NRDE2 Interacts with an Early Transcription Elongation Complex and Widely Impacts Gene Expression
Abstract
1. Introduction
2. Results
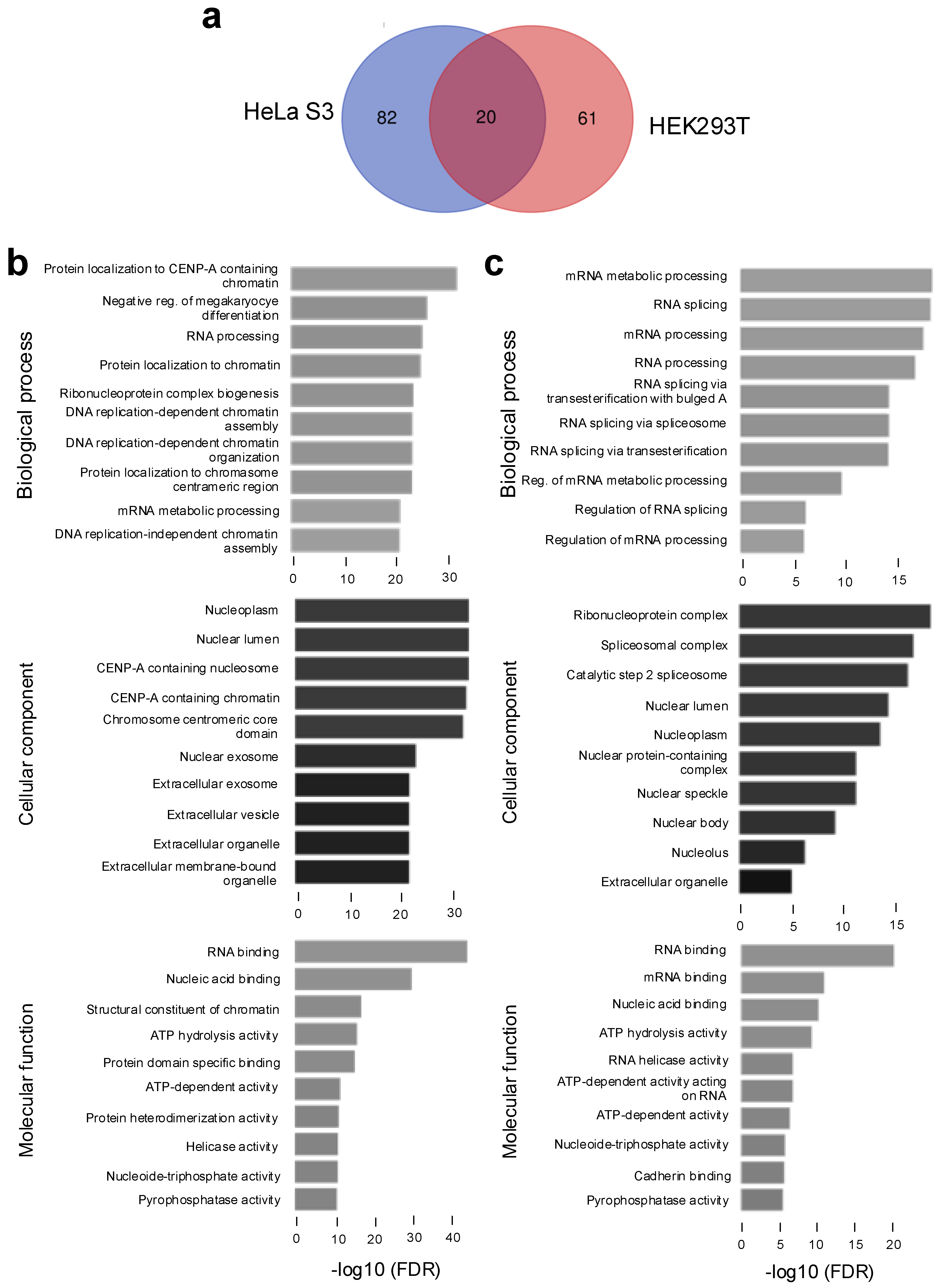
2.1. NRDE2 Interacts with Transcription-Associated Proteins and RNA Processing Factors
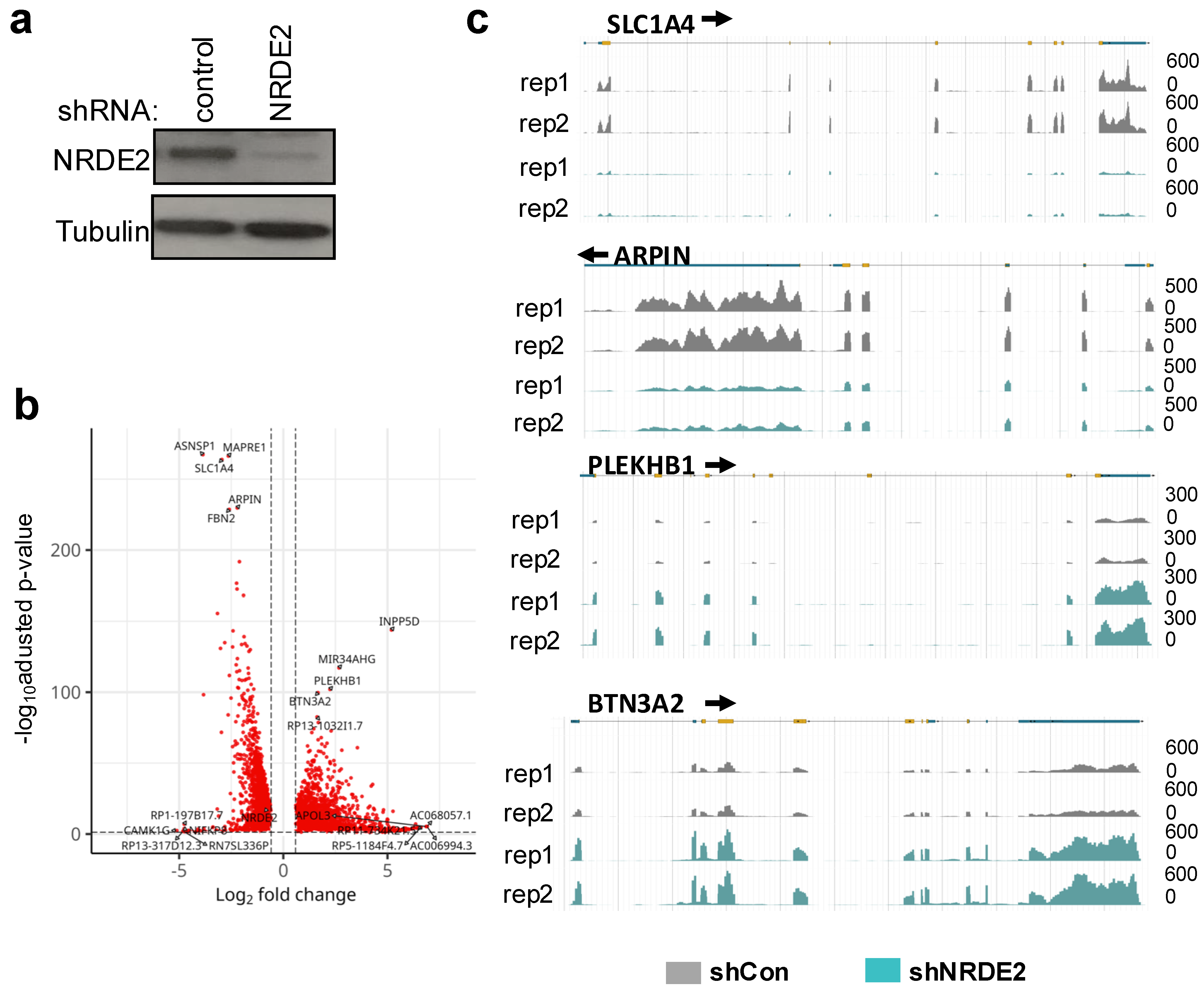
2.2. NRDE2 Affects Expression and mRNA Splicing at a Subset of Genes
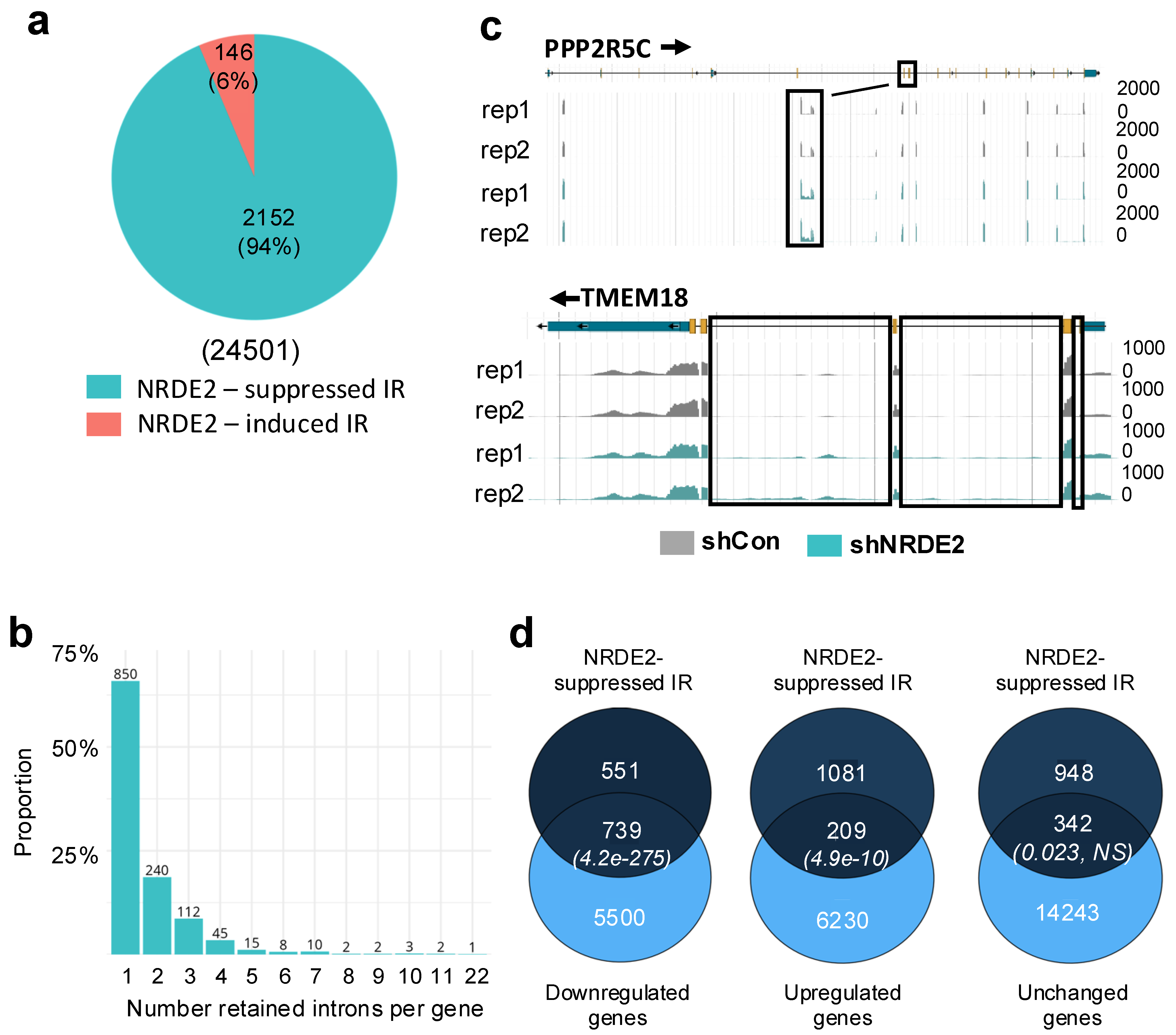
2.3. NRDE2 Regulates Intron Retention and Additional Splicing Events
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Antibodies
4.3. Generation of Fh-NRDE2-Expressing Cells
4.4. RNAi
4.5. NRDE2 Protein Complex Purification
4.6. Coimmunoprecipitation Analysis
4.7. RNA-Seq and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CDC73 | Cell division cycle 73 |
| DMEM | Dulbecco modified Eagle’s minimal essential medium |
| FDR | False discovery rate |
| IR | Intron retention |
| NRDE2 | Nuclear RNAi-defective 2 |
| PAF1 | Polymerase-associated Factor 1 |
| P-TEFb | Positive transcription elongation factor b |
| RNAPII | RNA polymerase II |
| shRNA | Short hairpin RNA |
References
- Guang, S.; Bochner, A.F.; Burkhart, K.B.; Burton, N.; Pavelec, D.M.; Kennedy, S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 2010, 465, 1097–1101. [Google Scholar] [CrossRef]
- Burton, N.O.; Burkhart, K.B.; Kennedy, S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2011, 108, 19683–19688. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, A.; Sarkies, P.; Simon, M.; Doebley, A.L.; Goldstein, L.D.; Hedges, A.; Ikegami, K.; Alvares, S.M.; Yang, L.; LaRocque, J.R.; et al. Caenorhabditis elegans RSD-2 and RSD-6 promote germ cell immortality by maintaining small interfering RNA populations. Proc. Natl. Acad. Sci. USA 2014, 111, E4323–E4331. