Glutaminase Reprogramming in Hepatocellular Carcinoma: Implications for Diagnosis, Prognosis, and Potential as a Novel Therapeutic Target
Abstract
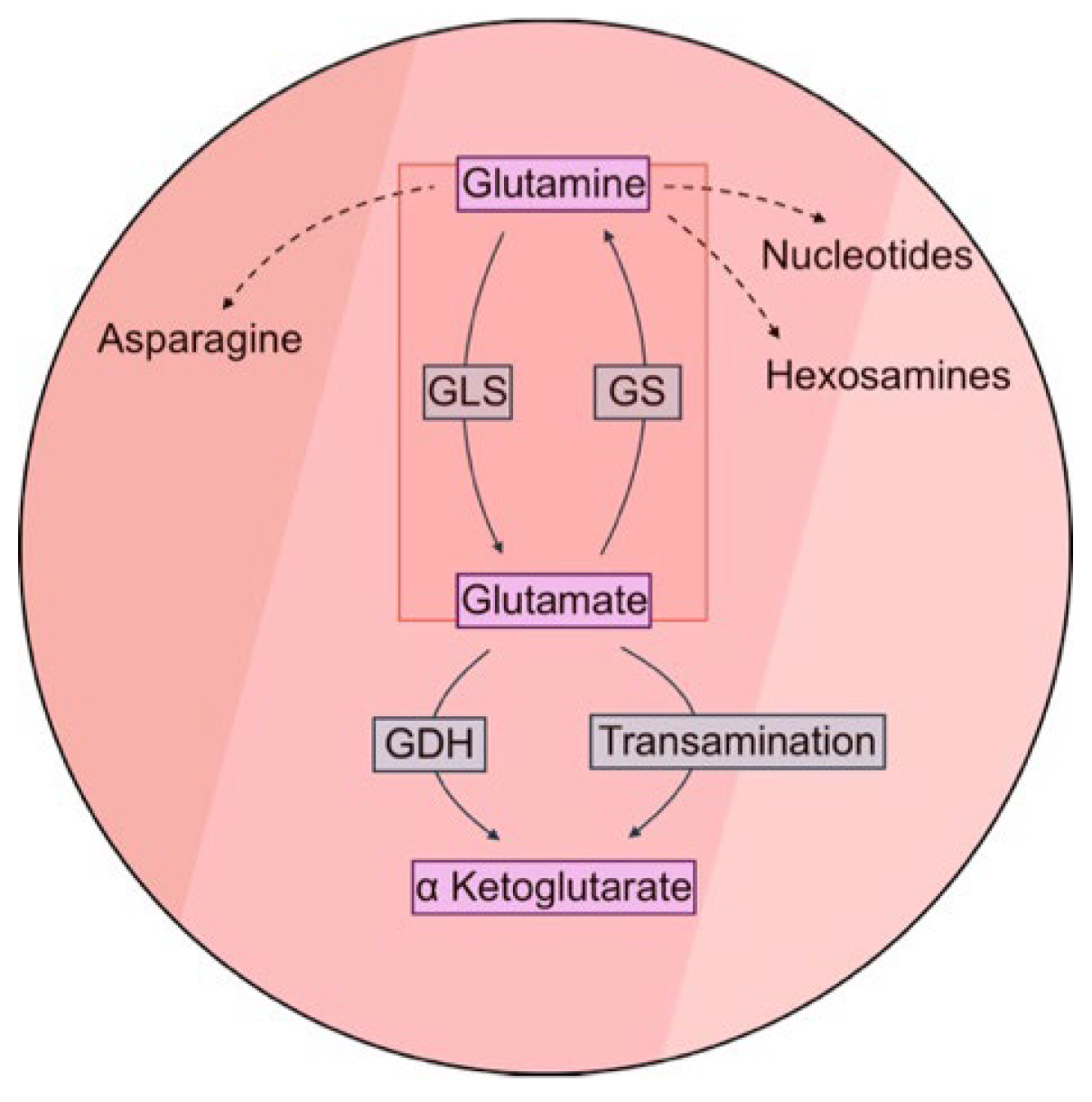
1. Introduction
2. Results
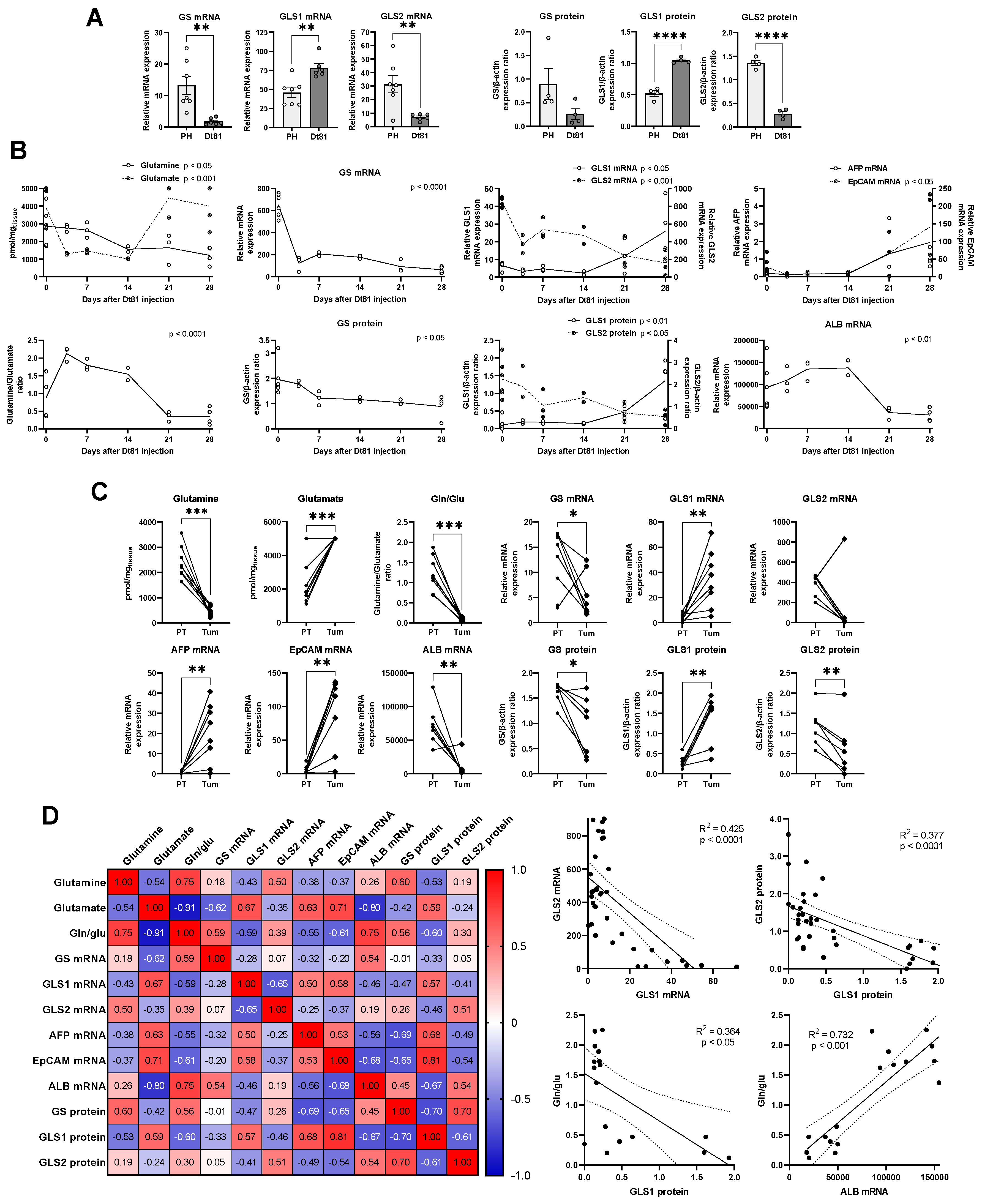
2.1. Rewiring of Hepatic Glutamine Metabolism Correlates with Development and Progression of Murine Hepatocellular Carcinoma
2.2. Discriminative Performance of Glutamine Metabolism Reprogramming in Human HCC Compared to Cirrhotic and Normal Livers
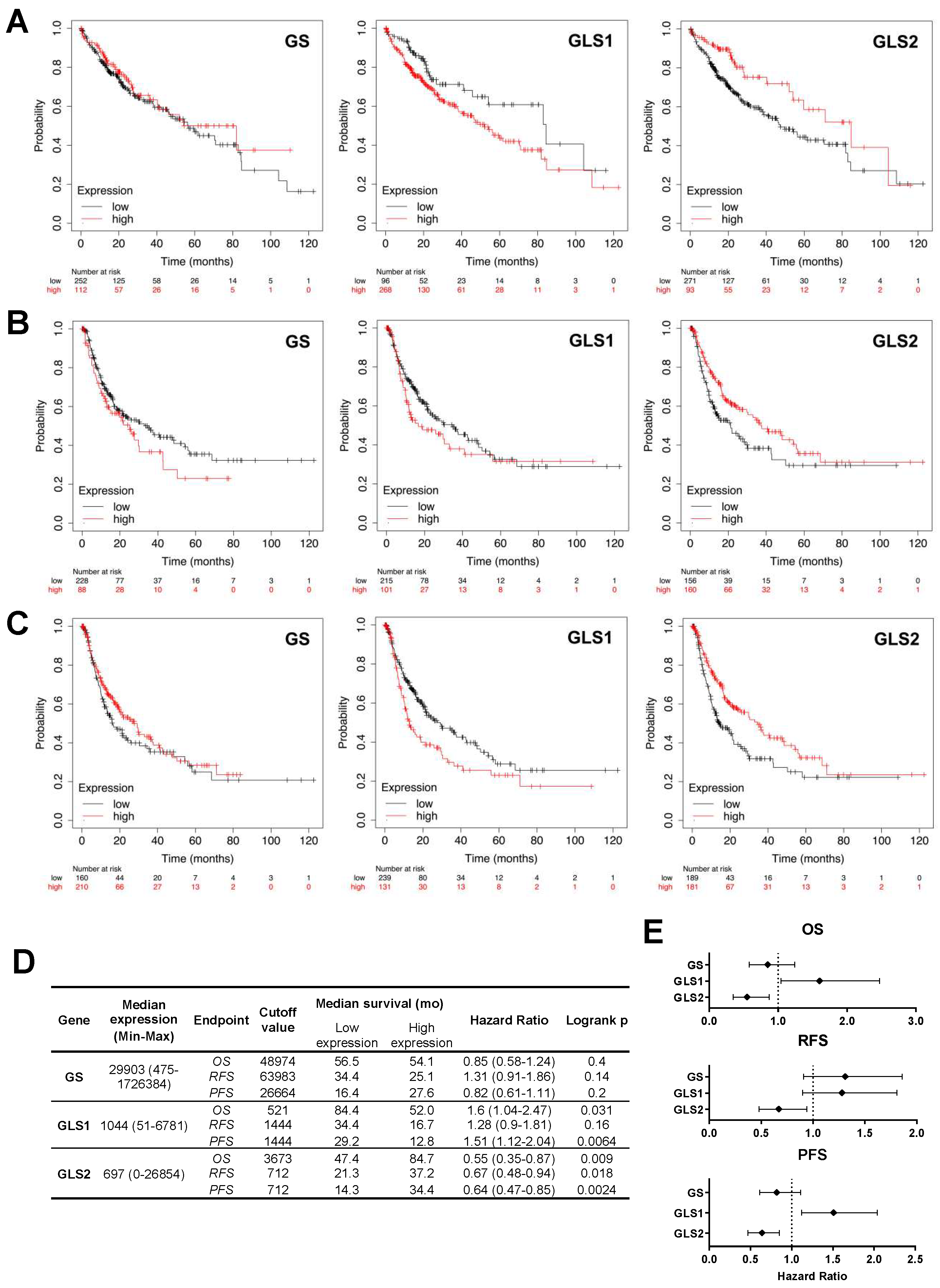
2.3. Alterations in the Expression of Tumoral Glutaminase and Glutamine Synthetase Expression Bear Significant Prognostic Implications in Patients with Hepatocellular Carcinoma
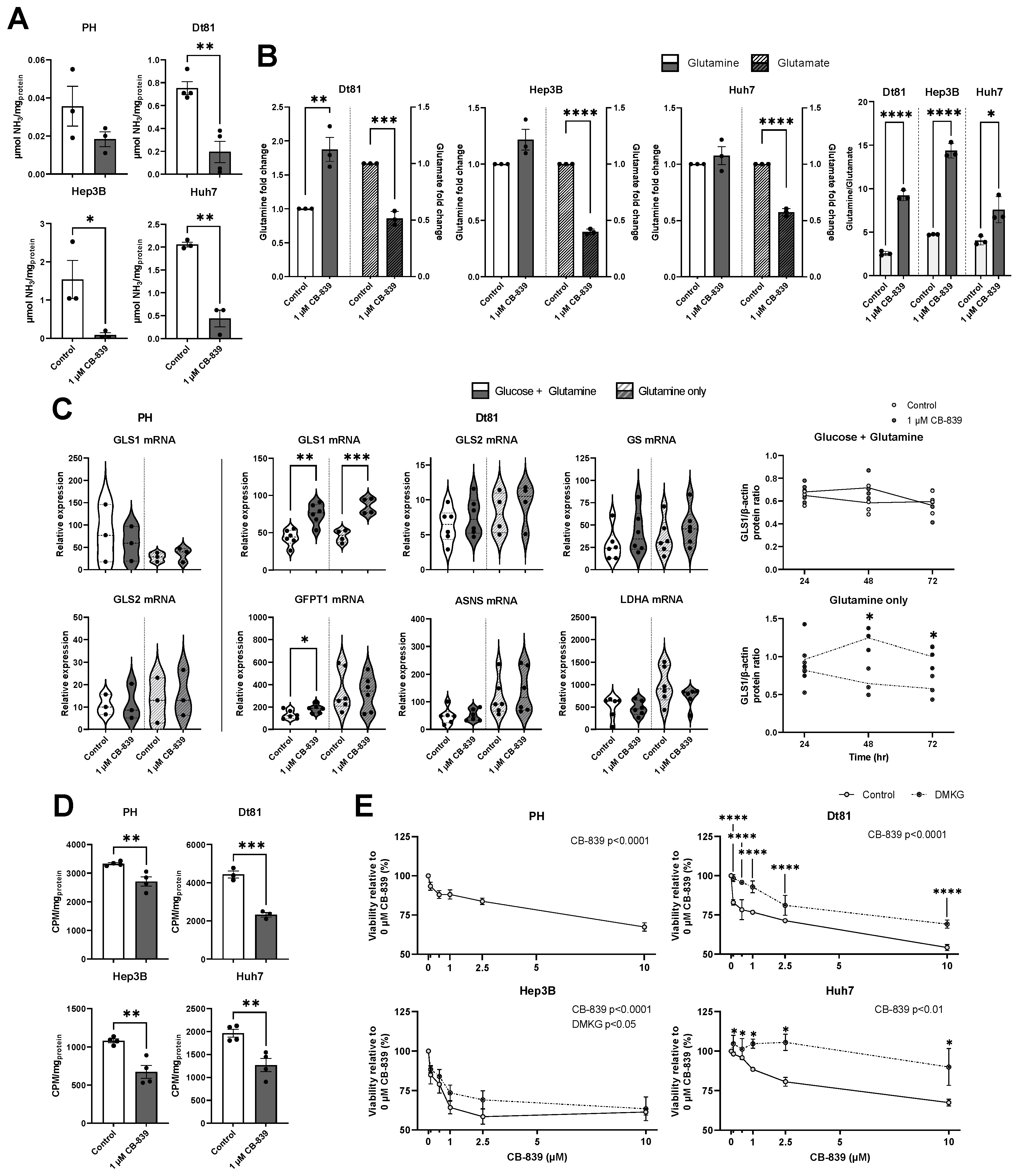
2.4. Pharmacological Targeting of Kidney Glutaminase Inhibits Glutaminolysis and Downstream Metabolism in Hepatocellular Carcinoma Cells
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Hepatocyte Isolation, and Culture Reagents
4.2. Animals and Experimental Hepatocarcinogenesis
4.3. Biobank Samples and Patient Cohort
4.4. Gene Expression Analysis
4.5. Western Blotting
4.6. Glutamine and Glutamate Quantification
4.7. Assessment of Cell Viability
4.8. H-Glutamine Uptake Assay
4.9. Ammonia Measurement
4.10. Survival Analysis of HCC Patients
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFP | Alpha-fetoprotein |
| ALB | Albumin |
| ANOVA | Analysis of variance |
| ASNS | Asparagine synthetase |
| AUROC | Area under receiver operating characteristic curve |
| CHUM | Centre hospitalier de l’Université de Montréal |
| DMEM | Dulbecco’s modified essential medium |
| DMKG | Dimethyl-α-ketoglutarate |
| Dt81 | Dt81Hepa1-6 |
| EpCAM | Epithelial cell adhesion molecule |
| GFPT1 | Glucosamine-fructose-6-phosphate aminotransferase |
| GLS | Glutaminase |
| GLS1 | Kidney glutaminase |
| GLS2 | Liver glutaminase |
| GS | Glutamine synthetase |
| HMBS | Hydroxymethylbilane synthase |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 |
| HRP | Horseradish peroxidase |
| H2AFZ | Histone H2A.Z variant |
| LC-MS | Liquid chromatography/mass spectrometry |
| LDHA | Lactate dehydrogenase |
| MTT | (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) tetrazolium salt |
| OS | Overall survival |
| PFS | Progression-free survival |
| PH | Primary hepatocyte |
