Nuclear Retention of mRNAs Through Paraspeckle Protein Binding to a Sequence Determinant in 3′UTR
Abstract
1. Introduction
2. Results
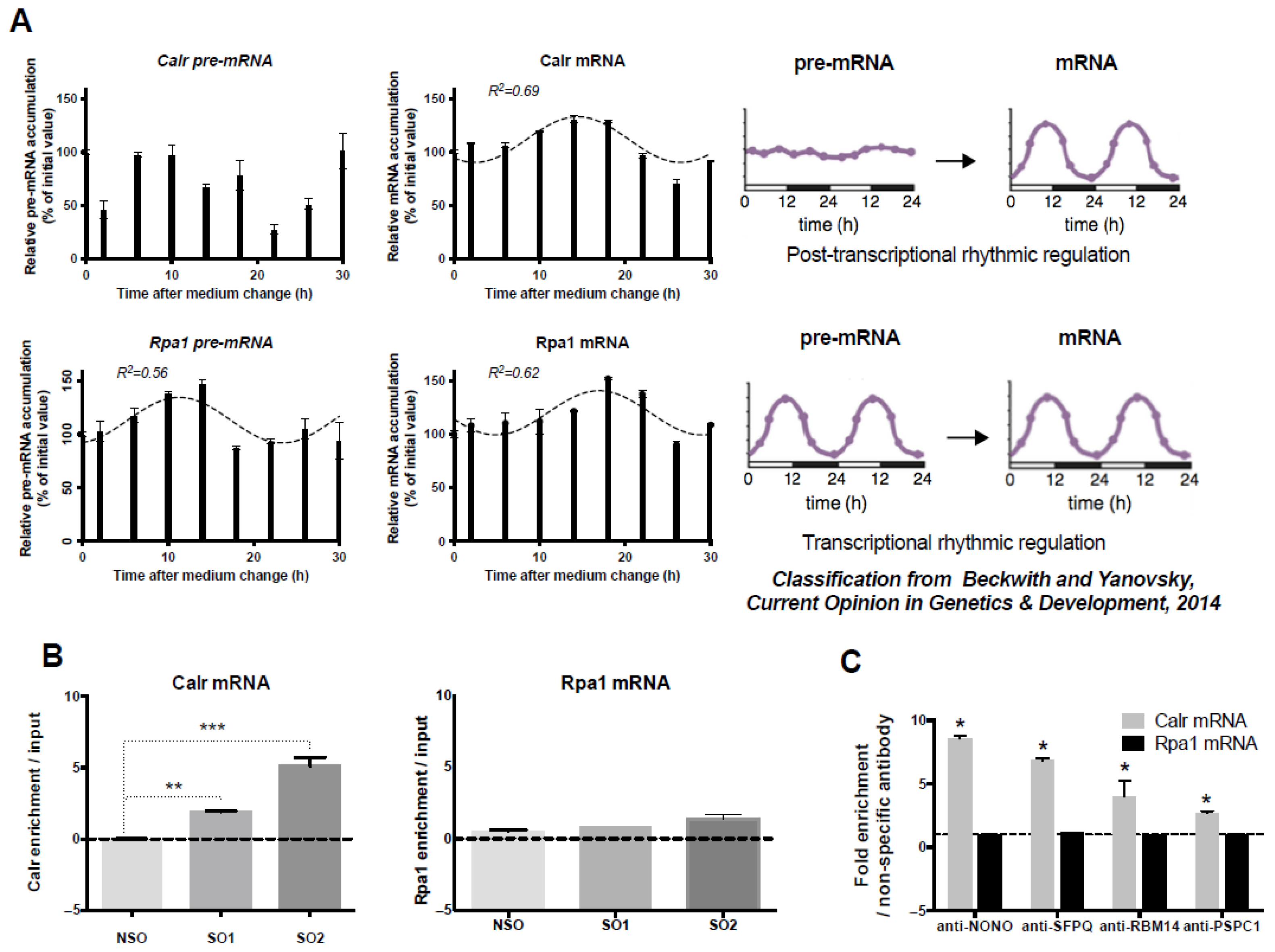
2.1. Post-Transcriptional Calr mRNA Circadian Expression
2.2. Association of Calr mRNA with Paraspeckles in GH4C1 Cells
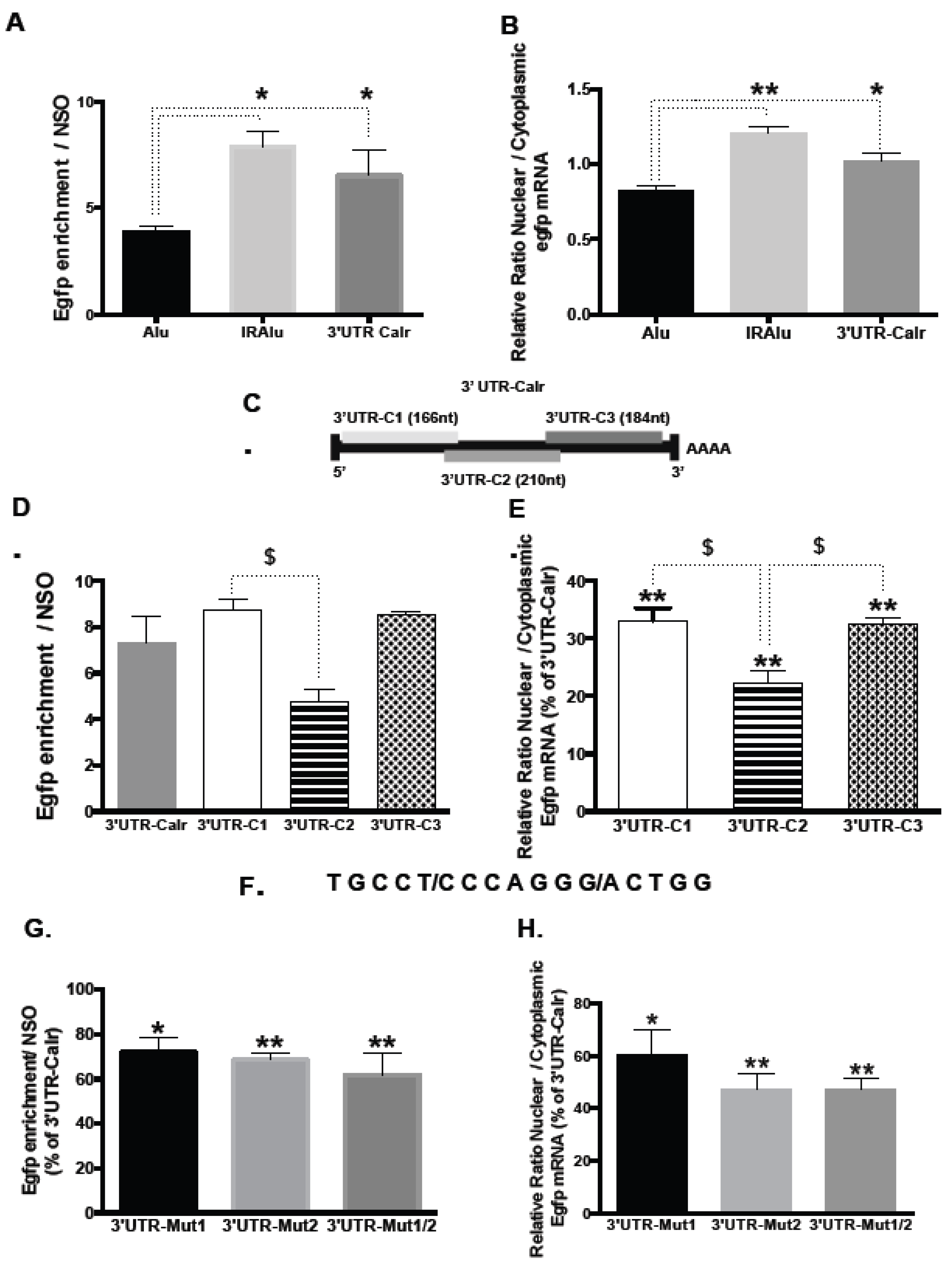
2.3. Involvement of 3′UTR in the Binding of Calr mRNA to Paraspeckles
2.4. Delineation of Sub-Regions in 3′UTR-Calr mRNA Engaged in Paraspeckle Binding
2.5. Involvement of a Tandem Sequence of 15 Nucleotides in 3′UTR-Calr mRNA in the Binding to Paraspeckles
2.6. Mass Spectrometry Analysis of the Proteins Bounded to the 15 Nucleotides Sequence and Its 5′ Flanked Region
2.7. Number of Occurrences of the 15-Nucleotide Sequence Surrounded in 5′ by a C Stretch in the 3′UTR of the Neat1 mRNA Targets
3. Discussion
4. Materials and Methods
4.1. Cell Line Culture and Preparation of Stably-Transfected Cell Lines
4.2. Plasmid Constructs
4.3. Mutagenesis
4.4. RNA Expression Analysis
4.5. RNA Immunoprecipitation (RIP) Experiments
4.6. Neat1 RNA Pull-Down
4.7. RNA-Protein Pull-Down
4.8. MS Analysis
4.9. Bioinformatics
4.9.1. Mass Spectrometry Data Analysis
4.9.2. Nucleotide Sequence Analysis
- Consensus motif
- 3′UTR sequences
- Consensus matrix
4.10. RNA-FISH
4.11. Cosinor and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| UTR | Untranslated region |
| IRAlu | inverted repeats of Alu sequences |
| Neat1 | nuclear-enriched abundant transcript one |
| DBHS | Drosophila Melanogaster Behavior Human Splicing |
| SO | specific oligonucleotides |
| NSO | Non-specific oligonucleotides |
| SINE | short interspersed nuclear elements |
| SIRLOIN | SINE-derived nuclear RNA LOcalizatIoN |
| RIP | RNA--immunoprecipitation |
| MEME | Multiple Expression motifs for Motif Elicitation |
| Amadeus | A Motif Algorithm for Detecting Enrichment in mUltiple Species |
| IntaRNA | RNA-RNA interaction prediction algorithms |
| HCD | high-energy collision dissociation |