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, K.B.; Guang, S.; Buckley, B.A.; Wong, L.; Bochner, A.F.; Kennedy, S. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 2011, 7, e1002249. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Yan, J.; Fei, Y.; Pagano, D.J.; Kennedy, S. A Conserved NRDE-2/MTR-4 Complex Mediates Nuclear RNAi in Caenorhabditis elegans. Genetics 2020, 216, 1071–1085. [Google Scholar] [CrossRef]
- Lee, N.N.; Chalamcharla, V.R.; Reyes-Turcu, F.; Mehta, S.; Zofall, M.; Balachandran, V.; Dhakshnamoorthy, J.; Taneja, N.; Yamanaka, S.; Zhou, M.; et al. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell 2013, 155, 1061–1074. [Google Scholar] [CrossRef]
- Cipakova, I.; Jurcik, M.; Rubintova, V.; Borbova, M.; Mikolaskova, B.; Jurcik, J.; Bellova, J.; Barath, P.; Gregan, J.; Cipak, L. Identification of proteins associated with splicing factors Ntr1, Ntr2, Brr2 and Gpl1 in the fission yeast Schizosaccharomyces pombe. Cell Cycle 2019, 18, 1532–1536. [Google Scholar] [CrossRef]
- Flemr, M.; Schwaiger, M.; Hess, D.; Iesmantavicius, V.; Ahel, J.; Tuck, A.C.; Mohn, F.; Buhler, M. Mouse nuclear RNAi-defective 2 promotes splicing of weak 5′ splice sites. RNA 2023, 29, 1140–1165. [Google Scholar] [CrossRef]
- Jiao, A.L.; Perales, R.; Umbreit, N.T.; Haswell, J.R.; Piper, M.E.; Adams, B.D.; Pellman, D.; Kennedy, S.; Slack, F.J. Human nuclear RNAi-defective 2 (NRDE2) is an essential RNA splicing factor. RNA 2019, 25, 352–363. [Google Scholar] [CrossRef]
