miR-106b-5p as a Central Regulator of Cancer Progression and Chemotherapy-Induced Cardiotoxicity: From Molecular Mechanisms to Clinical Translation
Abstract
1. Introduction
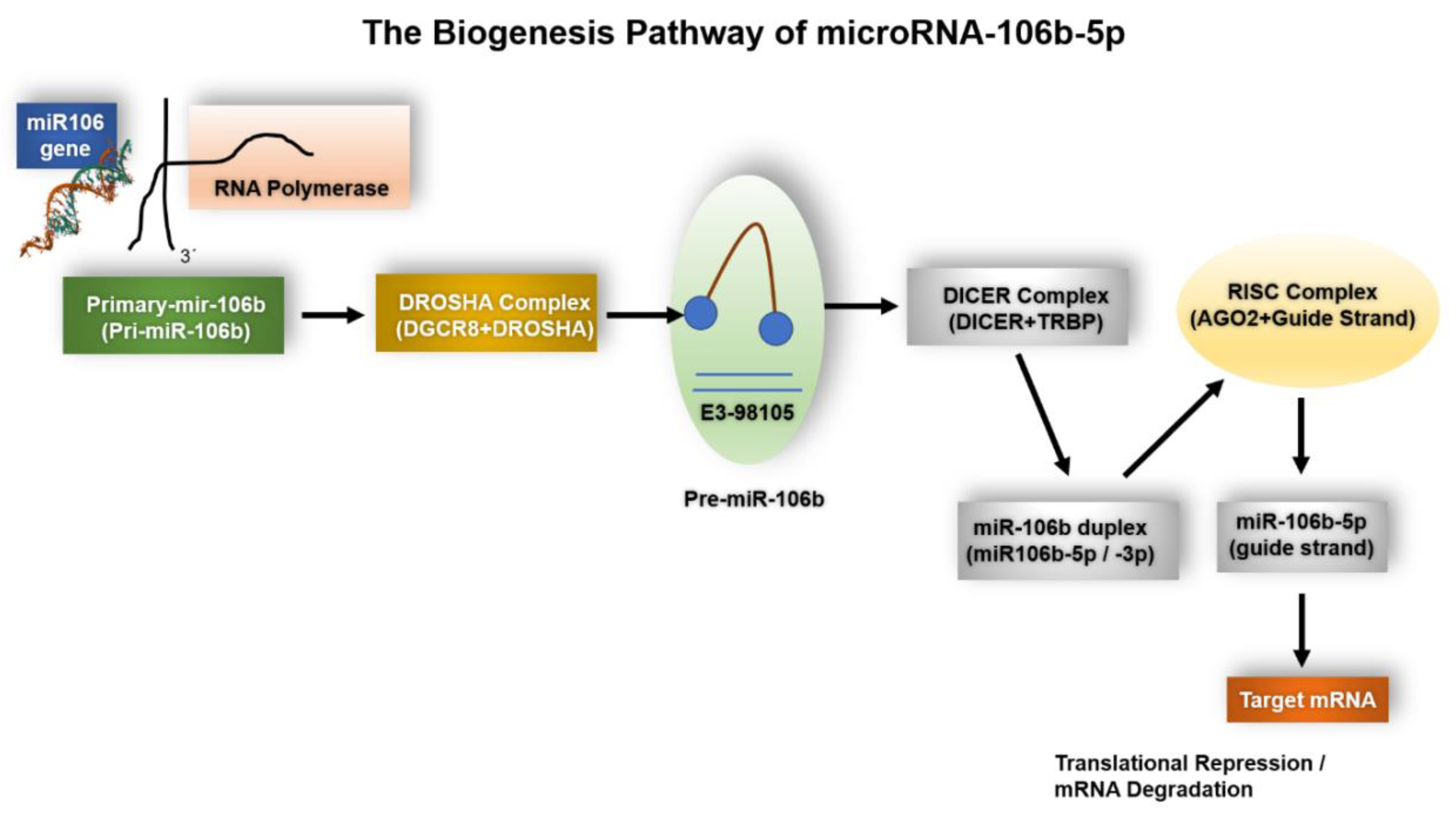
1.1. The Biogenesis of miR-106b-5p
1.1.1. Genomic Context and Transcriptional Regulation
1.1.2. Nuclear Processing and Modulation by Accessory Factors
1.1.3. Cytoplasmic Processing, Strand Selection and RISC Loading
1.1.4. Host Gene-Cluster Cooperation and Oncogenic Dysregulation
1.1.5. Contextual and Stress-Responsive Regulation—Implications for Experiments
1.1.6. Summary (Practical Takeaways)
- miR-106b-5p arises from the intronic miR-106b~25 cluster within MCM7 and is commonly co-expressed with its host gene, a relationship with strong relevance for tumor biology [17].
- Experimental designs should monitor host-gene expression and precursor/mature ratios, and consider RBP and stress-response states, to draw robust conclusions about miR-106b-5p function [18].
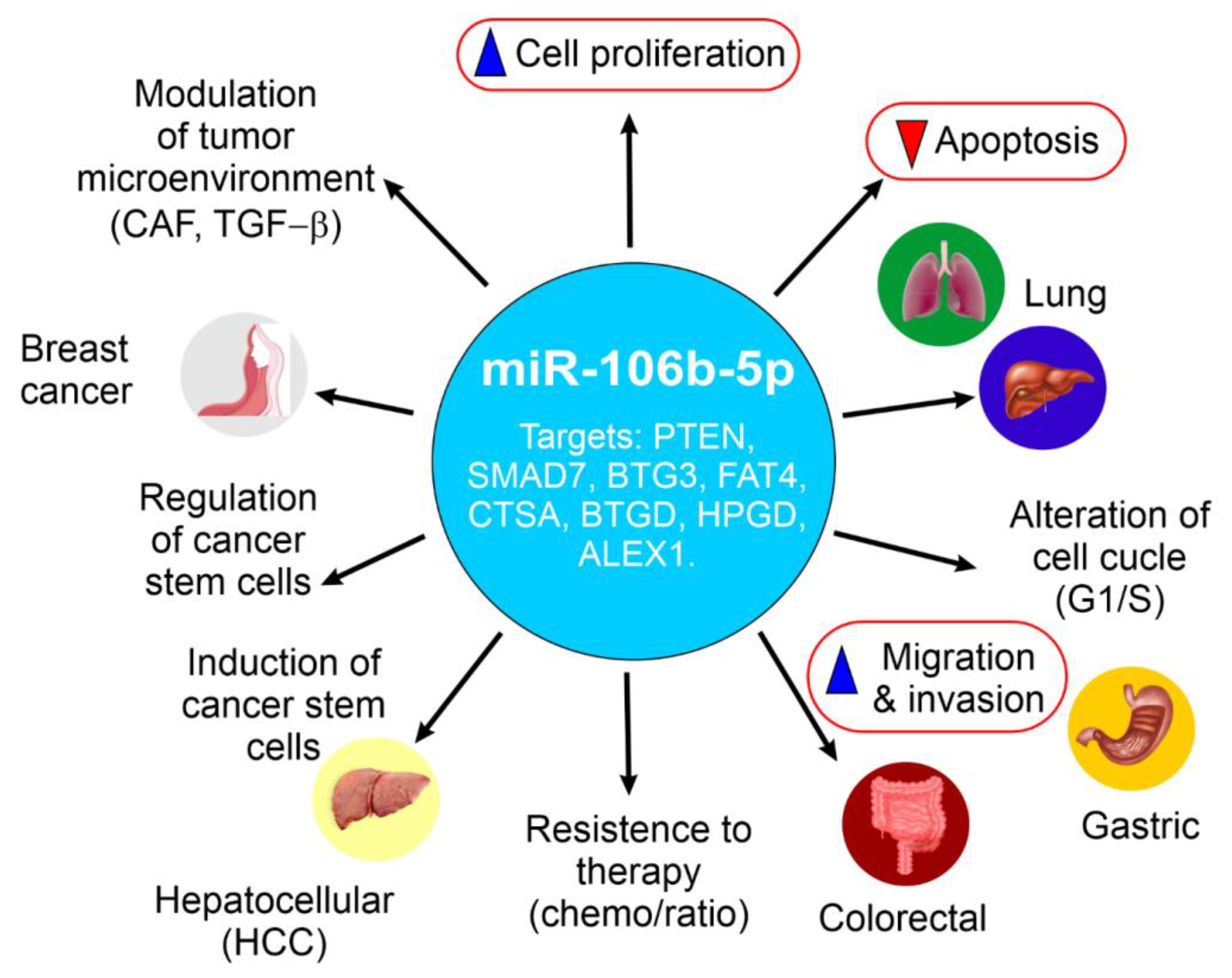
2. Elevated miR-106b-5p Expression and Tumorigenic Roles in Specific Cancers
2.1. Breast Cancer: Examining the Dual Nature of miR-106b-5p
2.2. Prostate Cancer: Oncogenic Functions and Target Genes
2.3. Lung Cancer (NSCLC): Promoting Proliferation and Inhibiting Apoptosis via BTG3
2.4. Gastric Cancer: Upregulation and Impact on Tumorigenesis
2.5. Colorectal Cancer: Context-Dependent Effects on Migration and Invasion, Targeting FAT4 and CTSA
