Serum Metabolomics Uncovers Immune and Lipid Pathway Alterations in Lambs Supplemented with Novel LAB-Bifidobacterium Cocktail
Abstract
1. Introduction
2. Results
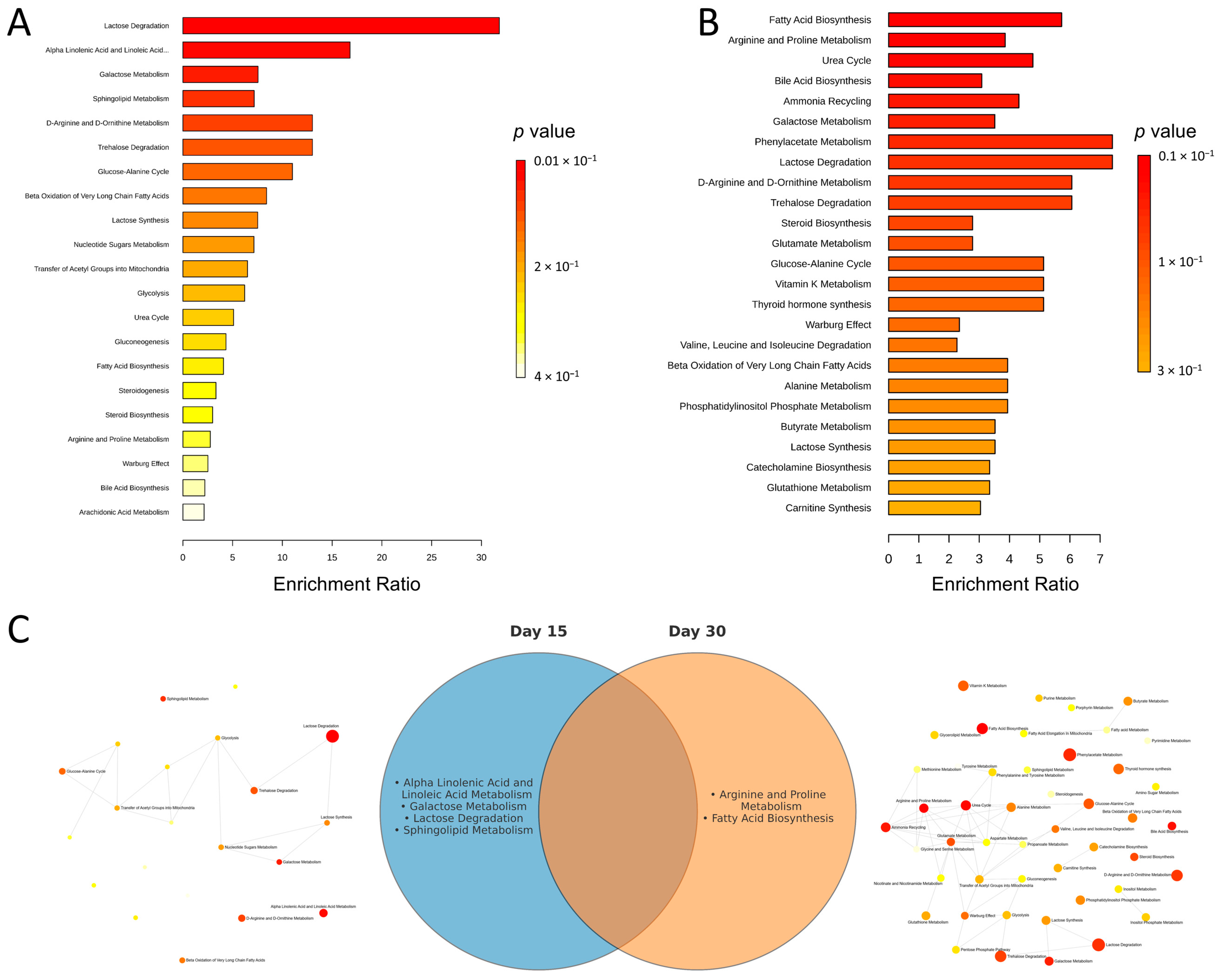
2.1. Metabolic Alterations After 15 Days of Supplementation
2.2. Metabolic Alterations After 30 Days of Supplementation
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Sample Collection
4.2. GC-TOF Compound Identification and Quantification
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mathew, A.G.; Cissell, R.; Liamthong, S. Antibiotic Resistance in Bacteria Associated with Food Animals: A United States Perspective of Livestock Production. Foodborne Pathog. Dis. 2007, 4, 115–133. [Google Scholar] [CrossRef]
- Nunes, J.O.; Bertechini, A.G.; de Brito, J.Á.G.; Fassani, É.J.; Mesquita, F.R.; Makiyama, L.; Meneghetti, C. Evaluation of the Use of Probiotic (Bacillus Subtilis C-3102) as Additive to Improve Performance in Broiler Chicken Diets. Rev. Bras. Zootec. 2012, 41, 2374–2378. [Google Scholar] [CrossRef]
- Teillant, A.; Brower, C.H.; Laxminarayan, R. Economics of Antibiotic Growth Promoters in Livestock. Annu. Rev. Resour. Econ. 2015, 7, 349–374. [Google Scholar] [CrossRef]
- Fischer, A.J.; Song, Y.; He, Z.; Haines, D.M.; Guan, L.L.; Steele, M.A. Effect of Delaying Colostrum Feeding on Passive Transfer and Intestinal Bacterial Colonization in Neonatal Male Holstein Calves. J. Dairy. Sci. 2018, 101, 3099–3109. [Google Scholar] [CrossRef]
- Faber, S.N.; Faber, N.E.; Mccauley, T.C.; Ax, R.L. Case Study: Effects Of Colostrum Ingestion on Lactational Performance1. Prof. Anim. Sci. 2005, 21, 420–425. [Google Scholar] [CrossRef]
- Bartels, C.J.M.; Holzhauer, M.; Jorritsma, R.; Swart, W.A.J.M.; Lam, T.J.G.M. Prevalence, Prediction and Risk Factors of Enteropathogens in Normal and Non-Normal Faeces of Young Dutch Dairy Calves. Prev. Vet. Med. 2010, 93, 162–169. [Google Scholar] [CrossRef]
- Millet, S.; Maertens, L. The European Ban on Antibiotic Growth Promoters in Animal Feed: From Challenges to Opportunities. Vet. J. 2011, 187, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Wallinga, D.; Smit, L.A.M.; Davis, M.F.; Casey, J.A.; Nachman, K.E. A Review of the Effectiveness of Current US Policies on Antimicrobial Use in Meat and Poultry Production. Curr. Environ. Health Rep. 2022, 9, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut Microbiota Functions: Metabolism of Nutrients and Other Food Components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Pessione, E. Lactic Acid Bacteria Contribution to Gut Microbiota Complexity: Lights and Shadows. Front. Cell Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef]