| PPIA | Peptidylprolyl isomerase A |
| RFS | Relapse-free survival |
| ROC | Receiver operating characteristic |
| S9 | 40S ribosomal protein S9 |
| TCGA | The Cancer Genome Atlas |
| TGCA-LIHC | The Cancer Genome Atlas—Liver Hepatocellular Carcinoma |
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Kulik, L.; El-Serag, H.B. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019, 156, 477–491.e471. [Google Scholar] [CrossRef] [PubMed]
- Chidambaranathan-Reghupaty, S.; Fisher, P.B.; Sarkar, D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv. Cancer Res. 2021, 149, 1–61. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Garrido, A.; Djouder, N. Cirrhosis: A Questioned Risk Factor for Hepatocellular Carcinoma. Trends Cancer 2021, 7, 29–36. [Google Scholar] [CrossRef]
- Toh, M.R.; Wong, E.Y.T.; Wong, S.H.; Ng, A.W.T.; Loo, L.H.; Chow, P.K.; Ngeow, J. Global Epidemiology and Genetics of Hepatocellular Carcinoma. Gastroenterology 2023, 164, 766–782. [Google Scholar] [CrossRef]
- Tambay, V.; Raymond, V.A.; Goossens, C.; Rousseau, L.; Turcotte, S.; Bilodeau, M. Metabolomics-Guided Identification of a Distinctive Hepatocellular Carcinoma Signature. Cancers 2023, 15, 3232. [Google Scholar] [CrossRef]
- He, M.J.; Pu, W.; Wang, X.; Zhong, X.; Zhao, D.; Zeng, Z.; Cai, W.; Liu, J.; Huang, J.; Tang, D.; et al. Spatial metabolomics on liver cirrhosis to hepatocellular carcinoma progression. Cancer Cell Int. 2022, 22, 366. [Google Scholar] [CrossRef]
- Hu, C.; Wang, T.; Zhuang, X.; Sun, Q.; Wang, X.; Lin, H.; Feng, M.; Zhang, J.; Cao, Q.; Jiang, Y. Metabolic analysis of early nonalcoholic fatty liver disease in humans using liquid chromatography-mass spectrometry. J. Transl. Med. 2021, 19, 152. [Google Scholar] [CrossRef]
- Jin, R.; Banton, S.; Tran, V.T.; Konomi, J.V.; Li, S.; Jones, D.P.; Vos, M.B. Amino Acid Metabolism is Altered in Adolescents with Nonalcoholic Fatty Liver Disease-An Untargeted, High Resolution Metabolomics Study. J. Pediatr. 2016, 172, 14–19.e15. [Google Scholar] [CrossRef]
- Beyoglu, D.; Idle, J.R. Metabolomic and Lipidomic Biomarkers for Premalignant Liver Disease Diagnosis and Therapy. Metabolites 2020, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Suciu, A.M.; Crisan, D.A.; Procopet, B.D.; Radu, C.I.; Socaciu, C.; Tantau, M.V.; Stefanescu, H.O.; Grigorescu, M. What’s in Metabolomics for Alcoholic Liver Disease? J. Gastrointest. Liver Dis. 2018, 27, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef]