- Richard, P.; Ogami, K.; Chen, Y.; Feng, S.; Moresco, J.J.; Yates, J.R., 3rd; Manley, J.L. NRDE-2, the human homolog of fission yeast Nrl1, prevents DNA damage accumulation in human cells. RNA Biol. 2018, 15, 868–876. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Zuo, X.; Wang, C.; Zhang, Z.; Zhang, H.; Zeng, T.; Chen, S.; Liu, M.; Chen, H.; et al. NRDE2 deficiency impairs homologous recombination repair and sensitizes hepatocellular carcinoma to PARP inhibitors. Cell Genom. 2024, 4, 100550. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Wu, G.; Zhang, H.; Du, X.; Chen, S.; Zhang, L.; Wang, K.; Fan, J.; Gao, S.; et al. NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction. Genes Dev. 2019, 33, 536–549. [Google Scholar] [CrossRef]
- Chiu, C.G.; Nakamura, Y.; Chong, K.K.; Huang, S.K.; Kawas, N.P.; Triche, T.; Elashoff, D.; Kiyohara, E.; Irie, R.F.; Morton, D.L.; et al. Genome-wide characterization of circulating tumor cells identifies novel prognostic genomic alterations in systemic melanoma metastasis. Clin. Chem. 2014, 60, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.C.; Devlin, A.M.; Kim, J.; Conrad, N.K. Kaposi’s Sarcoma-Associated Herpesvirus Fine-Tunes the Temporal Expression of Late Genes by Manipulating a Host RNA Quality Control Pathway. J. Virol. 2020, 94, e00287-20. [Google Scholar] [CrossRef] [PubMed]
- Contreras, X.; Salifou, K.; Sanchez, G.; Helsmoortel, M.; Beyne, E.; Bluy, L.; Pelletier, S.; Rousset, E.; Rouquier, S.; Kiernan, R. Nuclear RNA surveillance complexes silence HIV-1 transcription. PLoS Pathog. 2018, 14, e1006950. [Google Scholar] [CrossRef] [PubMed]