- Verschuere, L.; Rombaut, G.; Sorgeloos, P.; Verstraete, W. Probiotic Bacteria as Biological Control Agents in Aquaculture. Microbiol. Mol. Biol. Rev. 2000, 64, 655–671. [Google Scholar] [CrossRef]
- Liao, S.F.; Nyachoti, M. Using Probiotics to Improve Swine Gut Health and Nutrient Utilization. Anim. Nutr. 2017, 3, 331–343. [Google Scholar] [CrossRef]
- Weidenmaier, C.; Peschel, A. Teichoic Acids and Related Cell-Wall Glycopolymers in Gram-Positive Physiology and Host Interactions. Nat. Rev. Microbiol. 2008, 6, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Cazorla, S.I.; Maldonado-Galdeano, C.; Weill, R.; de Paula, J.; Perdigón, G.D.V. Oral Administration of Probiotics Increases Paneth Cells and Intestinal Antimicrobial Activity. Front. Microbiol. 2018, 9, 736. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, R.; Małaczewska, J.; Tobolski, D.; Miciński, J.; Kaczorek-Łukowska, E.; Zwierzchowski, G. The Effect of Orally Administered Multi-Strain Probiotic Formulation (Lactobacillus, Bifidobacterium) on the Phagocytic Activity and Oxidative Metabolism of Peripheral Blood Granulocytes and Monocytes in Lambs. Int. J. Mol. Sci. 2024, 25, 5068. [Google Scholar] [CrossRef]
- Daszkiewicz, T.; Miciński, J.; Wójcik, R.; Tobolski, D.; Zwierzchowski, G.; Kobzhassarov, T.; Ząbek, K.; Charkiewicz, K. The Effect of Probiotic Supplementation in Kamieniec Lambs on Meat Quality. Small Rumin. Res. 2025, 244, 107444. [Google Scholar] [CrossRef]
- Jiye, A.; Trygg, J.; Gullberg, J.; Johansson, A.I.; Jonsson, P.; Antti, H.; Marklund, S.L.; Moritz, T. Extraction and GC/MS Analysis of the Human Blood Plasma Metabolome. Anal. Chem. 2005, 77, 8086–8094. [Google Scholar] [CrossRef]
- Dervishi, E.; Zhang, G.; Dunn, S.M.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. GC–MS Metabolomics Identifies Metabolite Alterations That Precede Subclinical Mastitis in the Blood of Transition Dairy Cows. J. Proteome Res. 2016, 16, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Love, J.; Selker, R.; Marsman, M.; Jamil, T.; Dropmann, D.; Verhagen, J.; Ly, A.; Gronau, Q.F.; Smíra, M.; Epskamp, S.; et al. JASP: Graphical Statistical Software for Common Statistical Designs. J. Stat. Softw. 2019, 88, 1–17. [Google Scholar] [CrossRef]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational Biomarker Discovery in Clinical Metabolomics: An Introductory Tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef]