References
- Fox, A.H.; Lam, Y.W.; Leung, A.K.; Lyon, C.E.; Andersen, J.; Mann, M.; Lamond, A.I. Paraspeckles: A Novel Nuclear Domain. Curr. Biol. 2002, 12, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Bond, C.S.; Fox, A.H. Paraspeckles: Nuclear Bodies Built on Long Noncoding RNA. J. Cell Biol. 2009, 186, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Carmichael, G.G. Altered Nuclear Retention of mRNAs Containing Inverted Repeats in Human Embryonic Stem Cells: Functional Role of a Nuclear Noncoding RNA. Mol. Cell 2009, 35, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Guru, S.C.; Agarwal, S.K.; Manickam, P.; Olufemi, S.-E.; Crabtree, J.S.; Weisemann, J.M.; Kester, M.B.; Kim, Y.S.; Wang, Y.; Emmert-Buck, M.R.; et al. A Transcript Map for the 2.8-Mb Region Containing the Multiple Endocrine Neoplasia Type 1 Locus. Genome Res. 1997, 7, 725–735. [Google Scholar] [CrossRef]
- Sasaki, Y.T.; Hirose, T. How to Build a Paraspeckle. Genome Biol. 2009, 10, 227. [Google Scholar] [CrossRef]
- Naganuma, T.; Hirose, T. Paraspeckle Formation during the Biogenesis of Long Non-Coding RNAs. RNA Biol. 2013, 10, 456–461. [Google Scholar] [CrossRef]
- Nakagawa, S.; Hirose, T. Paraspeckle Nuclear Bodies—Useful Uselessness? Cell. Mol. Life Sci. 2012, 69, 3027–3036. [Google Scholar] [CrossRef]
- Prasanth, K.V.; Prasanth, S.G.; Xuan, Z.; Hearn, S.; Freier, S.M.; Bennett, C.F.; Zhang, M.Q.; Spector, D.L. Regulating Gene Expression through RNA Nuclear Retention. Cell 2005, 123, 249–263. [Google Scholar] [CrossRef]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.F.; Goshima, N.; Hirose, T. Alternative 3′-End Processing of Long Noncoding RNA Initiates Construction of Nuclear Paraspeckles: LncRNA Processing for Nuclear Body Architecture. EMBO J. 2012, 31, 4020–4034. [Google Scholar] [CrossRef]
- Torres, M.; Becquet, D.; Guillen, S.; Boyer, B.; Moreno, M.; Blanchard, M.-P.; Franc, J.-L.; François-Bellan, A.-M. RNA Pull-down Procedure to Identify RNA Targets of a Long Non-Coding RNA. JoVE 2018, 134, e57379. [Google Scholar] [CrossRef]
