Methods for Assessing MAGL Enzymatic Activity: An Extensive Review of Past and Emerging Approaches
Abstract
1. Introduction
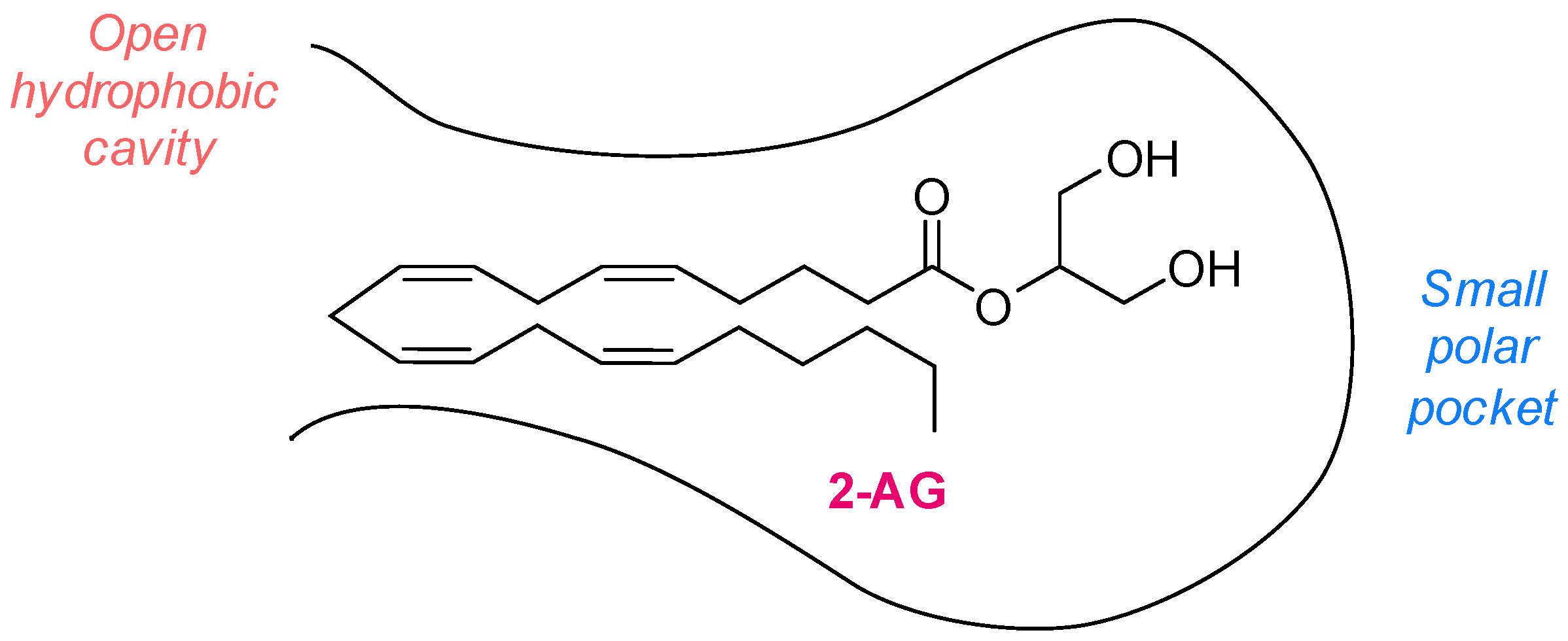
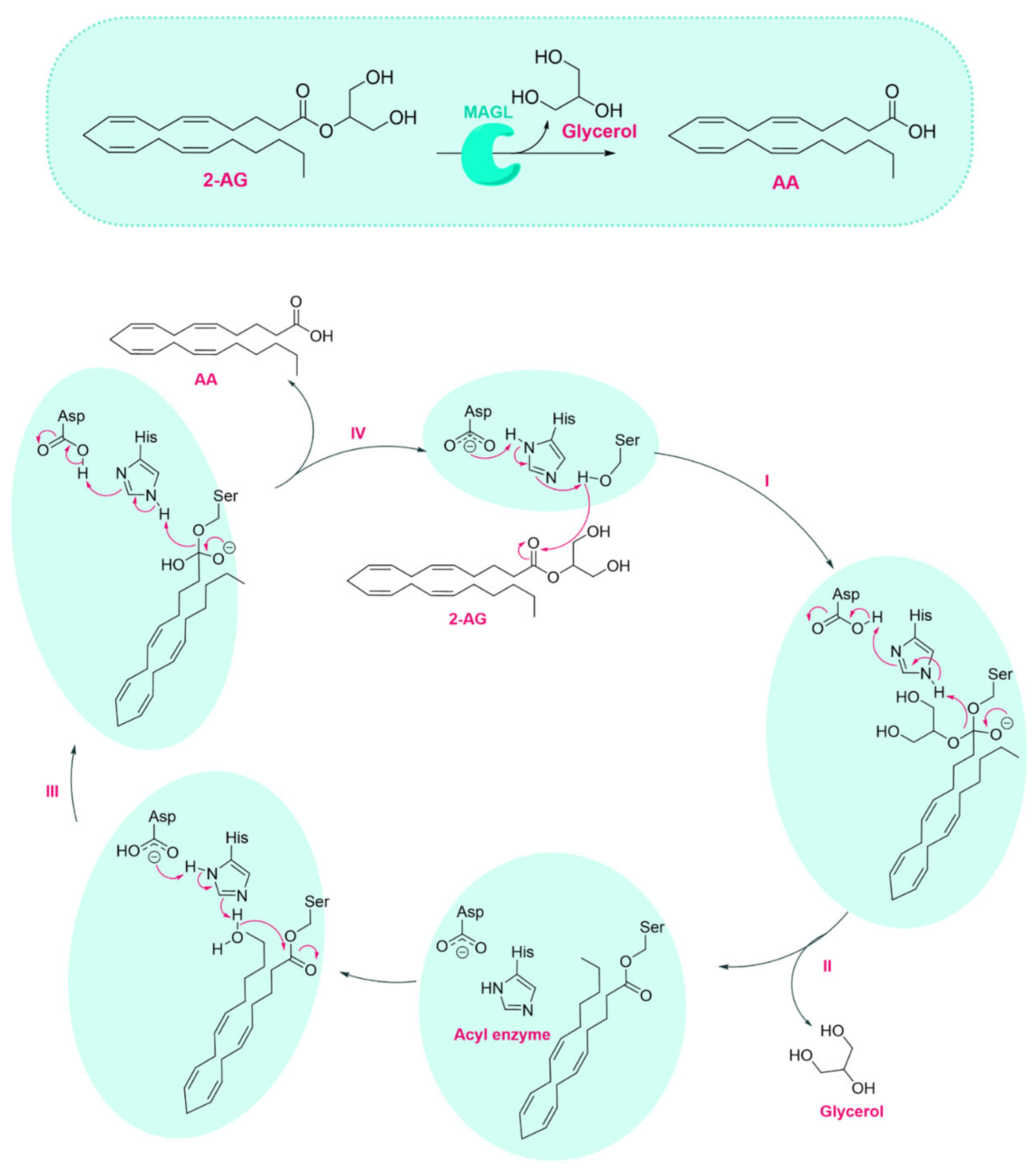
1.1. MAGL Structure and Catalytic Mechanism
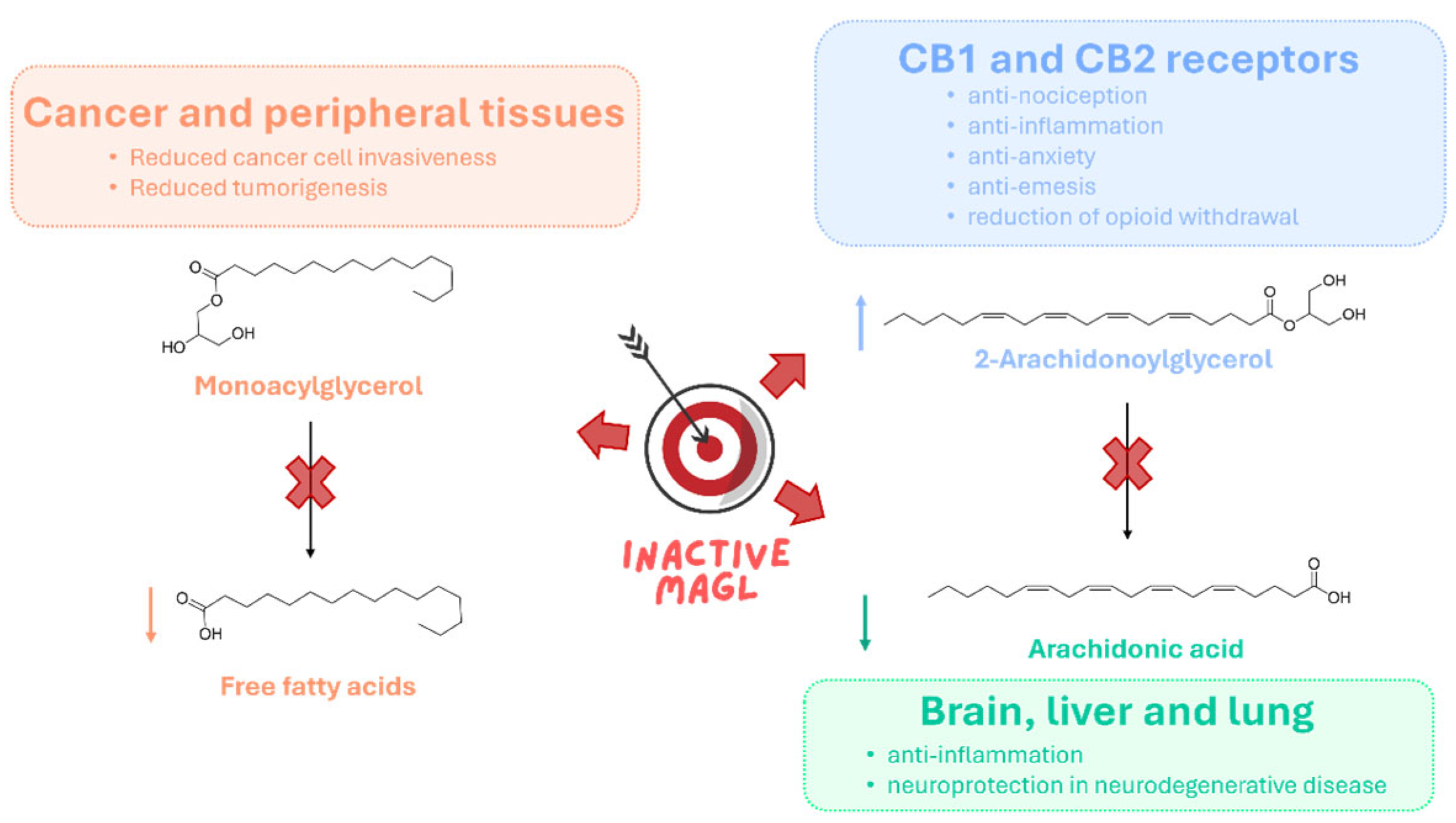
1.2. Therapeutic Potential of MAGL Inhibition
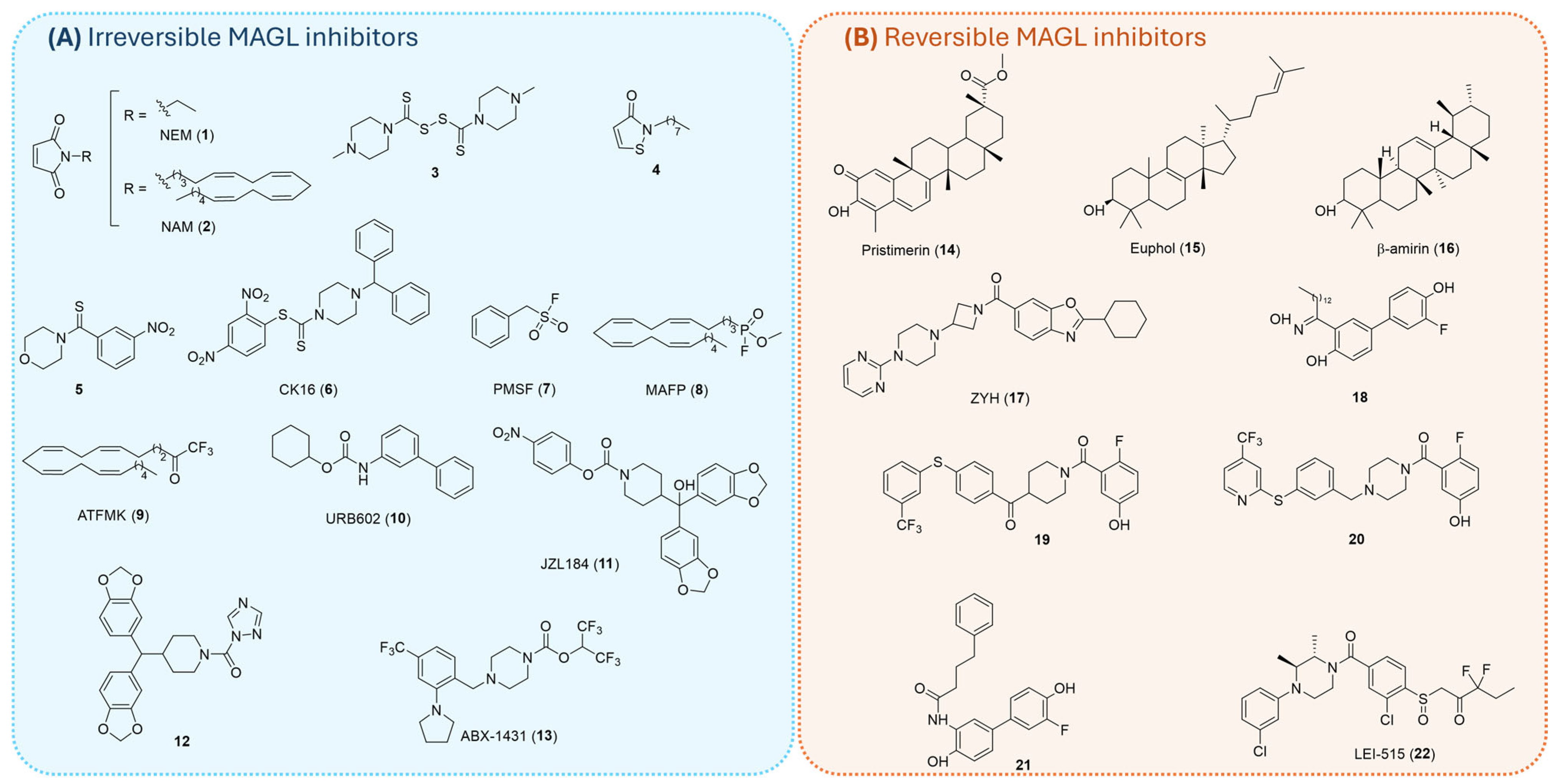
1.3. MAGL Inhibitors
1.3.1. Irreversible Inhibitors
1.3.2. Reversible Inhibitors
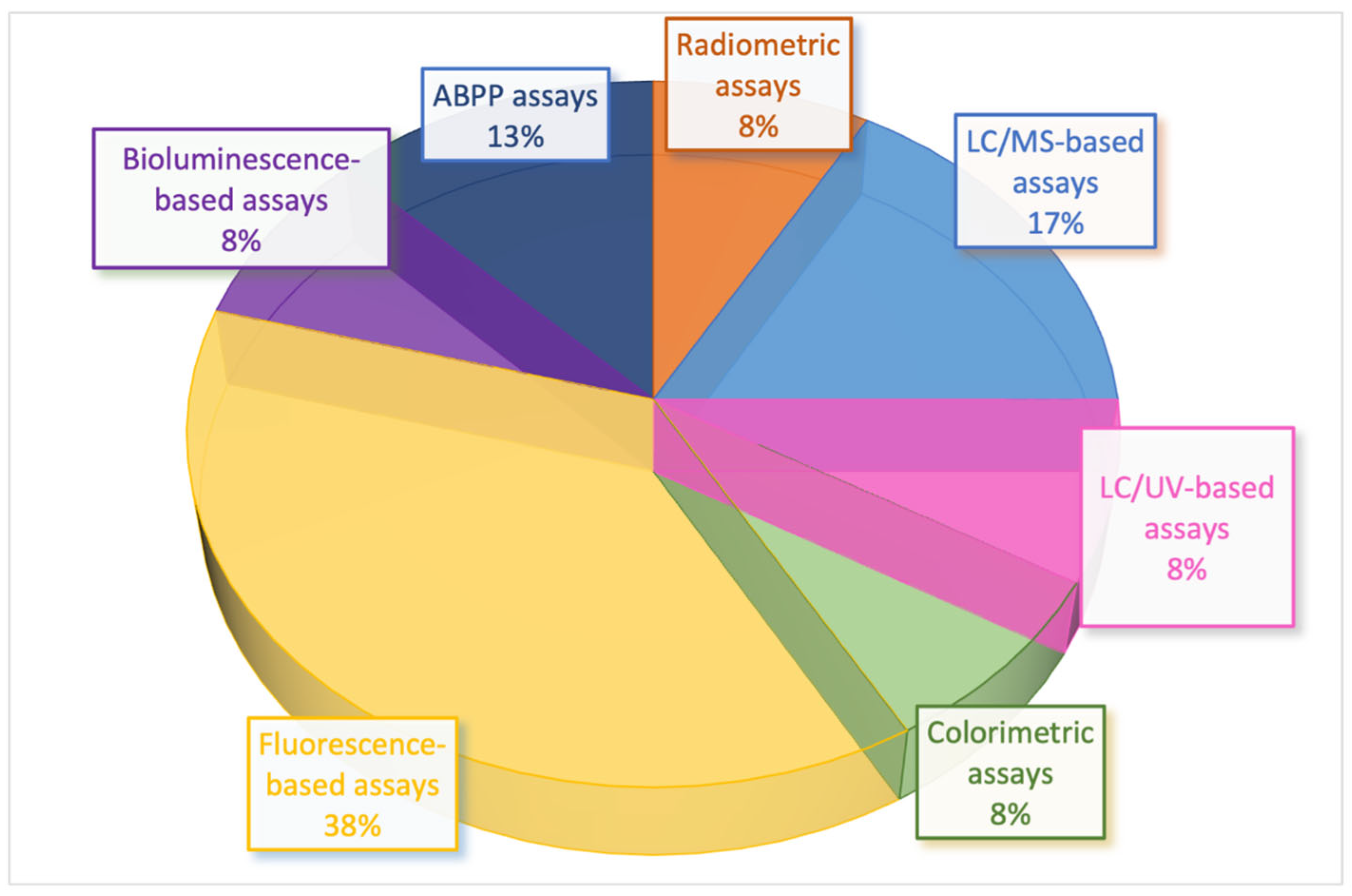
2. Biochemical Methods to Assess MAGL Activity: From Conventional to Novel Approaches
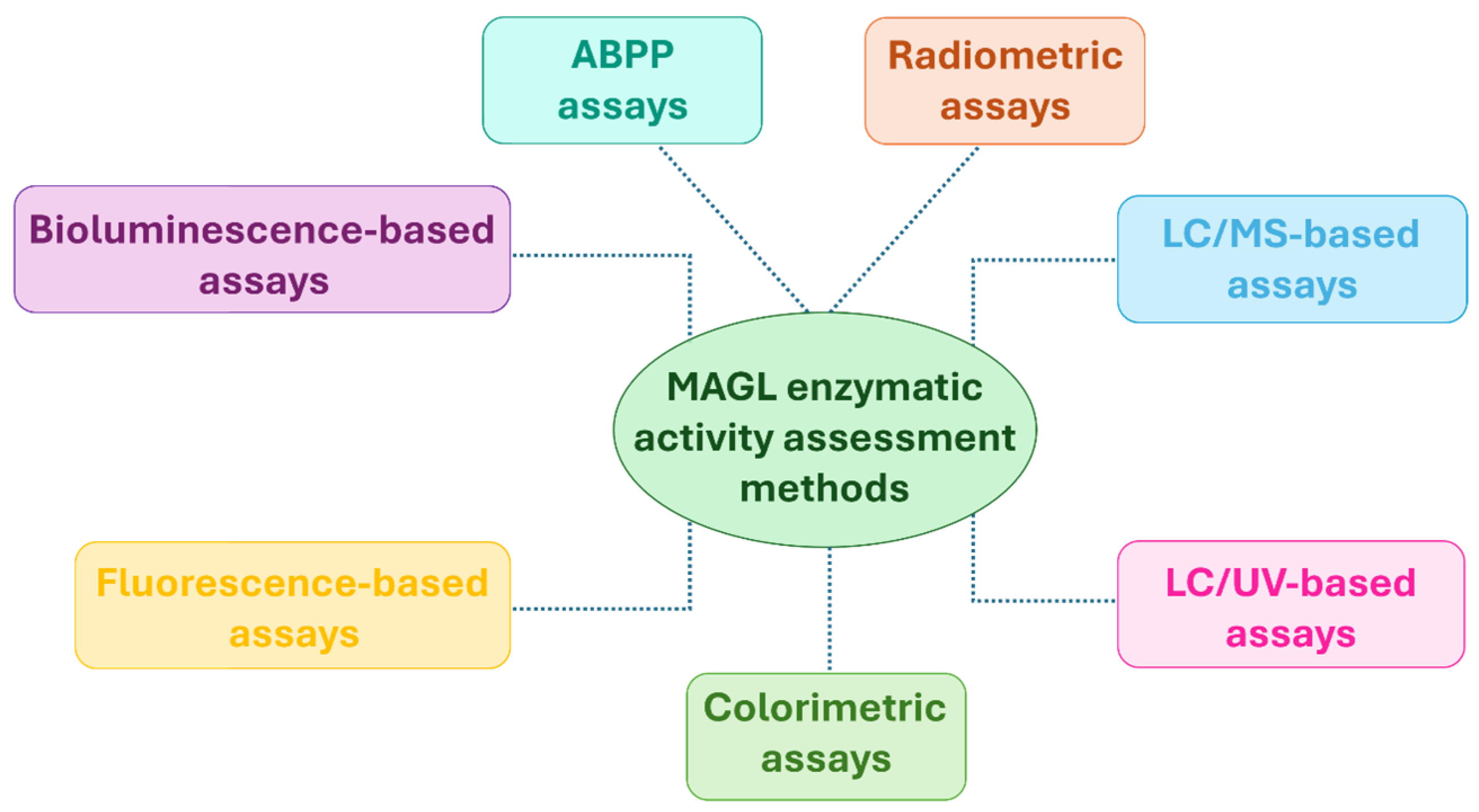
2.1. Radiometric Assays
2.2. Mass Spectrometry (MS)-Based Assays
2.3. Ultraviolet (UV)-Based Assays
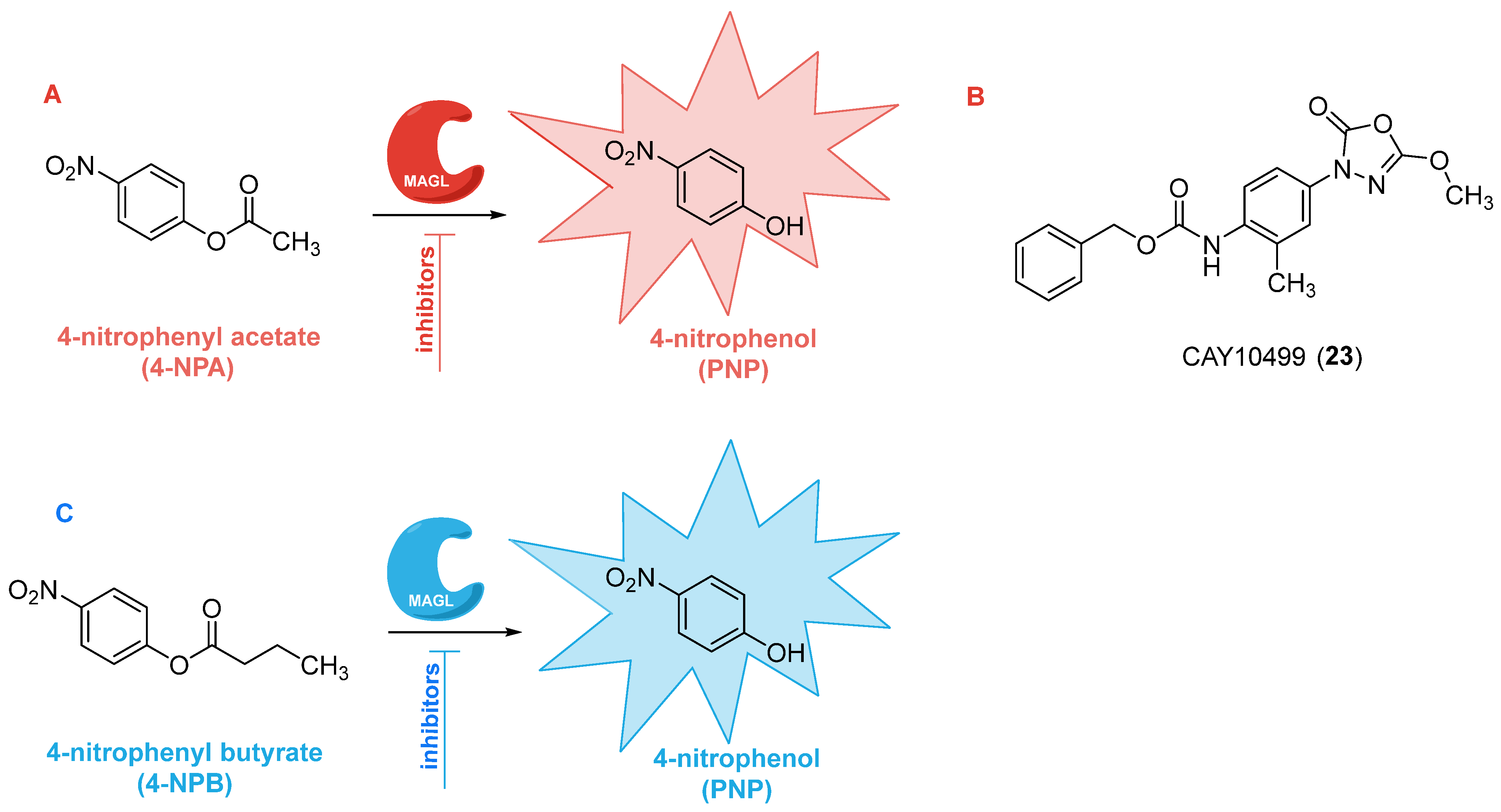
2.4. Colorimetric Assays
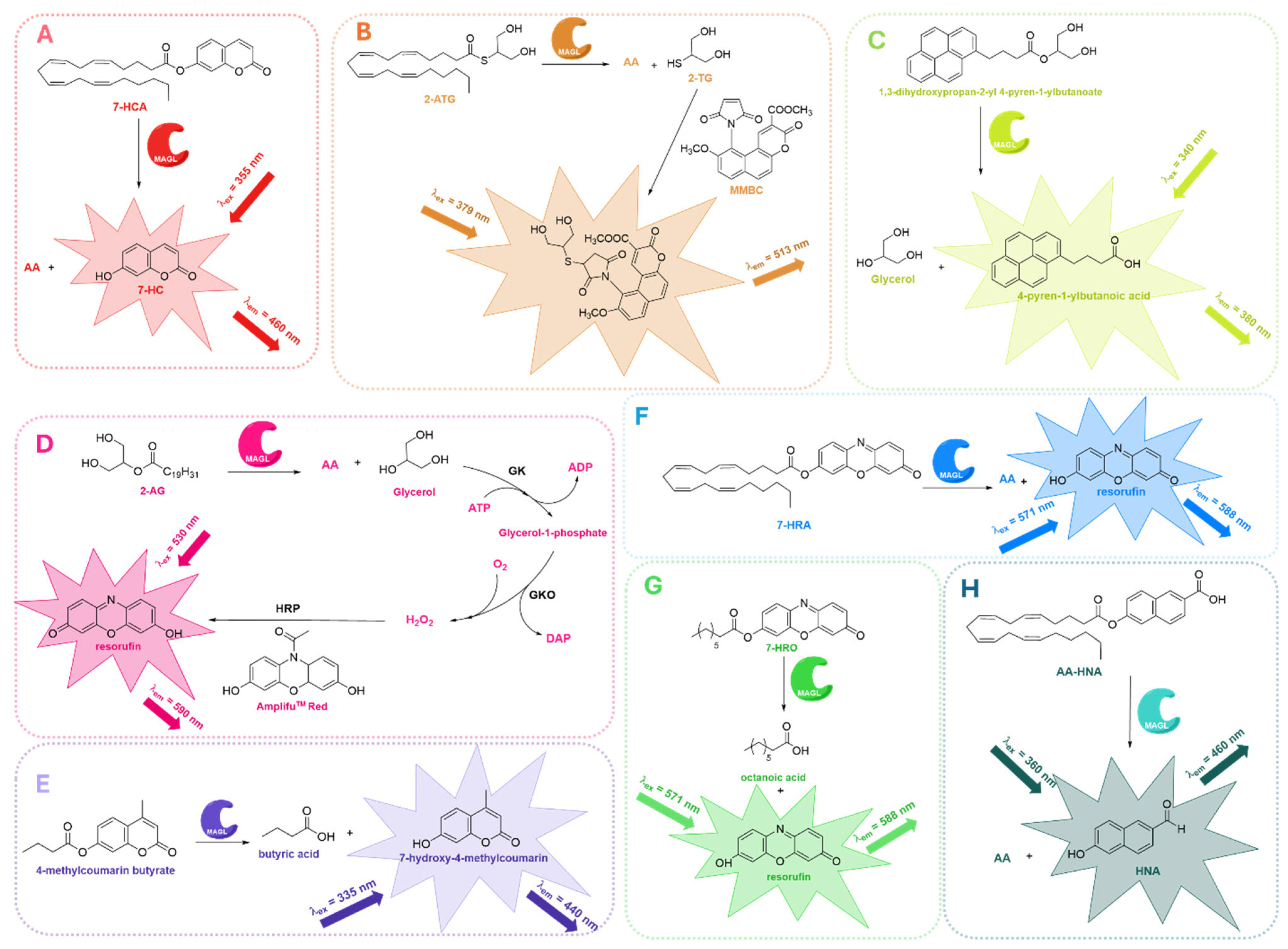
2.5. Fluorescence-Based Assays
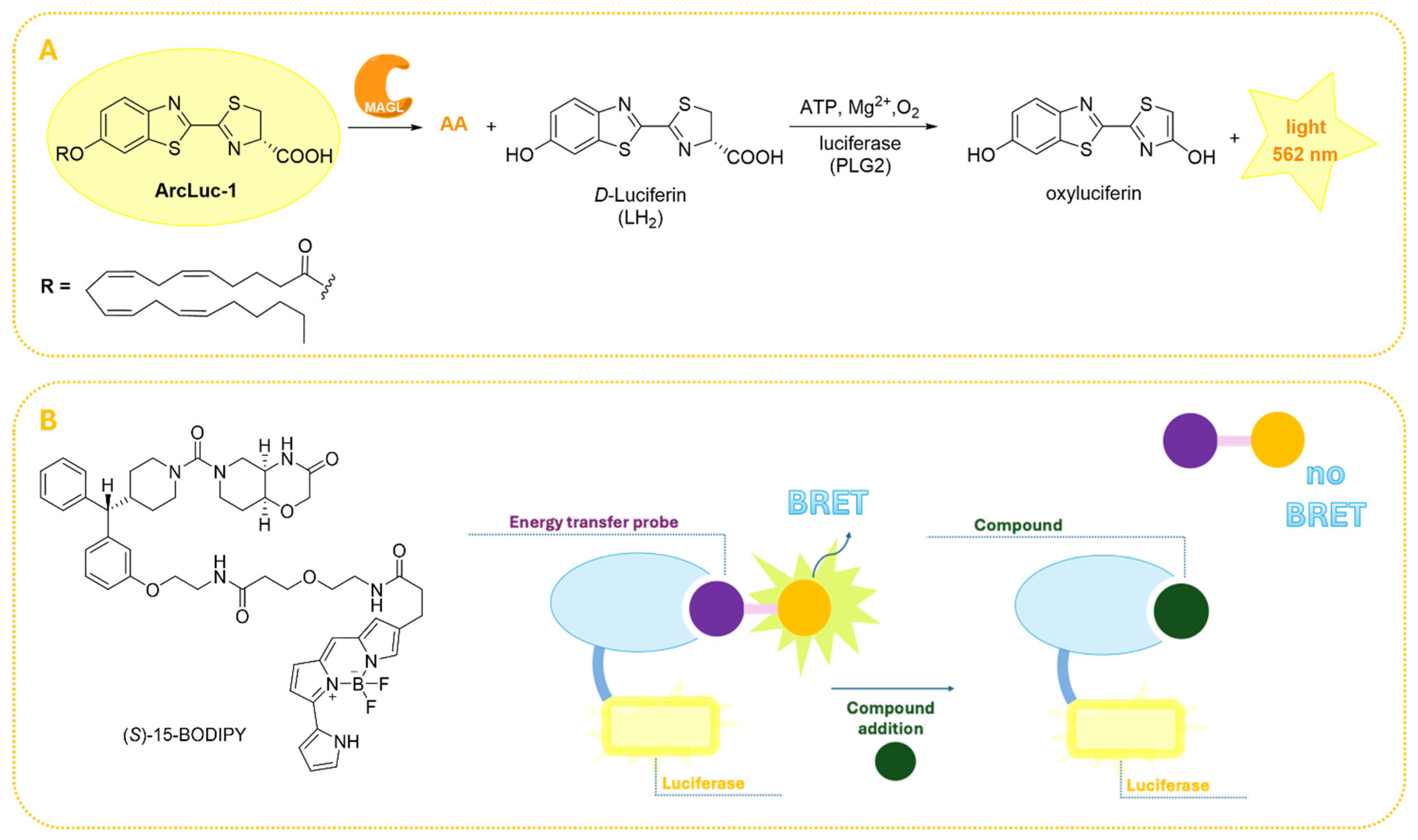
2.6. Bioluminescence-Based Assays
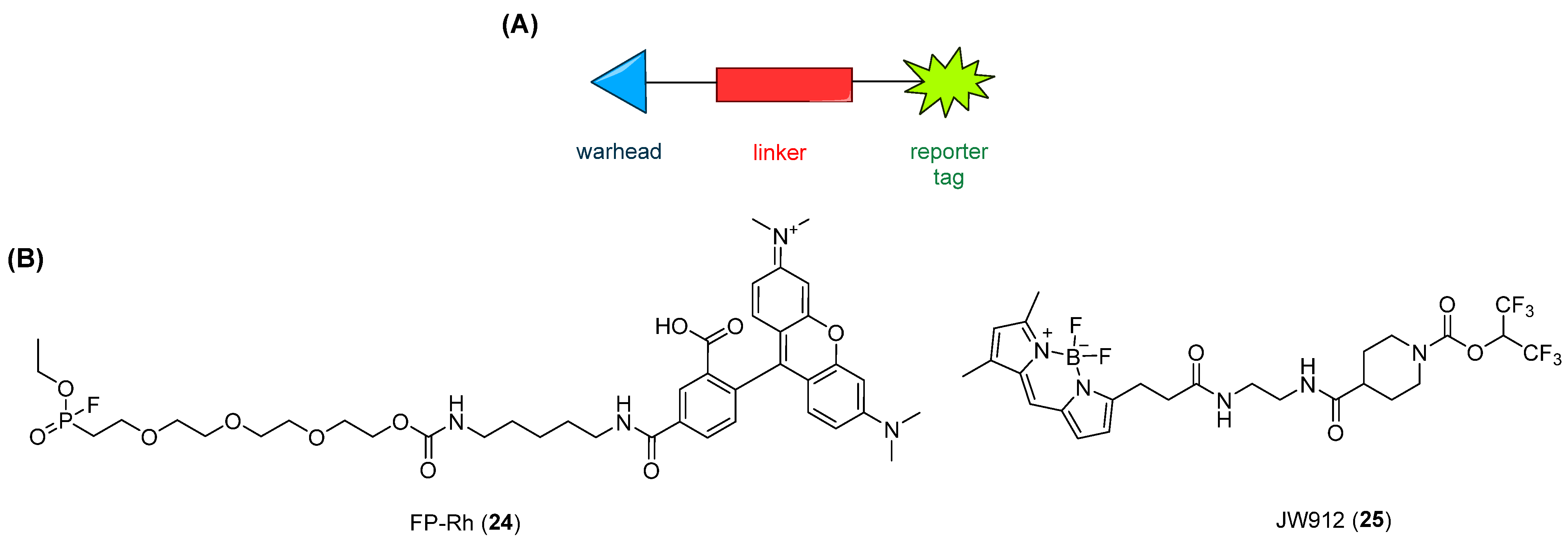
2.7. Activity-Based Protein Profiling (ABPP) Assays
3. Concluding Remarks
| Assessment Method | Substrate | Pros | Cons | Reference |
|---|---|---|---|---|
| Radiometric assays | Natural (2-AG or 2-OG) substrates | Very high sensitivity | - Complex experimental procedures (e.g., lipid extraction, fractioning by TLC, and radiolabeled substrates) - Costly - No real-time monitoring - Incompatibility with HTS | [64,65] |
| Chromatographic (UV- or MS-based) assays | Natural (2-AG or 2-OG) substrates | - High sensitivity and accuracy - Specific quantification of products | - Complex experimental procedures (e.g., lipid extraction and phase separation) - Costly - Low throughput - Not ideal for HTS - No real-time monitoring | [45,48,49,66,67,68] |
| Colorimetric assays | Surrogate substrates (4-NPA or 4-NPB) | - Cost-effective - Easy detection method (absorbance) - Simple setup - Real-time monitoring - HTS-compatible - No lipid extraction required | - Low sensitivity - Reduced accuracy due to the use of surrogate substrates | [69,70] |
| Fluorescence-based assays | Surrogate (7-HRA, 7-HRO, 4-methylcoumarin butyrate) and natural (glycerol) substrates | - Cost-effective - Easy detection method (fluorescence) - Real-time monitoring - HTS-compatible - No lipid extraction required - Possibility of using natural substrates under more physiological conditions | - Some compounds may interfere with fluorescence signal - Require optimized conditions - Less straightforward than colorimetric assays | [71,72,73,74,75,76,77,78,106] |
| Bioluminescence-based assays | Surrogate substrates (luciferase-coupled) | - Very high sensitivity - Broad dynamic range - Minimal background noise - Real-time monitoring - HTS-compatible - Amenable to automation | - Require specialized reagents (luciferase substrates) - Higher cost compared to colorimetric/fluorescent assays | [80,81] |
| ABPP assays | No requirement for substrates | - Proteome-wide selectivity profiling - Simultaneous assessment of enzymatic activity and inhibitor selectivity in a single experiment - Applicable to both in vitro and in vivo studies | - Availability of a suitable ABP - Potential probe-related artifacts (off-target labeling or incomplete proteome coverage) - Lower throughput compared to simple biochemical assays (gel-based ABPP assays) | [82,83,118] |
| MAGL Inhibitor | Inhibition Activity | Inhibition Type | Assay Method | Detection Substrate | Reference |
|---|---|---|---|---|---|
| NEM (1) | IC50 = 53 µM | Irreversible | Chromatographic UV-based assay | Natural substrate: 2-AG | [41] |
| NAM (2) | IC50 = 0.14 µM | Irreversible | Chromatographic UV-based assay | Natural substrate: 2-AG | [41] |
| 3 | pIC50 = 6.96 | Irreversible | Radiometric assay | Natural substrate: 2-OG | [44] |
| 4 | IC50 = 88 nM | Irreversible | Chromatographic MS-based assay | Natural substrate: 2-OG | [45] |
| 5 | pIC50 = 5.02 | Irreversible | Radiometric assay | Natural substrate: 2-OG | [46] |
| CK16 (6) | pIC50 = 6.45 | Irreversible | Radiometric assay | Natural substrate: 2-OG | [47] |
| PMSF (7) | IC50 = 155 µM | Irreversible | Chromatographic UV-based assay | Natural substrate: 2-AG | [48] |
| MAFP (8) | IC50 = 2.2 nM | Irreversible | Chromatographic UV-based assay | Natural substrate: 2-AG | [48] |
| ATFMK (9) | IC50 = 66 µM | Irreversible | Chromatographic UV-based assay | Natural substrate: 2-AG | [48] |
| URB602 (10) | IC50 = 223 µM | Irreversible | Chromatographic MS-based assay | Natural substrate: 2-OG | [49] |
| JZL184 (11) | IC50 = 8 nM | Irreversible | ABPP assay | No requirement for substrate | [50] |
| 12 | IC50 = 363 pM | Irreversible | Fluorescence-based assay | Natural substrate: 2-AG and GK | [51] |
| ABX-1341 (13) | IC50 = 0.014 µM | Irreversible | ABPP assay | No requirement for substrate | [52] |