2.6. Hepatocellular Carcinoma: Driving Proliferation, Migration, and Cell Cycle Progression Through PTEN/PI3K/Akt
2.7. Esophageal Squamous Cell Carcinomas: Contribution to Tumorigenesis
2.8. miR-106b-5p: A Key Driver of Renal Cell Carcinoma Aggressiveness via Wnt/β-Catenin Signaling
2.9. miR-106b-5p in Bladder Cancer
2.10. miR-106b-5p in Cutaneous Melanoma
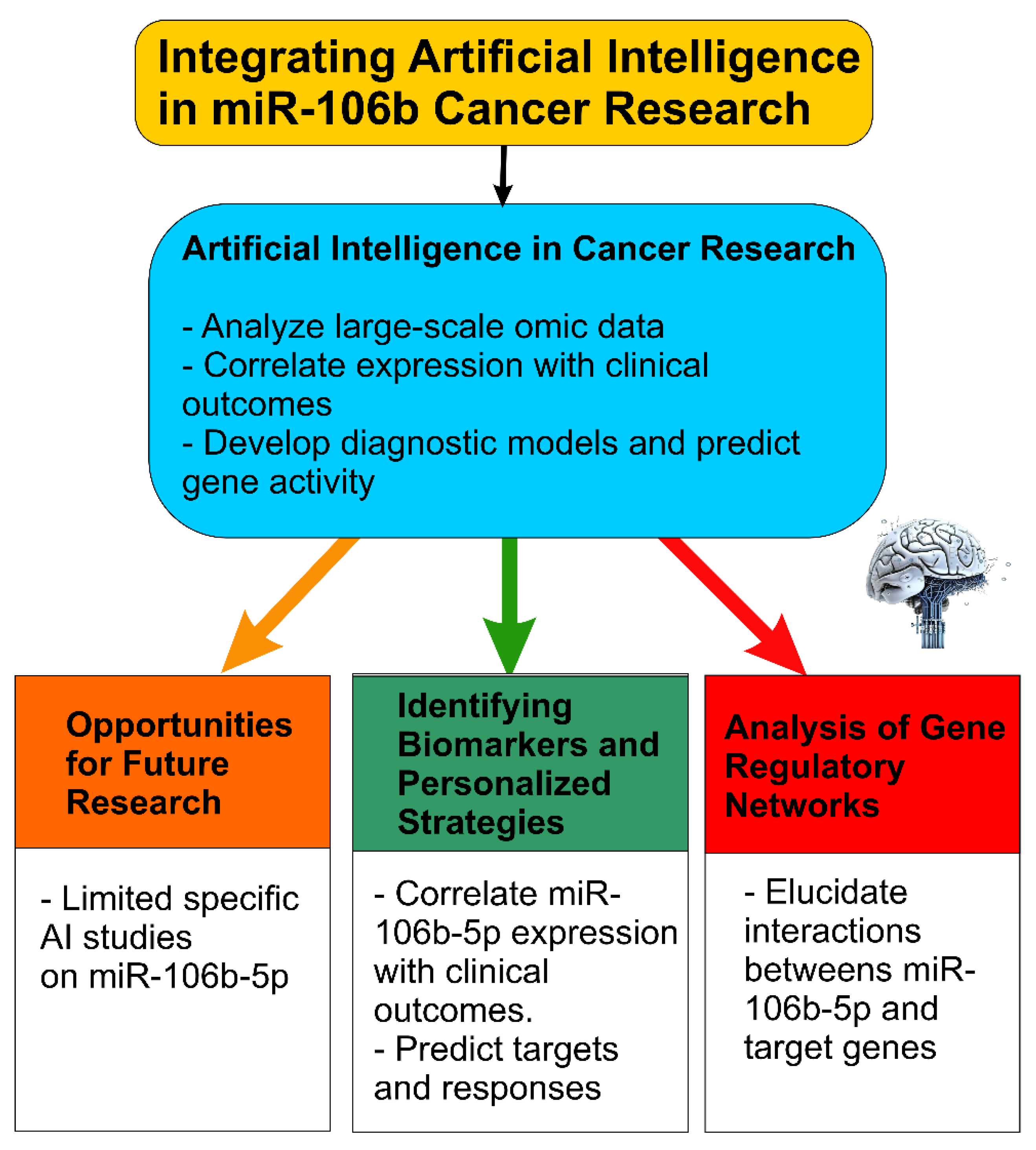
3. Integrating Artificial Intelligence into miR-106b-5p Cancer Research
3.1. Limited Specific AI Studies on miR-106b-5p: Opportunities for Future Research
3.2. Potential of AI in Identifying Biomarkers, Therapeutic Targets, and Personalized Strategies
3.3. AI-Driven Analysis of Gene Regulatory Networks Involving miR-106b-5p
- Combine predictive bioinformatic tools (TargetScan, miRDB, DIANA, StarBase) and experimentally validated databases (miRTarBase, TarBase) when building AI models, to reduce false positives.
- Use multi-omics datasets (miRNA, mRNA, lncRNA, proteome) plus clinical metadata (survival, response) to train AI/ML models for predicting biomarkers or prognostic signatures specific to miR-106b-5p.
- Apply network analysis (ceRNA, ceRNA-miRNA-mRNA, regulatory networks) with AI to identify hub genes and context-specific modules involving miR-106b-5p.
- Explore deep learning models tailored for target prediction (e.g., TargetNet) or for integrative tasks such as combining imaging + molecular data.
