Structures, Interactions, and Antimicrobial Activity of the Shortest Thanatin Peptide from Anasa tristis
Abstract
1. Introduction
2. Results
2.1. Antibacterial Activity of Ana-Thanatin
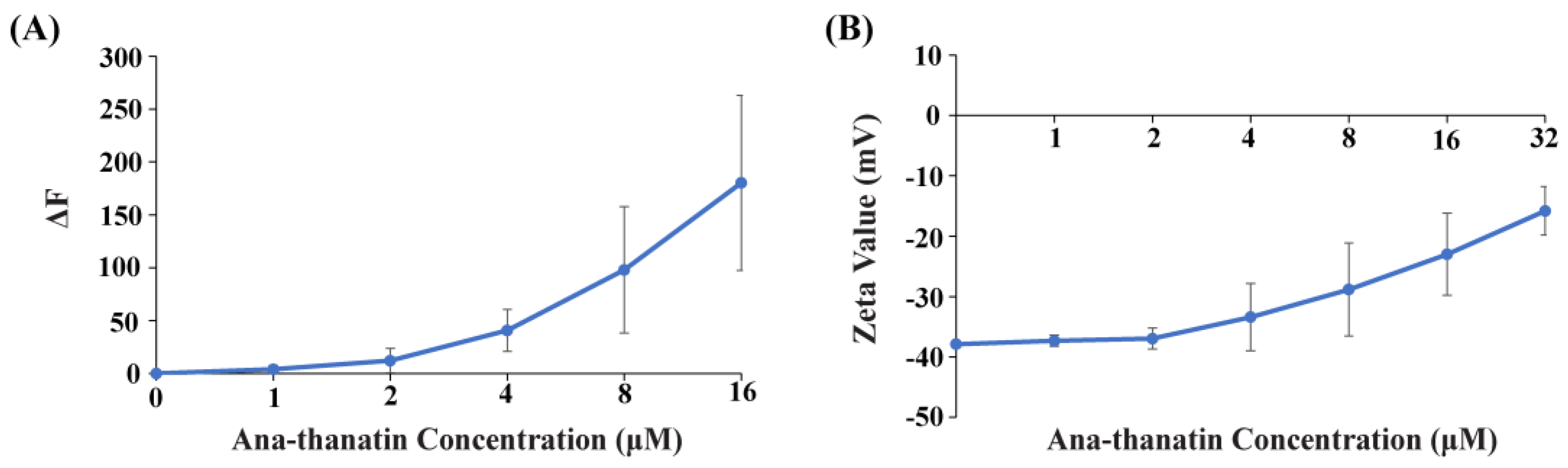
2.2. Ana-Thanatin-Mediated Outer Membrane Permeabilization and Bacterial Surface Charge Neutralization
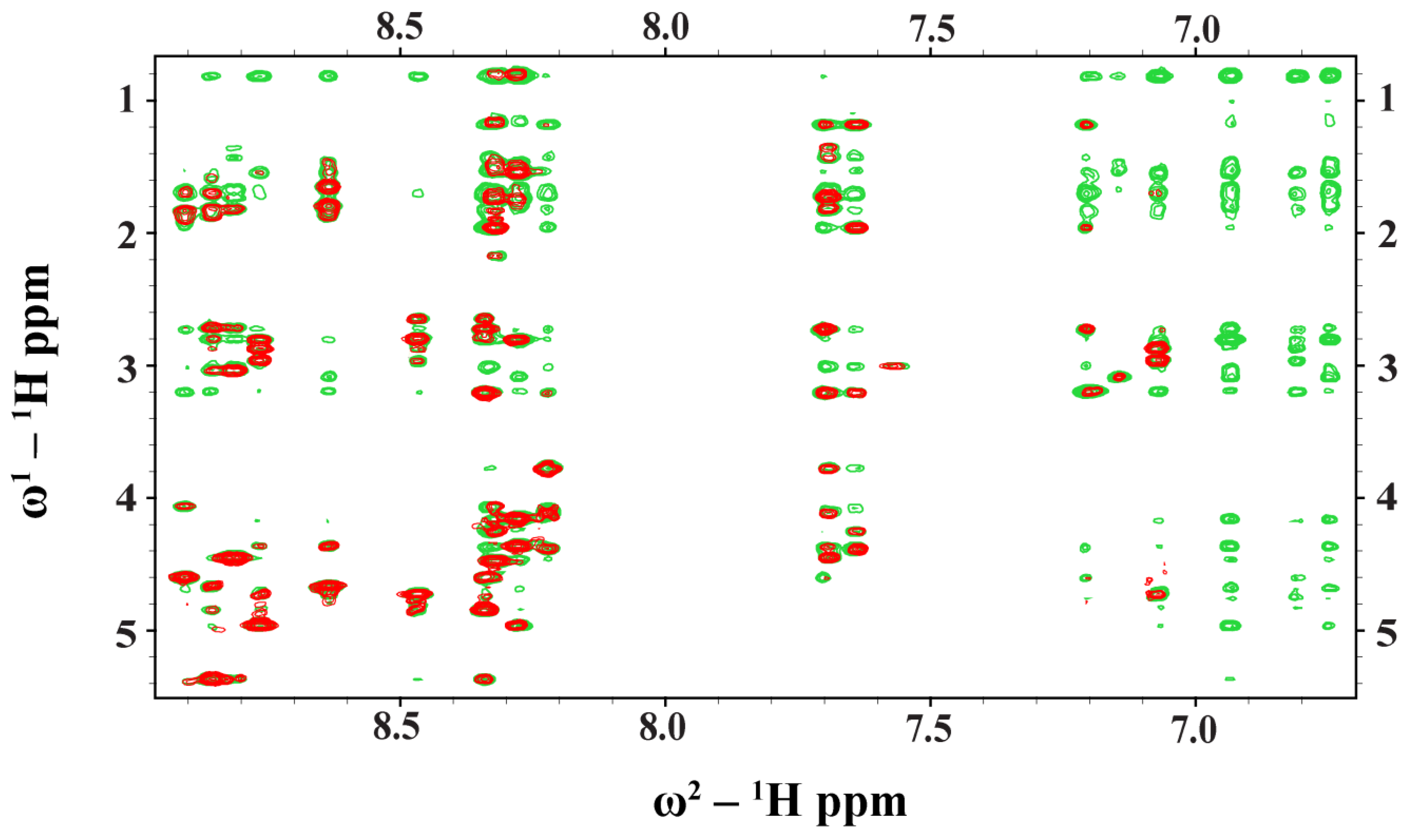
2.3. NMR Analyses of Ana-Thanatin in Free Solution and in Complex of the LPS Micelle
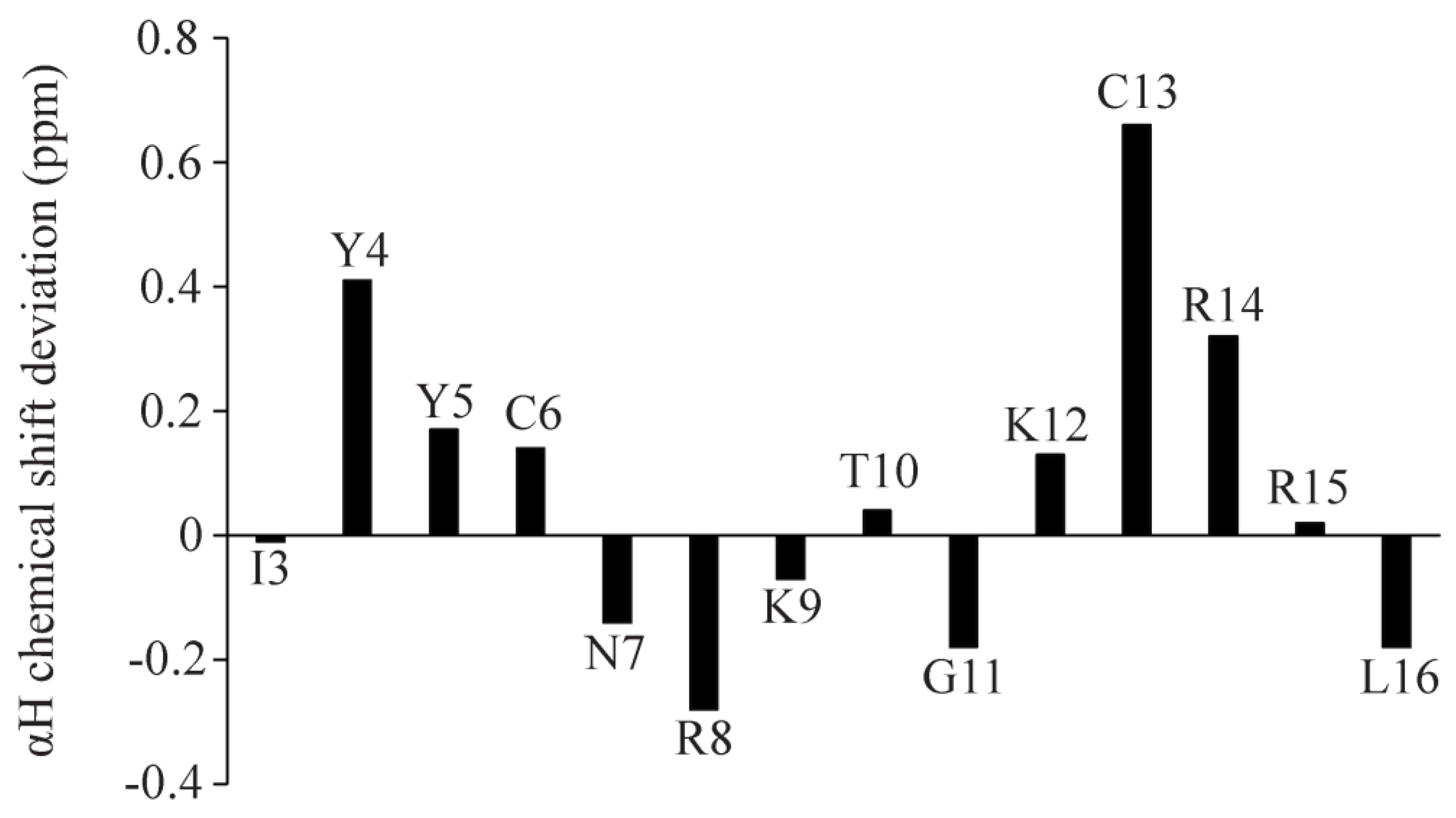