- Torres, M.; Becquet, D.; Blanchard, M.-P.; Guillen, S.; Boyer, B.; Moreno, M.; Franc, J.-L.; François-Bellan, A.-M. Circadian RNA Expression Elicited by 3′-UTR IRAlu-Paraspeckle Associated Elements. eLife 2016, 5, e14837. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Becquet, D.; Blanchard, M.-P.; Guillen, S.; Moreno, M.; Franc, J.-L.; François-Bellan, A.-M.; François-Bellan, A.-M. Paraspeckles as Rhythmic Nuclear mRNA Anchorages Responsible for Circadian Gene Expression. Nucleus 2017, 8, 249–254. [Google Scholar] [CrossRef]
- Chen, L.-L.; DeCerbo, J.N.; Carmichael, G.G. Alu Element-Mediated Gene Silencing. EMBO J. 2008, 27, 1694–1705. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Carmichael, G.G. Gene Regulation by SINES and Inosines: Biological Consequences of A-to-I Editing of Alu Element Inverted Repeats. Cell Cycle 2008, 7, 3294–3301. [Google Scholar] [CrossRef]
- Menet, J.S.; Rodriguez, J.; Abruzzi, K.C.; Rosbash, M. Nascent-Seq Reveals Novel Features of Mouse Circadian Transcriptional Regulation. Elife 2012, 1, e00011. [Google Scholar] [CrossRef]
- Beckwith, E.J.; Yanovsky, M.J. Circadian Regulation of Gene Expression: At the Crossroads of Transcriptional and Post-Transcriptional Regulatory Networks. Curr. Opin. Genet. Dev. 2014, 27, 35–42. [Google Scholar] [CrossRef]
- Guillaumond, F.; Becquet, D.; Blanchard, M.-P.; Attia, J.; Moreno, M.; Bosler, O.; François-Bellan, A.-M. Nocturnal Expression of Phosphorylated-ERK1/2 in Gastrin-Releasing Peptide Neurons of the Rat Suprachiasmatic Nucleus. J. Neurochem. 2007, 101, 1224–1235. [Google Scholar] [CrossRef]
- Yamazaki, T.; Hirose, T. The Building Process of the Functional Paraspeckle with Long Non-Coding RNAs. Front. Biosci. (Elite Ed.) 2015, 7, 1–41. [Google Scholar] [CrossRef][Green Version]
