, A.H.; Ogretmen, B. Targeting Sphingolipid Metabolism as a Therapeutic Strategy in Cancer Treatment. Cancers 2022, 14, 2183. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Ge, J.; He, B.; Zeng, B. Glycosphingolipids in Filamentous Fungi: Biological Roles and Potential Applications in Cosmetics and Health Foods. Front. Microbiol. 2021, 12, 690211. [Google Scholar] [CrossRef] [PubMed]
- Yong, T.L.; Zaman, R.; Rehman, N.; Tan, C.K. Ceramides and Skin Health: New Insights. Exp. Dermatol. 2025, 34, e70042. [Google Scholar] [CrossRef]
- Borodzicz, S.; Rudnicka, L.; Mirowska-Guzel, D.; Cudnoch-Jedrzejewska, A. The role of epidermal sphingolipids in dermatologic diseases. Lipids Health Dis. 2016, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Jennemann, R.; Sandhoff, R.; Langbein, L.; Kaden, S.; Rothermel, U.; Gallala, H.; Sandhoff, K.; Wiegandt, H.; Gröne, H.-J. Integrity and Barrier Function of the Epidermis Critically Depend on Glucosylceramide Synthesis. J. Biol. Chem. 2007, 282, 3083–3094. [Google Scholar] [CrossRef]
- Hülsmeier, A.J. Glycosphingolipids in neurodegeneration–Molecular mechanisms, cellular roles, and therapeutic perspectives. Neurobiol. Dis. 2025, 207, 106851. [Google Scholar] [CrossRef]
- Zhang, T.; de Waard, A.A.; Wuhrer, M.; Spaapen, R.M. The Role of Glycosphingolipids in Immune Cell Functions. Front. Immunol. 2019, 10, 90. [Google Scholar] [CrossRef]
- Ilan, Y. β-Glycosphingolipids as Mediators of Both Inflammation and Immune Tolerance: A Manifestation of Randomness in Biological Systems. Front. Immunol. 2019, 10, 1143. [Google Scholar] [CrossRef]
- Nakayama, H.; Nagafuku, M.; Suzuki, A.; Iwabuchi, K.; Inokuchi, J.-I. The regulatory roles of glycosphingolipid-enriched lipid rafts in immune systems. FEBS Lett. 2018, 592, 3921–3942. [Google Scholar] [CrossRef]
- Hua, Y.; Zhang, G.; Liu, Y.; Tian, X.; Zhang, X.; Song, G.; Tian, Q.; Yin, F. Regulation of sphingolipid metabolism in the immune microenvironment of gastric cancer: Current insights and future directions. Front. Oncol. 2025, 15, 1604227. [Google Scholar] [CrossRef]
- Soula, M.; Unlu, G.; Welch, R.; Chudnovskiy, A.; Uygur, B.; Shah, V.; Alwaseem, H.; Bunk, P.; Subramanyam, V.; Yeh, H.W.; et al. Glycosphingolipid synthesis mediates immune evasion in KRAS-driven cancer. Nature 2024, 633, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Gonzalez, M.; Angelia, J.; Piroozmand, S.; Jamehdor, S.; Behrooz, A.B.; Latifi-Navid, H.; Ahmadi, M.; Pecic, S. Lipidomics-driven drug discovery and delivery strategies in glioblastoma. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2025, 1871, 167637. [Google Scholar] [CrossRef]
- Zhakupova, A.; Zeinolla, A.; Kokabi, K.; Sergazy, S.; Aljofan, M. Drug Resistance: The Role of Sphingolipid Metabolism. Int. J. Mol. Sci. 2025, 26, 3716. [Google Scholar] [CrossRef] [PubMed]
- Wajapeyee, N.; Beamon, T.C.; Gupta, R. Roles and therapeutic targeting of ceramide metabolism in cancer. Mol. Metab. 2024, 83, 101936. [Google Scholar] [CrossRef]
