Computer-Aided Molecular Design Meets Network Toxicology and Molecular Docking: A Joint Strategy to Explore Common Molecular Mechanisms of Phthalates on Human Breast Cancer and Structure–Activity Relationship
Abstract
1. Introduction
2. Results
2.1. Common Target Genes of Three PAEs
2.2. Functional Enrichment Analysis
2.3. Protein–Protein Interaction Network (PPIN)
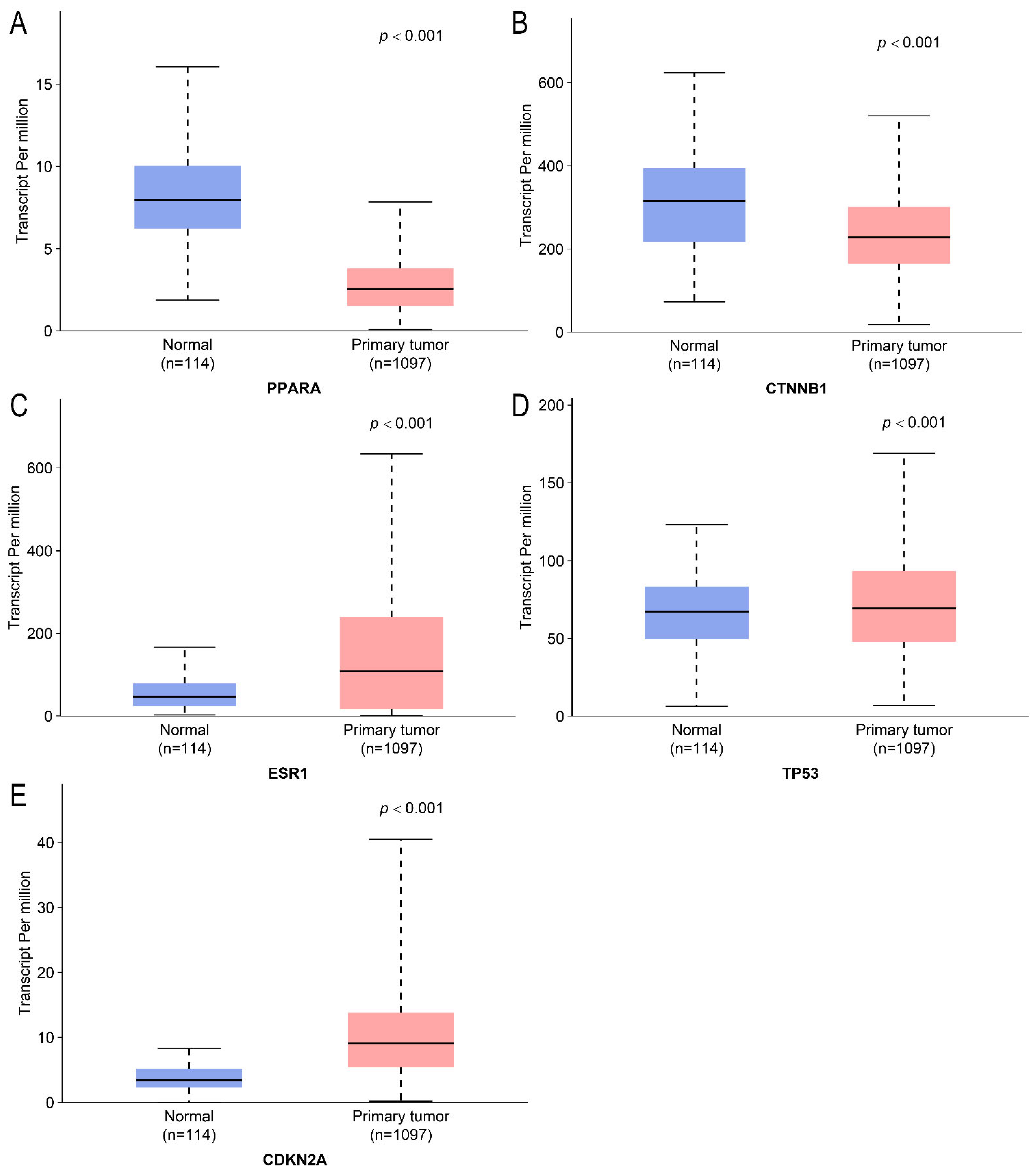
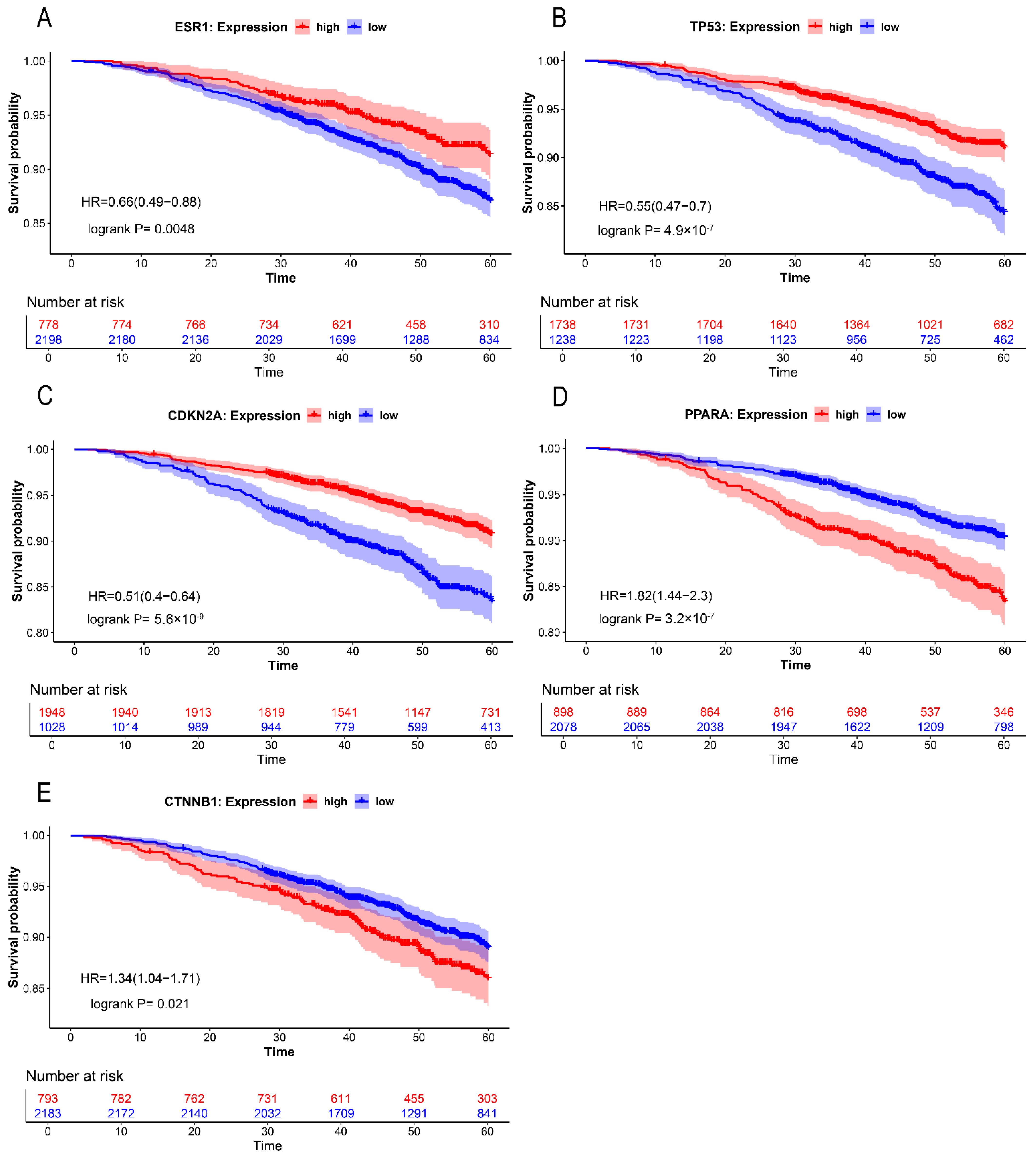
2.4. Differential Expression Analysis and Survival Analysis
2.5. Molecular Docking
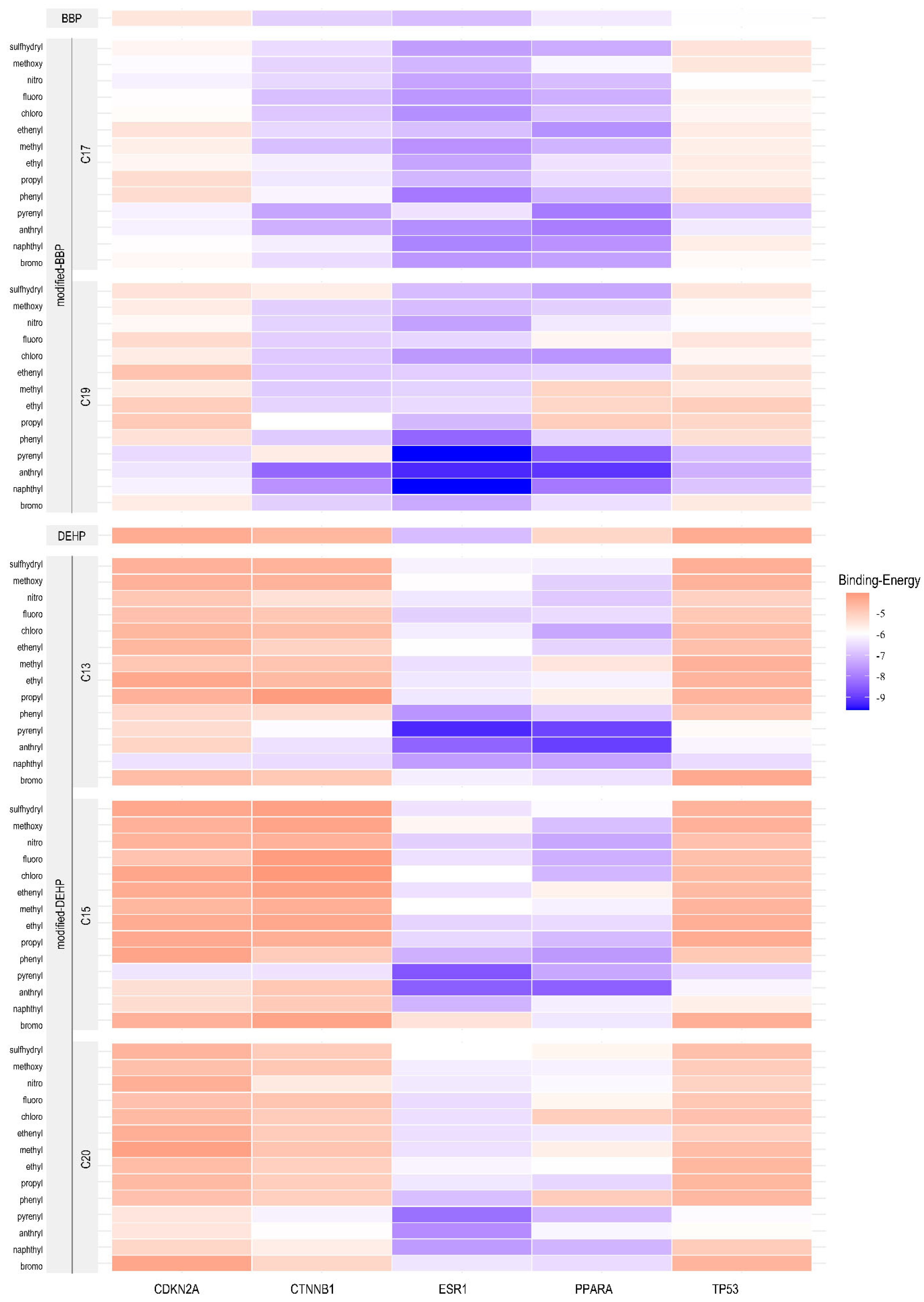
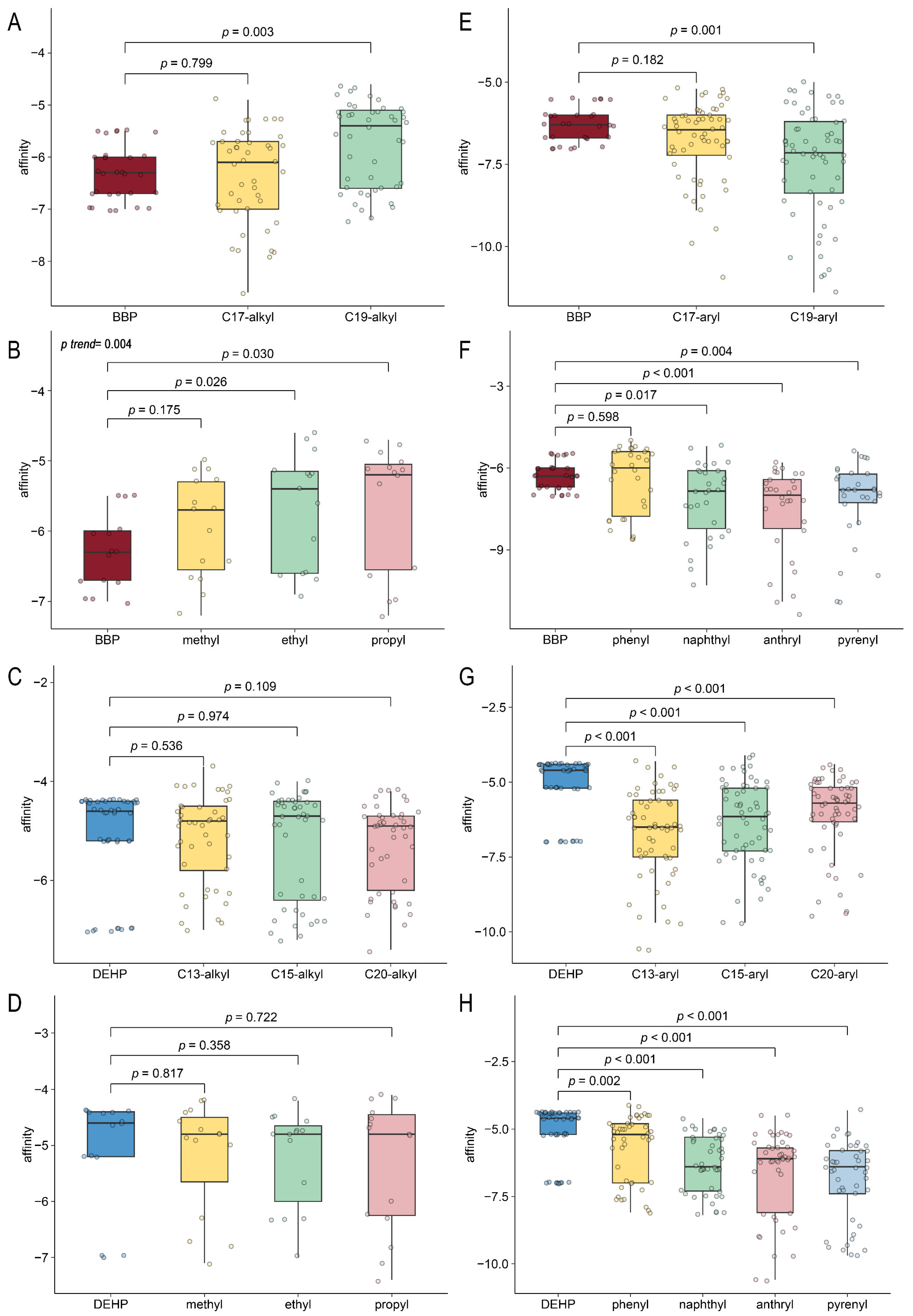
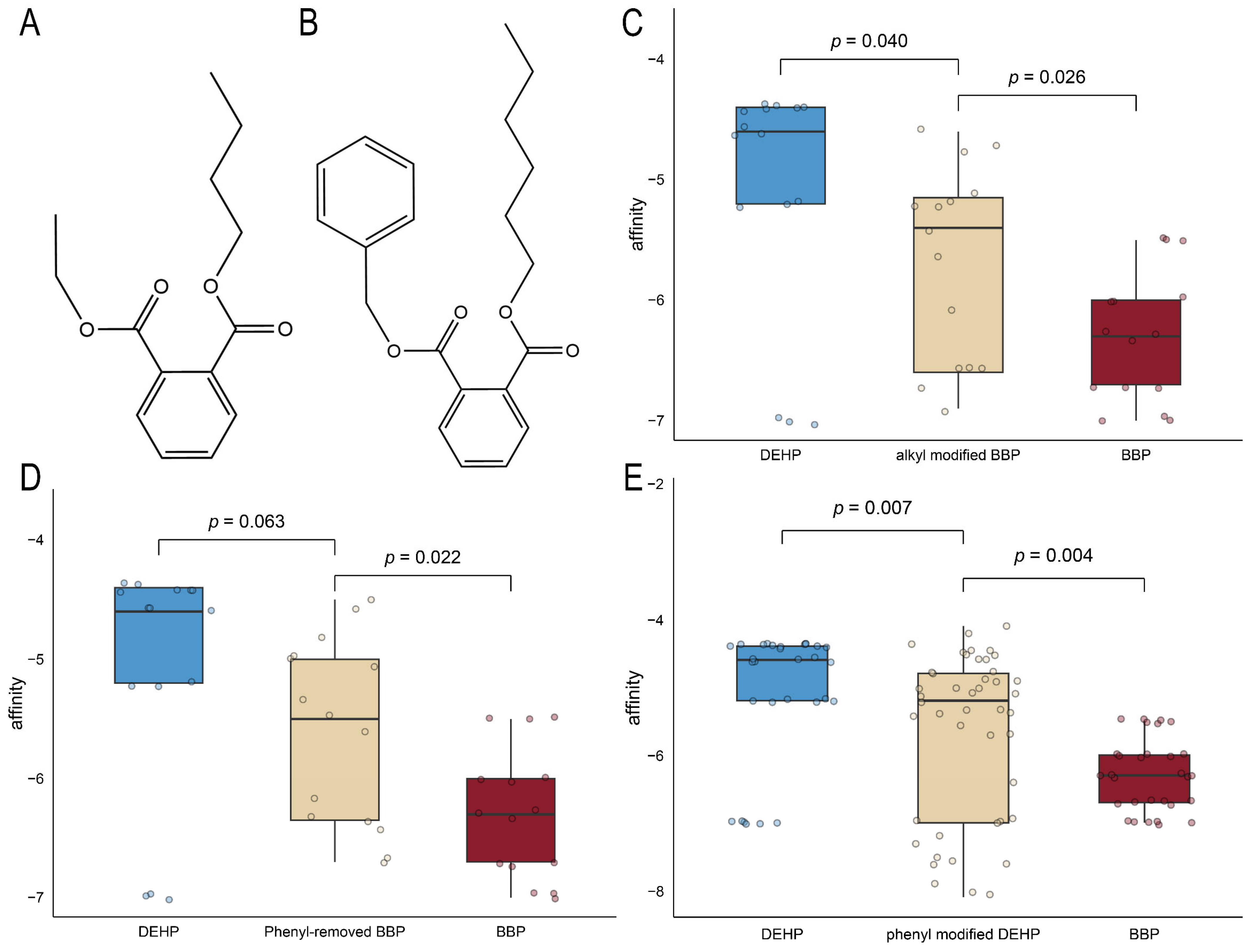
2.6. Computer-Aided Molecular Design and Molecular Docking for Validation
3. Discussion
4. Methods
4.1. Constructing Datasets and Identifying Interactive Genes