| Pristimerin (14) | IC50 = 93 nM | Reversible | Chromatographic MS-based assay | Natural substrate: 2-OG | [55] |
| Euphol (15) | IC50 = 315 nM | Reversible | Chromatographic MS-based assay | Natural substrate: 2-OG | [55] |
| β-Amirin (16) | IC50 = 2800 nM | Reversible | Chromatographic MS-based assay | Natural substrate: 2-OG | [56] |
| ZYH (17) | IC50 = 0.010 µM | Reversible | Fluorescence-based assay | Surrogate substrate: 4-methylcoumarin butyrate | [57] |
| 18 | IC50 = 0.68 µM | Reversible | Colorimetric assay | Surrogate substrate: 4-NPA | [59] |
| 19 | IC50 = 1.26 nM | Reversible | Colorimetric assay | Surrogate substrate: 4-NPA | [60] |
| 20 | IC50 = 5.2 nM | Reversible | Colorimetric assay | Surrogate substrate: 4-NPA | [61] |
| 21 | IC50 = 0.34 µM | Reversible | Colorimetric assay | Surrogate substrate: 4-NPA | [62] |
| LEI-515 (22) | IC50 = 25 nM | Reversible | ABPP assay | No requirement for substrate | [63] |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cristino, L.; Bisogno, T.; Di Marzo, V. Cannabinoids and the Expanded Endocannabinoid System in Neurological Disorders. Nat. Rev. Neurol. 2020, 16, 9–29. [Google Scholar] [CrossRef]
- Rezende, B.; Alencar, A.K.N.; de Bem, G.F.; Fontes-Dantas, F.L.; Montes, G.C. Endocannabinoid System: Chemical Characteristics and Biological Activity. Pharmaceuticals 2023, 16, 148. [Google Scholar] [CrossRef]
- Pertwee, R.G. Pharmacology of Cannabinoid CB1 and CB2 Receptors. Pharmacol. Ther. 1997, 74, 129–180. [Google Scholar] [CrossRef]
- Howlett, A.C.; Abood, M.E. CB1 and CB2 Receptor Pharmacology. Adv. Pharmacol. 2017, 80, 169–206. [Google Scholar] [CrossRef]
- Matsuda, L.A.; Lolait, S.J.; Brownstein, M.J.; Young, A.C.; Bonner, T.I. Structure of a Cannabinoid Receptor and Functional Expression of the Cloned CDNA. Nature 1990, 346, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Li, C.; Jaffe, A.E.; Shin, J.H.; Deep-Soboslay, A.; Yamin, R.; Weinberger, D.R.; Hyde, T.M.; Kleinman, J.E. Cannabinoid Receptor CNR1 Expression and DNA Methylation in Human Prefrontal Cortex, Hippocampus and Caudate in Brain Development and Schizophrenia. Transl. Psychiatry 2020, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Glass, M.; Faull, R.L.; Dragunow, M. Cannabinoid Receptors in the Human Brain: A Detailed Anatomical and Quantitative Autoradiographic Study in the Fetal, Neonatal and Adult Human Brain. Neuroscience 1997, 77, 299–318. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.; Thomas, K.L.; Abu-Shaar, M. Molecular Characterization of a Peripheral Receptor for Cannabinoids. Nature 1993, 365, 61–65. [Google Scholar] [CrossRef]
- Ye, L.; Cao, Z.; Wang, W.; Zhou, N. New Insights in Cannabinoid Receptor Structure and Signaling. Curr. Mol. Pharmacol. 2019, 12, 239–248. [Google Scholar] [CrossRef]
- Lu, H.C.; Mackie, K. An Introduction to the Endogenous Cannabinoid System. Biol. Psychiatry 2016, 79, 516–525. [Google Scholar] [CrossRef]
- Piette, C.; Cui, Y.; Gervasi, N.; Venance, L. Lights on Endocannabinoid-Mediated Synaptic Potentiation. Front. Mol. Neurosci. 2020, 13, 132. [Google Scholar] [CrossRef]
- Gil-Ordóñez, A.; Martín-Fontecha, M.; Ortega-Gutiérrez, S.; López-Rodríguez, M.L. Monoacylglycerol Lipase (MAGL) as a Promising Therapeutic Target. Biochem. Pharmacol. 2018, 157, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Mulvihill, M.M.; Mukhopadhyay, P.; Xu, H.; Erdélyi, K.; Hao, E.; Holovac, E.; Haskó, G.; Cravatt, B.F.; Nomura, D.K.; et al. Monoacylglycerol Lipase Controls Endocannabinoid and Eicosanoid Signaling and Hepatic Injury in Mice. Gastroenterology 2013, 144, 808–817.e15. [Google Scholar] [CrossRef]
- Ahn, K.; McKinney, M.K.; Cravatt, B.F. Enzymatic Pathways That Regulate Endocannabinoid Signaling in the Nervous System. Chem. Rev. 2008, 108, 1687–1707. [Google Scholar] [CrossRef]
- Tornqvist, H.; Belfrage, P. Purification and Some Properties of a Monoacylglycerol-Hydrolyzing Enzyme of Rat Adipose Tissue. J. Biol. Chem. 1976, 251, 813–819. [Google Scholar] [CrossRef]
- Karlsson, M.; Reue, K.; Xia, Y.-R.; Lusis, A.J.; Langin, D.; Tornqvist, H.; Holm, C. Exon–Intron Organization and Chromosomal Localization of the Mouse Monoglyceride Lipase Gene. Gene 2001, 272, 11–18. [Google Scholar] [CrossRef]
- Dinh, T.P.; Freund, T.F.; Piomelli, D. A Role for Monoglyceride Lipase in 2-Arachidonoylglycerol Inactivation. Chem. Phys. Lipids 2002, 121, 149–158. [Google Scholar] [CrossRef]
- Labar, G.; Bauvois, C.; Borel, F.; Ferrer, J.-L.; Wouters, J.; Lambert, D.M. Crystal Structure of the Human Monoacylglycerol Lipase, a Key Actor in Endocannabinoid Signaling. ChemBioChem 2010, 11, 218–227. [Google Scholar] [CrossRef]