- Jennemann, R.; Kaden, S.; Sandhoff, R.; Nordström, V.; Wang, S.; Volz, M.; Robine, S.; Amen, N.; Rothermel, U.; Wiegandt, H.; et al. Glycosphingolipids are essential for intestinal endocytic function. J. Biol. Chem. 2012, 287, 32598–32616. [Google Scholar] [CrossRef]
- Wang, H.; Jin, X.; Zhang, Y.; Wang, Z.; Zhang, T.; Xu, J.; Shen, J.; Zan, P.; Sun, M.; Wang, C.; et al. Inhibition of sphingolipid metabolism in osteosarcoma protects against CD151-mediated tumorigenicity. Cell Biosci. 2022, 12, 169. [Google Scholar] [CrossRef]
- Hughes, D.A.; Duke, V.M.; Baker, R.J.; Wright, F.; Foroni, L.; Atul, M.B. High Prevalence of B-Cell Clonality in Patients with Gaucher’s Disease. Blood 2004, 104, 2391. [Google Scholar] [CrossRef]
- Cox, T.M.; Rosenbloom, B.E.; Barker, R.A. Gaucher disease and comorbidities: B-cell malignancy and parkinsonism. Am. J. Hematol. 2015, 90 (Suppl. S1), S25–S28. [Google Scholar] [CrossRef]
- Ducatez, F.; Berger, M.G.; Pilon, C.; Plichet, T.; Lesueur, C.; Berger, J.; Belmatoug, N.; Marret, S.; Bekri, S.; Tebani, A. Deciphering metabolic shifts in Gaucher disease type 1: A multi-omics study. J. Mol. Med. 2025, 103, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Bonesteele, G.; Gargus, J.J.; Curtin, E.; Tang, M.; Rosenbloom, B.; Kimonis, V. Diffuse large B-cell non-Hodgkin’s lymphoma in Gaucher disease. Mol. Genet. Metab. Rep. 2020, 25, 100663. [Google Scholar] [CrossRef] [PubMed]
- Rosenbloom, B.E.; Weinreb, N.J.; Zimran, A.; Kacena, K.A.; Charrow, J.; Ward, E. Gaucher disease and cancer incidence: A study from the Gaucher Registry. Blood 2005, 105, 4569–4572. [Google Scholar] [CrossRef]
- Khamrui, E.; Banerjee, S.; Mukherjee, D.D.; Biswas, K. Emerging role of MAPK signaling in glycosphingolipid-associated tumorigenesis. Glycoconj. J. 2024, 41, 343–360. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Que, H.; Li, Q.; Wei, X. Wnt/β-catenin mediated signaling pathways in cancer: Recent advances, and applications in cancer therapy. Mol. Cancer 2025, 24, 171. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, T.; Sato, T.; Natsuka, S.; Kobayashi, Y.; Zhou, D.; Shinkai, T.; Hayakawa, S.; Furukawa, K. Involvement of murine β-1,4-galactosyltransferase V in lactosylceramide biosynthesis. Glycoconj. J. 2010, 27, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Kartal Yandim, M.; Apohan, E.; Baran, Y. Therapeutic potential of targeting ceramide/glucosylceramide pathway in cancer. Cancer Chemother. Pharmacol. 2013, 71, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Hill, R.A.; Li, Y.T. Ceramide glycosylation catalyzed by glucosylceramide synthase and cancer drug resistance. Adv. Cancer Res. 2013, 117, 59–89. [Google Scholar] [CrossRef] [PubMed]