4.2. Functional Enrichment Analysis (FEA)
4.3. Construction of Protein Interaction Network and Pathway Enrichment Analysis
4.4. Protein Structure Retrieval and Homology Modeling
4.5. Differential Expression Analysis and Survival Analysis
4.6. Molecular Docking
4.7. Computer-Aided Molecular Design
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hong, Y.; Xiao, S.; Naraginti, S.; Liao, W.; Feng, C.; Xu, D.; Guo, C.; Jin, X.; Xie, F. Freshwater water quality criteria for phthalate esters and recommendations for the revision of the water quality standards. Ecotoxicol. Environ. Saf. 2024, 279, 116517. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Guo, J.L.; Xue, J.C.; Bai, C.L.; Guo, Y. Phthalate metabolites: Characterization, toxicities, global distribution, and exposure assessment. Environ. Pollut. 2021, 291, 118106. [Google Scholar] [CrossRef] [PubMed]
- Kantor, E.D.; Romano, M.E. Phthalate Exposure From Prescription Medications and Breast Cancer Risk. J. Clin. Oncol. 2019, 37, 1775–1777. [Google Scholar] [CrossRef] [PubMed]
- Trasande, L.; Nelson, M.E.; Alshawabkeh, A.; Barrett, E.S.; Buckley, J.P.; Dabelea, D.; Dunlop, A.L.; Herbstman, J.B.; Meeker, J.D.; Naidu, M.; et al. Prenatal phthalate exposure and adverse birth outcomes in the USA: A prospective analysis of births and estimates of attributable burden and costs. Lancet Planet. Health 2024, 8, e74–e85. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M. JAMA Pediatrics patient page. Phthalate exposure and health risks. JAMA Pediatr. 2014, 168, 96. [Google Scholar] [CrossRef]
- The Commission of the European Communities. Commission Directive 2004/93/EC of 21 September 2004 Amending Council Directive 76/768/EEC for the Purpose of Adapting Its Annexes II and III to Technical Progress. Off. J. Eur. Union 2004, L 300, 13–41. Available online: http://data.europa.eu/eli/dir/2004/93/oj (accessed on 21 August 2025).
- The Commission of the European Communities. Directive 2005/84/EC of the European Parliament and of the Council 14 December 2005 Amending for the 22nd time Council Directive 76/769/EEC on the Approximation of the Laws, Regulations and Administrative Provisions of the Member States Relating to Restrictions on the Marketing and Use of Certain Dangerous Substances and Preparations (Phthalates in Toys and Childcare Articles). Off. J. Eur. Union 2005, L 344, 40–43. Available online: http://data.europa.eu/eli/dir/2005/84/oj (accessed on 21 August 2025).