- Bertrand, T.; Augé, F.; Houtmann, J.; Rak, A.; Vallée, F.; Mikol, V.; Berne, P.F.; Michot, N.; Cheuret, D.; Hoornaert, C.; et al. Structural Basis for Human Monoglyceride Lipase Inhibition. J. Mol. Biol. 2010, 396, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Casas-Godoy, L.; Duquesne, S.; Bordes, F.; Sandoval, G.; Marty, A. Lipases: An Overview. Methods Mol. Biol. 2012, 861, 3–30. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Li, W. Monoacylglycerol Lipase Inhibitors: Modulators for Lipid Metabolism in Cancer Malignancy, Neurological and Metabolic Disorders. Acta Pharm. Sin. B 2020, 10, 582–602. [Google Scholar] [CrossRef]
- Mulvihill, M.M.; Nomura, D.K. Therapeutic Potential of Monoacylglycerol Lipase Inhibitors. Life Sci. 2013, 92, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Nomura, D.K.; Morrison, B.E.; Blankman, J.L.; Long, J.Z.; Kinsey, S.G.; Marcondes, M.C.G.; Ward, A.M.; Hahn, Y.K.; Lichtman, A.H.; Conti, B.; et al. Endocannabinoid Hydrolysis Generates Brain Prostaglandins That Promote Neuroinflammation. Science 2011, 334, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Wise, L.E.; Chen, Y.; Gujjar, R.; Mahadevan, A.; Cravatt, B.F.; Lichtman, A.H. The Monoacylglycerol Lipase Inhibitor JZL184 Suppresses Inflammatory Pain in the Mouse Carrageenan Model. Life Sci. 2013, 92, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Brindisi, M.; Maramai, S.; Gemma, S.; Brogi, S.; Grillo, A.; Di Cesare Mannelli, L.; Gabellieri, E.; Lamponi, S.; Saponara, S.; Gorelli, B.; et al. Development and Pharmacological Characterization of Selective Blockers of 2-Arachidonoyl Glycerol Degradation with Efficacy in Rodent Models of Multiple Sclerosis and Pain. J. Med. Chem. 2016, 59, 2612–2632. [Google Scholar] [CrossRef]
- Pasquarelli, N.; Porazik, C.; Bayer, H.; Buck, E.; Schildknecht, S.; Weydt, P.; Witting, A.; Ferger, B. Contrasting Effects of Selective MAGL and FAAH Inhibition on Dopamine Depletion and GDNF Expression in a Chronic MPTP Mouse Model of Parkinson’s Disease. Neurochem. Int. 2017, 110, 14–24. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, M.; Teng, Z.; Tang, Y.-P.; Chen, C. Synaptic and Cognitive Improvements by Inhibition of 2-AG Metabolism Are through Upregulation of MicroRNA-188-3p in a Mouse Model of Alzheimer’s Disease. J. Neurosci. 2014, 34, 14919–14933. [Google Scholar] [CrossRef]
- Feliú, A.; Bonilla del Río, I.; Carrillo-Salinas, F.J.; Hernández-Torres, G.; Mestre, L.; Puente, N.; Ortega-Gutiérrez, S.; López-Rodríguez, M.L.; Grandes, P.; Mecha, M.; et al. 2-Arachidonoylglycerol Reduces Proteoglycans and Enhances Remyelination in a Progressive Model of Demyelination. J. Neurosci. 2017, 37, 8385–8398. [Google Scholar] [CrossRef]
- Pasquarelli, N.; Engelskirchen, M.; Hanselmann, J.; Endres, S.; Porazik, C.; Bayer, H.; Buck, E.; Karsak, M.; Weydt, P.; Ferger, B.; et al. Evaluation of Monoacylglycerol Lipase as a Therapeutic Target in a Transgenic Mouse Model of ALS. Neuropharmacology 2017, 124, 157–169. [Google Scholar] [CrossRef]
- Schurman, L.D.; Lichtman, A.H. Endocannabinoids: A Promising Impact for Traumatic Brain Injury. Front. Pharmacol. 2017, 8, 69. [Google Scholar] [CrossRef]
- Nomura, D.K.; Long, J.Z.; Niessen, S.; Hoover, H.S.; Ng, S.-W.; Cravatt, B.F. Monoacylglycerol Lipase Regulates a Fatty Acid Network That Promotes Cancer Pathogenesis. Cell 2010, 140, 49–61. [Google Scholar] [CrossRef]
- Baba, Y.; Funakoshi, T.; Mori, M.; Emoto, K.; Masugi, Y.; Ekmekcioglu, S.; Amagai, M.; Tanese, K. Expression of Monoacylglycerol Lipase as a Marker of Tumour Invasion and Progression in Malignant Melanoma. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 2038–2045. [Google Scholar] [CrossRef]
- Nomura, D.K.; Lombardi, D.P.; Chang, J.W.; Niessen, S.; Ward, A.M.; Long, J.Z.; Hoover, H.H.; Cravatt, B.F. Monoacylglycerol Lipase Exerts Dual Control over Endocannabinoid and Fatty Acid Pathways to Support Prostate Cancer. Chem. Biol. 2011, 18, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, B.; Seviour, E.G.; Tao, K.; Liu, X.; Ling, Y.; Chen, J.; Wang, G. Monoacylglycerol Lipase (MAGL) Knockdown Inhibits Tumor Cells Growth in Colorectal Cancer. Cancer Lett. 2011, 307, 6–17. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Z.; Lian, Z.; Liao, R.; Chen, Y.; Qin, Y.; Wang, J.; Jiang, Q.; Wang, X.; Gong, J. Monoacylglycerol Lipase: A Novel Potential Therapeutic Target and Prognostic Indicator for Hepatocellular Carcinoma. Sci. Rep. 2016, 6, 35784. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhao, Y.; Zhou, J.; Wang, X.; Pan, Q.; Zhang, N.; Wang, L.; Wang, M.; Zhan, D.; Liu, Z.; et al. Monoacylglycerol Lipase Promotes Progression of Hepatocellular Carcinoma via NF-ΚB-Mediated Epithelial-Mesenchymal Transition. J. Hematol. Oncol. 2016, 9, 127. [Google Scholar] [CrossRef]