- Moro, K.; Ichikawa, H.; Koyama, Y.; Abe, S.; Uchida, H.; Naruse, K.; Obata, Y.; Tsuchida, J.; Toshikawa, C.; Ikarashi, M.; et al. Oral Administration of Glucosylceramide Suppresses Tumor Growth by Affecting the Ceramide/Sphingosine-1-Phosphate Balance in Breast Cancer Tissue. World J. Oncol. 2023, 14, 430–437. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Han, T.Y.; Yu, J.Y.; Bitterman, A.; Le, A.; Giuliano, A.E.; Cabot, M.C. Oligonucleotides blocking glucosylceramide synthase expression selectively reverse drug resistance in cancer cells. J. Lipid Res. 2004, 45, 933–940. [Google Scholar] [CrossRef]
- Stathem, M.; Marimuthu, S.; O’Neal, J.; Rathmell, J.C.; Chesney, J.A.; Beverly, L.J.; Siskind, L.J. Glucose availability and glycolytic metabolism dictate glycosphingolipid levels. J. Cell Biochem. 2015, 116, 67–80. [Google Scholar] [CrossRef]
- Mostaq, M.S.; Kang, L.; Patwardhan, G.A.; Zhao, Y.; Shi, R.; Liu, Y.-Y. Glucosylceramide Synthase, a Key Enzyme in Sphingolipid Metabolism, Regulates Expression of Genes Accounting for Cancer Drug Resistance. Int. J. Mol. Sci. 2025, 26, 5112. [Google Scholar] [CrossRef]
- Varela, A.R.P.; Gonçalves da Silva, A.M.P.S.; Fedorov, A.; Futerman, A.H.; Prieto, M.; Silva, L.C. Effect of glucosylceramide on the biophysical properties of fluid membranes. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2013, 1828, 1122–1130. [Google Scholar] [CrossRef][Green Version]
- Raj, S.; Nazemidashtarjandi, S.; Kim, J.; Joffe, L.; Zhang, X.; Singh, A.; Mor, V.; Desmarini, D.; Djordjevic, J.; Raleigh, D.P.; et al. Changes in glucosylceramide structure affect virulence and membrane biophysical properties of Cryptococcus neoformans. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2224–2233. [Google Scholar] [CrossRef]
- Straus, A.J.; Mavodza, G.; Senkal, C.E. Glycosylation of ceramide synthase 6 is required for its activity. J. Lipid Res. 2025, 66, 100715. [Google Scholar] [CrossRef]
- Obeid, L.M.; Hannun, Y.A. Ceramide: A stress signal and mediator of growth suppression and apoptosis. J. Cell Biochem. 1995, 58, 191–198. [Google Scholar] [CrossRef]
- Zhu, X.F.; Liu, Z.C.; Xie, B.F.; Feng, G.K.; Zeng, Y.X. Ceramide induces cell cycle arrest and upregulates p27kip in nasopharyngeal carcinoma cells. Cancer Lett. 2003, 193, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Morjani, H.; Aouali, N.; Belhoussine, R.; Veldman, R.J.; Levade, T.; Manfait, M. Elevation of glucosylceramide in multidrug-resistant cancer cells and accumulation in cytoplasmic droplets. Int. J. Cancer 2001, 94, 157–165. [Google Scholar] [CrossRef]
- Lavie, Y.; Cao, H.; Bursten, S.L.; Giuliano, A.E.; Cabot, M.C. Accumulation of glucosylceramides in multidrug-resistant cancer cells. J. Biol. Chem. 1996, 271, 19530–19536. [Google Scholar] [CrossRef]
- van Vlerken, L.E.; Duan, Z.; Little, S.R.; Seiden, M.V.; Amiji, M.M. Augmentation of therapeutic efficacy in drug-resistant tumor models using ceramide coadministration in temporal-controlled polymer-blend nanoparticle delivery systems. AAPS J. 2010, 12, 171–180. [Google Scholar] [CrossRef]