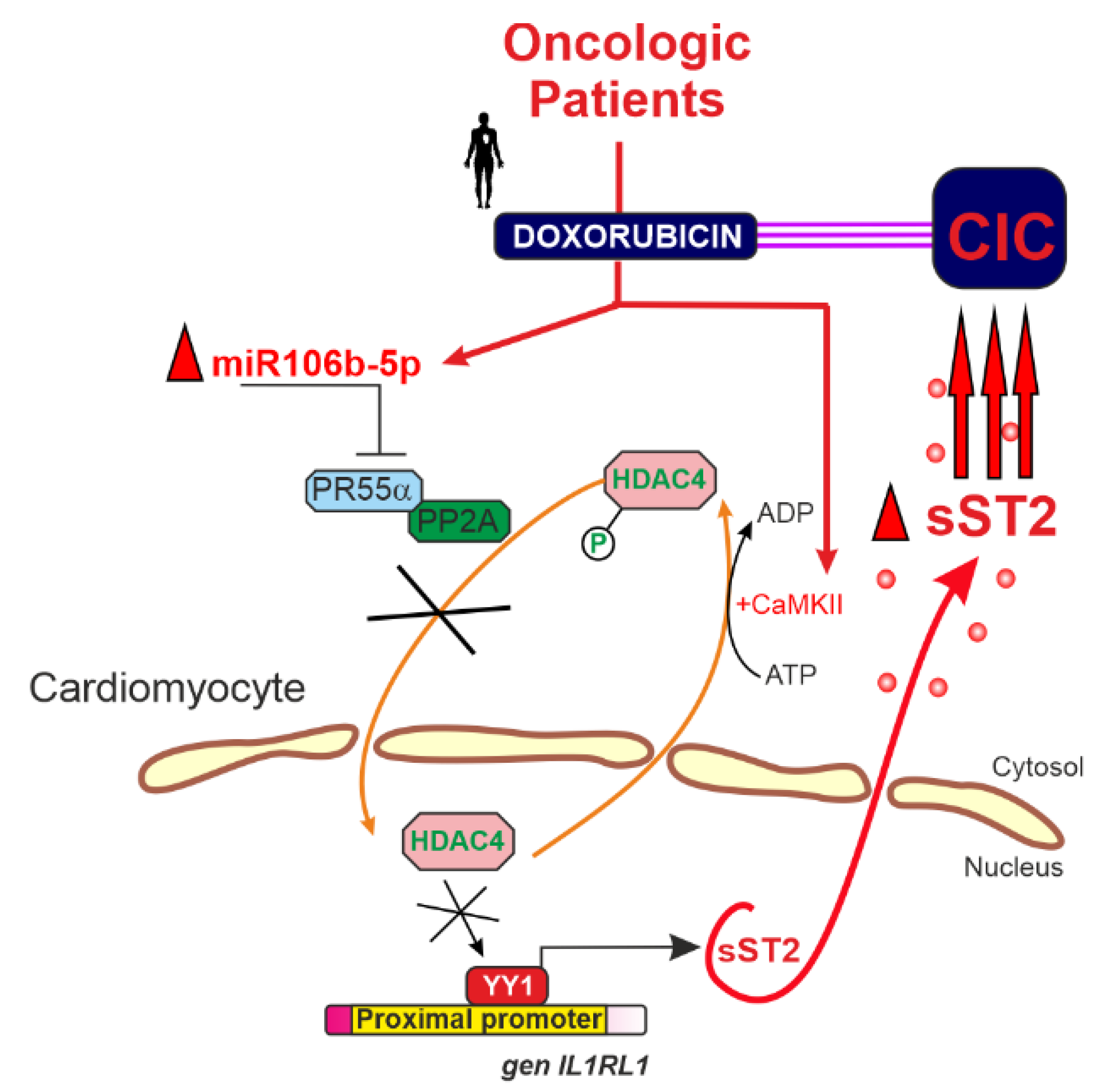
4. miR-106b-5p in Cancer Therapy-Related Cardiovascular Toxicity: A Key Mediator of Cardiac Dysfunction
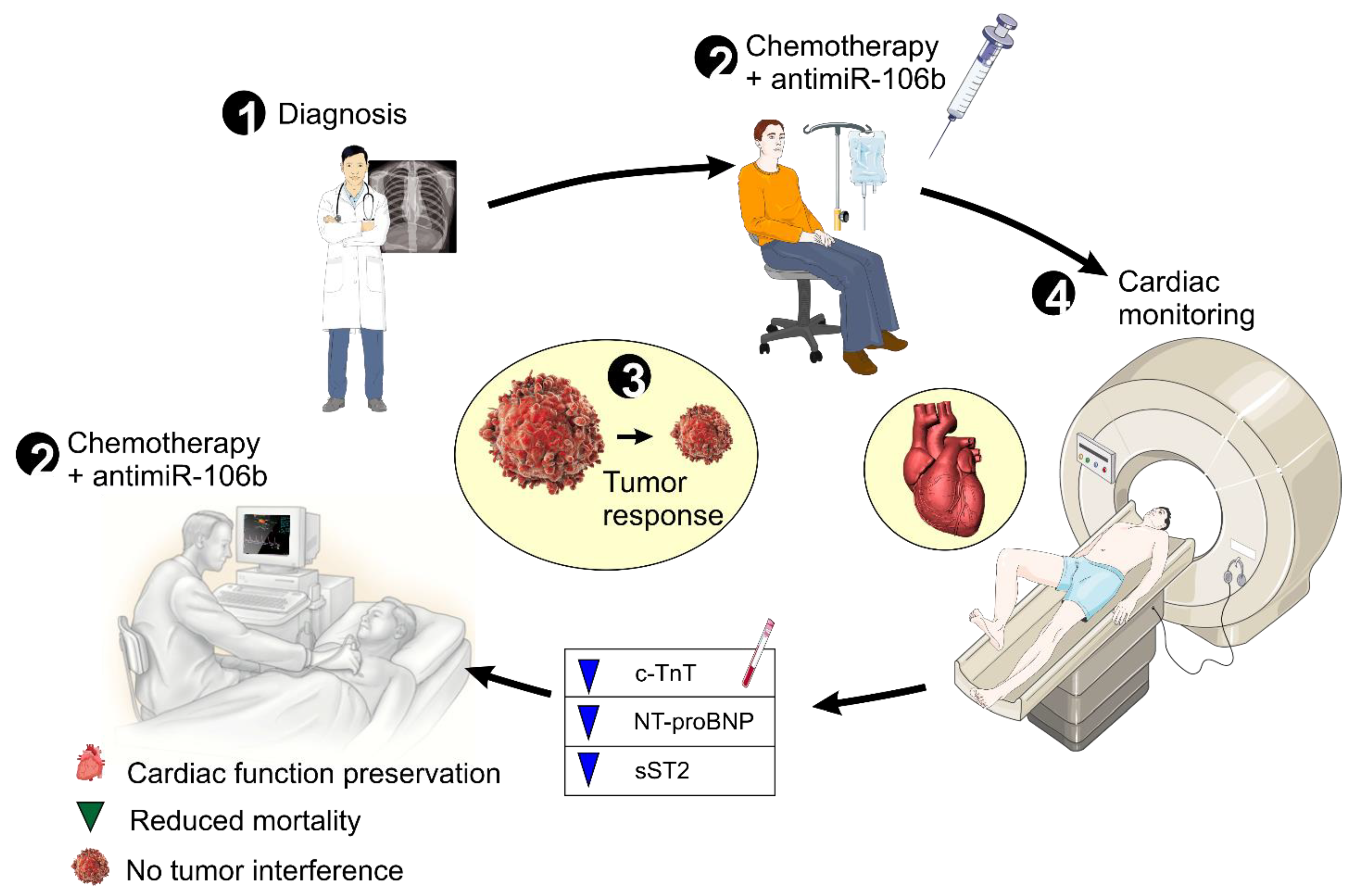
5. Clinical Implications and Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3’UTR | 3’ Untranslated Region |
| AKT | Protein kinase B |
| AM106 | AntimiR específico contra miR-106b-5p |
| BCL2 | B-cell lymphoma 2 |
| BNP | Brain Natriuretic Peptide |
| cTnT | Cardiac Troponin T |
| EF | Ejection Fraction |
| EMT | Epithelial-to-Mesenchymal Transition |
| GMP | Good Manufacturing Practice |
| HDAC4 | Histone Deacetylase 4 |
| NT-proBNP | N-terminal pro b-type Natriuretic Peptide |
| PI3K | Phosphoinositide 3-Kinase |
| PP2A | Protein Phosphatase 2A |
| PR55α | Regulatory subunit of PP2A |
| SMAD7 | Mothers against decapentaplegic homolog 7 |
| sST2 | Soluble Suppression of Tumorigenicity 2 |
| TGF-β | Transforming Growth Factor Beta |
| YY1 | Transcription factor Yin Yang 1 |
References
- Ho, P.T.B.; Clark, I.M.; Le, L.T.T. MicroRNA-Based Diagnosis and Therapy. Int. J. Mol. Sci. 2022, 23, 7167. [Google Scholar] [CrossRef]
- MicroRNA MiR-106b|Cancer Genetics Web. Available online: http://www.cancerindex.org/geneweb/MIR106B.htm (accessed on 9 April 2025).