- Contreras, X.; Depierre, D.; Akkawi, C.; Srbic, M.; Helsmoortel, M.; Nogaret, M.; LeHars, M.; Salifou, K.; Heurteau, A.; Cuvier, O.; et al. PAPgamma associates with PAXT nuclear exosome to control the abundance of PROMPT ncRNAs. Nat. Commun. 2023, 14, 6745. [Google Scholar] [CrossRef]
- Francette, A.M.; Tripplehorn, S.A.; Arndt, K.M. The Paf1 Complex: A Keystone of Nuclear Regulation Operating at the Interface of Transcription and Chromatin. J. Mol. Biol. 2021, 433, 166979. [Google Scholar] [CrossRef]
- Vos, S.M.; Farnung, L.; Boehning, M.; Wigge, C.; Linden, A.; Urlaub, H.; Cramer, P. Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 2018, 560, 607–612. [Google Scholar] [CrossRef]
- Yik, J.H.; Chen, R.; Nishimura, R.; Jennings, J.L.; Link, A.J.; Zhou, Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 2003, 12, 971–982. [Google Scholar] [CrossRef]
- Gardini, A.; Baillat, D.; Cesaroni, M.; Hu, D.; Marinis, J.M.; Wagner, E.J.; Lazar, M.A.; Shilatifard, A.; Shiekhattar, R. Integrator regulates transcriptional initiation and pause release following activation. Mol. Cell 2014, 56, 128–139. [Google Scholar] [CrossRef]
- Stadelmayer, B.; Micas, G.; Gamot, A.; Martin, P.; Malirat, N.; Koval, S.; Raffel, R.; Sobhian, B.; Severac, D.; Rialle, S.; et al. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat. Commun. 2014, 5, 5531. [Google Scholar] [CrossRef]
- Yamamoto, J.; Hagiwara, Y.; Chiba, K.; Isobe, T.; Narita, T.; Handa, H.; Yamaguchi, Y. DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nat. Commun. 2014, 5, 4263. [Google Scholar] [CrossRef]
- Middleton, R.; Gao, D.; Thomas, A.; Singh, B.; Au, A.; Wong, J.J.; Bomane, A.; Cosson, B.; Eyras, E.; Rasko, J.E.; et al. IRFinder: Assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017, 18, 51. [Google Scholar] [CrossRef]
