Integrative Methylome and Transcriptome Analysis Reveals Epigenetic Regulation of Pigmentation in Oujiang Color Common Carp
Abstract
1. Introduction
2. Results
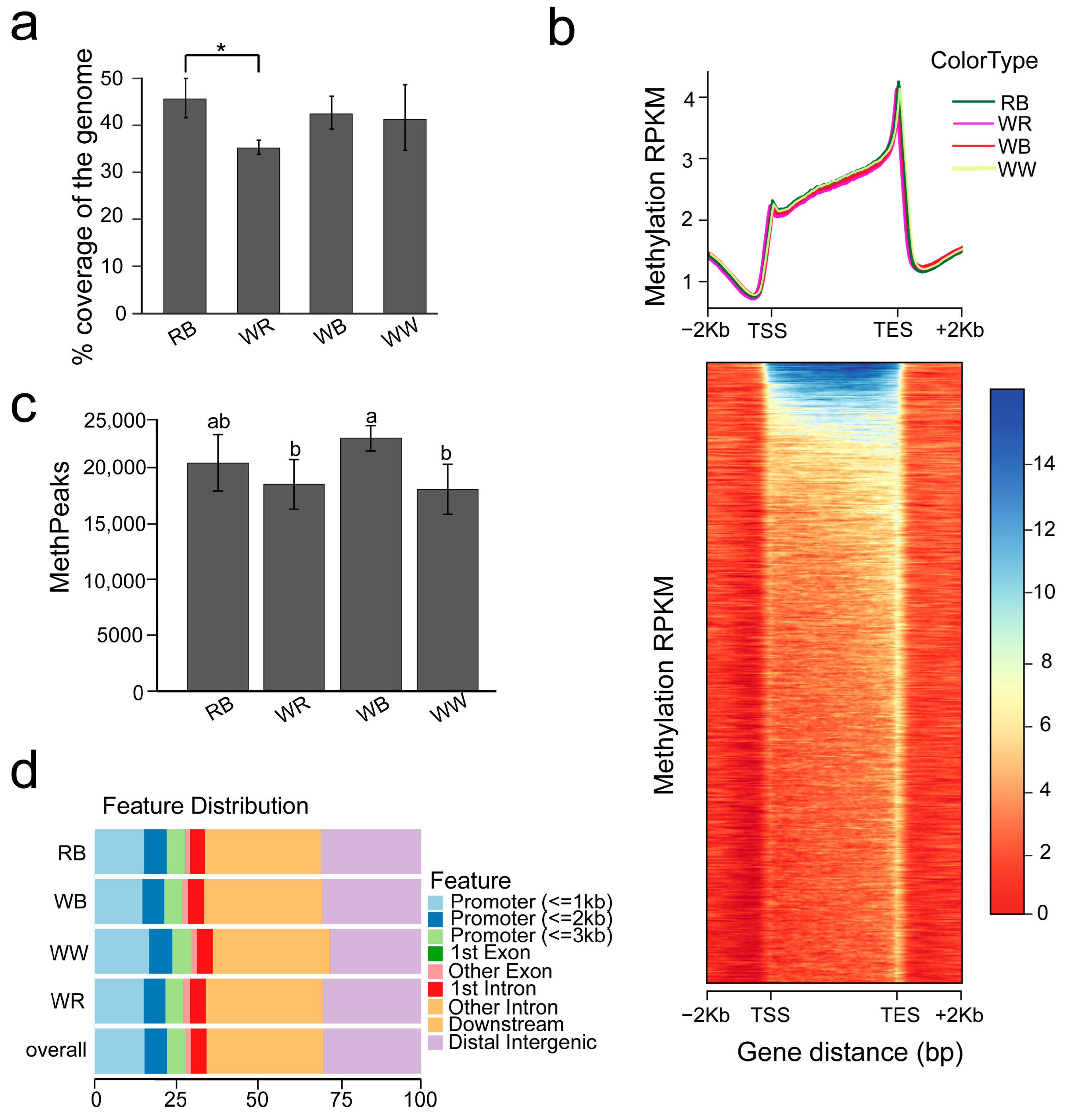
2.1. Whole Genome Methylation Profiles Among Four Common Color Carp Varieties
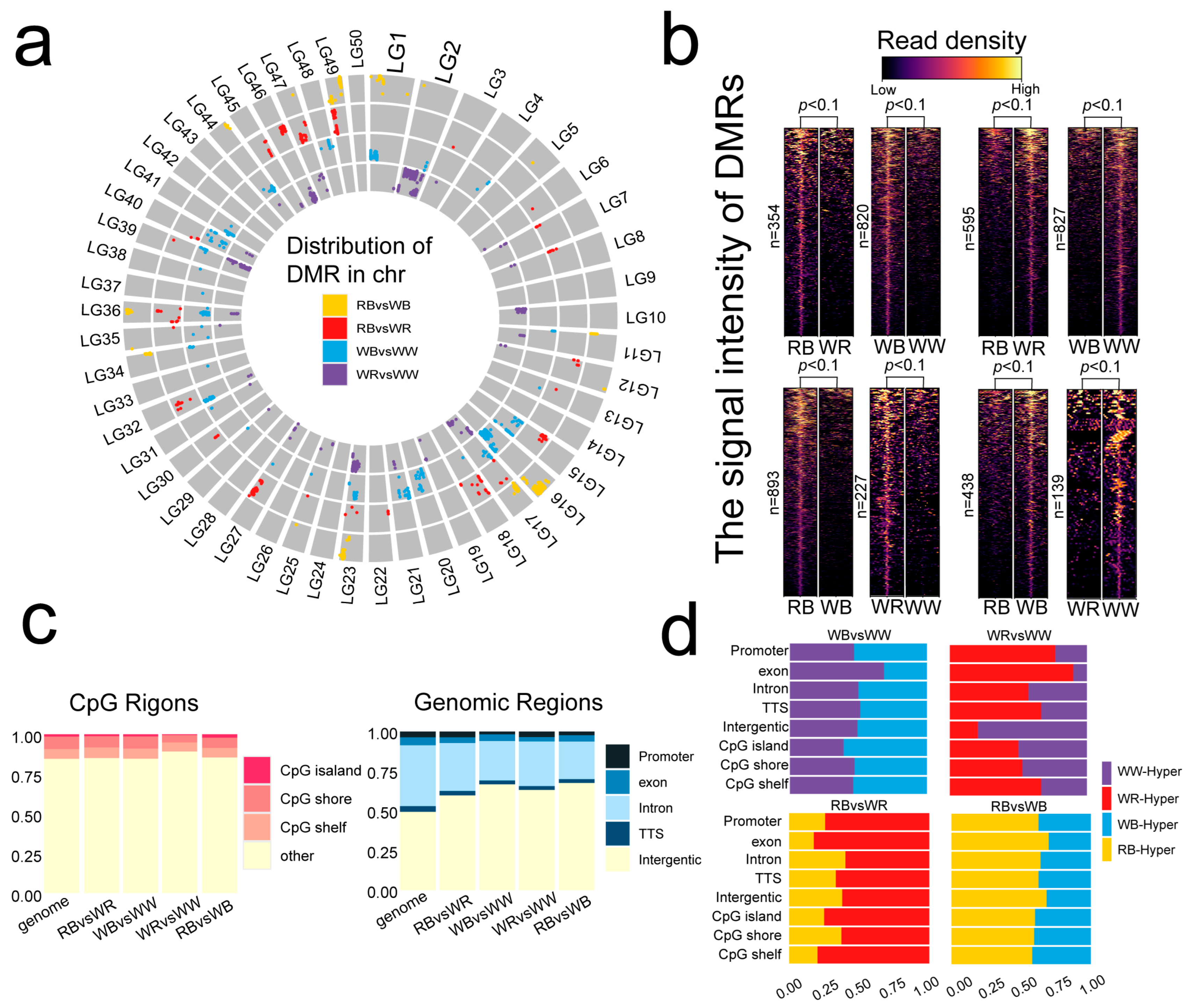
2.2. Differential Methylation Analysis Among the Four-Color Varieties
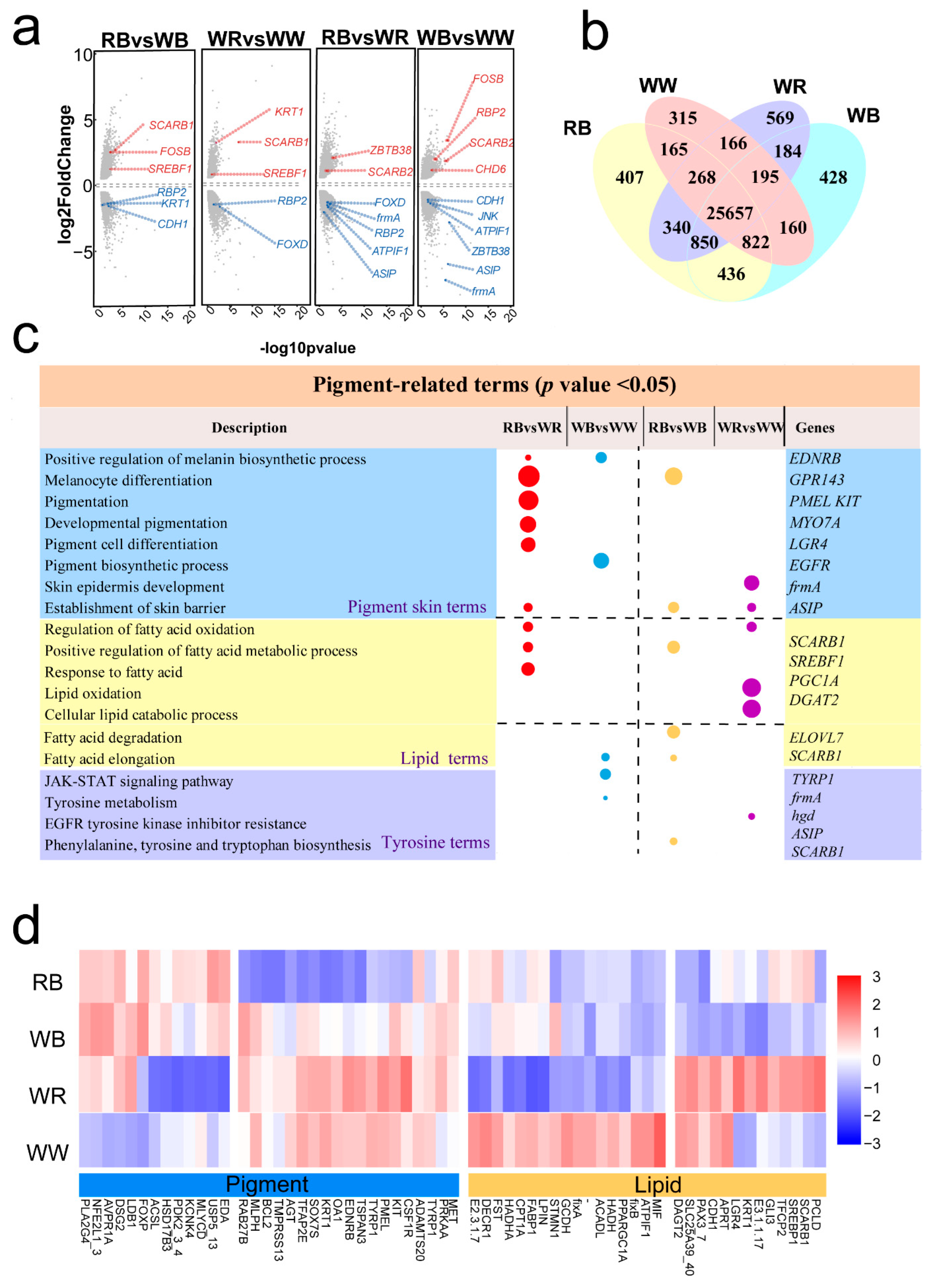
2.3. Different Expression Profiles of the Four-Color Varieties
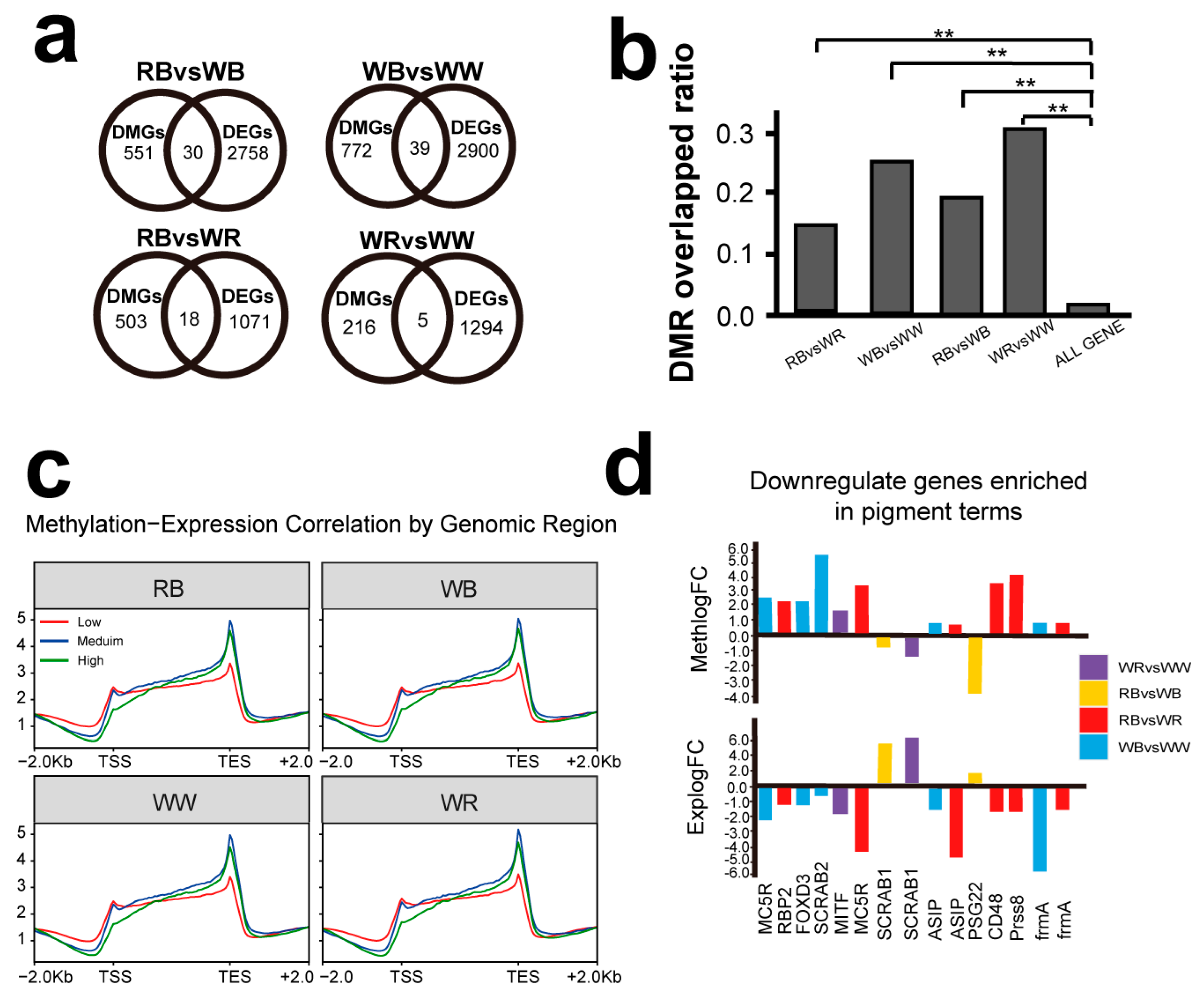
2.4. Integrated Transcriptome-Methylome Analysis
3. Discussion
3.1. Regulatory Effects of DNA Methylation on Gene Expression in Oujiang Color Common Carp
3.2. Epigenetic Differences Between Black-Spotted and Non-Black-Spotted Color Common Carp