- Hu, W.R.; Lian, Y.F.; Peng, L.X.; Lei, J.J.; Deng, C.C.; Xu, M.; Feng, Q.S.; Chen, L.Z.; Bei, J.X.; Zeng, Y.X. Monoacylglycerol Lipase Promotes Metastases in Nasopharyngeal Carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 3704–3713. [Google Scholar]
- Sticht, M.A.; Long, J.Z.; Rock, E.M.; Limebeer, C.L.; Mechoulam, R.; Cravatt, B.F.; Parker, L.A. Inhibition of Monoacylglycerol Lipase Attenuates Vomiting in Suncus Murinus and 2-arachidonoyl Glycerol Attenuates Nausea in Rats. Br. J. Pharmacol. 2012, 165, 2425–2435. [Google Scholar] [CrossRef]
- Mitra, S.; Gobira, P.H.; Werner, C.T.; Martin, J.A.; Iida, M.; Thomas, S.A.; Erias, K.; Miracle, S.; Lafargue, C.; An, C.; et al. A Role for the Endocannabinoid Enzymes Monoacylglycerol and Diacylglycerol Lipases in Cue-induced Cocaine Craving Following Prolonged Abstinence. Addict. Biol. 2021, 26, e13007. [Google Scholar] [CrossRef]
- Martínez-Rivera, A.; Fetcho, R.N.; Birmingham, L.; Xu, J.; Yang, R.; Foord, C.; Scala-Chávez, D.; Mekawy, N.; Pleil, K.; Pickel, V.M.; et al. Elevating Levels of the Endocannabinoid 2-Arachidonoylglycerol Blunts Opioid Reward but Not Analgesia. Sci. Adv. 2024, 10, eadq4779. [Google Scholar] [CrossRef] [PubMed]
- Saario, S.M.; Salo, O.M.H.; Nevalainen, T.; Poso, A.; Laitinen, J.T.; Järvinen, T.; Niemi, R. Characterization of the Sulfhydryl-Sensitive Site in the Enzyme Responsible for Hydrolysis of 2- Arachidonoyl-Glycerol in Rat Cerebellar Membranes. Chem. Biol. 2005, 12, 649–656. [Google Scholar] [CrossRef]
- Zvonok, N.; Pandarinathan, L.; Williams, J.; Johnston, M.; Karageorgos, I.; Janero, D.R.; Krishnan, S.C.; Makriyannis, A. Covalent Inhibitors of Human Monoacylglycerol Lipase: Ligand-Assisted Characterization of the Catalytic Site by Mass Spectrometry and Mutational Analysis. Chem. Biol. 2008, 15, 854–862. [Google Scholar] [CrossRef]
- Labar, G.; Bauvois, C.; Muccioli, G.G.; Wouters, J.; Lambert, D.M. Disulfiram Is an Inhibitor of Human Purified Monoacylglycerol Lipase, the Enzyme Regulating 2-Arachidonoylglycerol Signaling. ChemBioChem 2007, 8, 1293–1297. [Google Scholar] [CrossRef]
- Kapanda, C.N.; Muccioli, G.G.; Labar, G.; Poupaert, J.H.; Lambert, D.M. Bis(Dialkylaminethiocarbonyl)Disulfides as Potent and Selective Monoglyceride Lipase Inhibitors. J. Med. Chem. 2009, 52, 7310–7314. [Google Scholar] [CrossRef]
- King, A.; Lodola, A.; Carmi, C.; Fu, J.; Mor, M.; Piomelli, D. A Critical Cysteine Residue in Monoacylglycerol Lipase Is Targeted by a New Class of Isothiazolinone-based Enzyme Inhibitors. Br. J. Pharmacol. 2009, 157, 974–983. [Google Scholar] [CrossRef]
- Kapanda, C.N.; Muccioli, G.G.; Labar, G.; Draoui, N.; Lambert, D.M.; Poupaert, J.H. Search for Monoglyceride Lipase Inhibitors: Synthesis and Screening of Arylthioamides Derivatives. Med. Chem. Res. 2009, 18, 243–254. [Google Scholar] [CrossRef]
- Kapanda, C.N.; Masquelier, J.; Labar, G.; Muccioli, G.G.; Poupaert, J.H.; Lambert, D.M. Synthesis and Pharmacological Evaluation of 2,4-Dinitroaryldithiocarbamate Derivatives as Novel Monoacylglycerol Lipase Inhibitors. J. Med. Chem. 2012, 55, 5774–5783. [Google Scholar] [CrossRef] [PubMed]
- Saario, S.M.; Savinainen, J.R.; Laitinen, J.T.; Järvinen, T.; Niemi, R. Monoglyceride Lipase-like Enzymatic Activity Is Responsible for Hydrolysis of 2-Arachidonoylglycerol in Rat Cerebellar Membranes. Biochem. Pharmacol. 2004, 67, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- King, A.R.; Duranti, A.; Tontini, A.; Rivara, S.; Rosengarth, A.; Clapper, J.R.; Astarita, G.; Geaga, J.A.; Luecke, H.; Mor, M.; et al. URB602 Inhibits Monoacylglycerol Lipase and Selectively Blocks 2-Arachidonoylglycerol Degradation in Intact Brain Slices. Chem. Biol. 2007, 14, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Long, J.Z.; Li, W.; Booker, L.; Burston, J.J.; Kinsey, S.G.; Schlosburg, J.E.; Pavón, F.J.; Serrano, A.M.; Selley, D.E.; Parsons, L.H.; et al. Selective Blockade of 2-Arachidonoylglycerol Hydrolysis Produces Cannabinoid Behavioral Effects. Nat. Chem. Biol. 2009, 5, 37–44. [Google Scholar] [CrossRef]
- Aaltonen, N.; Savinainen, J.R.; Ribas, C.R.; Rönkkö, J.; Kuusisto, A.; Korhonen, J.; Navia-Paldanius, D.; Häyrinen, J.; Takabe, P.; Käsnänen, H.; et al. Piperazine and Piperidine Triazole Ureas as Ultrapotent and Highly Selective Inhibitors of Monoacylglycerol Lipase. Chem. Biol. 2013, 20, 379–390. [Google Scholar] [CrossRef]
- Cisar, J.S.; Weber, O.D.; Clapper, J.R.; Blankman, J.L.; Henry, C.L.; Simon, G.M.; Alexander, J.P.; Jones, T.K.; Ezekowitz, R.A.B.; O’Neill, G.P.; et al. Identification of ABX-1431, a Selective Inhibitor of Monoacylglycerol Lipase and Clinical Candidate for Treatment of Neurological Disorders. J. Med. Chem. 2018, 61, 9062–9084. [Google Scholar] [CrossRef]