- Trincado, J.L.; Entizne, J.C.; Hysenaj, G.; Singh, B.; Skalic, M.; Elliott, D.J.; Eyras, E. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018, 19, 40. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Qin, X.; Xu, H.; Li, L.; Wang, Z.; Li, F.; Xie, X.; Zhou, H.; Shen, Y.; Long, J. Structural insights into Paf1 complex assembly and histone binding. Nucleic Acids Res. 2013, 41, 10619–10629. [Google Scholar] [CrossRef] [PubMed]
- Rozenblatt-Rosen, O.; Nagaike, T.; Francis, J.M.; Kaneko, S.; Glatt, K.A.; Hughes, C.M.; LaFramboise, T.; Manley, J.L.; Meyerson, M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc. Natl. Acad. Sci. USA 2009, 106, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Mandal, S.S.; Pham, A.D.; Zheng, Y.; Erdjument-Bromage, H.; Batra, S.K.; Tempst, P.; Reinberg, D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005, 19, 1668–1673. [Google Scholar] [CrossRef]
- Salifou, K.; Burnard, C.; Basavarajaiah, P.; Grasso, G.; Helsmoortel, M.; Mac, V.; Depierre, D.; Franckhauser, C.; Beyne, E.; Contreras, X.; et al. Chromatin-associated MRN complex protects highly transcribing genes from genomic instability. Sci. Adv. 2021, 7, eabb2947. [Google Scholar] [CrossRef]
- Bejjani, F.; Segeral, E.; Mosca, K.; Lecourieux, A.; Bakail, M.; Hamoudi, M.; Emiliani, S. Overlapping and distinct functions of SPT6, PNUTS, and PCF11 in regulating transcription termination. Nucleic Acids Res. 2025, 53, gkaf179. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Ramirez, F.; Dundar, F.; Diehl, S.; Gruning, B.A.; Manke, T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–W191. [Google Scholar] [CrossRef]