3.3. Epigenetic Differences Between Red and White Related Color Common Carp
3.4. Pigmentation-Related Genes and Pathways
3.5. Molecular Mechanisms Underlying Skin Coloration in Oujiang Color Common Carp
3.6. Translational Relevance: Methylation Markers for Selective Breeding
4. Materials and Methods
4.1. Sample Collection
4.2. DNA and RNA Extraction
4.3. MBD-Seq Sequencing
4.4. Genome-Wide Methylome Profiling
4.5. RNA-Seq Sequencing and Data Analysis
4.6. RT-qPCR Verification of RNA-Seq Results
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cuthill, I.C.; Allen, W.L.; Arbuckle, K.; Caspers, B.; Chaplin, G.; Hauber, M.E.; Hill, G.E.; Jablonski, N.G.; Jiggins, C.D.; Kelber, A.; et al. The biology of color. Science 2017, 357, eaan0221. [Google Scholar] [CrossRef]
- Kelsh, R.N. Genetics and evolution of pigment patterns in fish. Pigment. Cell Res. 2004, 17, 326–336. [Google Scholar] [CrossRef]
- Parichy, D.M. Evolution of pigment cells and patterns: Recent insights from teleost fishes. Curr. Opin. Genet. Dev. 2021, 69, 88–96. [Google Scholar] [CrossRef]
- Patterson, L.B.; Parichy, D.M. Zebrafish pigment pattern formation: Insights into the development and evolution of adult form. Annu. Rev. Genet. 2019, 53, 505–530. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Lu, G.; Yin, H.; Wang, L.; Atuganile, M.; Dong, Z. Fish pigmentation and coloration: Molecular mechanisms and aquaculture perspectives. Rev. Aquac. 2021, 13, 2395–2412. [Google Scholar] [CrossRef]
- Braasch, I.; Schartl, M.; Volff, J.N. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol. Biol. 2007, 7, 74. [Google Scholar] [CrossRef]