- Hill, M.; Tran, N. MiRNA Interplay: Mechanisms and Consequences in Cancer. Dis. Model. Mech. 2021, 14, dmm047662. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Sen, A.; Saini, S.; Dwivedi, S.; Agrawal, R.; Bansal, A.; Shekhar, S. MicroRNA Significance in Cancer: An Updated Review on Diagnostic, Prognostic, and Therapeutic Perspectives. Ejifcc 2024, 35, 265–284. [Google Scholar]
- Sessa, F.; Salerno, M.; Esposito, M.; Cocimano, G.; Pomara, C. MiRNA Dysregulation in Cardiovascular Diseases: Current Opinion and Future Perspectives. Int. J. Mol. Sci. 2023, 24, 5192. [Google Scholar] [CrossRef]
- Guarnieri, A.L.; Towers, C.G.; Drasin, D.J.; Oliphant, M.U.J.; Andrysik, Z.; Hotz, T.J.; Vartuli, R.L.; Linklater, E.S.; Pandey, A.; Khanal, S.; et al. The miR-106b-25 Cluster Mediates Breast Tumor Initiation through Activation of NOTCH1 via Direct Repression of NEDD4L. Oncogene 2018, 37, 3879–3893. [Google Scholar] [CrossRef]
- Yang, C.; Dou, R.; Yin, T.; Ding, J. MiRNA-106b-5p in Human Cancers: Diverse Functions and Promising Biomarker. Biomed. Pharmacother. 2020, 127, 110211. [Google Scholar] [CrossRef]
- Pereira, J.; Santos, M.; Delabio, R.; Barbosa, M.; Smith, M.; Payão, S.; Rasmussen, L. Analysis of Gene Expression of MiRNA-106b-5p and TRAIL in the Apoptosis Pathway in Gastric Cancer. Genes 2020, 11, 393. [Google Scholar] [CrossRef]
- Melman, Y.F.; Shah, R.; Das, S. MicroRNAs in Heart Failure: Is the Picture Becoming Less MiRky? Circ. Heart Fail. 2014, 7, 203–214. [Google Scholar] [CrossRef]
- He, C.; Chen, H.; Liu, Y.; Li, X.; Zhang, C.; Qin, Q.; Pang, Q. MiR-106b-5p Promotes Cell Proliferation and Cell Cycle Progression by Directly Targeting CDKN1A in Osteosarcoma. Exp. Ther. Med. 2020, 19, 3203–3210. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Collazo-Lorduy, A.; Castillo-Martin, M.; Gong, Y.; Wang, L.; Oh, W.K.; Galsky, M.D.; Cordon-Cardo, C.; Zhu, J. Identification of MicroR-106b as a Prognostic Biomarker of P53-like Bladder Cancers by ActMiR. Oncogene 2018, 37, 5858–5872. [Google Scholar] [CrossRef] [PubMed]
- Mastropasqua, F.; Marzano, F.; Valletti, A.; Aiello, I.; Di Tullio, G.; Morgano, A.; Liuni, S.; Ranieri, E.; Guerrini, L.; Gasparre, G.; et al. TRIM8 Restores p53 Tumour Suppressor Function by Blunting N-MYC Activity in Chemo-Resistant Tumours. Mol. Cancer 2017, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Lax, A.; Soler, F.; Fernandez del Palacio, M.J.; Pascual-Oliver, S.; Ballester, M.R.; Fuster, J.J.; Pascual-Figal, D.; Asensio-Lopez, M. del C. Silencing of MicroRNA-106b-5p Prevents Doxorubicin-Mediated Cardiotoxicity Through Modulation of the PR55α/YY1/SST2 Signaling Axis. Mol. Ther. Nucleic Acids 2023, 32, 704–720. [Google Scholar] [CrossRef] [PubMed]
- US20240417727A1—Antagonist of the Mirna-106b-5p to Treat or Prevent the Cardiotoxicity That Is Developed in Oncologic Patients after Receiving Anthracycline-Based Chemotherapy—Google Patents. Available online: https://patents.google.com/patent/US20240417727A1/en?q=(microRNA106b-5p)&oq=microRNA106b-5p (accessed on 9 April 2025).
- Liu, X.; Fortin, K.; Mourelatos, Z. MicroRNAs: Biogenesis and Molecular Functions. Brain Pathol. 2008, 18, 113–121. [Google Scholar] [CrossRef]
- Hynes, C.; Kakumani, P.K. Regulatory Role of RNA-Binding Proteins in MicroRNA Biogenesis. Front. Mol. Biosci. 2024, 11, 1374843. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Salmena, L.; Riccardi, L.; Fornari, A.; Song, M.S.; Hobbs, R.M.; Sportoletti, P.; Varmeh, S.; Egia, A.; Fedele, G.; et al. Identification of the MiR-106b~25 MicroRNA Cluster as a Proto-Oncogenic PTEN-Targeting Intron That Cooperates with Its Host Gene MCM7 in Transformation. Sci. Signal. 2010, 3, ra29. [Google Scholar] [CrossRef]
- Gupta, S.; Read, D.E.; Deepti, A.; Cawley, K.; Gupta, A.; Oommen, D.; Verfaillie, T.; Matus, S.; Smith, M.A.; Mott, J.L.; et al. Perk-Dependent Repression of MiR-106b-25 Cluster Is Required for ER Stress-Induced Apoptosis. Cell Death Dis. 2012, 3, ra29. [Google Scholar] [CrossRef]
- Sagar, S.K. MiR-106b as an Emerging Therapeutic Target in Cancer. Genes Dis. 2021, 9, 889–899. [Google Scholar] [CrossRef]
- Daneshpour, M.; Ghadimi-Daresajini, A. Overview of MiR-106a Regulatory Roles: From Cancer to Aging. Bioengineering 2023, 10, 892. [Google Scholar] [CrossRef]
- Farré, P.L.; Duca, R.B.; Massillo, C.; Dalton, G.N.; Graña, K.D.; Gardner, K.; Lacunza, E.; De Siervi, A. Mir-106b-5p: A Master Regulator of Potential Biomarkers for Breast Cancer Aggressiveness and Prognosis. Int. J. Mol. Sci. 2021, 22, 11135. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.E.; Song, Y.S.; Cho, E.Y.; Lee, A. MiR-106b-5p and MiR-17-5p Could Predict Recurrence and Progression in Breast Ductal Carcinoma in Situ Based on the Transforming Growth Factor-Beta Pathway. Breast Cancer Res. Treat. 2019, 176, 119–130. [Google Scholar] [CrossRef]
- Lee, J.; Yim, K.; Kim, D.-M.; Song, Y.-S.; Lee, A. MiR-106b-5p, MiR-17-5p to Predict Recurrence and Progression in Breast DCIS Model Based on TGFβ Paradox. J. Clin. Oncol. 2017, 35, e23019. [Google Scholar] [CrossRef]
- Wang, Z.; Li, T.E.; Chen, M.; Pan, J.J.; Shen, K.W. MiR-106b-5p Contributes to the Lung Metastasis of Breast Cancer via Targeting CNN1 and Regulating Rho/ROCK1 Pathway. Aging 2020, 12, 1867–1887. [Google Scholar] [CrossRef]
- Tufail, M. PTEN-Mediated Resistance in Cancer: From Foundation to Future Therapies. Toxicol. Reports 2025, 14, 101987. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, Y.; Liu, Y.; Zhou, M.; Lu, Y.; Yuan, L.; Zhang, C.; Hong, M.; Wang, S.; Li, X. MiR-106b Induces Cell Radioresistance via the PTEN/PI3K/AKT Pathways and P21 in Colorectal Cancer. J. Transl. Med. 2015, 13, 252. [Google Scholar] [CrossRef]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt Signal Transduction for Cancer Therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Mohamed Mansoor, M.A.; Zhu, X.; Ashiqueali, S.A.; Alam, M.T.; Winiarska, H.; Pazdrowski, P.; Kaminski, F.; Copik, A.; Masternak, M.M.; Kuznar-Kaminska, B. Circulating MicroRNAs as a Prognostic Tool to Determine Treatment Efficacy in Lung Cancer Patients Undergoing Pembrolizumab PD-1 Blockade Immunotherapy. Cancers 2024, 16, 4202. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Pan, C.; Yao, G.; Liu, B.; Ma, T.; Xia, Y.; Jiang, W.; Chen, L.; Chen, Y. MiR-106b-5p Promotes Proliferation and Inhibits Apoptosis by Regulating BTG3 in Non-Small Cell Lung Cancer. Cell. Physiol. Biochem. 2017, 44, 1545–1558. [Google Scholar] [CrossRef] [PubMed]