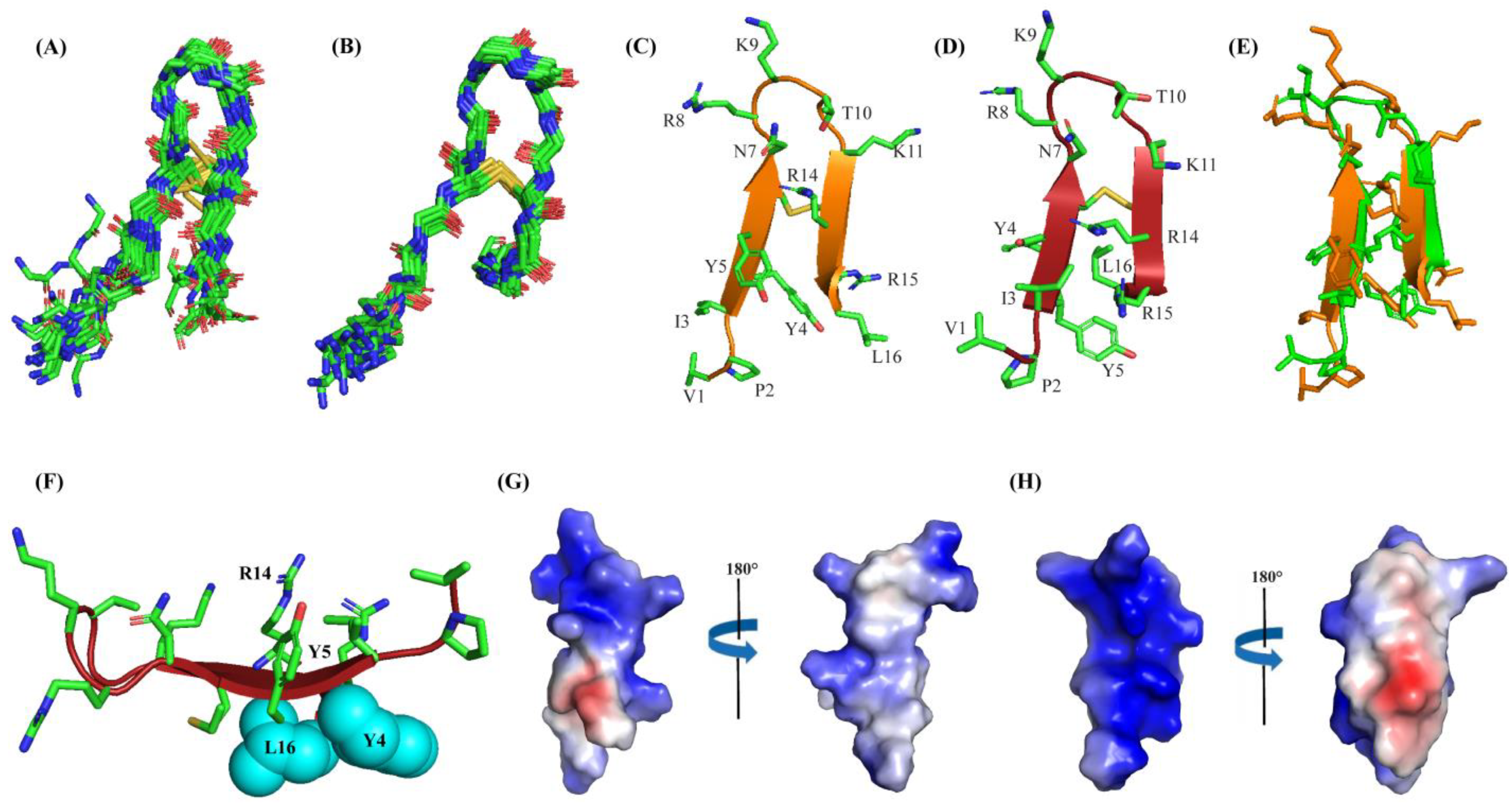
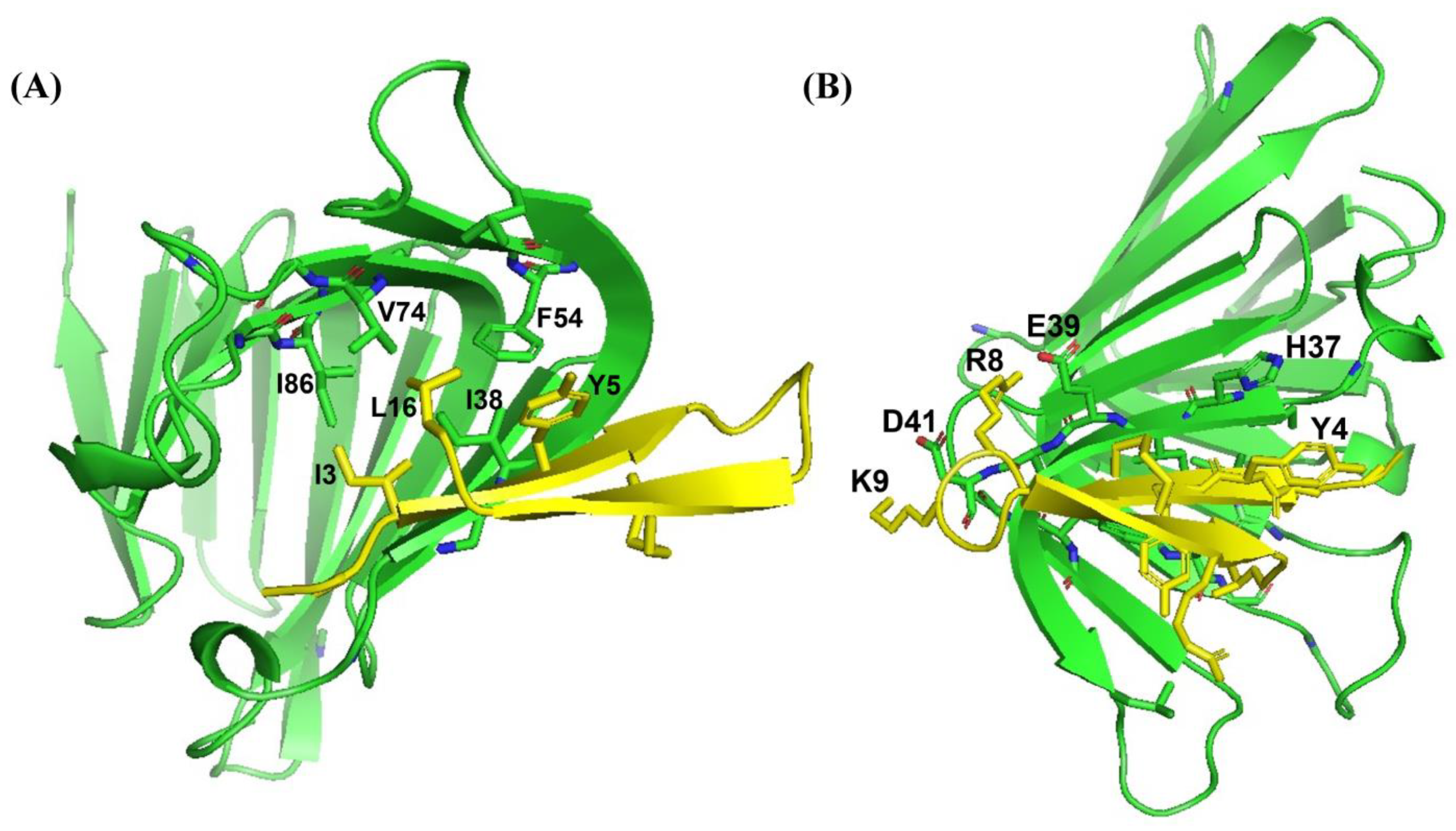
2.4. Atomic-Resolution Structures of Ana-Thanatin in Free Solution and in Complex with LPS Outer Membrane
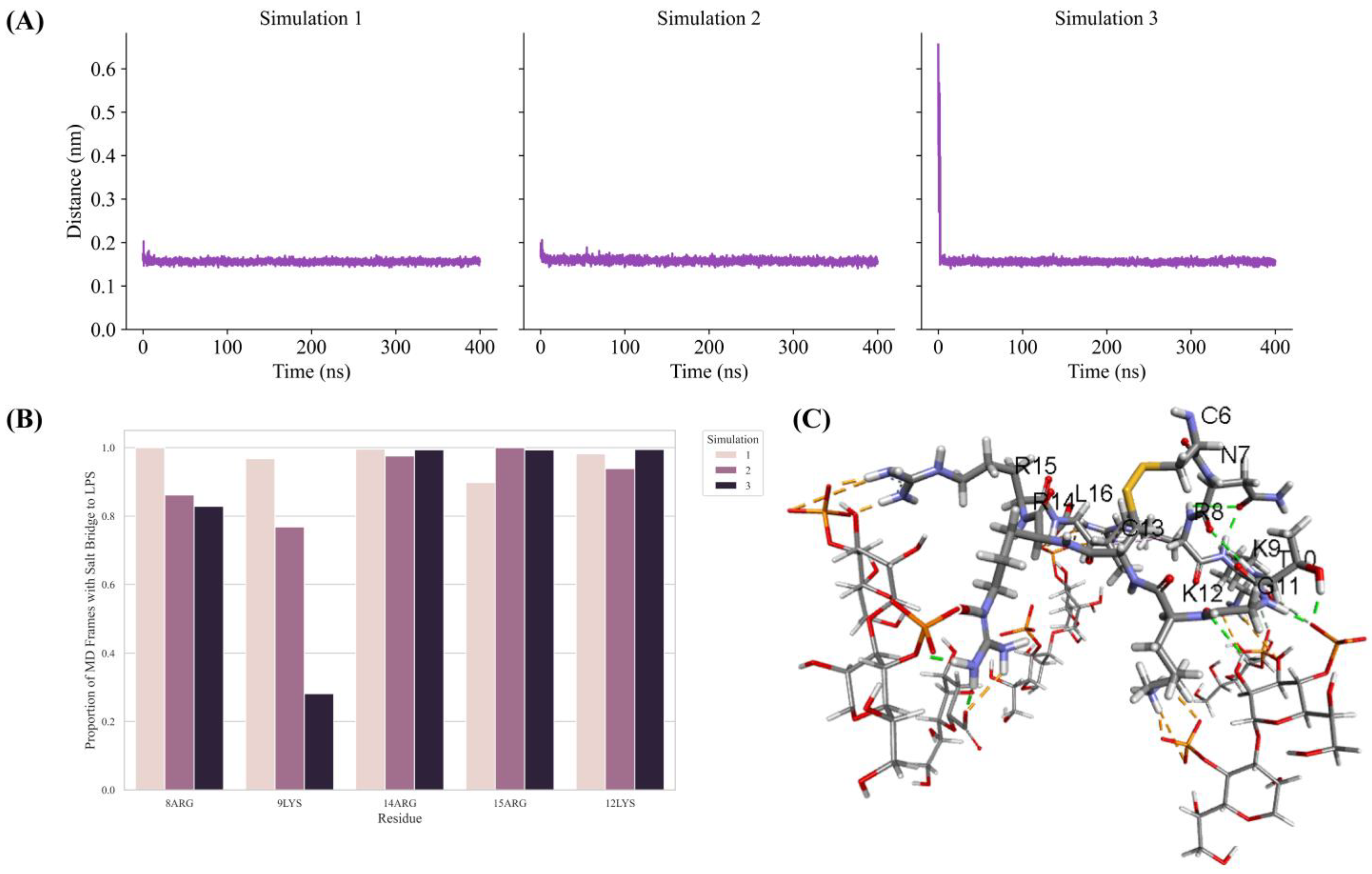
2.5. MD Simulation Studies of Ana-Thanatin with LPS Dipalmitoylphosphatidylethanolamine (DPPE) Lipid Bilayer
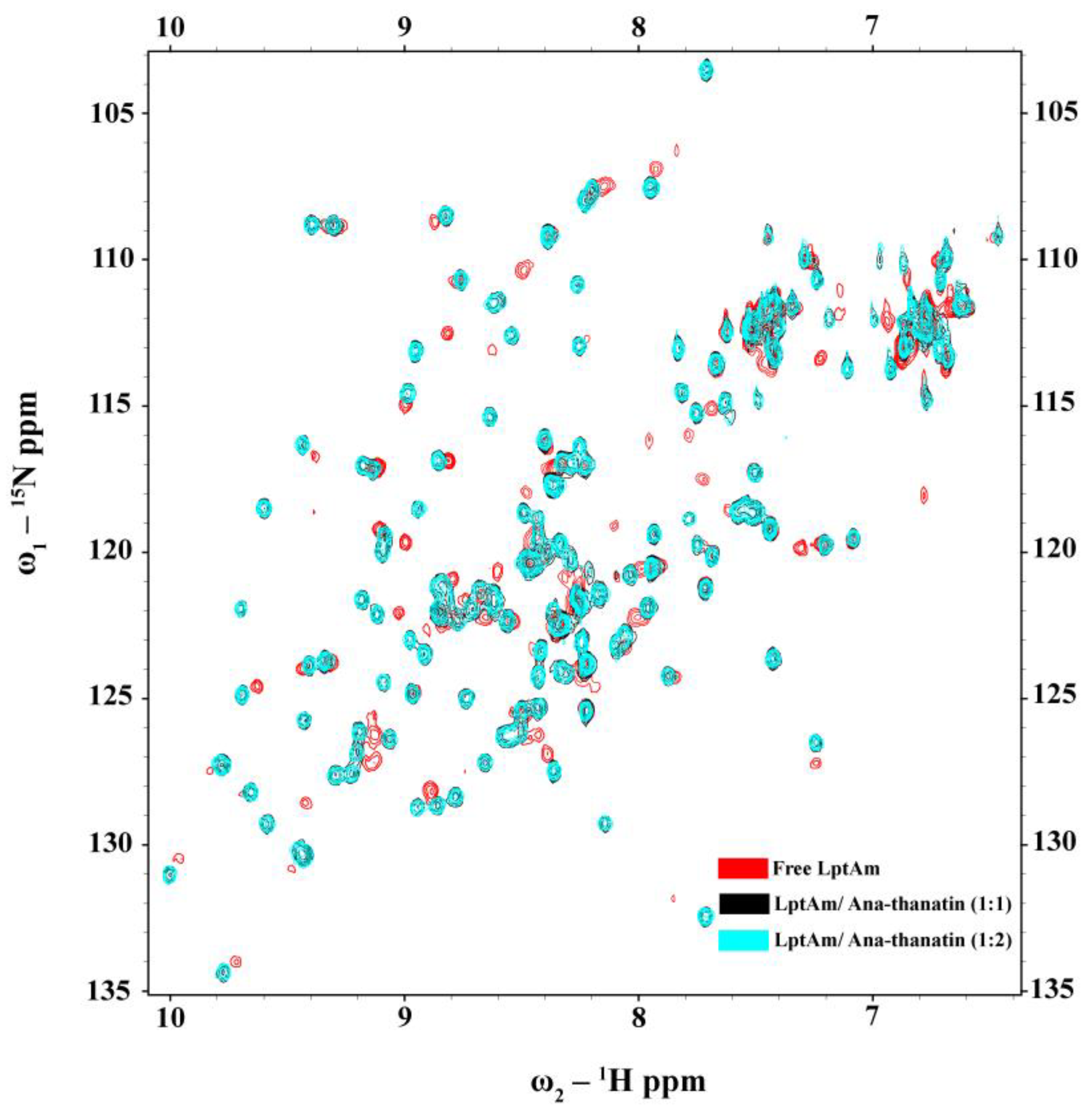
2.6. Binding of Ana-Thanatin with LptAm and Alpha-Fold3-Derived Structure of the Complex
3. Discussion
4. Materials and Methods