- Zhou, S.; Zeng, H.; Huang, J.; Lei, L.; Tong, X.; Li, S.; Zhou, Y.; Guo, H.; Khan, M.; Luo, L. Epigenetic regulation of melanogenesis. Ageing Res. Rev. 2021, 69, 101349. [Google Scholar] [CrossRef]
- Marcoli, R.; Jones, D.B.; Massault, C.; Harrison, P.J.; Cate, H.S.; Jerry, D.R. Barramundi (Lates calcarifer) rare coloration patterns: A multiomics approach to understand the “panda” phenotype. J. Fish Biol. 2024, 105, 1268–1279. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, J.; Fu, W.; Xu, W.; Zhang, H.; Chen, S.; Liu, W.; Peng, L.; Xiao, Y. Comparative Transcriptome and DNA methylation analyses of the molecular mechanisms underlying skin color variations in Crucian carp (Carassius carassius L.). BMC Genet. 2017, 18, 95. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Li, Z.; Li, L.; Ding, T.; Chen, S.; Duan, P.; Wang, X.; Qiu, Y.; Ding, X. DNA Methylation and Transcriptome Profiling Reveal the Role of the Antioxidant Pathway and Lipid Metabolism in Plectropomus leopardus Skin Color Formation. Antioxidants 2025, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Blakkan, D.; Lee, G.; Bashier, R.; Fernald, R.; Alvarado, S. DNA methylation of the endothelin receptor B makes blue fish yellow. bioRxiv 2022. bioRxiv:2022.09.27.509821. [Google Scholar] [CrossRef]
- Li, S.; Wang, C. Genetic diversity and selective breeding of red common carps in China. NAGA 2001, 24, 56–61. Available online: https://hdl.handle.net/20.500.12348/2352 (accessed on 10 September 2025).