- Morad, S.A.; Levin, J.C.; Shanmugavelandy, S.S.; Kester, M.; Fabrias, G.; Bedia, C.; Cabot, M.C. Ceramide--antiestrogen nanoliposomal combinations--novel impact of hormonal therapy in hormone-insensitive breast cancer. Mol. Cancer Ther. 2012, 11, 2352–2361. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Gupta, V.; Patwardhan, G.A.; Bhinge, K.; Zhao, Y.; Bao, J.; Mehendale, H.; Cabot, M.C.; Li, Y.T.; Jazwinski, S.M. Glucosylceramide synthase upregulates MDR1 expression in the regulation of cancer drug resistance through cSrc and β-catenin signaling. Mol. Cancer 2010, 9, 145. [Google Scholar] [CrossRef]
- La Monica, S.; Vacondio, F.; Eltayeb, K.; Lodola, A.; Volta, F.; Viglioli, M.; Ferlenghi, F.; Galvani, F.; Galetti, M.; Bonelli, M.; et al. Targeting glucosylceramide synthase induces antiproliferative and proapoptotic effects in osimertinib-resistant NSCLC cell models. Sci. Rep. 2024, 14, 6491. [Google Scholar] [CrossRef]
- Stefanovic, M.; Tutusaus, A.; Martinez-Nieto, G.A.; Bárcena, C.; de Gregorio, E.; Moutinho, C.; Barbero-Camps, E.; Villanueva, A.; Colell, A.; Marí, M.; et al. Targeting glucosylceramide synthase upregulation reverts sorafenib resistance in experimental hepatocellular carcinoma. Oncotarget 2016, 7, 8253–8267. [Google Scholar] [CrossRef] [PubMed]
- Tyler, A.; Johansson, A.; Karlsson, T.; Gudey, S.K.; Brännström, T.; Grankvist, K.; Behnam-Motlagh, P. Targeting glucosylceramide synthase induction of cell surface globotriaosylceramide (Gb3) in acquired cisplatin-resistance of lung cancer and malignant pleural mesothelioma cells. Exp. Cell Res. 2015, 336, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Medatwal, N.; Kumar, S.; Kar, A.; Komalla, V.; Yavvari, P.S.; Mishra, D.; Rizvi, Z.A.; Nandan, S.; Malakar, D.; et al. A Localized Chimeric Hydrogel Therapy Combats Tumor Progression through Alteration of Sphingolipid Metabolism. ACS Cent. Sci. 2019, 5, 1648–1662. [Google Scholar] [CrossRef] [PubMed]
- Aizaz, M.; Khan, A.; Khan, F.; Khan, M.; Musad Saleh, E.A.; Nisar, M.; Baran, N. The cross-talk between macrophages and tumor cells as a target for cancer treatment. Front. Oncol. 2023, 13, 1259034. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Uikey, B.N.; Rathore, S.S.; Gupta, P.; Kashyap, D.; Kumar, C.; Shukla, D.; Vijayamahantesh; Chandel, A.S.; Ahirwar, B.; et al. Role of cytokine in malignant T-cell metabolism and subsequent alternation in T-cell tumor microenvironment. Front. Oncol. 2023, 13, 1235711. [Google Scholar] [CrossRef]
- Zeng, W.; Li, F.; Jin, S.; Ho, P.-C.; Liu, P.-S.; Xie, X. Functional polarization of tumor-associated macrophages dictated by metabolic reprogramming. J. Exp. Clin. Cancer Res. 2023, 42, 245. [Google Scholar] [CrossRef]
- Wang, J.; He, Y.; Hu, F.; Hu, C.; Sun, Y.; Yang, K.; Yang, S. Metabolic Reprogramming of Immune Cells in the Tumor Microenvironment. Int. J. Mol. Sci. 2024, 25, 12223. [Google Scholar] [CrossRef]
- Pascual, G.; Benitah, S.A. Lipids in the tumor microenvironment: Immune modulation and metastasis. Front. Oncol. 2024, 14, 1435480. [Google Scholar] [CrossRef] [PubMed]