- Ling, B.; Liao, X.; Tang, Q.; Ye, G.; Bin, X.; Wang, J.; Pang, Y.; Qi, G. MicroRNA-106b-5p Inhibits Growth and Progression of Lung Adenocarcinoma Cells by Downregulating IGSF10. Aging 2021, 13, 18740–18756. [Google Scholar] [CrossRef]
- Mehlich, D.; Garbicz, F.; Włodarski, P.K. The Emerging Roles of the Polycistronic MiR-106b∼25 Cluster in Cancer—A Comprehensive Review. Biomed. Pharmacother. 2018, 107, 1183–1195. [Google Scholar] [CrossRef]
- Lucarini, V.; Nardozi, D.; Angiolini, V.; Benvenuto, M.; Focaccetti, C.; Carrano, R.; Besharat, Z.M.; Bei, R.; Masuelli, L. Tumor Microenvironment Remodeling in Gastrointestinal Cancer: Role of MiRNAs as Biomarkers of Tumor Invasion. Biomedicines 2023, 11, 1761. [Google Scholar] [CrossRef]
- Zhu, Z.; Yang, Q.; Zhang, B.; Wu, W.; Yuan, F.; Zhu, Z. MiR-106b Promotes Metastasis of Early Gastric Cancer by Targeting ALEX1 in Vitro and in Vivo. Cell. Physiol. Biochem. 2019, 52, 606–616. [Google Scholar] [CrossRef]
- Li, F.; Guo, Y.; Liu, J.; Zhang, R. The Significance of Elevated Plasma Expression of MicroRNA 106b~25 Clusters in Gastric Cancer. PLoS ONE 2017, 12, e0178427. [Google Scholar] [CrossRef]
- Pan, M.; Chen, Q.; Lu, Y.; Wei, F.; Chen, C.; Tang, G.; Huang, H. MiR-106b-5p Regulates the Migration and Invasion of Colorectal Cancer Cells by Targeting FAT4. Biosci. Rep. 2020, 40, BSR20200098. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Weng, W.; Xu, M.; Wang, Q.; Tan, C.; Sun, H.; Wang, L.; Huang, D.; Du, X.; Sheng, W. MiR-106b-5p Inhibits the Invasion and Metastasis of Colorectal Cancer by Targeting CTSA. Onco Targets Ther. 2018, 11, 3835–3845. [Google Scholar] [CrossRef]
- Shen, G.; Jia, H.; Tai, Q.; Li, Y.; Chen, D. MiR-106b Downregulates Adenomatous Polyposis Coli and Promotes Cell Proliferation in Human Hepatocellular Carcinoma. Carcinogenesis 2013, 34, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.A.; Wang, Z.; Yang, C. MicroRNA Regulation of the Small Rho GTPase Regulators—Complexities and Opportunities in Targeting Cancer Metastasis. Cancers 2020, 12, 1092. [Google Scholar] [CrossRef]
- MIR106B (MicroRNA 106b). Available online: https://atlasgeneticsoncology.org/gene/51084/mir106b-(microrna-106b) (accessed on 9 April 2025).
- Yang, F.; Sun, Z.; Wang, D.; Du, T. MiR-106b-5p Regulates Esophageal Squamous Cell Carcinoma Progression by Binding to HPGD. BMC Cancer 2022, 22, 308. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, I.; Ishige, F.; Iwatate, Y.; Gunji, H.; Shiratori, F.; Kuwayama, N.; Nabeya, Y.; Takeshita, N.; Matsubara, H. Usefulness of Serum MiR-1246/MiR-106b Ratio in Patients with Esophageal Squamous Cell Carcinoma. Oncol. Lett. 2020, 20, 350. [Google Scholar] [CrossRef]
- Lu, J.; Wei, J.-H.; Feng, Z.-H.; Chen, Z.-H.; Wang, Y.-Q.; Huang, Y.; Fang, Y.; Liang, Y.-P.; Cen, J.-J.; Pan, Y.-H.; et al. MiR-106b-5p Promotes Renal Cell Carcinoma Aggressiveness and Stem-Cell-like Phenotype by Activating Wnt/β-Catenin Signalling. Oncotarget 2017, 8, 21461–21471. [Google Scholar] [CrossRef]
- Xiang, W.; He, J.; Huang, C.; Chen, L.; Tao, D.; Wu, X.; Wang, M.; Luo, G.; Xiao, X.; Zeng, F.; et al. MiR-106b-5p Targets Tumor Suppressor Gene SETD2 to Inactive Its Function in Clear Cell Renal Cell Carcinoma. Oncotarget 2015, 6, 4066–4079. [Google Scholar] [CrossRef]
- Miao, L.J.; Yan, S.; Zhuang, Q.F.; Mao, Q.Y.; Xue, D.; He, X.Z.; Chen, J.P. MiR-106b Promotes Proliferation and Invasion by Targeting Capicua through MAPK Signaling in Renal Carcinoma Cancer. Onco Targets Ther. 2019, 12, 3595–3607. [Google Scholar] [CrossRef]
- de Sousa Neto, P.I.; Pinto, V.B.P.; Piancó, E.D.S.; Gomes, M.L.; Monteiro, S.C.M.; Vidal, F.C.B.; Nascimento, M.d.D.S.B.; Pinho, J.D.; Calixto, J.d.R.R.; de Andrade, M.S. The Role of MicroRNAs in Non-Invasive Diagnosis of Bladder Cancer: A Systematic Review. Einstein 2024, 22, eRW0611. [Google Scholar] [CrossRef]
- Chen, S.; Lu, C.; Lin, S.; Sun, C.; Wen, Z.; Ge, Z.; Chen, W.; Li, Y.; Zhang, P.; Wu, Y.; et al. A Panel Based on Three-MiRNAs as Diagnostic Biomarker for Prostate Cancer. Front. Genet. 2024, 15, 1371441. [Google Scholar] [CrossRef] [PubMed]
- Pavalean, M.I.; Dobre, M.; Pelisenco, I.A.; Madan, V.L.; Milanesi, E.; Hinescu, M.E. Association of MiRNA-17-92 Cluster with Muscle Invasion in Bladder Cancer. Int. J. Mol. Sci. 2025, 26, 7546. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.; Ding, Y.; Xi, H.; Ruan, H.; Lu, F.; Ma, S.; Wang, J. Exosomal MiR-106b-5p Derived from Melanoma Cell Promotes Primary Melanocytes Epithelial-Mesenchymal Transition through Targeting EphA4. J. Exp. Clin. Cancer Res. 2021, 40, 107. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gao, Y.; Cao, Y.; He, F.; Jiang, R.; Liu, H.; Cai, H.; Zan, T. Extracellular RNA in Melanoma: Advances, Challenges, and Opportunities. Front. Cell Dev. Biol. 2023, 11, 1141543. [Google Scholar] [CrossRef]
- Ye, Q.; Li, Z.; Li, Y.; Li, Y.; Zhang, Y.; Gui, R.; Cui, Y.; Zhang, Q.; Qian, L.; Xiong, Y.; et al. Exosome-Derived MicroRNA: Implications in Melanoma Progression, Diagnosis and Treatment. Cancers 2023, 15, 80. [Google Scholar] [CrossRef]
- Zhou, Q.; Hu, Q. Oncogenic MiR-106b-5p Promotes Cisplatin Resistance in Triple-Negative Breast Cancer by Targeting GDF11. Histol. Histopathol. 2024, 39, 533–541. [Google Scholar] [CrossRef]
- Mizielska, A.; Dziechciowska, I.; Szczepański, R.; Cisek, M.; Dąbrowska, M.; Ślężak, J.; Kosmalska, I.; Rymarczyk, M.; Wilkowska, K.; Jacczak, B.; et al. Doxorubicin and Cisplatin Modulate MiR-21, MiR-106, MiR-126, MiR-155 and MiR-199 Levels in MCF7, MDA-MB-231 and SK-BR-3 Cells That Makes Them Potential Elements of the DNA-Damaging Drug Treatment Response Monitoring in Breast Cancer Cells-A Preliminary Study. Genes 2023, 14, 702. [Google Scholar] [CrossRef]
- Hashemi, M.; Taheriazam, A.; Daneii, P.; Hassanpour, A.; Kakavand, A.; Rezaei, S.; Hejazi, E.S.; Aboutalebi, M.; Gholamrezaie, H.; Saebfar, H.; et al. Targeting PI3K/Akt Signaling in Prostate Cancer Therapy. J. Cell Commun. Signal. 2023, 17, 423–443. [Google Scholar] [CrossRef]
- Doghish, A.S.; Ismail, A.; El-Mahdy, H.A.; Elkady, M.A.; Elrebehy, M.A.; Sallam, A.A.M. A Review of the Biological Role of MiRNAs in Prostate Cancer Suppression and Progression. Int. J. Biol. Macromol. 2022, 197, 141–156, Erratum in Int. J. Biol. Macromol. 2025, 319, 146252. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Y.; Wang, Z.; Xia, Q. LncRNA PSMG3-AS1 Is Upregulated in Prostate Carcinoma and Downregulates MiR-106b through DNA Methylation. Syst. Biol. Reprod. Med. 2023, 69, 264–270. [Google Scholar] [CrossRef]