- Molder, F.; Jablonski, K.P.; Letcher, B.; Hall, M.B.; Tomkins-Tinch, C.H.; Sochat, V.; Forster, J.; Lee, S.; Twardziok, S.O.; Kanitz, A.; et al. Sustainable data analysis with Snakemake. F1000Research 2021, 10, 33. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srbic, M.; Belhaouari, C.; Raffel, R.; Lemaire, L.; Barbier, J.; Bossuyt, J.; Akkawi, C.; Contreras, X.; Kiernan, R. NRDE2 Interacts with an Early Transcription Elongation Complex and Widely Impacts Gene Expression. Int. J. Mol. Sci. 2025, 26, 9792. https://doi.org/10.3390/ijms26199792
Srbic M, Belhaouari C, Raffel R, Lemaire L, Barbier J, Bossuyt J, Akkawi C, Contreras X, Kiernan R. NRDE2 Interacts with an Early Transcription Elongation Complex and Widely Impacts Gene Expression. International Journal of Molecular Sciences. 2025; 26(19):9792. https://doi.org/10.3390/ijms26199792
Chicago/Turabian StyleSrbic, Marina, Chaïmaa Belhaouari, Raoul Raffel, Laurine Lemaire, Jerome Barbier, Julie Bossuyt, Charbel Akkawi, Xavier Contreras, and Rosemary Kiernan. 2025. "NRDE2 Interacts with an Early Transcription Elongation Complex and Widely Impacts Gene Expression" International Journal of Molecular Sciences 26, no. 19: 9792. https://doi.org/10.3390/ijms26199792
APA StyleSrbic, M., Belhaouari, C., Raffel, R., Lemaire, L., Barbier, J., Bossuyt, J., Akkawi, C., Contreras, X., & Kiernan, R. (2025). NRDE2 Interacts with an Early Transcription Elongation Complex and Widely Impacts Gene Expression. International Journal of Molecular Sciences, 26(19), 9792. https://doi.org/10.3390/ijms26199792