- The Commission of the European Communities. Commission Directive 2007/19/EC of 30 March 2007 Amending Directive 2002/72/EC Relating to Plastic Materials and Articles Intended to Come into Contact with Food and Council Directive 85/572/EEC Laying Down the List of Simulants to Be Used for Testing Migration of Constituents of Plastic Materials and Articles Intended to Come into Contact with Foodstuffs. Off. J. Eur. Union 2007, L 91, 17–36. Available online: http://data.europa.eu/eli/dir/2007/19/oj (accessed on 21 August 2025).
- Yu, L.; Shen, Y.; Gao, P.; Zhang, Q.; Hu, X.; Xu, Q. A novel molecularly imprinted photoelectrochemical aptasensor based on dual recognition mechanism for ultratrace detection of plasticizer dibutyl phthalate. Chem. Eng. J. 2023, 472, 144925. [Google Scholar] [CrossRef]
- Yang, N.; Zhang, Y.; Yang, N.; Men, C.; Zuo, J. Distribution characteristics and relationship of microplastics, phthalate esters, and bisphenol A in the Beiyun River basin of Beijing. J. Hazard. Mater. 2024, 480, 136190. [Google Scholar] [CrossRef]
- Johns, L.E.; Cooper, G.S.; Galizia, A.; Meeker, J.D. Exposure assessment issues in epidemiology studies of phthalates. Environ. Int. 2015, 85, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Chen, L.L.; Guo, Y.; Wu, Q.; Yang, M.; Wu, M.H.; Kannan, K. Phthalate diesters in airborne PM2.5 and PM10 in a suburban area of Shanghai: Seasonal distribution and risk assessment. Sci. Total Environ. 2014, 497–498, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Wormuth, M.; Scheringer, M.; Vollenweider, M.; Hungerbuhler, K. What are the sources of exposure to eight frequently used phthalic acid esters in Europeans? Risk Anal. 2006, 26, 803–824. [Google Scholar] [CrossRef]
- Massahi, T.; Omer, A.K.; Kiani, A.; Soleimani, H.; Fattahi, N.; Sharafi, K. Assessing the effect of sunlight exposure and reuse of polyethylene terephthalate bottles on phthalate migration. Sci. Total Environ. 2025, 962, 178480. [Google Scholar] [CrossRef]
- Crobeddu, B.; Ferraris, E.; Kolasa, E.; Plante, I. Di(2-ethylhexyl) phthalate (DEHP) increases proliferation of epithelial breast cancer cells through progesterone receptor dysregulation. Environ. Res. 2019, 173, 165–173. [Google Scholar] [CrossRef]
- Chen, F.P.; Chien, M.H. Lower concentrations of phthalates induce proliferation in human breast cancer cells. Climacteric 2014, 17, 377–384. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ahern, T.P.; Broe, A.; Lash, T.L.; Cronin-Fenton, D.P.; Ulrichsen, S.P.; Christiansen, P.M.; Cole, B.F.; Tamimi, R.M.; Sørensen, H.T.; Damkier, P. Phthalate Exposure and Breast Cancer Incidence: A Danish Nationwide Cohort Study. J. Clin. Oncol. 2019, 37, 1800–1809. [Google Scholar] [CrossRef]
- Yoon, K.; Kwack, S.J.; Kim, H.S.; Lee, B.M. Estrogenic endocrine-disrupting chemicals: Molecular mechanisms of actions on putative human diseases. J. Toxicol. Environ. Health B Crit. Rev. 2014, 17, 127–174. [Google Scholar] [CrossRef]
- Yager, J.D.; Davidson, N.E. Estrogen carcinogenesis in breast cancer. N. Engl. J. Med. 2006, 354, 270–282. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Tsai, C.F.; Hsu, C.Y.; Kuo, P.L.; Lee, J.N.; Chai, C.Y.; Wang, S.C.; Tsai, E.M. Phthalates induce proliferation and invasiveness of estrogen receptor-negative breast cancer through the AhR/HDAC6/c-Myc signaling pathway. FASEB J. 2012, 26, 778–787. [Google Scholar] [CrossRef]
- Wang, Y.C.; Tsai, C.F.; Chuang, H.L.; Chang, Y.C.; Chen, H.S.; Lee, J.N.; Tsai, E.M. Benzyl butyl phthalate promotes breast cancer stem cell expansion via SPHK1/S1P/S1PR3 signaling. Oncotarget 2016, 7, 29563–29576. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.H.; Tsai, C.F.; Hsu, C.Y.; Kuo, P.L.; Hsi, E.; Suen, J.L.; Hung, C.H.; Lee, J.N.; Chai, C.Y.; Wang, S.C.; et al. n-Butyl benzyl phthalate promotes breast cancer progression by inducing expression of lymphoid enhancer factor 1. PLoS ONE 2012, 7, e42750. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.N. In silico tools to aid risk assessment of endocrine disrupting chemicals. Toxicology 2004, 205, 43–53. [Google Scholar] [CrossRef]