- Liang, H.; Studach, L.; Hullinger, R.L.; Xie, J.; Andrisani, O.M. Down-Regulation of RE-1 Silencing Transcription Factor (REST) in Advanced Prostate Cancer by Hypoxia-Induced MiR-106b~25. Exp. Cell Res. 2014, 320, 188–199. [Google Scholar] [CrossRef]
- Xie, H.; Yao, J.; Wang, Y.; Ni, B. Exosome-Transmitted CircVMP1 Facilitates the Progression and Cisplatin Resistance of Non-Small Cell Lung Cancer by Targeting MiR-524-5p-METTL3/SOX2 Axis. Drug Deliv. 2022, 29, 1257–1271. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Li, S.; Li, Y.; Wan, M.; Fang, X.; Zhao, Y.; Zuo, W.; Long, D.; Xuan, Y. Regulation of BTG3 by MicroRNA-20b-5p in Non-Small Cell Lung Cancer. Oncol. Lett. 2019, 18, 137–144. [Google Scholar] [CrossRef] [PubMed]
- El-Aal, A.E.A.; Elshafei, A.; Ismail, M.Y.; El-Shafey, M.M. Identification of MiR-106b-5p, MiR-601, and MiR-760 Expression and Their Clinical Values in Non-Small Cell Lung Cancer (NSCLC) Patients’ Serum. Pathol. Res. Pract. 2023, 248, 154663. [Google Scholar] [CrossRef] [PubMed]
- Stolfi, C.; Troncone, E.; Marafini, I.; Monteleone, G. Role of TGF-Beta and Smad7 in Gut Inflammation, Fibrosis and Cancer. Biomolecules 2020, 11, 17. [Google Scholar] [CrossRef]
- Su, J.; Chen, D.; Ruan, Y.; Tian, Y.; Lv, K.; Zhou, X.; Ying, D.; Lu, Y. LncRNA MBNL1-AS1 Represses Gastric Cancer Progression via the TGF-β Pathway by Modulating MiR-424-5p/Smad7 Axis. Bioengineered 2022, 13, 6978–6995. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, K.; Yang, Y.; Guo, Z.; Wang, X.; Teng, B.; Zhao, Q.; Huang, C.; Qiu, Z. MEF2A-Mediated LncRNA HCP5 Inhibits Gastric Cancer Progression via MiR-106b-5p/P21 Axis. Int. J. Biol. Sci. 2021, 17, 623–634. [Google Scholar] [CrossRef]
- Wei, R.; Xiao, Y.; Song, Y.; Yuan, H.; Luo, J.; Xu, W. FAT4 Regulates the EMT and Autophagy in Colorectal Cancer Cells in Part via the PI3K-AKT Signaling Axis. J. Exp. Clin. Cancer Res. 2019, 38, 112. [Google Scholar] [CrossRef]
- Quan, L.; Stassen, A.P.M.; Ruivenkamp, C.A.L.; van Wezel, T.; Fijneman, R.J.A.; Hutson, A.; Kakarlapudi, N.; Hart, A.A.M.; Demant, P. Most Lung and Colon Cancer Susceptibility Genes Are Pair-Wise Linked in Mice, Humans and Rats. PLoS ONE 2011, 6, e14727. [Google Scholar] [CrossRef]
- Liu, S.; An, G.; Cao, Q.; Li, T.; Jia, X.; Lei, L. The MiR-106b/NR2F2-AS1/PLEKHO2 Axis Regulates Migration and Invasion of Colorectal Cancer through the MAPK Pathway. Int. J. Mol. Sci. 2021, 22, 5877. [Google Scholar] [CrossRef]
- Yao, J.; Wang, J.; Xu, Y.; Guo, Q.; Sun, Y.; Liu, J.; Li, S.; Guo, Y.; Wei, L. CDK9 Inhibition Blocks the Initiation of PINK1-PRKN-Mediated Mitophagy by Regulating the SIRT1-FOXO3-BNIP3 Axis and Enhances the Therapeutic Effects Involving Mitochondrial Dysfunction in Hepatocellular Carcinoma. Autophagy 2022, 18, 1879–1897. [Google Scholar] [CrossRef]
- Shilbayeh, S.A.R.; Khedr, N.F.; Alshabeeb, M.A.; Alsaleh, A.A.; Alanizi, A.H.; Abd El-Baset, O.A.; Werida, R.H. Genetic Polymorphisms as Predictors of the Response of Hepatocellular Carcinoma Patients to Doxorubicin Chemotherapy: A Genome-Wide Association Study. Front. Pharmacol. 2025, 16, 1604473. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Suo, C.; Zhang, T.; Shen, S.; Gu, X.; Qiu, S.; Zhang, P.; Wei, H.; Ma, W.; Yan, R.; et al. ENO1 Promotes Liver Carcinogenesis through YAP1-Dependent Arachidonic Acid Metabolism. Nat. Chem. Biol. 2023, 19, 1492–1503. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Liu, T.; Zheng, S.; Liu, Q.; Yang, C.; Zhou, J.; Chen, Y.; Sheyhidin, I.; Lu, X. MiR-106b Promotes Migration and Invasion through Enhancing EMT via Downregulation of Smad 7 in Kazakh’s Esophageal Squamous Cell Carcinoma. Tumour Biol. 2016, 37, 14595–14604. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Huang, Y.; Hou, Y.; Li, Q.; Wang, H. Circular RNA ITCH Is a Tumor Suppressor in Clear Cell Renal Cell Carcinoma Metastasis through MiR-106b-5p/PDCD4 Axis. J. Immunol. Res. 2021, 2021, 5524344. [Google Scholar] [CrossRef]
- Jin, N.; Jin, X.; Gu, X.; Na, W.; Zhang, M.; Zhao, R. Screening Biomarkers of Bladder Cancer Using Combined MiRNA and MRNA Microarray Analysis. Mol. Med. Rep. 2015, 12, 3170–3176. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, X.; Yang, Y.; Li, Z.; Du, L.; Dong, Z.; Qu, A.; Jiang, X.; Li, P.; Wang, C. Urinary Cell-Free MicroRNA-106b as a Novel Biomarker for Detection of Bladder Cancer. Med. Oncol. 2014, 31, 197. [Google Scholar] [CrossRef]
- Lin, N.; Zhou, Y.; Lian, X.; Tu, Y. Expression of MicroRNA-106b and Its Clinical Significance in Cutaneous Melanoma. Genet. Mol. Res. 2015, 14, 16379–16385. [Google Scholar] [CrossRef]
- Prasad, R.; Katiyar, S.K. Down-Regulation of MiRNA-106b Inhibits Growth of Melanoma Cells by Promoting G1-Phase Cell Cycle Arrest and Reactivation of P21/WAF1/Cip1 Protein. Oncotarget 2014, 5, 10636–10649. [Google Scholar] [CrossRef]
- Saleh, Y.; Abdelkarim, O.; Herzallah, K.; Abela, G.S. Anthracycline-Induced Cardiotoxicity: Mechanisms of Action, Incidence, Risk Factors, Prevention, and Treatment. Heart Fail. Rev. 2021, 26, 1159–1173. [Google Scholar] [CrossRef]
- Yerukala Sathipati, S.; Tsai, M.J.; Shukla, S.K.; Ho, S.Y. Artificial Intelligence-Driven Pan-Cancer Analysis Reveals MiRNA Signatures for Cancer Stage Prediction. Hum. Genet. Genom. Adv. 2023, 4, 100190. [Google Scholar] [CrossRef]
- Mullany, L.E.; Wolff, R.K.; Slattery, M.L. Effectiveness and Usability of Bioinformatics Tools to Analyze Pathways Associated with MiRNA Expression. Cancer Inform. 2015, 14, 121–130. [Google Scholar] [CrossRef]
- Rincón-Riveros, A.; Morales, D.; Rodríguez, J.A.; Villegas, V.E.; López-Kleine, L. Bioinformatic Tools for the Analysis and Prediction of NcRNA Interactions. Int. J. Mol. Sci. 2021, 22, 11397. [Google Scholar] [CrossRef]
- Das, S.S.; James, M.; Paul, S.; Chakravorty, N. MiRnalyze: An Interactive Database Linking Tool to Unlock Intuitive MicroRNA Regulation of Cell Signaling Pathways. Database 2017, 2017, bax015. [Google Scholar] [CrossRef]
- AI Tool “Sees” Cancer Gene Signatures in Biopsy Images. Available online: https://med.stanford.edu/news/all-news/2024/11/cancer-gene-biopsy.html (accessed on 9 April 2025).