- Martinez-Reyes, I.; Chandel, N.S. Cancer metabolism: Looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine reliance in cell metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. [Google Scholar] [CrossRef]
- Aledo, J.C.; Gomez-Fabre, P.M.; Olalla, L.; Marquez, J. Identification of two human glutaminase loci and tissue-specific expression of the two related genes. Mamm. Genome 2000, 11, 1107–1110. [Google Scholar] [CrossRef]
- Gebhardt, R.; Matz-Soja, M. Liver zonation: Novel aspects of its regulation and its impact on homeostasis. World J. Gastroenterol. 2014, 20, 8491–8504. [Google Scholar] [CrossRef]
- Häussinger, D.; Lamers, W.H.; Moorman, A.F. Hepatocyte heterogeneity in the metabolism of amino acids and ammonia. Enzyme 1992, 46, 72–93. [Google Scholar] [CrossRef]
- Goel, C.; Monga, S.P.; Nejak-Bowen, K. Role and Regulation of Wnt/beta-Catenin in Hepatic Perivenous Zonation and Physiological Homeostasis. Am. J. Pathol. 2022, 192, 4–17. [Google Scholar] [CrossRef]
- Yang, L.; Venneti, S.; Nagrath, D. Glutaminolysis: A Hallmark of Cancer Metabolism. Annu. Rev. Biomed. Eng. 2017, 19, 163–194. [Google Scholar] [CrossRef]
- Groenewald, W.; Lund, A.H.; Gay, D.M. The Role of WNT Pathway Mutations in Cancer Development and an Overview of Therapeutic Options. Cells 2023, 12, 990. [Google Scholar] [CrossRef]
- Nguyen, T.B.; Roncalli, M.; Di Tommaso, L.; Kakar, S. Combined use of heat-shock protein 70 and glutamine synthetase is useful in the distinction of typical hepatocellular adenoma from atypical hepatocellular neoplasms and well-differentiated hepatocellular carcinoma. Mod. Pathol. 2016, 29, 283–292. [Google Scholar] [CrossRef]
- Di Tommaso, L.; Franchi, G.; Park, Y.N.; Fiamengo, B.; Destro, A.; Morenghi, E.; Montorsi, M.; Torzilli, G.; Tommasini, M.; Terracciano, L.; et al. Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology 2007, 45, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Curthoys, N.P.; Watford, M. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 1995, 15, 133–159. [Google Scholar] [CrossRef] [PubMed]