- Wang, B.; Ji, P.; Wang, J.; Sun, J.; Wang, C.; Xu, P.; Sun, X. The complete mitochondrial genome of the Oujiang color carp, Cyprinus carpio var. color (Cypriniformes, Cyprinidae). Mitochondrial DNA 2013, 24, 19–21. [Google Scholar] [CrossRef]
- Wang, C.-H.; Li, S.-F.; Xiang, S.-P.; Wang, J.; Liu, Z.-G.; Pang, Z.-Y.; Duan, J.-P.; Xu, Z.-B. Genetic parameter estimates for growth-related traits in Oujiang color common carp (Cyprinus carpio var. color). Aquaculture 2006, 259, 103–107. [Google Scholar] [CrossRef]
- Du, J.; Chen, X.; Wang, J.; Chen, H.; Yue, W.; Lu, G.; Wang, C. Comparative skin transcriptome of two Oujiang color common carp (Cyprinus carpio var. color) varieties. Fish Physiol. Biochem. 2019, 45, 177–185. [Google Scholar] [CrossRef]
- Wang, C.; Wachholtz, M.; Wang, J.; Liao, X.; Lu, G. Analysis of the skin transcriptome in two oujiang color varieties of common carp. PLoS ONE 2014, 9, e90074. [Google Scholar] [CrossRef] [PubMed]
- Orteu, A.; Jiggins, C.D. The genomics of coloration provides insights into adaptive evolution. Nat. Rev. Genet. 2020, 21, 461–475, Correction in Nat. Rev. Genet. 2020, 21, 503. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Zhao, F.; Xiong, T.; Lai, L.; Li, H.; Lin, W.; Xiao, T.; Lin, W. Genetic mapping identifies SNP mutations in MITF-M promoter associated with melanin formation in Putian black duck. Poult. Sci. 2024, 103, 103191. [Google Scholar] [CrossRef] [PubMed]
- Dolinoy, D.C.; Weidman, J.R.; Waterland, R.A.; Jirtle, R.L. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 2006, 114, 567–572. [Google Scholar] [CrossRef]
- Yuan, B.; Qi, Y.; Zhang, X.; Hu, J.; Fan, Y.; Ji, X. The relationship of MITF gene expression and promoter methylation with plumage colour in quail. Br. Poult. Sci. 2024, 65, 259–264. [Google Scholar] [CrossRef]
- Yuan, C.; Mao, J.; Sun, H.; Wang, Y.; Guo, M.; Wang, X.; Tian, Y.; Hao, Z.; Ding, J.; Chang, Y. Genome-wide DNA methylation profile changes associated with shell colouration in the Yesso scallop (Patinopecten yessoensis) as measured by whole-genome bisulfite sequencing. Bmc Genom. 2021, 22, 740. [Google Scholar] [CrossRef]
- Kaluscha, S.; Domcke, S.; Wirbelauer, C.; Stadler, M.B.; Durdu, S.; Burger, L.; Schübeler, D. Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation. Nat. Genet. 2022, 54, 1895–1906. [Google Scholar] [CrossRef]
- Bogan, S.N.; Strader, M.E.; Hofmann, G.E. Associations between DNA methylation and gene regulation depend on chromatin accessibility during transgenerational plasticity. BMC Biol. 2023, 21, 149. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.S.; Chen, Y.; Ge, J.; Wilkening, A.N.; Hou, Y.; Lee, H.J.; Choi, Y.R.; Lowdon, R.F.; Xing, X.; Li, D. Epigenetic dynamics shaping melanophore and iridophore cell fate in zebrafish. Genome Biol. 2021, 22, 282. [Google Scholar] [CrossRef] [PubMed]
- Ahi, E.P.; Lecaudey, L.A.; Ziegelbecker, A.; Steiner, O.; Glabonjat, R.; Goessler, W.; Hois, V.; Wagner, C.; Lass, A.; Sefc, K.M. Comparative transcriptomics reveals candidate carotenoid color genes in an East African cichlid fish. BMC Genom. 2020, 21, 54. [Google Scholar] [CrossRef]
- Granneman, J.G.; Kimler, V.A.; Zhang, H.; Ye, X.; Luo, X.; Postlethwait, J.H.; Thummel, R. Lipid droplet biology and evolution illuminated by the characterization of a novel perilipin in teleost fish. Elife 2017, 6, e21771. [Google Scholar] [CrossRef] [PubMed]