- Borgmästars, E.; De Weerd, H.A.; Lubovac-Pilav, Z.; Sund, M. MiRFA: An Automated Pipeline for MicroRNA Functional Analysis with Correlation Support from TCGA and TCPA Expression Data in Pancreatic Cancer. BMC Bioinform. 2019, 20, 393. [Google Scholar] [CrossRef] [PubMed]
- Shakyawar, S.; Southekal, S.; Guda, C. MintRULS: Prediction of MiRNA-MRNA Target Site Interactions Using Regularized Least Square Method. Genes 2022, 13, 1528. [Google Scholar] [CrossRef] [PubMed]
- Min, S.; Lee, B.; Yoon, S. TargetNet: Functional MicroRNA Target Prediction with Deep Neural Networks. Bioinformatics 2022, 38, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, L.; Wei, X.; Zhao, C.; Li, S.; Li, J.; Le, T.D. Pan-Cancer Characterization of NcRNA Synergistic Competition Uncovers Potential Carcinogenic Biomarkers. PLoS Comput. Biol. 2023, 19, e1011308. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Vlachos, I.S.; Karagkouni, D.; Georgakilas, G.; Kanellos, I.; Vergoulis, T.; Zagganas, K.; Tsanakas, P.; Floros, E.; Dalamagas, T.; et al. DIANA-LncBase v2: Indexing MicroRNA Targets on Non-Coding Transcripts. Nucleic Acids Res. 2016, 44, D231–D238. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. StarBase v2.0: Decoding MiRNA-CeRNA, MiRNA-NcRNA and Protein–RNA Interaction Networks from Large-Scale CLIP-Seq Data. Nucleic Acids Res. 2013, 42, D92–D97. [Google Scholar] [CrossRef]
- Linders, A.N.; Dias, I.B.; López Fernández, T.; Tocchetti, C.G.; Bomer, N.; Van der Meer, P. A Review of the Pathophysiological Mechanisms of Doxorubicin-Induced Cardiotoxicity and Aging. NPJ Aging 2024, 10, 9. [Google Scholar] [CrossRef]
- Asensio-Lopez, M.C.; Lax, A.; Fernandez del Palacio, M.J.; Sassi, Y.; Hajjar, R.J.; Januzzi, J.L.; Bayes-Genis, A.; Pascual-Figal, D.A. Yin-Yang 1 Transcription Factor Modulates ST2 Expression during Adverse Cardiac Remodeling Post-Myocardial Infarction. J. Mol. Cell. Cardiol. 2019, 130, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Asensio-Lopez, M.C.; Sassi, Y.; Soler, F.; Fernandez del Palacio, M.J.; Pascual-Figal, D.; Lax, A. The MiRNA199a/SIRT1/P300/Yy1/SST2 Signaling Axis Regulates Adverse Cardiac Remodeling Following MI. Sci. Rep. 2021, 11, 3915. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.E.; Chen, P.; Chen, S.S.; Ma, T.; Shi, G.; Zhou, Y.; Li, J.; Sheng, L. MiR-106b-5p Promotes Cell Cycle Progression of Malignant Melanoma by Targeting PTEN. Oncol. Rep. 2018, 39, 331–337. [Google Scholar] [CrossRef]
- Li, N.; Miao, Y.; Shan, Y.; Liu, B.; Li, Y.; Zhao, L.; Jia, L. MiR-106b and MiR-93 Regulate Cell Progression by Suppression of PTEN via PI3K/Akt Pathway in Breast Cancer. Cell Death Dis. 2017, 8, e2796. [Google Scholar] [CrossRef]
- Zhang, G.J.; Li, J.S.; Zhou, H.; Xiao, H.X.; Li, Y.; Zhou, T. MicroRNA-106b Promotes Colorectal Cancer Cell Migration and Invasion by Directly Targeting DLC1. J. Exp. Clin. Cancer Res. 2015, 34, 73. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, D.; Zhu, X.; Yang, G.; Ren, M. MiR-106a Associated with Diabetic Peripheral Neuropathy Through the Regulation of 12/15-LOX-Meidiated Oxidative/Nitrative Stress. Curr. Neurovasc. Res. 2017, 14, 117–124. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, H.; Tao, Y.; Xu, Z.; Lai, H. Relationship between Serum MiR-106 and MYL4 Levels and the Prevalence, Risk Stratification, and Prognosis of Atrial Fibrillation. J. Immunol. Res. 2022, 2022, 1069866. [Google Scholar] [CrossRef] [PubMed]
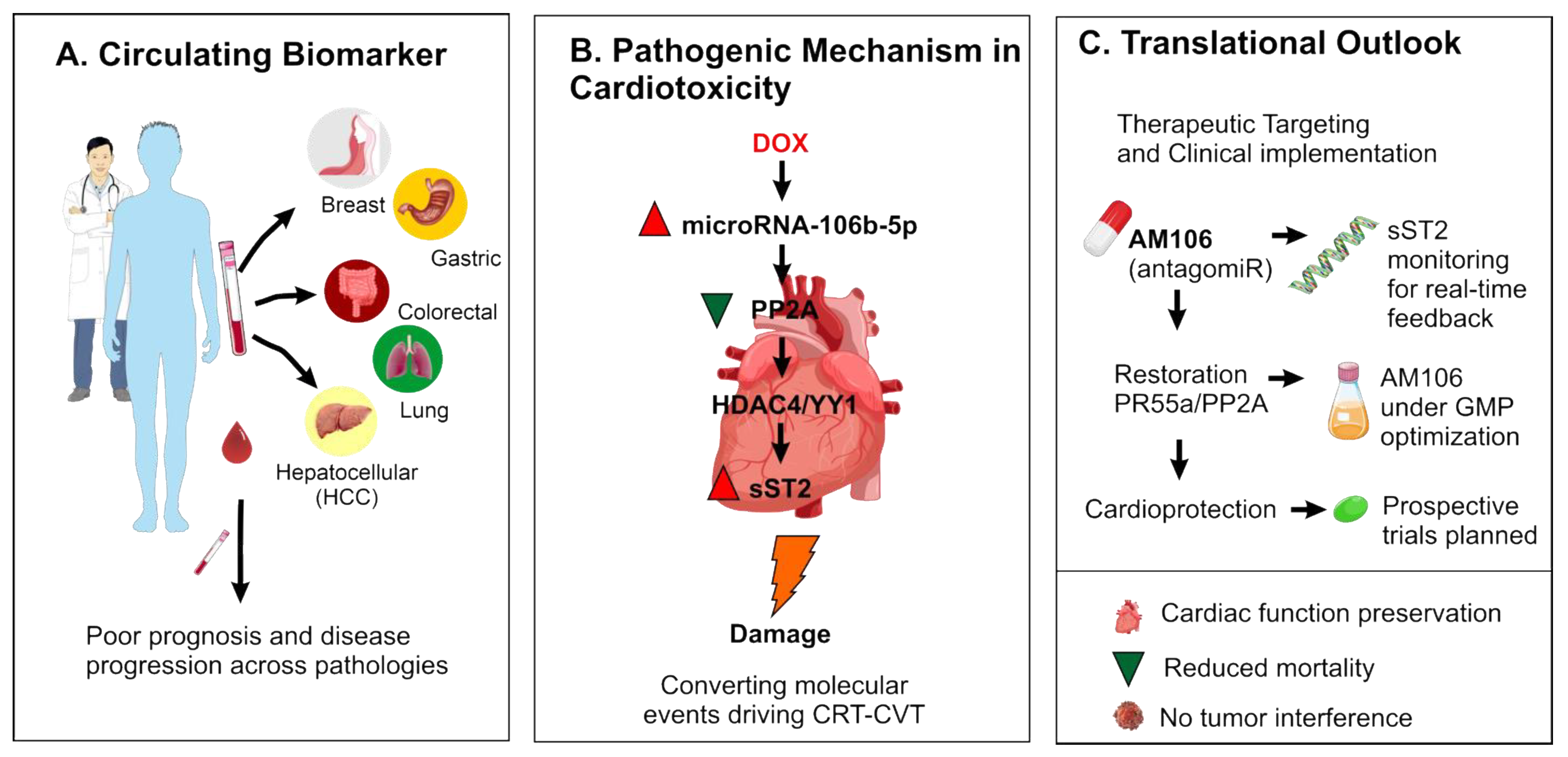
| Cancer Type | Role of microRNA-106b-5p | Key Target Genes | Affected Signaling Pathways | Key References |
|---|---|---|---|---|
| Breast Cancer | Oncogenic/Tumor Suppressor (context-dependent) | PTEN, CNN1, GAB1, GNG12, HBP1, SESN1 | PI3K/AKT, Rho/ROCK1, TGF-β | [21,24,51,52] |
| Prostate Cancer | Oncogenic | PTEN, p21, Caspase-7, adhesion molecules | PI3K/AKT, cell cycle regulation | [53,54,55,56] |
| Non-Small Cell Lung Cancer (NSCLC) | Oncogenic | BTG3, PKD2, IGSF10 (context-dependent) | Proliferation, apoptosis, cisplatin resistance | [57,58,59] |
| Gastric Cancer | Oncogenic | SMAD7, PTEN, ALEX1 | TGF-β inhibition, migration/invasion | [60,61,62] |
| Colorectal Cancer | Oncogenic/Tumor Suppressor (context-dependent) | FAT4, CTSA, DLC-1 | Migration, invasion, metastasis, radioresistance | [63,64,65] |