- Wegner, M.S.; Schömel, N.; Olzomer, E.M.; Trautmann, S.; Olesch, C.; Byrne, F.L.; Brüne, B.; Gurke, R.; Ferreirós, N.; Weigert, A.; et al. Increased glucosylceramide production leads to decreased cell energy metabolism and lowered tumor marker expression in non-cancerous liver cells. Cell Mol. Life Sci. 2021, 78, 7025–7041. [Google Scholar] [CrossRef]
- Netea-Maier, R.T.; Smit, J.W.A.; Netea, M.G. Metabolic changes in tumor cells and tumor-associated macrophages: A mutual relationship. Cancer Lett. 2018, 413, 102–109. [Google Scholar] [CrossRef]
- Jin, H.-R.; Wang, J.; Wang, Z.-J.; Xi, M.-J.; Xia, B.-H.; Deng, K.; Yang, J.-L. Lipid metabolic reprogramming in tumor microenvironment: From mechanisms to therapeutics. J. Hematol. Oncol. 2023, 16, 103. [Google Scholar] [CrossRef]
- Broadfield, L.A.; Pane, A.A.; Talebi, A.; Swinnen, J.V.; Fendt, S.-M. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Dev. Cell 2021, 56, 1363–1393. [Google Scholar] [CrossRef]
- Zhang, M.; Wei, T.; Zhang, X.; Guo, D. Targeting lipid metabolism reprogramming of immunocytes in response to the tumor microenvironment stressor: A potential approach for tumor therapy. Front. Immunol. 2022, 13, 937406. [Google Scholar] [CrossRef]
- Weber, C.E.; Kuo, P.C. The tumor microenvironment. Surg. Oncol. 2012, 21, 172–177. [Google Scholar] [CrossRef]
- Huang, R.; Kang, T.; Chen, S. The role of tumor-associated macrophages in tumor immune evasion. J. Cancer Res. Clin. Oncol. 2024, 150, 238. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.H.; Beury, D.W.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cells: Critical Cells Driving Immune Suppression in the Tumor Microenvironment. Adv. Cancer Res. 2015, 128, 95–139. [Google Scholar] [CrossRef]
- Li, K.; Shi, H.; Zhang, B.; Ou, X.; Ma, Q.; Chen, Y.; Shu, P.; Li, D.; Wang, Y. Myeloid-derived suppressor cells as immunosuppressive regulators and therapeutic targets in cancer. Signal Transduct. Target. Ther. 2021, 6, 362. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, M.; Do, M.H.; Chou, C.; Stamatiades, E.G.; Nixon, B.G.; Shi, W.; Zhang, X.; Li, P.; Gao, S. Cancer immunotherapy via targeted TGF-β signalling blockade in TH cells. Nature 2020, 587, 121–125. [Google Scholar] [CrossRef]
- Aguilera, K.Y.; Rivera, L.B.; Hur, H.; Carbon, J.G.; Toombs, J.E.; Goldstein, C.D.; Dellinger, M.T.; Castrillon, D.H.; Brekken, R.A. Collagen signaling enhances tumor progression after anti-VEGF therapy in a murine model of pancreatic ductal adenocarcinoma. Cancer Res. 2014, 74, 1032–1044. [Google Scholar] [CrossRef]
- Veglia, F.; Perego, M.; Gabrilovich, D. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 2018, 19, 108–119. [Google Scholar] [CrossRef]
- Salah, N.Y. Vascular endothelial growth factor (VEGF), tissue inhibitors of metalloproteinase-1 (TIMP-1) and nail fold capillaroscopy changes in children and adolescents with Gaucher disease; relation to residual disease severity. Cytokine 2020, 133, 155120. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Sig. Transduct. Target Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Nakamura, K.; Smyth, M.J. Myeloid immunosuppression and immune checkpoints in the tumor microenvironment. Cell. Mol. Immunol. 2020, 17, 1–12. [Google Scholar] [CrossRef]