- Botman, D.; Tigchelaar, W.; Van Noorden, C.J. Determination of phosphate-activated glutaminase activity and its kinetics in mouse tissues using metabolic mapping (quantitative enzyme histochemistry). J. Histochem. Cytochem. 2014, 62, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Tanaka, T.; Poyurovsky, M.V.; Nagano, H.; Mayama, T.; Ohkubo, S.; Lokshin, M.; Hosokawa, H.; Nakayama, T.; Suzuki, Y.; et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl. Acad. Sci. USA 2010, 107, 7461–7466. [Google Scholar] [CrossRef]
- Suzuki, S.; Venkatesh, D.; Kanda, H.; Nakayama, A.; Hosokawa, H.; Lee, E.; Miki, T.; Stockwell, B.R.; Yokote, K.; Tanaka, T.; et al. GLS2 Is a Tumor Suppressor and a Regulator of Ferroptosis in Hepatocellular Carcinoma. Cancer Res. 2022, 82, 3209–3222. [Google Scholar] [CrossRef]
- Xu, L.; Zhou, D.; Li, F.; Ji, L. Glutaminase 2 functions as a tumor suppressor gene in gastric cancer. Transl. Cancer Res. 2020, 9, 4906–4913. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Zhao, Y.; Yue, X.; Zhu, Y.; Wang, X.; Wu, H.; Blanco, F.; Li, S.; Bhanot, G.; et al. Glutaminase 2 is a novel negative regulator of small GTPase Rac1 and mediates p53 function in suppressing metastasis. eLife 2016, 5, e10727. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341e1323. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Lin, M.; Zhu, W.; Liang, Y.; Hong, X.; Zhao, Y.; Young, K.H.; Hu, W.; Feng, Z. Glutaminase 2 negatively regulates the PI3K/AKT signaling and shows tumor suppression activity in human hepatocellular carcinoma. Oncotarget 2014, 5, 2635–2647. [Google Scholar] [CrossRef] [PubMed]
- Tambay, V.; Raymond, V.A.; Bilodeau, M. MYC Rules: Leading Glutamine Metabolism toward a Distinct Cancer Cell Phenotype. Cancers 2021, 13, 4484. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, F.; Fan, N.; Zhou, C.; Li, D.; Macvicar, T.; Dong, Q.; Bruns, C.J.; Zhao, Y. Targeting Glutaminolysis: New Perspectives to Understand Cancer Development and Novel Strategies for Potential Target Therapies. Front. Oncol. 2020, 10, 589508. [Google Scholar] [CrossRef]
- Xi, J.; Sun, Y.; Zhang, M.; Fa, Z.; Wan, Y.; Min, Z.; Xu, H.; Xu, C.; Tang, J. GLS1 promotes proliferation in hepatocellular carcinoma cells via AKT/GSK3beta/CyclinD1 pathway. Exp. Cell Res. 2019, 381, 1–9. [Google Scholar] [CrossRef]
- Li, B.; Cao, Y.; Meng, G.; Qian, L.; Xu, T.; Yan, C.; Luo, O.; Wang, S.; Wei, J.; Ding, Y.; et al. Targeting glutaminase 1 attenuates stemness properties in hepatocellular carcinoma by increasing reactive oxygen species and suppressing Wnt/beta-catenin pathway. EBioMedicine 2019, 39, 239–254. [Google Scholar] [CrossRef]