- Jiang, M.; van der Stelt, M. Activity-Based Protein Profiling Delivers Selective Drug Candidate ABX-1431, a Monoacylglycerol Lipase Inhibitor, To Control Lipid Metabolism in Neurological Disorders. J. Med. Chem. 2018, 61, 9059–9061. [Google Scholar] [CrossRef] [PubMed]
- Schlosburg, J.E.; Blankman, J.L.; Long, J.Z.; Nomura, D.K.; Pan, B.; Kinsey, S.G.; Nguyen, P.T.; Ramesh, D.; Booker, L.; Burston, J.J.; et al. Chronic Monoacylglycerol Lipase Blockade Causes Functional Antagonism of the Endocannabinoid System. Nat. Neurosci. 2010, 13, 1113–1119. [Google Scholar] [CrossRef]
- King, A.R.; Dotsey, E.Y.; Lodola, A.; Jung, K.M.; Ghomian, A.; Qiu, Y.; Fu, J.; Mor, M.; Piomelli, D. Discovery of Potent and Reversible Monoacylglycerol Lipase Inhibitors. Chem. Biol. 2009, 16, 1045–1052. [Google Scholar] [CrossRef]
- Chicca, A.; Marazzi, J.; Gertsch, J. The Antinociceptive Triterpene Β-amyrin Inhibits 2-arachidonoylglycerol (2-AG) Hydrolysis without Directly Targeting Cannabinoid Receptors. Br. J. Pharmacol. 2012, 167, 1596–1608. [Google Scholar] [CrossRef]
- Chevalier, K.M.; Dax, S.L.; Flores, C.M.; Liu, L.; Macielag, M.J.; McDonnel, M.E.; Nelen, M.I.; Prouty, S.; Todd, M.; Zhang, S.; et al. Heteroaromatic and Aromatic Piperazinyl Azetidinyl Amides as Monoacylglycerol Lipase Inhibitors. WO2010124121, 28 October 2010. [Google Scholar]
- Schalk-Hihi, C.; Schubert, C.; Alexander, R.; Bayoumy, S.; Clemente, J.C.; Deckman, I.; DesJarlais, R.L.; Dzordzorme, K.C.; Flores, C.M.; Grasberger, B.; et al. Crystal Structure of a Soluble Form of Human Monoglyceride Lipase in Complex with an Inhibitor at 1.35 Å Resolution. Protein Sci. 2011, 20, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Bononi, G.; Granchi, C.; Lapillo, M.; Giannotti, M.; Nieri, D.; Fortunato, S.; El Boustani, M.; Caligiuri, I.; Poli, G.; Carlson, K.E.; et al. Discovery of Long-Chain Salicylketoxime Derivatives as Monoacylglycerol Lipase (MAGL) Inhibitors. Eur. J. Med. Chem. 2018, 157, 817–836. [Google Scholar] [CrossRef]
- Bononi, G.; Tonarini, G.; Poli, G.; Barravecchia, I.; Caligiuri, I.; Macchia, M.; Rizzolio, F.; Demontis, G.C.; Minutolo, F.; Granchi, C.; et al. Monoacylglycerol Lipase (MAGL) Inhibitors Based on a Diphenylsulfide-Benzoylpiperidine Scaffold. Eur. J. Med. Chem. 2021, 223, 113679. [Google Scholar] [CrossRef]
- Di Stefano, M.; Masoni, S.; Bononi, G.; Poli, G.; Galati, S.; Gado, F.; Manzi, S.; Vagaggini, C.; Brai, A.; Caligiuri, I.; et al. Design, Synthesis, ADME and Biological Evaluation of Benzylpiperidine and Benzylpiperazine Derivatives as Novel Reversible Monoacylglycerol Lipase (MAGL) Inhibitors. Eur. J. Med. Chem. 2024, 263, 115916. [Google Scholar] [CrossRef] [PubMed]
- Bononi, G.; Gado, F.; Masoni, S.; Scalabrini, D.; Della Vedova, L.; Di Stefano, M.; Piazza, L.; Poles, C.; Lonzi, C.; Landucci, E.; et al. Discovery of Reversible Monoacylglycerol Lipase (MAGL) Inhibitors Based on Ortho-Hydroxyanilide Scaffold. Bioorg. Chem. 2025, 163, 108698. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Huizenga, M.C.W.; Wirt, J.L.; Paloczi, J.; Amedi, A.; van den Berg, R.J.B.H.N.; Benz, J.; Collin, L.; Deng, H.; Di, X.; et al. A Monoacylglycerol Lipase Inhibitor Showing Therapeutic Efficacy in Mice without Central Side Effects or Dependence. Nat. Commun. 2023, 14, 8039. [Google Scholar] [CrossRef]
- Dinh, T.P.; Carpenter, D.; Leslie, F.M.; Freund, T.F.; Katona, I.; Sensi, S.L.; Kathuria, S.; Piomelli, D. Brain Monoglyceride Lipase Participating in Endocannabinoid Inactivation. Proc. Natl. Acad. Sci. USA 2002, 99, 10819–10824, Erratum in: Proc. Natl. Acad. Sci. USA 2002, 99, 13961. [Google Scholar] [CrossRef]
- Brengdahl, J.; Fowler, C.J. A Novel Assay for Monoacylglycerol Hydrolysis Suitable for High-Throughput Screening. Anal. Biochem. 2006, 359, 40–44. [Google Scholar] [CrossRef]
- Del Carlo, S.; Manera, C.; Chicca, A.; Arena, C.; Bertini, S.; Burgalassi, S.; Tampucci, S.; Gertsch, J.; Macchia, M.; Saccomanni, G. Development of an HPLC/UV Assay for the Evaluation of Inhibitors of Human Recombinant Monoacylglycerol Lipase. J. Pharm. Biomed. Anal. 2015, 108, 113–121. [Google Scholar] [CrossRef]
- Blankman, J.L.; Simon, G.M.; Cravatt, B.F. A Comprehensive Profile of Brain Enzymes That Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem. Biol. 2007, 14, 1347–1356. [Google Scholar] [CrossRef]
- Kayacelebi, A.A.; Schauerte, C.; Kling, K.; Herbers, J.; Beckmann, B.; Engeli, S.; Jordan, J.; Zoerner, A.A.; Tsikas, D. Cross-Validated Stable-Isotope Dilution GC–MS and LC–MS/MS Assays for Monoacylglycerol Lipase (MAGL) Activity by Measuring Arachidonic Acid Released from the Endocannabinoid 2-Arachidonoyl Glycerol. J. Chromatogr. B 2017, 1047, 151–159. [Google Scholar] [CrossRef]
- Muccioli, G.G.; Labar, G.; Lambert, D.M. CAY10499, a Novel Monoglyceride Lipase Inhibitor Evidenced by an Expeditious MGL Assay. ChemBioChem 2008, 9, 2704–2710. [Google Scholar] [CrossRef]