- Hassan, A.; Gado, H.; Anele, U.Y.; Berasain, M.A.M.; Salem, A.Z.M. Influence of Dietary Probiotic Inclusion on Growth Performance, Nutrient Utilization, Ruminal Fermentation Activities and Methane Production in Growing Lambs. Anim. Biotechnol. 2020, 31, 365–372. [Google Scholar] [CrossRef]
- Armstrong, L.E.; Casa, D.J.; Belval, L.N. Metabolism, Bioenergetics and Thermal Physiology: Influences of the Human Intestinal Microbiota. Nutr. Res. Rev. 2019, 32, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Belury, M.A. Inhibition of Carcinogenesis by Conjugated Linoleic Acid: Potential Mechanisms of Action. J. Nutr. 2002, 132, 2995–2998. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wu, L.; Chen, J.; Dong, L.; Chen, C.; Wen, Z.; Hu, J.; Fleming, I.; Wang, D.W. Metabolism Pathways of Arachidonic Acids: Mechanisms and Potential Therapeutic Targets. Signal Transduct. Target. Ther. 2021, 6, 94. [Google Scholar] [CrossRef]
- Quaranta, A.; Revol-Cavalier, J.; Wheelock, C.E. The Octadecanoids: An Emerging Class of Lipid Mediators. Biochem. Soc. Trans. 2022, 50, 1569–1582. [Google Scholar] [CrossRef]
- Lv, L.; Mu, D.; Du, Y.; Yan, R.; Jiang, H. Mechanism of the Immunomodulatory Effect of the Combination of Live Bifidobacterium, Lactobacillus, Enterococcus, and Bacillus on Immunocompromised Rats. Front. Immunol. 2021, 12, 694344. [Google Scholar] [CrossRef]
- Lubbadeh, W.; Haddadin, M.S.Y.; Al-Tamimi, M.A.; Robinson, R.K. Effect on the Cholesterol Content of Fresh Lamb of Supplementing the Feed of Awassi Ewes and Lambs with Lactobacillus acidophilus. Meat Sci. 1999, 52, 381–385. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, T.; Zhang, Y.; Yang, L.; Duan, Y.; Su, L.; Tian, J.; Sun, L.; Wang, B.; Jin, Y. Impact of Feeding Probiotics on Blood Parameters, Tail Fat Metabolites, and Volatile Flavor Components of Sunit Sheep. Foods 2022, 11, 2644. [Google Scholar] [CrossRef]