- Pandey, M.K.; Grabowski, G.A. Cytology of Gaucher Disease; Taylor & Francis: Abingdon, UK, 2013. [Google Scholar]
- Campeau, P.M.; Rafei, M.; Boivin, M.N.; Sun, Y.; Grabowski, G.A.; Galipeau, J. Characterization of Gaucher disease bone marrow mesenchymal stromal cells reveals an altered inflammatory secretome. Blood 2009, 114, 3181–3190. [Google Scholar] [CrossRef] [PubMed]
- Boven, L.A.; van Meurs, M.; Boot, R.G.; Mehta, A.; Boon, L.; Aerts, J.M.; Laman, J.D. Gaucher cells demonstrate a distinct macrophage phenotype and resemble alternatively activated macrophages. Am. J. Clin. Pathol. 2004, 122, 359–369. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dubot, P.; Astudillo, L.; Therville, N.; Carrié, L.; Pettazzoni, M.; Cheillan, D.; Stirnemann, J.; Levade, T.; Andrieu-Abadie, N.; Sabourdy, F. Potential Role of Sphingolipidoses-Associated Lysosphingolipids in Cancer. Cancers 2022, 14, 4858. [Google Scholar] [CrossRef]
- Wang, M.; Xiao, Y.; Miao, J.; Zhang, X.; Liu, M.; Zhu, L.; Liu, H.; Shen, X.; Wang, J.; Xie, B.; et al. Oxidative Stress and Inflammation: Drivers of Tumorigenesis and Therapeutic Opportunities. Antioxidants 2025, 14, 735. [Google Scholar] [CrossRef]
- Neganova, M.; Liu, J.; Aleksandrova, Y.; Klochkov, S.; Fan, R. Therapeutic Influence on Important Targets Associated with Chronic Inflammation and Oxidative Stress in Cancer Treatment. Cancers 2021, 13, 6062. [Google Scholar] [CrossRef] [PubMed]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Augustin, R.C.; Delgoffe, G.M.; Najjar, Y.G. Characteristics of the Tumor Microenvironment That Influence Immune Cell Functions: Hypoxia, Oxidative Stress, Metabolic Alterations. Cancers 2020, 12, 3802. [Google Scholar] [CrossRef]
- Mistry, P.K.; Taddei, T.; vom Dahl, S.; Rosenbloom, B.E. Gaucher disease and malignancy: A model for cancer pathogenesis in an inborn error of metabolism. Crit. Rev. Oncog. 2013, 18, 235–246. [Google Scholar] [CrossRef]
- Choy, F.Y.; Campbell, T.N. Gaucher disease and cancer: Concept and controversy. Int. J. Cell Biol. 2011, 2011, 150450. [Google Scholar] [CrossRef]
- Revel-Vilk, S.; Zimran, A.; Istaiti, M.; Azani, L.; Shalev, V.; Chodick, G.; Manor, O.; Paltiel, O. Cancer Risk in Patients with Gaucher Disease Using Real-World Data. J. Clin. Med. 2023, 12, 7707. [Google Scholar] [CrossRef] [PubMed]
- Jennemann, R.; Federico, G.; Mathow, D.; Rabionet, M.; Rampoldi, F.; Popovic, Z.V.; Volz, M.; Hielscher, T.; Sandhoff, R.; Gröne, H.J. Inhibition of hepatocellular carcinoma growth by blockade of glycosphingolipid synthesis. Oncotarget 2017, 8, 109201–109216. [Google Scholar] [CrossRef]