- Schmeisser, J.; Verlhac-Trichet, V.; Madaro, A.; Lall, S.P.; Torrissen, O.; Olsen, R.E. Molecular mechanism involved in carotenoid metabolism in post-smolt Atlantic salmon: Astaxanthin metabolism during flesh pigmentation and its antioxidant properties. Mar. Biotechnol. 2021, 23, 653–670. [Google Scholar] [CrossRef]
- Robinson, C.L.; Evans, R.D.; Sivarasa, K.; Ramalho, J.S.; Briggs, D.A.; Hume, A.N. The adaptor protein melanophilin regulates dynamic myosin-Va: Cargo interaction and dendrite development in melanocytes. Mol. Biol. Cell 2019, 30, 742–752. [Google Scholar] [CrossRef]
- Saldana-Caboverde, A.; Kos, L. Roles of endothelin signaling in melanocyte development and melanoma. Pigment. Cell Melanoma Res. 2010, 23, 160–170. [Google Scholar] [CrossRef]
- Kubic, J.D.; Young, K.P.; Plummer, R.S.; Ludvik, A.E.; Lang, D. Pigmentation PAX-ways: The role of Pax3 in melanogenesis, melanocyte stem cell maintenance, and disease. Pigment. Cell Melanoma Res. 2008, 21, 627–645. [Google Scholar] [CrossRef]
- Toomey, M.B.; Lopes, R.J.; Araújo, P.M.; Johnson, J.D.; Gazda, M.A.; Afonso, S.; Mota, P.G.; Koch, R.E.; Hill, G.E.; Corbo, J.C. High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc. Natl. Acad. Sci. USA 2017, 114, 5219–5224. [Google Scholar] [CrossRef]
- Harris, C.A.; Haas, J.T.; Streeper, R.S.; Stone, S.J.; Kumari, M.; Yang, K.; Han, X.; Brownell, N.; Gross, R.W.; Zechner, R. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J. Lipid Res. 2011, 52, 657–667. [Google Scholar] [CrossRef]
- Liang, K. Mitochondrial CPT1A: Insights into structure, function, and basis for drug development. Front. Pharmacol. 2023, 14, 1160440. [Google Scholar] [CrossRef]
- Wang, X.; Shen, X.; Yan, Y.; Li, H. Pyruvate dehydrogenase kinases (PDKs): An overview toward clinical applications. Biosci. Rep. 2021, 41, BSR20204402. [Google Scholar] [CrossRef] [PubMed]
- Jackson, P.J.; Douglas, N.R.; Chai, B.; Binkley, J.; Sidow, A.; Barsh, G.S.; Millhauser, G.L. Structural and molecular evolutionary analysis of Agouti and Agouti-related proteins. Chem. Biol. 2006, 13, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Tiirikka, T.; Kantanen, J. A genome-wide scan study identifies a single nucleotide substitution in ASIP associated with white versus non-white coat-colour variation in sheep (Ovis aries). Heredity 2014, 112, 122–131. [Google Scholar] [CrossRef]
- Michaud, E.J.; Van Vugt, M.; Bultman, S.J.; Sweet, H.O.; Davisson, M.T.; Woychik, R.P. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 1994, 8, 1463–1472. [Google Scholar] [CrossRef]
- Cal, L.; Suarez-Bregua, P.; Comesaña, P.; Owen, J.; Braasch, I.; Kelsh, R.; Cerdá-Reverter, J.M.; Rotllant, J. Countershading in zebrafish results from an Asip1 controlled dorsoventral gradient of pigment cell differentiation. Sci. Rep. 2019, 9, 3449. [Google Scholar] [CrossRef]
- Blewitt, M.E.; Vickaryous, N.K.; Paldi, A.; Koseki, H.; Whitelaw, E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2006, 2, e49. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Chon, J.; Field, M.S.; Stover, P.J. Alcohol dehydrogenase 5 is a source of formate for de novo purine biosynthesis in HepG2 cells. J. Nutr. 2017, 147, 499–505. [Google Scholar] [CrossRef]
- Wu, J.; Chen, J.; Zhang, Y.; Dai, W.; Lin, X.; Yang, X.; Chen, M. ADH2 is downregulated by methylation and acts as a novel biomarker for breast carcinoma prognosis. Ann. Clin. Lab. Sci. 2021, 51, 12–21. [Google Scholar] [PubMed]