- Zhou, M.; Yang, J.; Li, Y. A model for phthalic acid esters’ biodegradability and biotoxicity multi-effect pharmacophore and its application in molecular modification. J. Environ. Sci. Health A Toxic Hazard. Subst. Environ. Eng. 2021, 56, 361–378. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, S.; Lu, H.; Xia, M.; Wang, S.; Li, X.; Li, H.; Wang, Y.; Ge, R.S.; Liu, Y. Enhanced inhibition of human and rat aromatase activity by benzene ring substitutions in bisphenol A: QSAR structure-activity relationship and in silico docking analysis. J. Hazard. Mater. 2024, 465, 133252. [Google Scholar] [CrossRef]
- Qiu, Y.; Jiang, L.; Zhang, S.; Li, Y. Molecular design of lower-oestrogen-activity phthalate acid esters with high fluorescence intensity using pharmacophore model. Int. J. Environ. Anal. Chem. 2021, 101, 895–910. [Google Scholar] [CrossRef]
- Gou, Y.Y.; Lin, S.; Que, D.E.; Tayo, L.L.; Lin, D.Y.; Chen, K.C.; Chen, F.A.; Chiang, P.C.; Wang, G.S.; Hsu, Y.C.; et al. Estrogenic effects in the influents and effluents of the drinking water treatment plants. Environ. Sci. Pollut. Res. Int. 2016, 23, 8518–8528. [Google Scholar] [CrossRef]
- Romero-Franco, M.; Hernandez-Ramirez, R.U.; Calafat, A.M.; Cebrian, M.E.; Needham, L.L.; Teitelbaum, S.; Wolff, M.S.; Lopez-Carrillo, L. Personal care product use and urinary levels of phthalate metabolites in Mexican women. Environ. Int. 2011, 37, 867–871. [Google Scholar] [CrossRef]
- Panagiotou, G.; Taboureau, O. The impact of network biology in pharmacology and toxicology. SAR QSAR Environ. Res. 2012, 23, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Hoeng, J.; Talikka, M.; Martin, F.; Sewer, A.; Yang, X.; Iskandar, A.; Schlage, W.K.; Peitsch, M.C. Case study: The role of mechanistic network models in systems toxicology. Drug Discov. Today 2014, 19, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Audouze, K.; Taboureau, O. Emerging Bioinformatics Methods and Resources in Drug Toxicology. Methods Mol. Biol. 2022, 2425, 133–146. [Google Scholar] [CrossRef]
- He, N.; Zhang, J.; Liu, M.; Yin, L. Elucidating the mechanism of plasticizers inducing breast cancer through network toxicology and molecular docking analysis. Ecotoxicol. Environ. Saf. 2024, 284, 116866. [Google Scholar] [CrossRef]
- He, J.; Zhu, X.; Xu, K.; Li, Y.; Zhou, J. Network toxicological and molecular docking to investigate the mechanisms of toxicity of agricultural chemical Thiabendazole. Chemosphere 2024, 363, 142711. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Y.; Sun, Y.; Wei, Z.; Wang, D.; Chen, S.; Yang, F.; Wang, J.; Kang, X. Assessing the toxicological impact of PET-MPs exposure on IVDD: Insights from network toxicology and molecular docking. J. Environ. Manag. 2025, 373, 123830. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Hsu, C.Y.; Yang, P.J.; Chiu, C.C.; Liang, S.S.; Ou-Yang, F.; Kan, J.Y.; Hou, M.F.; Wang, T.N.; Tsai, E.M. DEHP mediates drug resistance by directly targeting AhR in human breast cancer. Biomed. Pharmacother. 2022, 145, 112400. [Google Scholar] [CrossRef]
- Picard, K.; Lhuguenot, J.C.; Lavier-Canivenc, M.C.; Chagnon, M.C. Estrogenic activity and metabolism of n-butyl benzyl phthalate in vitro: Identification of the active molecule(s). Toxicol. Appl. Pharmacol. 2001, 172, 108–118. [Google Scholar] [CrossRef]
- Yang, P.J.; Hou, M.F.; Ou-Yang, F.; Hsieh, T.H.; Lee, Y.J.; Tsai, E.M.; Wang, T.N. Association between recurrent breast cancer and phthalate exposure modified by hormone receptors and body mass index. Sci. Rep. 2022, 12, 2858. [Google Scholar] [CrossRef]
- Hamid, N.; Junaid, M.; Manzoor, R.; Jia, P.P.; Pei, D.S. Prioritizing phthalate esters (PAEs) using experimental in vitro/vivo toxicity assays and computational in silico approaches. J. Hazard. Mater. 2020, 398, 122851. [Google Scholar] [CrossRef]
- Venkata, N.G.; Robinson, J.A.; Cabot, P.J.; Davis, B.; Monteith, G.R.; Roberts-Thomson, S.J. Mono(2-ethylhexyl)phthalate and mono-n-butyl phthalate activation of peroxisome proliferator activated-receptors alpha and gamma in breast. Toxicol. Lett. 2006, 163, 224–234. [Google Scholar] [CrossRef]
- Ozcan, G. PTCH1 and CTNNB1 emerge as pivotal predictors of resistance to neoadjuvant chemotherapy in ER+/HER2- breast cancer. Front. Oncol. 2023, 13, 1216438. [Google Scholar] [CrossRef]