- Cassim, S.; Raymond, V.A.; Lapierre, P.; Bilodeau, M. From in vivo to in vitro: Major metabolic alterations take place in hepatocytes during and following isolation. PLoS ONE 2017, 12, e0190366. [Google Scholar] [CrossRef]
- Jin, H.; Wang, S.; Zaal, E.A.; Wang, C.; Wu, H.; Bosma, A.; Jochems, F.; Isima, N.; Jin, G.; Lieftink, C.; et al. A powerful drug combination strategy targeting glutamine addiction for the treatment of human liver cancer. eLife 2020, 9, e56749. [Google Scholar] [CrossRef]
- Lacoste, B.; Raymond, V.A.; Cassim, S.; Lapierre, P.; Bilodeau, M. Highly tumorigenic hepatocellular carcinoma cell line with cancer stem cell-like properties. PLoS ONE 2017, 12, e0171215. [Google Scholar] [CrossRef]
- Musallam, L.; Ethier, C.; Haddad, P.S.; Bilodeau, M. Role of EGF receptor tyrosine kinase activity in antiapoptotic effect of EGF on mouse hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 280, G1360–G1369. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Menyhárt, O.; Nagy, Á.; Győrffy, B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R. Soc. Open Sci. 2018, 5, 181006. [Google Scholar] [CrossRef]

| HCC (n = 32) | Cirrhosis (n = 20) | Control (n = 20) | ||
|---|---|---|---|---|
| Demographics | Male | 26 (81.3%) | 18 (90.0%) | 14 (70.0%) |
| Median age (mean), years | 68 (63.8) | 68 (63.3) | 60 (60.5) | |
| Cirrhosis | 25 (78.1%) | 20 (100.0%) | 0 (0.0%) | |
| Etiology | Alcohol | 1 (3.1%) | 1 (5.0%) | - |
| HBV | 5 (15.6%) | 4 (20.0%) | - | |
| HCV | 4 (12.5%) | 4 (20.0%) | - | |
| MASLD | 6 (18.8%) | 6 (30.0%) | - | |
| Other/Unknown | 16 (50%) | 5 (25.0%) | - | |
| Tumor biology | Solitary, n (%) | 12 (71.9%) | - | - |
| Greatest tumor dimension, mean (cm) | 7.1 | - | - | |
| Vascular invasion | Microvascular | 8 (25.0%) | - | - |
| Macrovascular | 8 (25.0%) | - | - | |
| None | 14 (43.8%) | - | - | |
| Unknown | 4 (12.5%) | - | - | |
| Grade | I (n, %) | 3 (9.4%) | - | - |
| II (n, %) | 17 (53.1%) | - | - | |
| III (n, %) | 6 (18.8%) | - | - | |
| IV (n, %) | 2 (6.3%) | - | - | |
| Unknown (n, %) | 4 (12.5%) | - | - | |
| TNM staging | T1 | 12 (37.5%) | - | - |
| T2 | 12 (37.5%) | - | - | |
| T3 | 5 (15.6%) | - | - | |