- Wu, T.; Wang, G.; Xiong, Z.; Xia, Y.; Song, X.; Zhang, H.; Wu, Y.; Ai, L. Probiotics Interact with Lipids Metabolism and Affect Gut Health. Front. Nutr. 2022, 9, 917043. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Shataer, D.; Yan, H.; Dong, X.; Zhang, M.; Qin, Y.; Cui, J.; Wang, L. Probiotics and Non-Alcoholic Fatty Liver Disease: Unveiling the Mechanisms of Lactobacillus plantarum and Bifidobacterium bifidum in Modulating Lipid Metabolism, Inflammation, and Intestinal Barrier Integrity. Foods 2024, 13, 2992. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Ho, C.L. Recent Development of Probiotic Bifidobacteria for Treating Human Diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar] [CrossRef]
- Kelly, B.; Pearce, E.L. Amino Assets: How Amino Acids Support Immunity. Cell Metab. 2020, 32, 154–175. [Google Scholar] [CrossRef]
- Li, P.; Wu, G. Important Roles of Amino Acids in Immune Responses. Br. J. Nutr. 2022, 127, 398–402. [Google Scholar] [CrossRef]
- Aguayo-Cerón, K.A.; Sánchez-Muñoz, F.; Gutierrez-Rojas, R.A.; Acevedo-Villavicencio, L.N.; Flores-Zarate, A.V.; Huang, F.; Giacoman-Martinez, A.; Villafaña, S.; Romero-Nava, R. Glycine: The Smallest Anti-Inflammatory Micronutrient. Int. J. Mol. Sci. 2023, 24, 11236. [Google Scholar] [CrossRef]
- Chen, H.; Guo, B.; Yang, M.; Luo, J.; Hu, Y.; Qu, M.; Song, X. Response of Growth Performance, Blood Biochemistry Indices, and Rumen Bacterial Diversity in Lambs to Diets Containing Supplemental Probiotics and Chinese Medicine Polysaccharides. Front. Vet. Sci. 2021, 8, 681389. [Google Scholar] [CrossRef]
- Pintarič, M.; Langerholc, T. Probiotic Mechanisms Affecting Glucose Homeostasis: A Scoping Review. Life 2022, 12, 1187. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.-L.; Zhang, A.-H.; Kong, L.; Wang, X.-J. Advances in Mass Spectrometry-Based Metabolomics for Investigation of Metabolites. RSC Adv. 2018, 8, 22335–22350. [Google Scholar] [CrossRef] [PubMed]
- Bubnov, R.V.; Babenko, L.P.; Lazarenko, L.M.; Mokrozub, V.V.; Demchenko, O.A.; Nechypurenko, O.V.; Spivak, M.Y. Comparative Study of Probiotic Effects of Lactobacillus and Bifidobacteria Strains on Cholesterol Levels, Liver Morphology and the Gut Microbiota in Obese Mice. EPMA J. 2017, 8, 357–376. [Google Scholar] [CrossRef]