| Hepatocellular Carcinoma | Oncogenic | PTEN, APC, BTG3, BIM | PI3K/AKT, cell cycle, EMT | [66,67,68] |
| Esophageal Squamous Cell Carcinoma | Oncogenic | PTEN, SMAD-7, HPGD | EMT, proliferation, invasion | [40,69] |
| Renal Cell Carcinoma (RCC) | Oncogenic | SETD2, CIC, Wnt inhibitors (indirect), β-catenin targets (Cyclin D1, MYC, CD44, MMP7) | Wnt/β-catenin activation, MAPK signaling, epigenetic regulation | [42,44,70] |
| Bladder Cancer | Prognostic biomarker/Oncogenic | TP53-related networks, cell cycle regulators | p53-like tumor biology, apoptosis, recurrence risk | [11,71,72] |
| Melanoma | Oncogenic | PTEN (context-dependent), EphA4 | PI3K/AKT, ERK/MAPK, EMT via exosomes | [48,73,74,75] |
| Study Type | Experimental Model | Major Findings | Clinical/Translational Implications | Key References |
|---|---|---|---|---|
| In Vitro | Cancer cell lines (breast, prostate, NSCLC) | miR-106b-5p modulates proliferation, apoptosis, migration, and invasion by targeting PTEN, BTG3, and others | Potential diagnostic and prognostic biomarker; target for therapeutic intervention | [52,92] |
| In Vivo | Animal models of doxorubicin-induced cardiotoxicity | Upregulation of miR-106b-5p linked to cardiac dysfunction; inhibition restored PP2A function and improved outcomes | Provides proof-of-concept for anti-miR therapy (AM106) to prevent cardiotoxicity | [13,93] |
| Clinical | Analysis of circulating miRNA levels in cancer patients | Elevated miR-106b-5p correlates with poor outcomes and higher risk of cardiotoxicity when using anthracyclines | Supports the use of miR-106b-5p as a stratification biomarker and guides personalized therapy strategies | [20,94] |
| Disease Context | miR-106b-5p Function | Key Target Genes | Affected Signaling Pathways |
|---|---|---|---|
| Osteosarcoma | Oncogenic | CDKN1A/p21 | Cell cycle |
| Non-Small Cell Lung Cancer | Oncogenic | BTG3 | Proliferation, Apoptosis |
| Colorectal Cancer | Tumor Suppressor/Oncogenic | FAT4, SLAIN2 | Migration, Invasion, Metastasis |
| Hepatocellular Carcinoma | Oncogenic | PTEN, BTG3, BIM | PI3K/AKT, Proliferation, Apoptosis, Cell cycle |
| Malignant Melanoma | Oncogenic | PTEN | Akt/ERK, Cell cycle |
| Prostate Cancer | Oncogenic | PTEN, p21 | Viability, Migration, Cell cycle |
| Clear Cell Renal Cell Carcinoma | Oncogenic | SETD2 | Cell cycle, Apoptosis |
| Breast Cancer | Oncogenic/Tumor Suppressor | PTEN, CNN1 | PI3K/AKT, Rho/ROCK1, TGF-beta |
| Doxorubicin-Induced Cardiotoxicity | Pro-cardiotoxic | FOG2, sST2 | Apoptosis, Oxidative stress |
| Oxidative Stress in Neonatal Mouse Cardiomyocytes | Cardioprotective | Unknown | Antioxidant, Inflammatory |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asensio Lopez, M.d.C.; Ruiz Ballester, M.; Bastida Nicolas, F.J.; Soler Pardo, F.; Alonso-Romero, J.L.; Caro-Martinez, C.; Pascual Figal, D.; Lax, A. miR-106b-5p as a Central Regulator of Cancer Progression and Chemotherapy-Induced Cardiotoxicity: From Molecular Mechanisms to Clinical Translation. Int. J. Mol. Sci. 2025, 26, 10002. https://doi.org/10.3390/ijms262010002
Asensio Lopez MdC, Ruiz Ballester M, Bastida Nicolas FJ, Soler Pardo F, Alonso-Romero JL, Caro-Martinez C, Pascual Figal D, Lax A. miR-106b-5p as a Central Regulator of Cancer Progression and Chemotherapy-Induced Cardiotoxicity: From Molecular Mechanisms to Clinical Translation. International Journal of Molecular Sciences. 2025; 26(20):10002. https://doi.org/10.3390/ijms262010002
Chicago/Turabian StyleAsensio Lopez, Maria del Carmen, Miriam Ruiz Ballester, Francisco Jose Bastida Nicolas, Fernando Soler Pardo, Jose Luis Alonso-Romero, Cesar Caro-Martinez, Domingo Pascual Figal, and Antonio Lax. 2025. "miR-106b-5p as a Central Regulator of Cancer Progression and Chemotherapy-Induced Cardiotoxicity: From Molecular Mechanisms to Clinical Translation" International Journal of Molecular Sciences 26, no. 20: 10002. https://doi.org/10.3390/ijms262010002
APA StyleAsensio Lopez, M. d. C., Ruiz Ballester, M., Bastida Nicolas, F. J., Soler Pardo, F., Alonso-Romero, J. L., Caro-Martinez, C., Pascual Figal, D., & Lax, A. (2025). miR-106b-5p as a Central Regulator of Cancer Progression and Chemotherapy-Induced Cardiotoxicity: From Molecular Mechanisms to Clinical Translation. International Journal of Molecular Sciences, 26(20), 10002. https://doi.org/10.3390/ijms262010002