- Huang, F.; Cai, F.; Dahabieh, M.S.; Gunawardena, K.; Talebi, A.; Dehairs, J.; El-Turk, F.; Park, J.Y.; Li, M.; Goncalves, C.; et al. Peroxisome disruption alters lipid metabolism and potentiates antitumor response with MAPK-targeted therapy in melanoma. J. Clin. Investig. 2023, 133, e166644. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shen, Y.; Chen, C.; Sui, X.; Yang, J.; Wang, L.; Zhou, J. The crosstalk between autophagy and ferroptosis: What can we learn to target drug resistance in cancer? Cancer Biol. Med. 2019, 16, 630–646. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Tang, D. Autophagy and Ferroptosis-What’s the Connection? Curr. Pathobiol. Rep. 2017, 5, 153–159. [Google Scholar] [CrossRef]
- Lee, S.; Hwang, N.; Seok, B.G.; Lee, S.; Lee, S.J.; Chung, S.W. Autophagy mediates an amplification loop during ferroptosis. Cell Death Dis. 2023, 14, 464. [Google Scholar] [CrossRef]
- Eskander, G.; Abdelhamid, S.G.; Wahdan, S.A.; Radwan, S.M. Insights on the crosstalk among different cell death mechanisms. Cell Death Discov. 2025, 11, 56. [Google Scholar] [CrossRef]
- Bleicher, R.J.; Cabot, M.C. Glucosylceramide synthase and apoptosis. Biochim. Biophys. Acta 2002, 1585, 172–178. [Google Scholar] [CrossRef]
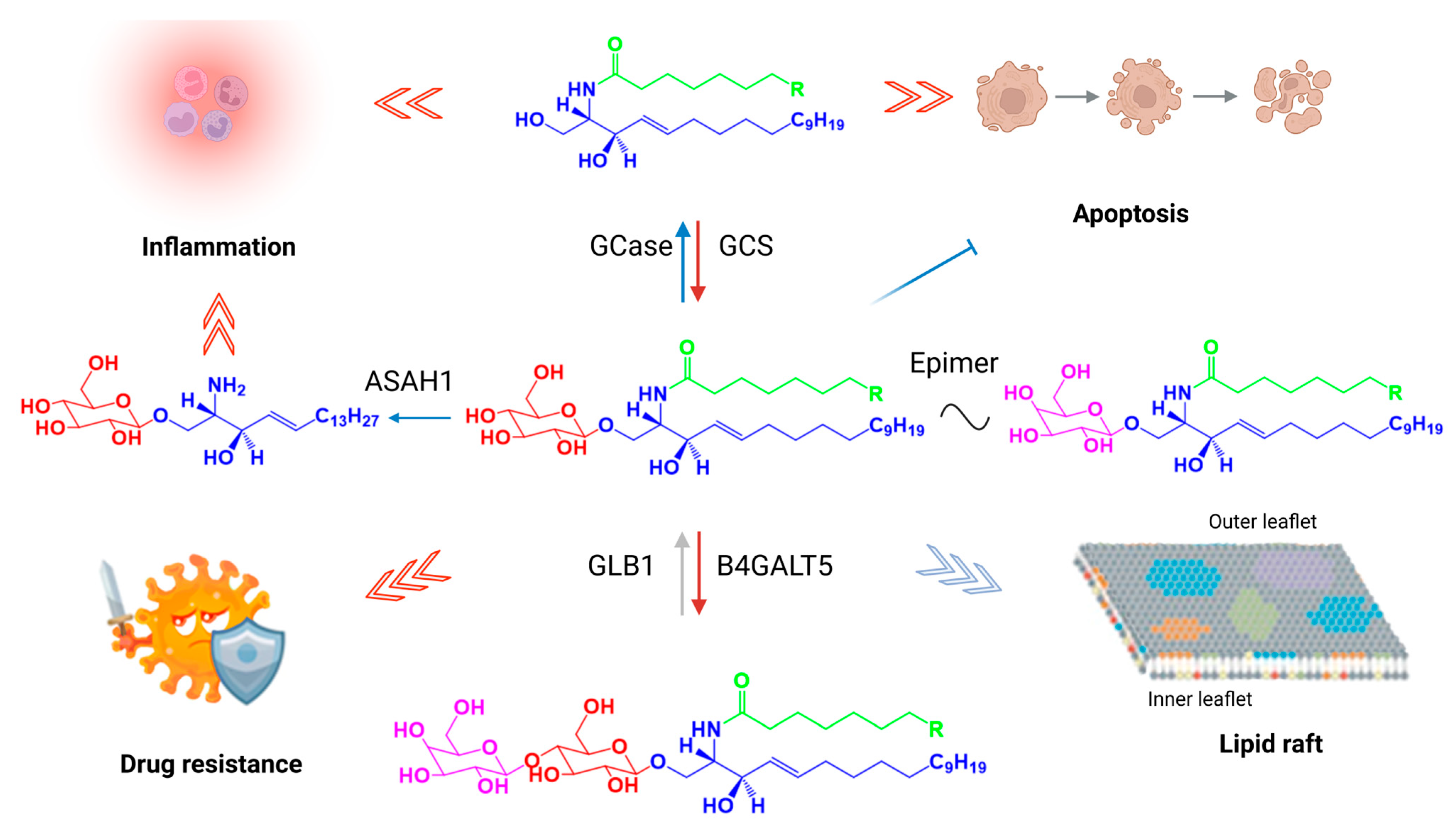
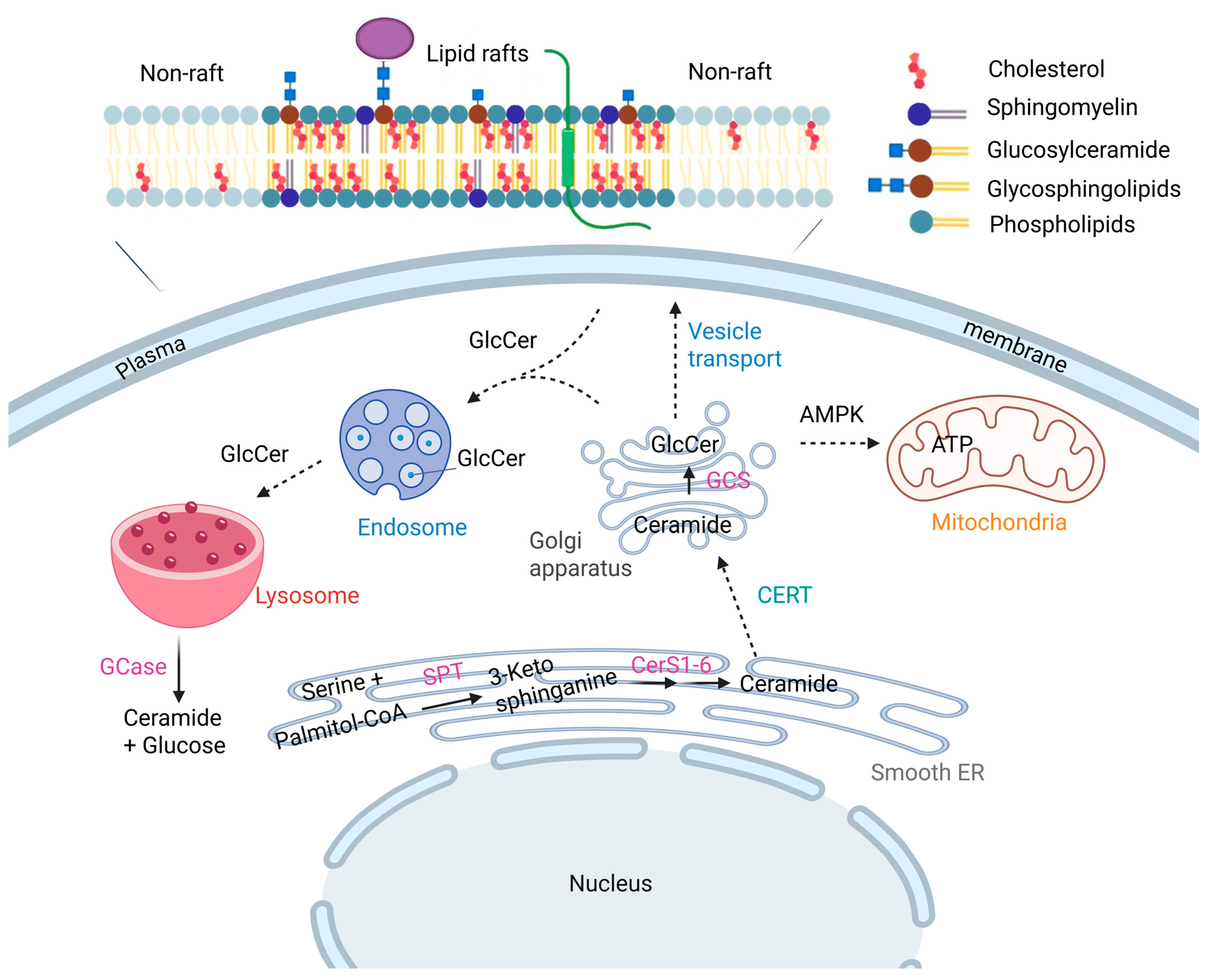
| Enzyme | UniProt ID (Human) | Mammalian Gene | Biochemical Function | References |
|---|---|---|---|---|
| Ceramide glucosyltranferase | Q16739 (EC 2.4.1.80) | UGCG (GCS) | Catalyzes the first glycosylation step in glycosphingolipid biosynthesis by transferring glucose from UDP-glucose to ceramide to form GlcCer. | [17,18] |
| Glucocerebrosidase (glucosylceramidase) | P04062 (EC 3.2.1.45) | GBA1 | Hydrolyzes the β-glucosidic linkage of GlcCer to yield glucose and ceramide. | [19,20,21,22,23,24] |
| Acid ceramidase | Q13510 (EC 3.5.1.23) | ASAH1 | Hydrolyzes ceramide into sphingosine and free fatty acids, thereby regulating sphingolipid homeostasis. | [24,25,26,27] |
| Galactosyltransferase orβ-1,4-galactosyltransferase 5 | Q9UBV7 (EC 2.4.1.38) | B4GALT5 | Transfers galactose from UDP-galactose to GlcCer to generate lactosylceramide (LacCer), a precursor for complex glycosphingolipids. | [28,29] |
| β-galactosidase | P16278 (EC 3.2.1.23) | GLB1 | Lysosomal hydrolase that cleaves terminal β-galactose residues from glycoconjugates including LacCer and ganglioside GM1. | [24,30,31,32,33] |
| GlcCer Species | Sample Types | Signaling Pathway/Metabolic Dysregulation | Cancer Type | References |
|---|---|---|---|---|
| C16:0, C18:1, C18:0, total | Breast cancer tissues and cells | Glutamine metabolism, NF-κB and Wnt/β-catenin pathway | Breast cancer | [75,76] |
| C16:0, C24:1 | Leukemic B-cell, human plasma | Pro-proliferation, mTOR, E2F1 | Chronic lymphocytic leukemia | [77,78] |
| C24:1 | Human plasma | Facilitate cancer cell survival | Oral squamous cell carcinoma (OSCC) | [79] |
| C18:0 | Human liver cancer cells and tissue | Activation of the Wnt/β-catenin pathway | Liver cancer | [80] |
| Very-long-chain (C ≥ 22) | Mouse colon epithelial cell | Activation of the β-catenin pathway | Colon cancer | [81,82,83] |
| Total | Human cervical cancer tissue and cell line | Activation of the PI3K/AKT pathway | Cervical cancer | [84] |