- Snyman, M.; Walsdorf, R.E.; Wix, S.N.; Gill, J.G. The metabolism of melanin synthesis—From melanocytes to melanoma. Pigment. Cell Melanoma Res. 2024, 37, 438–452. [Google Scholar] [CrossRef]
- Zhang, J.; Ye, Z.-W.; Bräutigam, L.; Chakraborty, P.; Luo, Z.; Culpepper, J.; Aslam, M.; Zhang, L.; Johansson, K.; Haeggström, J.Z. A role for microsomal glutathione transferase 1 in melanin biosynthesis and melanoma progression. J. Biol. Chem. 2023, 299, 104920. [Google Scholar] [CrossRef]
- Clarke, S.; Caulton, A.; McRae, K.; Brauning, R.; Couldrey, C.; Dodds, K. Beyond the genome: A perspective on the use of DNA methylation profiles as a tool for the livestock industry. Anim. Front. 2021, 11, 90–94. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, T.; Gao, D. Genetic and epigenetic regulation of growth, reproduction, disease resistance and stress responses in aquaculture. Front. Genet. 2022, 13, 994471. [Google Scholar] [CrossRef]
- Browne, D.J.; Brady, J.L.; Waardenberg, A.J.; Loiseau, C.; Doolan, D.L. An analytically and diagnostically sensitive RNA extraction and RT-qPCR protocol for peripheral blood mononuclear cells. Front. Immunol. 2020, 11, 402. [Google Scholar] [CrossRef]
- Aberg, K.A.; Chan, R.F.; Xie, L.; Shabalin, A.A.; van den Oord, E.J. Methyl-CpG-binding domain sequencing: MBD-seq. In DNA Methylation Protocols; Springer: New York, NY, USA, 2017; pp. 171–189. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, F.; Dündar, F.; Diehl, S.; Grüning, B.A.; Manke, T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–W191. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; He, Q.-Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Lienhard, M.; Grimm, C.; Morkel, M.; Herwig, R.; Chavez, L. MEDIPS: Genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 2014, 30, 284–286. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nie, H.; Jiang, K.; Li, N.; Jahan, K.; Jiang, L.; Huo, Z.; Yan, X. Transcriptome analysis reveals the pigmentation-related genes in two shell color strains of the Manila clam Ruditapes philippinarum. Anim. Biotechnol. 2021, 32, 439–450. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, W.; Chen, X.; Arif, A.; Guo, Z.; Kanika, N.H.; Song, Y.; Wang, J.; Wang, C. Integrative Methylome and Transcriptome Analysis Reveals Epigenetic Regulation of Pigmentation in Oujiang Color Common Carp. Int. J. Mol. Sci. 2025, 26, 10001. https://doi.org/10.3390/ijms262010001
Zhao W, Chen X, Arif A, Guo Z, Kanika NH, Song Y, Wang J, Wang C. Integrative Methylome and Transcriptome Analysis Reveals Epigenetic Regulation of Pigmentation in Oujiang Color Common Carp. International Journal of Molecular Sciences. 2025; 26(20):10001. https://doi.org/10.3390/ijms262010001
Chicago/Turabian StyleZhao, Wenqi, Xiaowen Chen, Ayesha Arif, Zhaoyang Guo, Nusrat Hasan Kanika, Yuehan Song, Jun Wang, and Chenghui Wang. 2025. "Integrative Methylome and Transcriptome Analysis Reveals Epigenetic Regulation of Pigmentation in Oujiang Color Common Carp" International Journal of Molecular Sciences 26, no. 20: 10001. https://doi.org/10.3390/ijms262010001
APA StyleZhao, W., Chen, X., Arif, A., Guo, Z., Kanika, N. H., Song, Y., Wang, J., & Wang, C. (2025). Integrative Methylome and Transcriptome Analysis Reveals Epigenetic Regulation of Pigmentation in Oujiang Color Common Carp. International Journal of Molecular Sciences, 26(20), 10001. https://doi.org/10.3390/ijms262010001