- Childs, C.E.; Röytiö, H.; Alhoniemi, E.; Fekete, A.A.; Forssten, S.D.; Hudjec, N.; Lim, Y.N.; Steger, C.J.; Yaqoob, P.; Tuohy, K.M.; et al. Xylo-Oligosaccharides Alone or in Synbiotic Combination with Bifidobacterium animalis subsp. lactis Induce Bifidogenesis and Modulate Markers of Immune Function in Healthy Adults: A Double-Blind, Placebo-Controlled, Randomised, Factorial Cross-over Study. Br. J. Nutr. 2014, 111, 1945–1956. [Google Scholar] [CrossRef]
- Fuentes, M.C.; Lajo, T.; Carrión, J.M.; Cuñé, J. Cholesterol-Lowering Efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in Hypercholesterolaemic Adults. Br. J. Nutr. 2013, 109, 1866–1872. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, Z.; Bath, C.; Marett, L.; Pryce, J.; Rochfort, S. Optimised Method for Short-Chain Fatty Acid Profiling of Bovine Milk and Serum. Molecules 2022, 27, 436. [Google Scholar] [CrossRef] [PubMed]
- Facchin, S.; Bertin, L.; Bonazzi, E.; Lorenzon, G.; de Barba, C.; Barberio, B.; Zingone, F.; Maniero, D.; Scarpa, M.; Ruffolo, C.; et al. Short-Chain Fatty Acids and Human Health: From Metabolic Pathways to Current Therapeutic Implications. Life 2024, 14, 559. [Google Scholar] [CrossRef] [PubMed]
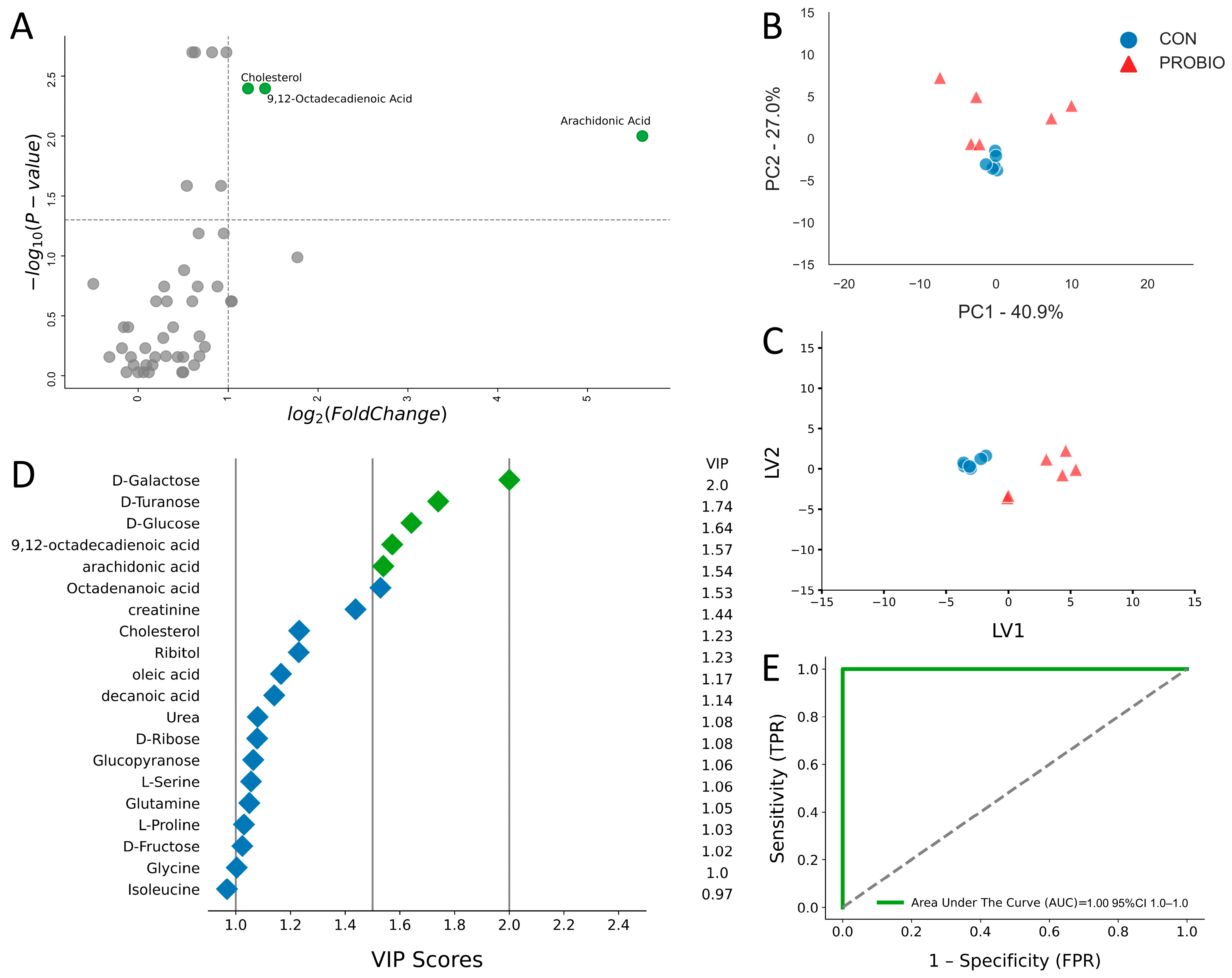
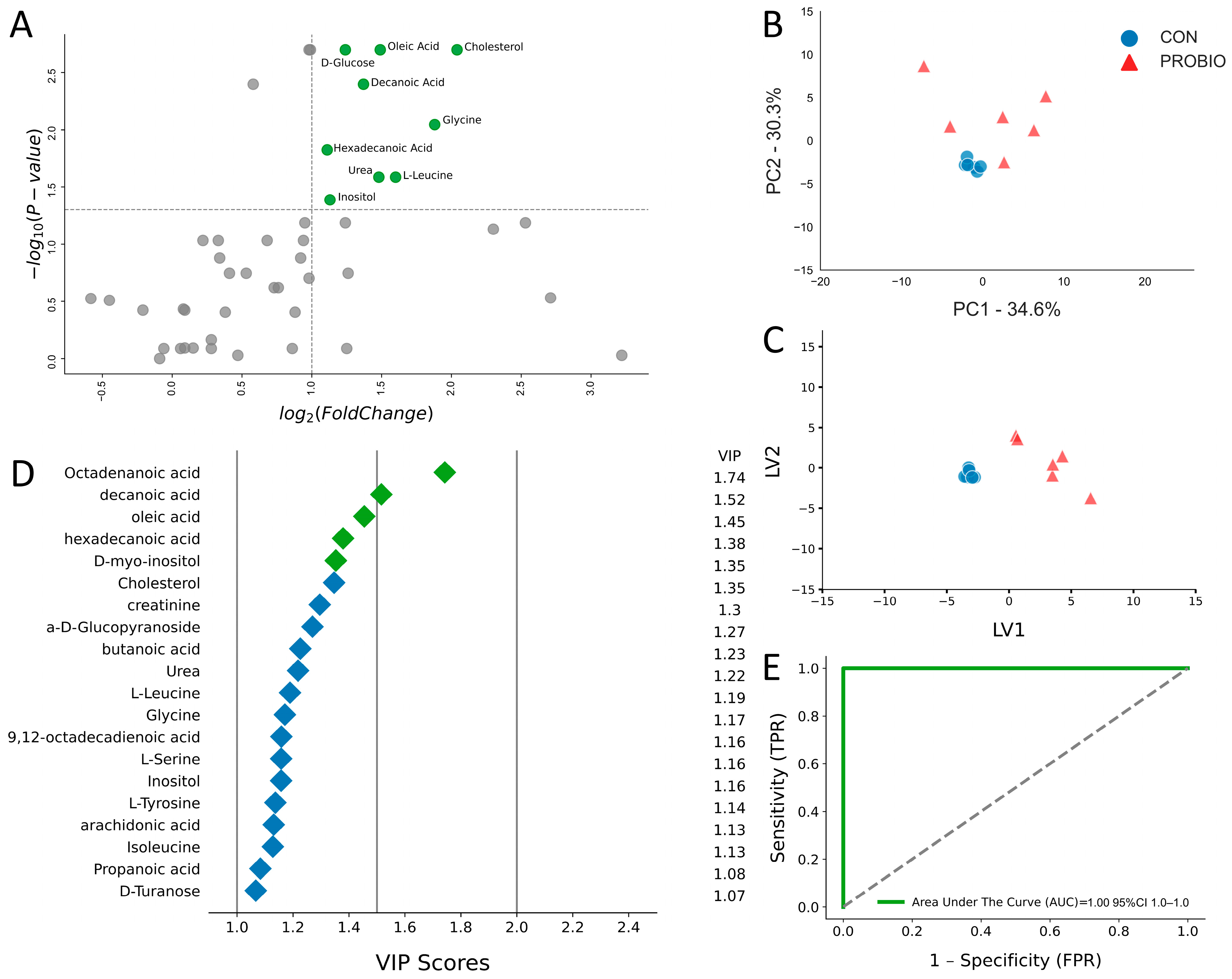
| Metabolite, μM | PROBIO | CON | p-Value | Fold Change | log2 (FC) | PROBIO/CON |
|---|---|---|---|---|---|---|
| Number of cases | 6 | 6 | - | - | - | - |
| 9,12-Octadecadienoic Acid | 0.60 (0.30) | 0.23 (0.03) | <0.01 | 2.66 | 1.41 | UP |
| Acetic Acid | 0.07 (0.04) | 0.08 (0.05) | 0.59 | 0.88 | −0.18 | DOWN |
| Alanine | 0.55 (0.16) | 0.49 (0.06) | 0.82 | 1.12 | 0.16 | UP |
| Arachidonic Acid | 0.05 (0.03) | < 0.01 | 0.01 | 48.76 | 5.61 | UP |
| B-Alanine | 0.02 (0.02) | 0.03 (0.01) | 0.94 | 0.91 | −0.13 | DOWN |
| Butanedioic Acid | 0.03 (0.01) | 0.03 (0.01) | 0.70 | 0.95 | −0.08 | DOWN |
| Butanoic Acid | 0.95 (0.62) | 0.68 (0.18) | 0.94 | 1.41 | 0.49 | UP |
| Cholesterol | 7.42 (4.37) | 3.19 (0.39) | <0.01 | 2.33 | 1.22 | UP |
| Creatinine | 0.61 (0.12) | 0.42 (0.09) | 0.03 | 1.45 | 0.54 | UP |
| Decanoic Acid | 0.1 (0.05) | 0.05 (0.02) | 0.03 | 1.89 | 0.92 | UP |
| D-Fructose | 0.49 (0.32) | 0.27 (0.05) | 0.18 | 1.84 | 0.88 | UP |
| D-Galactose | 0.10 (0.01) | 0.05 (0.01) | <0.01 | 1.76 | 0.82 | UP |
| D-Glucose | 8.63 (1.76) | 5.57 (0.34) | <0.01 | 1.55 | 0.63 | UP |
| Dihydroxybutanoic Acid | 0.02 (0.02) | 0.02 (0.01) | 0.94 | 1.04 | 0.06 | UP |
| D-Myo-Inositol | 1.84 (1.00) | 1.21 (0.13) | 0.24 | 1.52 | 0.60 | UP |
| D-Ribose | 0.18 (0.06) | 0.14 (0.03) | 0.48 | 1.22 | 0.28 | UP |