| T4 | 1 (3.1%) | - | - | |
| TX | 2 (6.3%) | - | - | |
| N0 | 4 (12.5%) | - | - | |
| N1 | 1 (3.1%) | - | - | |
| NX | 27 (84.4%) | - | - | |
| MX | 32 (100%) | - | - |
| NCT # | Title | Status | Conditions | Intervention | Sponsor (Collaborators) | Phase |
|---|---|---|---|---|---|---|
| 04250545 | Testing of the Anti Cancer Drugs CB-839 HCl (Telaglenastat) and MLN0128 (Sapanisertib) in Advanced Stage Non-small Cell Lung Cancer | Active, not recruiting | Leptomeningeal neoplasm, non-small cell lung cancer, metastatic malignant brain neoplasm | Telaglenastat HCl, sapanisertib | NCI | I/Ib |
| 03965845 | A Study of Telaglenastat (CB-839) in Combination With Palbociclib in Patients With Solid Tumors | Completed | Non-small cell lung cancer, colorectal carcinoma | Telaglenastat, palbociclib | Calithera Biosciences, Inc. | Ib/II |
| 04824937 | Telaglenastat + Talazoparib In Prostate Cancer | Unknown (12 September 2025) | Metastatic prostate cancer | Telaglenastat, talazoparib | Massachusetts General Hospital (Calithera Biosciences, Inc.; Pfizer; Prostate Cancer Foundation) | II |
| 03875313 | Study of CB-839 (Telaglenastat) in Combination With Talazoparib in Patients With Solid Tumors | Terminated | Renal cell carcinoma, triple-negative breast cancer, colorectal cancer | Telaglenastat, talazoparib | Calithera Biosciences, Inc. | I/II |
| 03528642 | Telaglenastat With Radiation Therapy and Temozolomide in Treating Patients With IDH-Mutated Diffuse Astrocytoma or Anaplastic Astrocytoma | Active, not recruiting | Astrocytoma | Telaglenastat, temozolomide, radiation therapy | NCI | I |
| 03872427 | Testing Whether Cancers With Specific Mutations Respond Better to Glutaminase Inhibitor, Telaglenastat Hydrochloride, Anti-Cancer Treatment, BeGIN Study | Active, not recruiting | Advanced/unresectable/metastatic malignant solid neoplasm, NF1-mutant malignant peripheral nerve sheath tumor | Telaglenastat HCl | NCI | II |
| 03163667 | CB-839 With Everolimus vs. Placebo With Everolimus in Participants With Renal Cell Carcinoma (RCC) (ENTRATA) | Completed | Clear-cell renal cell carcinoma | Telaglenastat everolimus | Calithera Biosciences, Inc. | II |
| 04265534 | KEAPSAKE: A Study of Telaglenastat (CB-839) With Standard-of-Care Chemoimmunotherapy in 1L KEAP1/NRF2-Mutated, Nonsquamous NSCLC (KEAPSAKE) | Terminated | Non-small cell lung cancer | Telaglenastat, carboplatin, pemetrexed, pembrolizumab | Calithera Biosciences, Inc. | II |