4.1. Peptide and Chemicals
4.2. Determination of Antibacterial Activity of Ana-Thanatin
4.3. LPS Outer Membrane (LPS-OM) Permeabilization Assay
4.4. Zeta (ζ) Potential Measurements
4.5. Recombinant Expression and Purification of LptAm
4.6. NMR Studies of Ana-Thanatin Peptide
4.7. Structure Calculations of Ana-Thanatin
4.8. MD Simulations of Ana-Thanatin in LPS-DPPE Lipid Bilayer
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ho, C.S.; Wong, C.T.H.; Aung, T.T.; Lakshminarayanan, R.; Mehta, J.S.; Rauz, S.; McNally, A.; Kintses, B.; Peacock, S.J.; de la Fuente-Nunez, C.; et al. Antimicrobial resistance: A concise update. Lancet Microbe 2025, 6, 100947. [Google Scholar] [CrossRef]
- Roope, L.S.J.; Smith, R.D.; Pouwels, K.B.; Buchanan, J.; Abel, L.; Eibich, P.; Butler, C.C.; Tan, P.S.; Walker, A.S.; Robotham, J.V.; et al. The challenge of antimicrobial resistance: What economics can contribute. Science 2019, 364, eaau4679. [Google Scholar] [CrossRef]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 2023, 21, 280–295, Erratum in Nat Rev Microbiol. 2024, 22, 255. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655, Erratum in Lancet 2022, 400, 1102. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling drug-resistant infections globally: Final report and recommendations. In Review on Antimicrobial Resistance; Wellcome Trust and HM Government: London, UK, 2016. [Google Scholar]
- World Health Organization. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 21 November 2023).