| D-Turanose | 0.64 (0.09) | 0.42 (0.05) | <0.01 | 1.51 | 0.60 | UP |
| Galactoric Acid | 0.08 (0.05) | 0.06 (0.02) | 0.70 | 1.36 | 0.44 | UP |
| Glucopyranose | 0.03 (0.03) | 0.01 (0.01) | 0.10 | 3.42 | 1.77 | UP |
| Glucopyranoside | 0.02 (0.02) | 0.01 (0.00) | 0.57 | 1.67 | 0.74 | UP |
| Glutamine | 0.83 (0.21) | 0.78 (0.10) | 0.82 | 1.07 | 0.09 | UP |
| Glycine | 0.74 (0.40) | 1.05 (0.32) | 0.18 | 0.70 | −0.51 | DOWN |
| Heptadecanoic Acid | 0.05 (0.04) | 0.04 (0.01) | 0.69 | 1.24 | 0.31 | UP |
| Hexadecanoic Acid | 1.17 (0.85) | 0.74 (0.07) | 0.18 | 1.58 | 0.66 | UP |
| Hexanoic Acid | 0.02 (0.01) | 0.02 (0.01) | 0.70 | 0.80 | −0.32 | DOWN |
| Inositol | 0.12 (0.04) | 0.10 (0.02) | 0.70 | 1.14 | 0.19 | UP |
| Isoleucine | 1.03 (0.53) | 0.67 (0.05) | 0.82 | 1.53 | 0.62 | UP |
| L-Asparagine | 0.03 (0.03) | 0.02 (0.01) | 0.47 | 1.60 | 0.68 | UP |
| L-Leucine | 3.79 (2.96) | 1.84 (0.36) | 0.24 | 2.06 | 1.04 | UP |
| L-Methionine | 1.35 (0.84) | 1.03 (0.14) | 0.39 | 1.31 | 0.39 | UP |
| L-Ornithine | 0.33 (0.25) | 0.33 (0.06) | 0.94 | 1.00 | 0.00 | DOWN |
| L-Proline | 0.55 (0.12) | 0.45 (0.06) | 0.18 | 1.22 | 0.29 | UP |
| L-Serine | 0.76 (0.18) | 0.66 (0.09) | 0.24 | 1.15 | 0.20 | UP |
| L-Threonine | 0.72 (0.20) | 0.67 (0.14) | 0.94 | 1.09 | 0.12 | UP |
| L-Tyrosine | 0.46 (0.16) | 0.44 (0.05) | 0.59 | 1.06 | 0.08 | UP |
| Malic Acid | 0.05 (0.02) | 0.05 (0.02) | 0.82 | 0.96 | −0.05 | DOWN |
| Octadenanoic Acid | 1.06 (0.30) | 0.54 (0.22) | <0.01 | 1.98 | 0.98 | UP |
| Oleic Acid | 1.13 (0.46) | 0.71 (0.27) | 0.06 | 1.59 | 0.67 | UP |
| Pentanedioic Acid | 0.11 (0.08) | 0.06 (0.03) | 0.24 | 2.04 | 1.03 | UP |
| Phosphoric Acid | 0.12 (0.08) | 0.13 (0.02) | 0.39 | 0.93 | −0.11 | DOWN |
| Propanoic Acid | 3.50 (2.62) | 2.48 (0.45) | 0.70 | 1.41 | 0.50 | UP |
| Ribitol | 0.09 (0.03) | 0.07 (0.01) | 0.13 | 1.43 | 0.51 | UP |
| Serotonin | 0.12 (0.09) | 0.14 (0.03) | 0.39 | 0.89 | −0.16 | DOWN |
| Tryptophan | 0.07 (0.07) | 0.04 (0.01) | 0.69 | 1.60 | 0.68 | UP |
| Urea | 6.87 (4.13) | 3.57 (0.52) | 0.06 | 1.93 | 0.95 | UP |
| Valine | 0.81 (0.52) | 0.57 (0.08) | 0.94 | 1.42 | 0.50 | UP |
| Xylitol | 0.07 (0.02) | 0.06 (0.02) | 0.24 | 1.25 | 0.32 | UP |
| Metabolite, μM | PROBIO | CON | p-Value | Fold Change | log2 (FC) | PROBIO/CON |
|---|---|---|---|---|---|---|
| Number of cases | 6 | 6 | - | - | - | - |