- Simko, E.A.J.; Liu, H.; Zhang, T.; Velasquez, A.; Teli, S.; Haeusler, A.R.; Wang, J. G-Quadruplexes Offer a Conserved Structural Motif for NONO Recruitment to NEAT1 Architectural lncRNA. Nucleic Acids Res. 2020, 48, gkaa475. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Linhart, C.; Halperin, Y.; Shamir, R. Transcription Factor and microRNA Motif Discovery: The Amadeus Platform and a Compendium of Metazoan Target Sets. Genome Res. 2008, 18, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Lubelsky, Y.; Ulitsky, I. Sequences Enriched in Alu Repeats Drive Nuclear Localization of Long RNAs in Human Cells. Nature 2018, 555, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and Customizable Prediction of RNA–RNA Interactions. Nucleic Acids Res. 2017, 45, W435–W439. [Google Scholar] [CrossRef]
- Maticzka, D.; Lange, S.J.; Costa, F.; Backofen, R. GraphProt: Modeling Binding Preferences of RNA-Binding Proteins. Genome Biol. 2014, 15, R17. [Google Scholar] [CrossRef]
- Choi, H.S.; Hwang, C.K.; Song, K.Y.; Law, P.-Y.; Wei, L.-N.; Loh, H.H. Poly(C)-Binding Proteins as Transcriptional Regulators of Gene Expression. Biochem. Biophys. Res. Commun. 2009, 380, 431–436. [Google Scholar] [CrossRef]
- Paziewska, A.; Wyrwicz, L.S.; Bujnicki, J.M.; Bomsztyk, K.; Ostrowski, J. Cooperative Binding of the hnRNP K Three KH Domains to mRNA Targets. FEBS Lett. 2004, 577, 134–140. [Google Scholar] [CrossRef]
- Nakamoto, M.Y.; Lammer, N.C.; Batey, R.T.; Wuttke, D.S. hnRNPK Recognition of the B Motif of Xist and Other Biological RNAs. Nucleic Acids Res. 2020, 1, 16. [Google Scholar] [CrossRef]
- Yao, J.; Tu, Y.; Shen, C.; Zhou, Q.; Xiao, H.; Jia, D.; Sun, Q. Nuclear Import Receptors and hnRNPK Mediates Nuclear Import and Stress Granule Localization of SIRLOIN. Cell. Mol. Life Sci. 2021, 78, 7617–7633. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Kavčič, P.; Rojc, B.; Dolenc-Grošelj, L.; Claustrat, B.; Fujs, K.; Poljak, M. The Impact of Sleep Deprivation and Nighttime Light Exposure on Clock Gene Expression in Humans. Croat. Med. J. 2011, 52, 594–603. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.-K.J.; Esterly, N.; Klaassen, C.D.; Wan, Y.-J.Y. Gender Disparity of Hepatic Lipid Homoeostasis Regulated by the Circadian Clock. J. Biochem. 2009, 145, 609–623. [Google Scholar] [CrossRef]