- GBD 2021 Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U. Global antimicrobial resistance in Gram-negative pathogens and clinical need. Curr. Opin. Microbiol. 2017, 39, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Platforms for Antibiotic Discovery. Nat. Rev. Drug Discovery 2013, 12, 371–387. [Google Scholar] [CrossRef]
- Brown, D. Antibiotic resistance breakers: Can repurposed drugs fill the antibiotic discovery void? Nat. Rev. Drug Discov. 2015, 14, 821–832, Erratum in Nat. Rev. Drug Discov. 2016, 15, 143. [Google Scholar] [CrossRef]
- Imai, Y.; Meyer, K.J.; Iinishi, A.; Favre-Godal, Q.; Green, R.; Manuse, S.; Caboni, M.; Mori, M.; Niles, S.; Ghiglieri, M.; et al. A new antibiotic selectively kills Gram-negative pathogens. Nature 2019, 576, 459–464, Erratum in Nature 2020, 580, 7802. [Google Scholar] [CrossRef]
- Zampaloni, C.; Mattei, P.; Bleicher, K.; Winther, L.; Thäte, C.; Bucher, C.; Adam, J.M.; Alanine, A.; Amrein, K.E.; Baidin, V.; et al. A novel antibiotic class targeting the lipopolysaccharide transporter. Nature 2024, 625, 566–571, Erratum in Nature 2024, 631, 8022. [Google Scholar] [CrossRef]
- Feng, J.; Zheng, Y.; Ma, W.; Weng, D.; Peng, D.; Xu, Y.; Wang, Z.; Wang, X. A synthetic antibiotic class with a deeply-optimized design for overcoming bacterial resistance. Nat. Commun. 2024, 15, 6040. [Google Scholar] [CrossRef]
- Chen, N.; Jiang, C. Antimicrobial peptides: Structure, mechanism, and modification. Eur. J. Med. Chem. 2023, 255, 115377. [Google Scholar] [CrossRef]
- Bhattacharjya, S.; Mohid, S.A.; Bhunia, A. Atomic-Resolution Structures and Mode of Action of Clinically Relevant Antimicrobial Peptides. Int. J. Mol. Sci. 2022, 23, 4558. [Google Scholar] [CrossRef]
- Starr, C.G.; Ghimire, J.; Guha, S.; Hoffmann, J.P.; Wang, Y.; Sun, L.; Landreneau, B.N.; Kolansky, Z.D.; Kilanowski-Doroh, I.M.; Sammarco, M.C.; et al. Synthetic molecular evolution of host cell-compatible, antimicrobial peptides effective against drug-resistant, biofilm-forming bacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 8437–8448. [Google Scholar] [CrossRef] [PubMed]
- Etayash, H.; Alford, M.; Akhoundsadegh, N.; Drayton, M.; Straus, S.K.; Hancock, R.E.W. Multifunctional Antibiotic-Host Defense Peptide Conjugate Kills Bacteria, Eradicates Biofilms, and Modulates the Innate Immune Response. J. Med. Chem. 2021, 64, 16854–16863, Erratum in J. Med. Chem. 2022, 65, 2710–2711. [Google Scholar] [CrossRef] [PubMed]
- Raheem, N.; Straus, S.K. Mechanisms of Action for Antimicrobial Peptides with Antibacterial and Antibiofilm Functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.; Zourou, A.C.; Cotten, M.L.; Pastor, R.W. A unified model of transient poration induced by antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2025, 122, e2510294122. [Google Scholar] [CrossRef]
- Wimley, W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef]
- Chen, E.H.; Wang, C.H.; Liao, Y.T.; Chan, F.Y.; Kanaoka, Y.; Uchihashi, T.; Kato, K.; Lai, L.; Chang, Y.W.; Ho, M.C.; et al. Visualizing the membrane disruption action of antimicrobial peptides by cryo-electron tomography. Nat. Commun. 2023, 14, 5464. [Google Scholar] [CrossRef]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar]
- Matsuzaki, K. Membrane Permeabilization Mechanisms. Adv. Exp. Med. Biol. 2019, 1117, 9–16. [Google Scholar]
- Bhattacharjya, S. NMR Structures and Interactions of Antimicrobial Peptides with Lipopolysaccharide: Connecting Structures to Functions. Curr. Top. Med. Chem. 2016, 16, 4–15. [Google Scholar] [PubMed]
- Bhunia, A.; Domadia, P.N.; Torres, J.; Hallock, K.J.; Ramamoorthy, A.; Bhattacharjya, S. NMR structure of pardaxin, a pore-forming antimicrobial peptide, in lipopolysaccharide micelles: Mechanism of outer membrane permeabilization. J. Biol. Chem. 2010, 285, 3883–3895. [Google Scholar] [PubMed]