- Rodrigues, M.S.; Ferreira, C.; Dias, C.; Pliássova, A.; Souza, L.; Ledo, A.; Laranjinha, J.; Cunha, R.A.; Köfalvi, A. An Optimized Spectrophotometric Assay Reveals Increased Activity of Enzymes Involved in 2-arachidonoyl Glycerol Turnover in the Cerebral Cortex of a Rat Model of Alzheimer’s Disease. Eur. J. Neurosci. 2022, 55, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chanda, P.; Jones, P.G.; Kennedy, J.D. A Fluorescence-Based Assay for Monoacylglycerol Lipase Compatible with Inhibitor Screening. Assay Drug Dev. Technol. 2008, 6, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Holtfrerich, A.; Makharadze, T.; Lehr, M. High-Performance Liquid Chromatography Assay with Fluorescence Detection for the Evaluation of Inhibitors against Human Recombinant Monoacylglycerol Lipase. Anal. Biochem. 2010, 399, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Casida, J.E.; Gulevich, A.G.; Sarpong, R.; Bunnelle, E.M. S-Arachidonoyl-2-Thioglycerol Synthesis and Use for Fluorimetric and Colorimetric Assays of Monoacylglycerol Lipase. Bioorg. Med. Chem. 2010, 18, 1942–1947. [Google Scholar] [CrossRef]
- Savinainen, J.R.; Yoshino, M.; Minkkilä, A.; Nevalainen, T.; Laitinen, J.T. Characterization of Binding Properties of Monoglyceride Lipase Inhibitors by a Versatile Fluorescence-Based Technique. Anal. Biochem. 2010, 399, 132–134. [Google Scholar] [CrossRef]
- Clemente, J.C.; Nulton, E.; Nelen, M.; Todd, M.J.; Maguire, D.; Schalk-Hihi, C.; Kuo, L.C.; Zhang, S.-P.; Flores, C.M.; Kranz, J.K. Screening and Characterization of Human Monoglyceride Lipase Active Site Inhibitors Using Orthogonal Binding and Functional Assays. SLAS Discov. 2012, 17, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Lauria, S.; Casati, S.; Ciuffreda, P. Synthesis and Characterization of a New Fluorogenic Substrate for Monoacylglycerol Lipase and Application to Inhibition Studies. Anal. Bioanal. Chem. 2015, 407, 8163–8167. [Google Scholar] [CrossRef] [PubMed]
- Miceli, M.; Casati, S.; Ottria, R.; Di Leo, S.; Eberini, I.; Palazzolo, L.; Parravicini, C.; Ciuffreda, P. Set-Up and Validation of a High Throughput Screening Method for Human Monoacylglycerol Lipase (MAGL) Based on a New Red Fluorescent Probe. Molecules 2019, 24, 2241. [Google Scholar] [CrossRef]
- Deng, H.; Zhang, Q.; Lei, Q.; Yang, N.; Yang, K.; Jiang, J.; Yu, Z. Discovering Monoacylglycerol Lipase Inhibitors by a Combination of Fluorogenic Substrate Assay and Activity-Based Protein Profiling. Front. Pharmacol. 2022, 13, 941522. [Google Scholar] [CrossRef]
- Ciuffreda, P.; Xynomilakis, O.; Casati, S.; Ottria, R. Fluorescence-Based Enzyme Activity Assay: Ascertaining the Activity and Inhibition of Endocannabinoid Hydrolytic Enzymes. Int. J. Mol. Sci. 2024, 25, 7693. [Google Scholar] [CrossRef]
- Miceli, M.; Casati, S.; Allevi, P.; Berra, S.; Ottria, R.; Rota, P.; Branchini, B.R.; Ciuffreda, P. A New Ultrasensitive Bioluminescence-Based Method for Assaying Monoacylglycerol Lipase. Int. J. Mol. Sci. 2021, 22, 6148. [Google Scholar] [CrossRef]
- Gazzi, T.; Brennecke, B.; Olikauskas, V.; Hochstrasser, R.; Wang, H.; Keen Chao, S.; Atz, K.; Mostinski, Y.; Topp, A.; Heer, D.; et al. Development of a Highly Selective NanoBRET Probe to Assess MAGL Inhibition in Live Cells. ChemBioChem 2025, 26, e202400704. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Wright, A.T.; Kozarich, J.W. Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu. Rev. Biochem. 2008, 77, 383–414. [Google Scholar] [CrossRef] [PubMed]
- Niphakis, M.J.; Cravatt, B.F. Enzyme Inhibitor Discovery by Activity-Based Protein Profiling. Annu. Rev. Biochem. 2014, 83, 341–377. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Lei, Q.; Wu, Y.; He, Y.; Li, W. Activity-Based Protein Profiling: Recent Advances in Medicinal Chemistry. Eur. J. Med. Chem. 2020, 191, 112151. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri, N.; Tiger, G.; Razdan, R.K.; Mahadevan, A.; Pertwee, R.G.; Martin, B.R.; Fowler, C.J. Inhibition of Monoacylglycerol Lipase and Fatty Acid Amide Hydrolase by Analogues of 2-arachidonoylglycerol. Br. J. Pharmacol. 2004, 143, 774–784. [Google Scholar] [CrossRef]
- Vandevoorde, S.; Jonsson, K.; Labar, G.; Persson, E.; Lambert, D.M.; Fowler, C.J. Lack of Selectivity of URB602 for 2-oleoylglycerol Compared to Anandamide Hydrolysis in Vitro. Br. J. Pharmacol. 2007, 150, 186–191. [Google Scholar] [CrossRef]
- Muccioli, G.G.; Xu, C.; Odah, E.; Cudaback, E.; Cisneros, J.A.; Lambert, D.M.; López Rodríguez, M.L.; Bajjalieh, S.; Stella, N. Identification of a Novel Endocannabinoid-Hydrolyzing Enzyme Expressed by Microglial Cells. J. Neurosci. 2007, 27, 2883–2889. [Google Scholar] [CrossRef]