| Number of 3′UTR | Number of Sequences | Number of Genes | Number of Sequences/Genes | |
|---|---|---|---|---|
| Neat1 RNA targets | 3347 | 2190 | 1330 | 1.65 |
| Background | 19,290 | 10,575 | 6386 | 1.65 |
| (total RNAs) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacq, A.; Becquet, D.; Boyer, B.; Guillen, S.; Bello-Goutierrez, M.-M.; Blanchard, M.-P.; Villard, C.; Belghazi, M.; Torres, M.; Franc, J.-L.; et al. Nuclear Retention of mRNAs Through Paraspeckle Protein Binding to a Sequence Determinant in 3′UTR. Int. J. Mol. Sci. 2025, 26, 6488. https://doi.org/10.3390/ijms26136488
Jacq A, Becquet D, Boyer B, Guillen S, Bello-Goutierrez M-M, Blanchard M-P, Villard C, Belghazi M, Torres M, Franc J-L, et al. Nuclear Retention of mRNAs Through Paraspeckle Protein Binding to a Sequence Determinant in 3′UTR. International Journal of Molecular Sciences. 2025; 26(13):6488. https://doi.org/10.3390/ijms26136488
Chicago/Turabian StyleJacq, Audrey, Denis Becquet, Bénédicte Boyer, Séverine Guillen, Maria-Montserrat Bello-Goutierrez, Marie-Pierre Blanchard, Claude Villard, Maya Belghazi, Manon Torres, Jean-Louis Franc, and et al. 2025. "Nuclear Retention of mRNAs Through Paraspeckle Protein Binding to a Sequence Determinant in 3′UTR" International Journal of Molecular Sciences 26, no. 13: 6488. https://doi.org/10.3390/ijms26136488
APA StyleJacq, A., Becquet, D., Boyer, B., Guillen, S., Bello-Goutierrez, M.-M., Blanchard, M.-P., Villard, C., Belghazi, M., Torres, M., Franc, J.-L., & François-Bellan, A.-M. (2025). Nuclear Retention of mRNAs Through Paraspeckle Protein Binding to a Sequence Determinant in 3′UTR. International Journal of Molecular Sciences, 26(13), 6488. https://doi.org/10.3390/ijms26136488