- Ilyas, H.; Kim, J.; Lee, D.; Malmsten, M.; Bhunia, A. Structural insights into the combinatorial effects of antimicrobial peptides reveal a role of aromatic-aromatic interactions in antibacterial synergism. J. Biol. Chem. 2019, 294, 14615–14633. [Google Scholar] [PubMed]
- Sandín, D.; Valle, J.; Chaves-Arquero, B.; Prats-Ejarque, G.; Larrosa, M.N.; González-López, J.J.; Jiménez, M.Á.; Boix, E.; Andreu, D.; Torrent, M. Rationally Modified Antimicrobial Peptides from the N-Terminal Domain of Human RNase 3 Show Exceptional Serum Stability. J. Med. Chem. 2021, 64, 11472–11482. [Google Scholar] [CrossRef]
- Oddo, A.; Hansen, P.R. Hemolytic Activity of Antimicrobial Peptides. Methods Mol. Biol. 2017, 1548, 427–435. [Google Scholar] [CrossRef]
- Wang, T.; Zou, C.; Wen, N.; Liu, X.; Meng, Z.; Feng, S.; Zheng, Z.; Meng, Q.; Wang, C. The effect of structural modification of antimicrobial peptides on their antimicrobial activity, hemolytic activity, and plasma stability. J. Pept. Sci. 2021, 27, e3306. [Google Scholar] [CrossRef]
- Verma, N.K.; Dewangan, R.P.; Harioudh, M.K.; Ghosh, J.K. Introduction of a β-leucine residue instead of leucine9 and glycine10 residues in Temporin L for improved cell selectivity, stability and activity against planktonic and biofilm of methicillin resistant S. aureus. Bioorg. Chem. 2023, 134, 106440. [Google Scholar] [CrossRef]
- Luther, A.; Moehle, K.; Chevalier, E.; Dale, G.; Obrecht, D. Protein epitope mimetic macrocycles as biopharmaceuticals. Curr. Opin. Chem. Biol. 2017, 38, 45–51. [Google Scholar] [CrossRef]
- Srinivas, N.; Jetter, P.; Ueberbacher, B.J.; Werneburg, M.; Zerbe, K.; Steinmann, J.; Van der Meijden, B.; Bernardini, F.; Lederer, A.; Dias, R.L.; et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 2010, 327, 1010–1013. [Google Scholar] [CrossRef]
- Dash, R.; Bhattacharjya, S. Thanatin: An Emerging Host Defense Antimicrobial Peptide with Multiple Modes of Action. Int. J. Mol. Sci. 2021, 22, 1522. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, Q.; Xu, T.; Malakar, P.K.; Zhu, Y.; Liu, J.; Zhao, Y.; Zhang, Z. Thanatin: A Promising Antimicrobial Peptide Targeting the Achilles’ Heel of Multidrug-Resistant Bacteria. Int. J. Mol. Sci. 2024, 25, 9496. [Google Scholar] [CrossRef] [PubMed]
- Panteleev, P.V.; Teplovodskaya, J.S.; Utkina, A.D.; Smolina, A.A.; Kruglikov, R.N.; Safronova, V.N.; Bolosov, I.A.; Korobova, O.V.; Borzilov, A.I.; Ovchinnikova, T.V. Discovery of Novel Thanatin-like Antimicrobial Peptides from Bean Bug Riptortus pedestris. Pharmaceutics 2024, 16, 1453. [Google Scholar] [CrossRef] [PubMed]
- Vetterli, S.U.; Zerbe, K.; Müller, M.; Urfer, M.; Mondal, M.; Wang, S.Y.; Moehle, K.; Zerbe, O.; Vitale, A.; Pessi, G.; et al. Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Sci. Adv. 2018, 4, eaau2634. [Google Scholar] [CrossRef]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-β-lactamase. Nat. Commun. 2019, 10, 3517. [Google Scholar] [CrossRef]
- Fehlbaum, P.; Bulet, P.; Chernysh, S.; Briand, J.P.; Roussel, J.P.; Letellier, L.; Hetru, C.; Hoffmann, J.A. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proc. Natl. Acad. Sci. USA 1996, 93, 1221–1225. [Google Scholar] [CrossRef]
- Sinha, S.; Zheng, L.; Mu, Y.; Ng, W.J.; Bhattacharjya, S. Structure and Interactions of A Host Defense Antimicrobial Peptide Thanatin in Lipopolysaccharide Micelles Reveal Mechanism of Bacterial Cell Agglutination. Sci. Rep. 2017, 7, 17795. [Google Scholar] [CrossRef]
- Moura, E.C.C.M.; Baeta, T.; Romanelli, A.; Laguri, C.; Martorana, A.M.; Erba, E.; Simorre, J.P.; Sperandeo, P.; Polissi, A. Thanatin Impairs Lipopolysaccharide Transport Complex Assembly by Targeting LptC-LptA Interaction and Decreasing LptA Stability. Front. Microbiol. 2020, 11, 909. [Google Scholar] [CrossRef]
- Sinha, S.; Ng, W.J.; Bhattacharjya, S. NMR structure and localization of the host defense antimicrobial peptide thanatin in zwitterionic dodecylphosphocholine micelle: Implications in antimicrobial activity. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183432. [Google Scholar] [CrossRef]