| 03831932 | Telaglenastat Hydrochloride and Osimertinib in Treating Patients With EGFR-Mutated Stage IV Non-small Cell Lung Cancer | Active, not recruiting | Advanced/metastatic non-small cell lung cancer | Telaglenastat HCl, osimertinib | NCI | I/II |
| 02771626 | Study CB-839 in Combination With Nivolumab in Patients With Melanoma, Clear Cell Renal Cell Carcinoma (ccRCC) and Non-Small Cell Lung Cancer (NSCLC) | Terminated | Clear-cell renal cell carcinoma, melanoma, non-small cell lung cancer | Telaglenastat, nivolumab | Calithera Biosciences, Inc. | I/II |
| 03428217 | CANTATA: CB-839 With Cabozantinib vs. Cabozantinib With Placebo in Patients With Metastatic Renal Cell Carcinoma (CANTATA) | Completed | Advanced/metastatic renal cell carcinoma | Telaglenastat, cabozantinib | Calithera Biosciences, Inc. | II |
| 04540965 | Impact of a Histamine H2 Receptor Antagonist (H2RA) on the Pharmacokinetics (PK) of Telaglenastat in Healthy Subjects | Completed | Drug interaction | Telaglenastat, famotidine | Calithera Biosciences, Inc. (Novotech (Australia) Pty Limited) | I |
| 03057600 | Study of CB-839 in Combination w/Paclitaxel in Participants of African Ancestry and Non-African Ancestry With Advanced Triple Negative Breast Cancer (TNBC) | Completed | Triple-negative breast cancer | Telaglenastat, paclitaxel | Calithera Biosciences, Inc. | II |
| 03263429 | Novel PET/CT Imaging Biomarkers of CB-839 in Combination With Panitumumab and Irinotecan in Patients With Metastatic and Refractory RAS Wildtype Colorectal Cancer | Completed | Colorectal cancer | Telaglenastat, panitumumab, irinotecan | Vanderbilt-Ingram Cancer Center (NCI, Calithera Biosciences, Inc.) | I/II |
| 03798678 | CB-839 HCl in Combination With Carfilzomib and Dexamethasone in Treating Patients With Recurrent or Refractory Multiple Myeloma | Active, not recruiting | Recurrent multiple myeloma | Telaglenastat, dexamethasone, carfilzomib | NCI | I |
| 02071888 | Study of the Glutaminase Inhibitor CB-839 in Hematological Tumors | Completed | Non-Hodgkin’s lymphoma | Telaglenastat, dexamethasone, pomalidomide | Calithera Biosciences, Inc. | I |
| 02071927 | Study of the Glutaminase Inhibitor CB-839 in Leukemia | Completed | Acute myeloid leukemia, acute lymphoid leukemia | Telaglenastat, azacitidine | Calithera Biosciences, Inc. | I |