- Song, P.; Lv, D.; Yang, L.; Zhou, J.; Yan, X.; Liu, Z.; Ma, K.; Yu, Y.; Liu, X.; Dong, Q. Di-(2-ethylhexyl) phthalate promotes benign prostatic hyperplasia through KIF11-Wnt/beta-catenin signaling pathway. Ecotoxicol. Environ. Saf. 2024, 281, 116602. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.X.; Yin, Y.C.; Xu, P.; Wei, M.; Zhang, W. Network toxicology and cell experiments reveal the mechanism of DEHP-induced diabetic nephropathy via Wnt signaling pathway. Toxicol. Appl. Pharmacol. 2024, 493, 117144. [Google Scholar] [CrossRef] [PubMed]
- Singer, C.F.; Holst, F.; Steurer, S.; Burandt, E.C.; Lax, S.F.; Jakesz, R.; Rudas, M.; Stöger, H.; Greil, R.; Sauter, G.; et al. Estrogen Receptor Alpha Gene Amplification Is an Independent Predictor of Long-Term Outcome in Postmenopausal Patients with Endocrine-Responsive Early Breast Cancer. Clin. Cancer Res. 2022, 28, 4112–4120. [Google Scholar] [CrossRef]
- Holst, F.; Stahl, P.R.; Ruiz, C.; Hellwinkel, O.; Jehan, Z.; Wendland, M.; Lebeau, A.; Terracciano, L.; Al-Kuraya, K.; Jänicke, F.; et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat. Genet. 2007, 39, 655–660. [Google Scholar] [CrossRef]
- Clurman, B.E.; Groudine, M. The CDKN2A tumor-suppressor locus—A tale of two proteins. N. Engl. J. Med. 1998, 338, 910–912. [Google Scholar] [CrossRef]
- Marvalim, C.; Datta, A.; Lee, S.C. Role of p53 in breast cancer progression: An insight into p53 targeted therapy. Theranostics 2023, 13, 1421–1442. [Google Scholar] [CrossRef]
- Aftab, A.; Shahzad, S.; Hussain, H.M.J.; Khan, R.; Irum, S.; Tabassum, S. CDKN2A/P16INK4A variants association with breast cancer and their in-silico analysis. Breast Cancer 2019, 26, 11–28. [Google Scholar] [CrossRef]
- Xu, T.; Zhao, H.; Wang, M.; Chow, A.; Fang, M. Metabolomics and In Silico Docking-Directed Discovery of Small-Molecule Enzyme Targets. Anal. Chem. 2021, 93, 3072–3081. [Google Scholar] [CrossRef]
- Böckers, M.; Paul, N.W.; Efferth, T. Butyl octyl phthalate interacts with estrogen receptor α in MCF-7 breast cancer cells to promote cancer development. World Acad. Sci. J. 2021, 3, 21. [Google Scholar] [CrossRef]
- Buteau-Lozano, H.; Velasco, G.; Cristofari, M.; Balaguer, P.; Perrot-Applanat, M. Xenoestrogens modulate vascular endothelial growth factor secretion in breast cancer cells through an estrogen receptor-dependent mechanism. J. Endocrinol. 2008, 196, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Hurst, C.H.; Waxman, D.J. Activation of PPARAlpha and PPARgamma by environmental phthalate monoesters. Toxicol. Sci. 2003, 74, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yao, X.; Jiang, N.; Zhang, J.; Liu, G.; Li, X.; Wang, C.; Yang, Z.; Wang, J.; Zhu, L.; et al. Environmentally relevant concentrations of butyl benzyl phthalate triggered oxidative stress and apoptosis in adult zebrafish (Danio rerio) liver: Combined analysis at physiological and molecular levels. Sci. Total Environ. 2023, 858 Pt 3, 160109. [Google Scholar] [CrossRef]
- Güner, O.F. History and evolution of the pharmacophore concept in computer-aided drug design. Curr. Top. Med. Chem. 2002, 2, 1321–1332. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Pu, Q.; Sun, P.; Zhao, W.; Liu, M.; Li, Y. Aquatic toxicity of tire microplastics on marine and freshwater organisms: An in silico approach. Chemosphere 2023, 313, 137523. [Google Scholar] [CrossRef]
- Li, X.; Mo, J.; Zhu, Q.; Ni, C.; Wang, Y.; Li, H.; Lin, Z.K.; Ge, R.S. The structure-activity relationship (SAR) for phthalate-mediated developmental and reproductive toxicity in males. Chemosphere 2019, 223, 504–513. [Google Scholar] [CrossRef]
- La Merrill, M.A.; Vandenberg, L.N.; Smith, M.T.; Goodson, W.; Browne, P.; Patisaul, H.B.; Guyton, K.Z.; Kortenkamp, A.; Cogliano, V.J.; Woodruff, T.J.; et al. Consensus on the key characteristics of endocrine-disrupting chemicals as a basis for hazard identification. Nat. Rev. Endocrinol. 2020, 16, 45–57. [Google Scholar] [CrossRef]
- Lovekamp-Swan, T.; Davis, B.J. Mechanisms of phthalate ester toxicity in the female reproductive system. Environ. Health Perspect. 2003, 111, 139–145. [Google Scholar] [CrossRef]
- Xiao, M.; Zhang, Y.; Zhang, X.; Zhang, G.; Jin, C.; Yang, J.; Wu, S.; Lu, X. Bisphenol A and Di(2-Ethylhexyl) Phthalate promote pulmonary carcinoma in female rats via estrogen receptor beta: In vivo and in silico analysis. Ecotoxicol. Environ. Saf. 2023, 250, 114496. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 15 February 2025).