| 9,12-Octadecadienoic Acid | 0.41 (0.26) | 0.21 (0.09) | 0.20 | 1.97 | 0.98 | UP |
| Acetic Acid | 0.10 (0.10) | 0.04 (0.02) | 0.82 | 2.37 | 1.25 | UP |
| Alanine | 4.25 (9.01) | 0.46 (0.03) | 0.94 | 9.29 | 3.22 | UP |
| Arachidonic Acid | 0.06 (0.06) | 0.01 (0.01) | 0.29 | 6.57 | 2.71 | UP |
| B-Alanine | 0.02 (0.02) | 0.02 (0.00) | 1.00 | 0.94 | −0.09 | DOWN |
| Butanedioic Acid | 0.04 (0.02) | 0.02 (0.01) | 0.24 | 1.69 | 0.76 | UP |
| Butanoic Acid | 1.52 (0.80) | 0.64 (0.15) | 0.06 | 2.37 | 1.24 | UP |
| Cholesterol | 9.32 (5.37) | 2.27 (1.15) | <0.01 | 4.11 | 2.04 | UP |
| Creatinine | 0.69 (0.18) | 0.46 (0.07) | <0.01 | 1.49 | 0.58 | UP |
| Decanoic Acid | 0.09 (0.03) | 0.03 (0.01) | <0.01 | 2.58 | 1.37 | UP |
| D-Fructose | 0.19 (0.06) | 0.20 (0.05) | 0.82 | 0.96 | −0.06 | DOWN |
| D-Galactose | 0.09 (0.05) | 0.05 (0.01) | 0.24 | 1.66 | 0.73 | UP |
| D-Glucose | 12.38 (9.99) | 5.26 (0.59) | <0.01 | 2.35 | 1.24 | UP |
| Dihydroxybutanoic Acid | 0.02 (0.02) | 0.02 (0.01) | 0.38 | 0.86 | −0.21 | DOWN |
| D-Myo-Inositol | 1.91 (0.73) | 0.97 (0.09) | <0.01 | 1.98 | 0.98 | UP |
| D-Ribose | 0.21 (0.10) | 0.13 (0.03) | 0.09 | 1.61 | 0.68 | UP |
| D-Turanose | 0.63 (0.21) | 0.43 (0.08) | 0.18 | 1.45 | 0.53 | UP |
| Galactoric Acid | 0.09 (0.08) | 0.07 (0.02) | 0.94 | 1.38 | 0.47 | UP |
| Glucopyranose | 0.01 (0.01) | 0.01 (0.00) | 0.37 | 1.06 | 0.08 | UP |
| Glucopyranoside | 0.01 (0.01) | <0.01 | 0.07 | 4.94 | 2.30 | UP |
| Glutamine | 0.83 (0.20) | 0.66 (0.11) | 0.09 | 1.26 | 0.33 | UP |
| Glycine | 3.10 (3.84) | 0.84 (0.16) | <0.01 | 3.68 | 1.88 | UP |
| Heptadecanoic Acid | 0.06 (0.06) | 0.06 (0.06) | 0.81 | 1.11 | 0.15 | UP |
| Hexadecanoic Acid | 1.92 (0.71) | 0.89 (0.30) | 0.01 | 2.16 | 1.11 | UP |
| Hexanoic Acid | 0.03 (0.03) | 0.03 (0.02) | 0.81 | 1.06 | 0.09 | UP |
| Inositol | 0.15 (0.09) | 0.07 (0.02) | 0.04 | 2.19 | 1.13 | UP |
| Isoleucine | 1.24 (0.66) | 0.65 (0.09) | 0.09 | 1.92 | 0.94 | UP |
| L-Asparagine | 0.02 (0.02) | 0.02 (0.01) | 0.29 | 0.67 | −0.58 | DOWN |
| L-Leucine | 5.17 (3.42) | 1.71 (0.43) | 0.03 | 3.03 | 1.60 | UP |
| L-Methionine | 1.54 (0.96) | 1.18 (0.28) | 0.39 | 1.30 | 0.38 | UP |
| L-Ornithine | 0.23 (0.19) | 0.31 (0.09) | 0.31 | 0.73 | −0.45 | DOWN |
| L-Proline | 0.58 (0.10) | 0.50 (0.06) | 0.09 | 1.17 | 0.22 | UP |
| L-Serine | 0.89 (0.23) | 0.70 (0.05) | 0.13 | 1.26 | 0.34 | UP |
| L-Threonine | 0.64 (0.24) | 0.53 (0.09) | 0.82 | 1.21 | 0.28 | UP |