| 02861300 | CB-839 + Capecitabine in Solid Tumors and Fluoropyrimidine Resistant PIK3CA Mutant Colorectal Cancer | Completed | Colorectal cancer | Telaglenastat, capecitabine | David Bajor, Case Comprehensive Cancer Center | I/II |
| 02071862 | Study of Glutaminase Inhibitor CB-839 in Solid Tumors | Completed | Triple-negative breast cancer, non-small cell lung cancer, renal cell carcinoma, mesothelioma, gastrointestinal stromal tumors | Telaglenastat, paclitaxel, everolimus, erlotnib, docetaxel, cabozantinib | Calithera Biosciences, Inc. | I |
| 03047993 | Glutaminase Inhibitor CB-839 and Azacitidine in Treating Patients with Advanced Myelodysplastic Syndrome | Completed | Acute myeloid leukemia, chronic myeloid leukemia | Telaglenastat, azacitidine | M. D. Anderson Cancer Center | I/II |
| 03944902 | CB-839 in Combination with Niraparib in Platinum-Resistant BRCA-Wild-type Ovarian Cancer | Terminated | Ovarian cancer | Telaglenastat, Niraparib | University of Alabama at Birmingham | I |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tambay, V.; Raymond, V.-A.; Turcotte, S.; Bilodeau, M. Glutaminase Reprogramming in Hepatocellular Carcinoma: Implications for Diagnosis, Prognosis, and Potential as a Novel Therapeutic Target. Int. J. Mol. Sci. 2025, 26, 9653. https://doi.org/10.3390/ijms26199653
Tambay V, Raymond V-A, Turcotte S, Bilodeau M. Glutaminase Reprogramming in Hepatocellular Carcinoma: Implications for Diagnosis, Prognosis, and Potential as a Novel Therapeutic Target. International Journal of Molecular Sciences. 2025; 26(19):9653. https://doi.org/10.3390/ijms26199653
Chicago/Turabian StyleTambay, Vincent, Valérie-Ann Raymond, Simon Turcotte, and Marc Bilodeau. 2025. "Glutaminase Reprogramming in Hepatocellular Carcinoma: Implications for Diagnosis, Prognosis, and Potential as a Novel Therapeutic Target" International Journal of Molecular Sciences 26, no. 19: 9653. https://doi.org/10.3390/ijms26199653
APA StyleTambay, V., Raymond, V.-A., Turcotte, S., & Bilodeau, M. (2025). Glutaminase Reprogramming in Hepatocellular Carcinoma: Implications for Diagnosis, Prognosis, and Potential as a Novel Therapeutic Target. International Journal of Molecular Sciences, 26(19), 9653. https://doi.org/10.3390/ijms26199653