| C12:0 | Human liver tissue | Early-stage cancer biomarker | Intrahepatic Cholangiocarcinoma | [85] |
| Total | Mouse liver tissue | mTOR pathway | Hepatocellular carcinoma (HCC) | [86] |
| Total | Gaucher cells | Systemic inflammation with infiltration of immune cells | Melanoma in GD | [87,88] |
| GCS Inhibitor | Co-Treatment | Disease Treated | GlcCer Level | Disease Phenotype Change | References |
|---|---|---|---|---|---|
| GENZ-123346 | None | Colon Cancer | Reduced | Significant lower tumor incidence in Genz-treated mice | [90] |
| threo-PPMP | Cisplatin | Head and neck cancer | Substrate ceramide increased | Sensitizes HNC preclinical tumor xenograft mouse model to cisplatin treatment | [91] |
| D-PDMP | None | Renal cancer | GlcCer increased and LacCer reduced | Marked reduction in tumor volume | [92] |
| Sinbaglustat | None | GM1 Gangliosidosis | Reduced in periphery | Decreased axonal damage and astrogliosis | [93] |
| Pyrazole Urea | None | Parkinson’s Disease | Reduced in CNS | Rescued lysosomal activity deficit and increased lysosomal hydrolysis activity | [94] |
| Eliglustat tartrate | None | GD | Reduced | Prevents GD-associated B-cell malignancy | [95] |
| GCS Inhibitor | Co-Treatment | Disease Treated | GlcCer Level | Disease Phenotype Change | References |
|---|---|---|---|---|---|
| Miglustat | None | Colon cancer | Reduced | Marked arrest of the cell cycle in human colon carcinoma cells | [90] |
| threo-PPMP | Cisplatin | Head and neck cancer | N/A (substrate ceramide increased) | Increased cisplatin-induced cell death in HNC cells | [91] |
| GENZ-123346 | Aripiprazole/cytostatic drugs | Hepatocellular carcinoma (HCC) | Reduced | Sensitizes HCC cells in therapy | [96] |
| Eliglustat | LAI | Melanoma | Reduced | Significantly inhibited tumor growth | [97] |
| Eliglustat | Anti-PD-1 antibody | Hematological malignancies and solid tumors | Reduced | Restore HLA antigen presentation | [98] |
| Eliglustat | None | Diffuse midline glioma (DMG) | N/A | Inhibited the proliferation of primary DMG cells | [99] |
| PDMP | SphK2 inhibitor | Lung cancer | N/A | Sensitize lung cancer cells to treatment | [100] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Pandey, M.K. Central Roles of Glucosylceramide in Driving Cancer Pathogenesis. Int. J. Mol. Sci. 2025, 26, 9879. https://doi.org/10.3390/ijms26209879
Zhao X, Pandey MK. Central Roles of Glucosylceramide in Driving Cancer Pathogenesis. International Journal of Molecular Sciences. 2025; 26(20):9879. https://doi.org/10.3390/ijms26209879
Chicago/Turabian StyleZhao, Xueheng, and Manoj Kumar Pandey. 2025. "Central Roles of Glucosylceramide in Driving Cancer Pathogenesis" International Journal of Molecular Sciences 26, no. 20: 9879. https://doi.org/10.3390/ijms26209879
APA StyleZhao, X., & Pandey, M. K. (2025). Central Roles of Glucosylceramide in Driving Cancer Pathogenesis. International Journal of Molecular Sciences, 26(20), 9879. https://doi.org/10.3390/ijms26209879