- Ortar, G.; Cascio, M.G.; Moriello, A.S.; Camalli, M.; Morera, E.; Nalli, M.; Di Marzo, V. Carbamoyl Tetrazoles as Inhibitors of Endocannabinoid Inactivation: A Critical Revisitation. Eur. J. Med. Chem. 2008, 43, 62–72. [Google Scholar] [CrossRef]
- Bisogno, T.; Ortar, G.; Petrosino, S.; Morera, E.; Palazzo, E.; Nalli, M.; Maione, S.; Di Marzo, V. Development of a Potent Inhibitor of 2-Arachidonoylglycerol Hydrolysis with Antinociceptive Activity in Vivo. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2009, 1791, 53–60. [Google Scholar] [CrossRef]
- Prescott, M.; Peek, K.; Veitch, D.P.; Daniel, R.M. The Use of Phenyl-Sepharose for the Affinity Purification of Proteinases. J. Biochem. Biophys. Methods 1993, 26, 51–60. [Google Scholar] [CrossRef]
- Mintzer, R.J.; Appell, K.C.; Cole, A.; Johns, A.; Pagila, R.; Polokoff, M.A.; Tabas, I.; Snider, R.M.; Meurer-Ogden, J.A. A Novel High-Throughput Screening Format to Identify Inhibitors of Secreted Acid Sphingomyelinase. SLAS Discov. 2005, 10, 225–234. [Google Scholar] [CrossRef]
- Jung, K.-M.; Piomelli, D. Assay of Monoacylglycerol Lipase Activity. Methods Mol. Biol. 2023, 2576, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Speers, A.E.; Cravatt, B.F. Activity-Based Protein Profiling (ABPP) and Click Chemistry (CC)–ABPP by MudPIT Mass Spectrometry. Curr. Protoc. Chem. Biol. 2009, 1, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Minkkilä, A.; Myllymäki, M.J.; Saario, S.M.; Castillo-Melendez, J.A.; Koskinen, A.M.P.; Fowler, C.J.; Leppänen, J.; Nevalainen, T. The Synthesis and Biological Evaluation of Para-Substituted Phenolic N-Alkyl Carbamates as Endocannabinoid Hydrolyzing Enzyme Inhibitors. Eur. J. Med. Chem. 2009, 44, 2994–3008. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-W.; Chen, L.; Xu, C.; Li, Z.; Zhang, H.; Zhang, X.-B.; Tan, W. Recent Progresses in Small-Molecule Enzymatic Fluorescent Probes for Cancer Imaging. Chem. Soc. Rev. 2018, 47, 7140–7180. [Google Scholar] [CrossRef]
- Wang, R.; Chen, J.; Gao, J.; Chen, J.-A.; Xu, G.; Zhu, T.; Gu, X.; Guo, Z.; Zhu, W.-H.; Zhao, C. A Molecular Design Strategy toward Enzyme-Activated Probes with near-Infrared I and II Fluorescence for Targeted Cancer Imaging. Chem. Sci. 2019, 10, 7222–7227. [Google Scholar] [CrossRef]
- Fernández-Ruiz, R. TXRF Spectrometry in the Bioanalytical Sciences: A Brief Review. X-Ray Spectrom. 2022, 51, 279–293. [Google Scholar] [CrossRef]
- Hua, L.; Wang, D.; Wang, K.; Wang, Y.; Gu, J.; Zhang, Q.; You, Q.; Wang, L. Design of Tracers in Fluorescence Polarization Assay for Extensive Application in Small Molecule Drug Discovery. J. Med. Chem. 2023, 66, 10934–10958. [Google Scholar] [CrossRef]
- Fang, B.; Shen, Y.; Peng, B.; Bai, H.; Wang, L.; Zhang, J.; Hu, W.; Fu, L.; Zhang, W.; Li, L.; et al. Small-Molecule Quenchers for Förster Resonance Energy Transfer: Structure, Mechanism, and Applications. Angew. Chemie Int. Ed. 2022, 61, e202207188. [Google Scholar] [CrossRef]
- Kundu, S.; Das, S.; Patra, A. Fluorescence Correlation Spectroscopy and Fluorescence Lifetime Imaging Microscopy for Deciphering the Morphological Evolution of Supramolecular Self-Assembly. Chem. Commun. 2023, 59, 8017–8031. [Google Scholar] [CrossRef]
- Millar, D.P. Time-Resolved Fluorescence Spectroscopy. Curr. Opin. Struct. Biol. 1996, 6, 637–642. [Google Scholar] [CrossRef]
- Krichevsky, O.; Bonnet, G. Fluorescence Correlation Spectroscopy: The Technique and Its Applications. Reports Prog. Phys. 2002, 65, 251–297. [Google Scholar] [CrossRef]
- Kask, P.; Palo, K.; Ullmann, D.; Gall, K. Fluorescence-Intensity Distribution Analysis and Its Application in Biomolecular Detection Technology. Proc. Natl. Acad. Sci. USA 1999, 96, 13756–13761. [Google Scholar] [CrossRef]
- Kanaoka, Y.; Sekine, T.; Machida, M.; Soma, Y.; Tanizawa, K.; Ban, Y. Studies on Protein-Sulfhydryl Reagents. I. Synthesis of Benzimidazole Derivatives of Maleimide; Fluorescent Labeling of Maleimide. Chem. Pharm. Bull. 1964, 12, 127–134. [Google Scholar] [CrossRef]
- Yang, J.; Langmuir, M.E. Synthesis and Properties of a Maleimide Fluorescent Thiol Reagent Derived from a Naphthopyranone. J. Heterocycl. Chem. 1991, 28, 1177–1180. [Google Scholar] [CrossRef]
- Navia-Paldanius, D.; Savinainen, J.R.; Laitinen, J.T. Biochemical and Pharmacological Characterization of Human α/β-Hydrolase Domain Containing 6 (ABHD6) and 12 (ABHD12). J. Lipid Res. 2012, 53, 2413–2424. [Google Scholar] [CrossRef]
- Ottria, R.; Casati, S.; Xynomilakis, O.; Veselinović, A.; Ciuffreda, P. Discovery of MAGL Inhibition by Lophine Derivatives: An Unexpected Finding from Chemiluminescent Assay Development. Molecules 2025, 30, 1605. [Google Scholar] [CrossRef] [PubMed]