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Gyorffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021, 19, 4101–4109. [Google Scholar] [CrossRef]
- Guo, D.; Jin, J.; Liu, J.; Wang, Y.; Li, D.; He, Y. Baicalein Inhibits the Progression and Promotes Radiosensitivity of Esophageal Squamous Cell Carcinoma by Targeting HIF-1A. Drug Des. Dev. Ther. 2022, 16, 2423–2436. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Liu, X.; Ouyang, S.; Yu, B.; Liu, Y.; Huang, K.; Gong, J.; Zheng, S.; Li, Z.; Li, H.; Jiang, H. PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010, 38, W609–W614. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef]


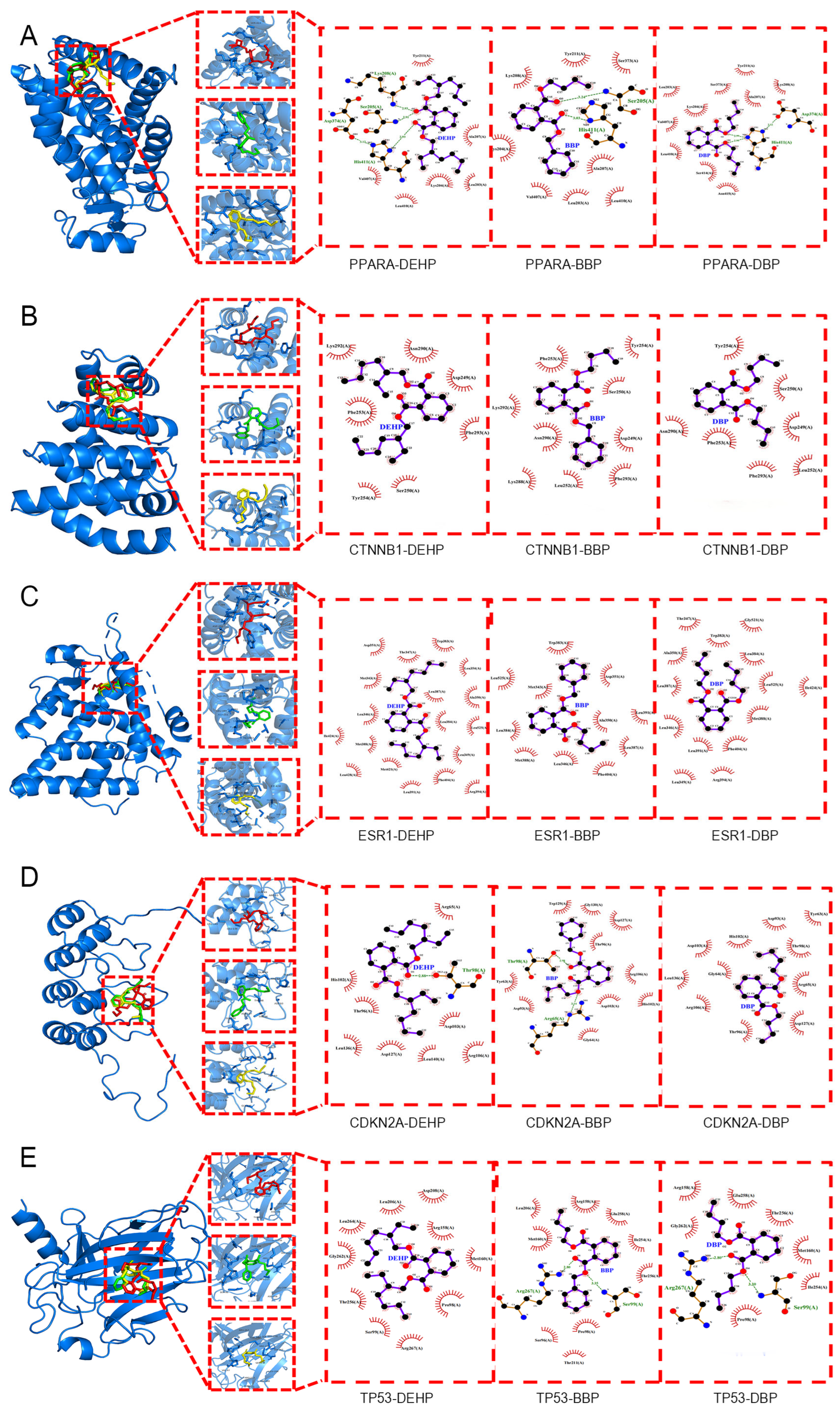
| Affinity (kcal/mol) | Hub Genes | |||||
|---|---|---|---|---|---|---|
| PPARA | CTNNB1 | ESR1 | CDKN2A | TP53 | ||
| PAE | DEHP | −5.2 | −4.6 | −7 | −4.4 | −4.4 |
| DBP | −5.5 | −5.3 | −6.6 | −5 | −4.8 | |
| BBP | −6.3 | −6.7 | −7 | −5.5 | −6 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Bai, Z.; Yan, X.; Zhou, Y.; Zhong, C.; Wu, J. Computer-Aided Molecular Design Meets Network Toxicology and Molecular Docking: A Joint Strategy to Explore Common Molecular Mechanisms of Phthalates on Human Breast Cancer and Structure–Activity Relationship. Int. J. Mol. Sci. 2025, 26, 9878. https://doi.org/10.3390/ijms26209878
Yang X, Bai Z, Yan X, Zhou Y, Zhong C, Wu J. Computer-Aided Molecular Design Meets Network Toxicology and Molecular Docking: A Joint Strategy to Explore Common Molecular Mechanisms of Phthalates on Human Breast Cancer and Structure–Activity Relationship. International Journal of Molecular Sciences. 2025; 26(20):9878. https://doi.org/10.3390/ijms26209878
Chicago/Turabian StyleYang, Xinyu, Zijun Bai, Xiaoyun Yan, Yu Zhou, Caiyun Zhong, and Jieshu Wu. 2025. "Computer-Aided Molecular Design Meets Network Toxicology and Molecular Docking: A Joint Strategy to Explore Common Molecular Mechanisms of Phthalates on Human Breast Cancer and Structure–Activity Relationship" International Journal of Molecular Sciences 26, no. 20: 9878. https://doi.org/10.3390/ijms26209878
APA StyleYang, X., Bai, Z., Yan, X., Zhou, Y., Zhong, C., & Wu, J. (2025). Computer-Aided Molecular Design Meets Network Toxicology and Molecular Docking: A Joint Strategy to Explore Common Molecular Mechanisms of Phthalates on Human Breast Cancer and Structure–Activity Relationship. International Journal of Molecular Sciences, 26(20), 9878. https://doi.org/10.3390/ijms26209878