| L-Tyrosine | 0.65 (0.35) | 0.34 (0.08) | 0.06 | 1.93 | 0.95 | UP |
| Malic Acid | 0.08 (0.06) | 0.03 (0.01) | 0.18 | 2.39 | 1.26 | UP |
| Octadenanoic Acid | 1.39 (0.24) | 0.70 (0.07) | <0.01 | 1.99 | 0.99 | UP |
| Oleic Acid | 1.93 (0.79) | 0.69 (0.16) | <0.01 | 2.81 | 1.49 | UP |
| Pentanedioic Acid | 0.15 (0.12) | 0.08 (0.02) | 0.82 | 1.82 | 0.86 | UP |
| Phosphoric Acid | 0.10 (0.13) | 0.09 (0.02) | 0.38 | 1.07 | 0.09 | UP |
| Propanoic Acid | 11.20 (14.29) | 1.93 (0.51) | 0.06 | 5.80 | 2.53 | UP |
| Ribitol | 0.11 (0.09) | 0.06 (0.01) | 0.13 | 1.90 | 0.92 | UP |
| Serotonin | 0.23 (0.08) | 0.17 (0.02) | 0.18 | 1.33 | 0.41 | UP |
| Tryptophan | 0.03 (0.04) | 0.03 (0.02) | 0.69 | 1.22 | 0.28 | UP |
| Urea | 10.13 (6.30) | 3.64 (0.88) | 0.03 | 2.78 | 1.48 | UP |
| Valine | 0.95 (0.64) | 0.52 (0.05) | 0.39 | 1.84 | 0.88 | UP |
| Xylitol | 0.04 (0.02) | 0.04 (0.01) | 0.82 | 1.04 | 0.06 | UP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wójcik, R.; Król-Grzymała, A.; Tobolski, D.; Paritova, A.; García-Calvo, E.; Miciński, J.; Zwierzchowski, G. Serum Metabolomics Uncovers Immune and Lipid Pathway Alterations in Lambs Supplemented with Novel LAB-Bifidobacterium Cocktail. Int. J. Mol. Sci. 2025, 26, 9808. https://doi.org/10.3390/ijms26199808
Wójcik R, Król-Grzymała A, Tobolski D, Paritova A, García-Calvo E, Miciński J, Zwierzchowski G. Serum Metabolomics Uncovers Immune and Lipid Pathway Alterations in Lambs Supplemented with Novel LAB-Bifidobacterium Cocktail. International Journal of Molecular Sciences. 2025; 26(19):9808. https://doi.org/10.3390/ijms26199808
Chicago/Turabian StyleWójcik, Roman, Angelika Król-Grzymała, Dawid Tobolski, Assel Paritova, Estefanía García-Calvo, Jan Miciński, and Grzegorz Zwierzchowski. 2025. "Serum Metabolomics Uncovers Immune and Lipid Pathway Alterations in Lambs Supplemented with Novel LAB-Bifidobacterium Cocktail" International Journal of Molecular Sciences 26, no. 19: 9808. https://doi.org/10.3390/ijms26199808
APA StyleWójcik, R., Król-Grzymała, A., Tobolski, D., Paritova, A., García-Calvo, E., Miciński, J., & Zwierzchowski, G. (2025). Serum Metabolomics Uncovers Immune and Lipid Pathway Alterations in Lambs Supplemented with Novel LAB-Bifidobacterium Cocktail. International Journal of Molecular Sciences, 26(19), 9808. https://doi.org/10.3390/ijms26199808