- Branchini, B.R.; Southworth, T.L. A Highly Sensitive Biosensor for ATP Using a Chimeric Firefly Luciferase. Methods Enzymol. 2017, 589, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Niwa, K.; Ichino, Y.; Kumata, S.; Nakajima, Y.; Hiraishi, Y.; Kato, D.; Viviani, V.R.; Ohmiya, Y. Quantum Yields and Kinetics of the Firefly Bioluminescence Reaction of Beetle Luciferases. Photochem. Photobiol. 2010, 86, 1046–1049. [Google Scholar] [CrossRef]
- Ando, Y.; Niwa, K.; Yamada, N.; Enomoto, T.; Irie, T.; Kubota, H.; Ohmiya, Y.; Akiyama, H. Firefly Bioluminescence Quantum Yield and Colour Change by PH-Sensitive Green Emission. Nat. Photonics 2008, 2, 44–47. [Google Scholar] [CrossRef]
- Luker, K.E.; Smith, M.C.P.; Luker, G.D.; Gammon, S.T.; Piwnica-Worms, H.; Piwnica-Worms, D. Kinetics of Regulated Protein–Protein Interactions Revealed with Firefly Luciferase Complementation Imaging in Cells and Living Animals. Proc. Natl. Acad. Sci. USA 2004, 101, 12288–12293. [Google Scholar] [CrossRef]
- Badr, C.E.; Tannous, B.A. Bioluminescence Imaging: Progress and Applications. Trends Biotechnol. 2011, 29, 624–633. [Google Scholar] [CrossRef]
- Yuan, M.; Ma, X.; Jiang, T.; Zhang, C.; Chen, H.; Gao, Y.; Yang, X.; Du, L.; Li, M. A Novel Coelenterate Luciferin-Based Luminescent Probe for Selective and Sensitive Detection of Thiophenols. Org. Biomol. Chem. 2016, 14, 10267–10274. [Google Scholar] [CrossRef]
- Dale, N.C.; Johnstone, E.K.M.; White, C.W.; Pfleger, K.D.G. NanoBRET: The Bright Future of Proximity-Based Assays. Front. Bioeng. Biotechnol. 2019, 7, 56. [Google Scholar] [CrossRef]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; et al. Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef]
- Bell, C.; Benz, J.; Gobbi, L.; Grether, U.; Groebke Zbinden, K.; Hansen, D.J.; Hornsperger, B.; Kocer, B.; Kroll, C.; Kuhn, B.; et al. Oxazine Monoacylglycerol Lipase (MAGL) Inhibitors. WO2019180185, 26 September 2019. [Google Scholar]
- He, Y.; Schild, M.; Grether, U.; Benz, J.; Leibrock, L.; Heer, D.; Topp, A.; Collin, L.; Kuhn, B.; Wittwer, M.; et al. Development of High Brain-Penetrant and Reversible Monoacylglycerol Lipase PET Tracers for Neuroimaging. J. Med. Chem. 2022, 65, 2191–2207. [Google Scholar] [CrossRef]
- Chang, J.W.; Cognetta, A.B.; Niphakis, M.J.; Cravatt, B.F. Proteome-Wide Reactivity Profiling Identifies Diverse Carbamate Chemotypes Tuned for Serine Hydrolase Inhibition. ACS Chem. Biol. 2013, 8, 1590–1599. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wu, Y.; Hou, J.; Wang, P.; Peng, X.; Ge, G. Coumarin-Based near-Infrared Fluorogenic Probes: Recent Advances, Challenges and Future Perspectives. Coord. Chem. Rev. 2023, 480, 215020. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, F.; Hao, Y.; Chen, L.; Zhou, Q.; Zeng, H.; Song, Y.; Guo, Z.; Peng, X.; Ge, G. Fluorogenic Probes for Functional Imaging of Endoplasmic Reticulum-Resident Proteins: From Molecular Engineering to Biomedical Applications. Adv. Funct. Mater. 2025, 35, 2416057. [Google Scholar] [CrossRef]
- Hentsch, A.; Guberman, M.; Radetzki, S.; Kaushik, S.; Huizenga, M.; Paul, J.; Schippers, M.; Benz, J.; Kuhn, B.; Heer, D.; et al. A Highly Selective and Versatile Probe Platform for Visualization of Monoacylglycerol Lipase. Angew. Chemie Int. Ed. 2025, 64, e202413405. [Google Scholar] [CrossRef]
- Hentsch, A.; Guberman, M.; Radetzki, S.; Kaushik, S.; Huizenga, M.; He, Y.; Contzen, J.; Kuhn, B.; Benz, J.; Schippers, M.; et al. Highly Specific Miniaturized Fluorescent Monoacylglycerol Lipase Probes Enable Translational Research. J. Am. Chem. Soc. 2025, 147, 10188–10202. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bononi, G.; Landucci, E.; Di Stefano, M.; Piazza, L.; Bertini, S.; Macchia, M.; Granchi, C. Methods for Assessing MAGL Enzymatic Activity: An Extensive Review of Past and Emerging Approaches. Int. J. Mol. Sci. 2025, 26, 9829. https://doi.org/10.3390/ijms26199829
Bononi G, Landucci E, Di Stefano M, Piazza L, Bertini S, Macchia M, Granchi C. Methods for Assessing MAGL Enzymatic Activity: An Extensive Review of Past and Emerging Approaches. International Journal of Molecular Sciences. 2025; 26(19):9829. https://doi.org/10.3390/ijms26199829
Chicago/Turabian StyleBononi, Giulia, Eva Landucci, Miriana Di Stefano, Lisa Piazza, Simone Bertini, Marco Macchia, and Carlotta Granchi. 2025. "Methods for Assessing MAGL Enzymatic Activity: An Extensive Review of Past and Emerging Approaches" International Journal of Molecular Sciences 26, no. 19: 9829. https://doi.org/10.3390/ijms26199829
APA StyleBononi, G., Landucci, E., Di Stefano, M., Piazza, L., Bertini, S., Macchia, M., & Granchi, C. (2025). Methods for Assessing MAGL Enzymatic Activity: An Extensive Review of Past and Emerging Approaches. International Journal of Molecular Sciences, 26(19), 9829. https://doi.org/10.3390/ijms26199829