- Schuster, M.; Brabet, E.; Oi, K.K.; Desjonquères, N.; Moehle, K.; Le Poupon, K.; Hell, S.; Gable, S.; Rithié, V.; Dillinger, S.; et al. Peptidomimetic antibiotics disrupt the lipopolysaccharide transport bridge of drug-resistant Enterobacteriaceae. Sci. Adv. 2023, 9, eadg3683. [Google Scholar] [CrossRef]
- Huynh, K.; Kibrom, A.; Donald, B.R.; Zhou, P. Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA. J. Struct. Biol. X 2023, 8, 100091. [Google Scholar] [CrossRef]
- Lee, J.; Cha, W.H.; Lee, D.-W. Multiple Precursor Proteins of Thanatin Isoforms, an Antimicrobial Peptide Associated with the Gut Symbiont of Riptortus pedestris. Front. Microbiol. 2022, 12, 796548. [Google Scholar] [CrossRef]
- Saad, A.; Bechinger, B. Solid-state NMR spectroscopy for structural studies of polypeptides and lipids in extended physiological membranes. Biochim. Biophys. Acta Biomembr. 2024, 1866, 184162. [Google Scholar] [CrossRef] [PubMed]
- Resende, J.M.; Verly, R.M.; Aisenbrey, C.; Cesar, A.; Bertani, P.; Piló-Veloso, D.; Bechinger, B. Membrane interactions of phylloseptin-1, -2, and -3 peptides by oriented solid-state NMR spectroscopy. Biophys. J. 2014, 107, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Mani, R.; Cady, S.D.; Tang, M.; Waring, A.J.; Lehrer, R.I.; Hong, M. Membrane-dependent oligomeric structure and pore formation of a beta-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. USA 2006, 103, 16242–16247. [Google Scholar] [CrossRef] [PubMed]
- Domadia, P.N.; Bhunia, A.; Ramamoorthy, A.; Bhattacharjya, S. Structure, interactions, and antibacterial activities of MSI-594 derived mutant peptide MSI-594F5A in lipopolysaccharide micelles: Role of the helical hairpin conformation in outer-membrane permeabilization. J. Am. Chem. Soc. 2010, 132, 18417–18428. [Google Scholar] [CrossRef]
- Nishiyama, Y.; Hou, G.; Agarwal, V.; Su, Y.; Ramamoorthy, A. Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy: Advances in Methodology and Applications. Chem. Rev. 2023, 123, 918–988. [Google Scholar] [CrossRef]
- Mielke, S.P.; Krishnan, V.V. Characterization of protein secondary structure from NMR chemical shifts. Prog. Nucl. Magn. Reson. Spectrosc. 2009, 54, 141–165. [Google Scholar] [CrossRef]
- Fiorentino, F.; Sauer, J.B.; Qiu, X.; Corey, R.A.; Cassidy, C.K.; Mynors-Wallis, B.; Mehmood, S.; Bolla, J.R.; Stansfeld, P.J.; Robinson, C.V. Dynamics of an LPS translocon induced by substrate and an antimicrobial peptide. Nat. Chem. Biol. 2021, 17, 187–195. [Google Scholar] [CrossRef]
- Sinha, S.; Dhanabal, V.B.; Sperandeo, P.; Polissi, A.; Bhattacharjya, S. Linking dual mode of action of host defense antimicrobial peptide thanatin: Structures, lipopolysaccharide and LptAm binding of designed analogs. Biochim. Biophys. Acta Biomembr. 2022, 1864, 183839. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.J.; Yan, B.T.S.; Palanivelu, N.; Dhanabal, V.B.; Bifani, J.P.; Bhattacharjya, S. Outer-Membrane Permeabilization, LPS Transport Inhibition: Activity, Interactions, and Structures of Thanatin Derived Antimicrobial Peptides. Int. J. Mol. Sci. 2024, 25, 2122. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.J.; Mu, Y.; Bhattacharjya, S. Structures, Interactions and Activity of the N-Terminal Truncated Variants of Antimicrobial Peptide Thanatin. Antibiotics 2024, 13, 74. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.J.; Guan, J.S.; Mu, Y.; Bhattacharjya, S. Single Disulfide Bond in Host Defense Thanatin Analog Peptides: Antimicrobial Activity, Atomic-Resolution Structures and Target Interactions. Int. J. Mol. Sci. 2024, 26, 51. [Google Scholar] [CrossRef]
- Sperandeo, P.; Martorana, A.M.; Polissi, A. The lipopolysaccharide transport (Lpt) machinery: A nonconventional transporter for lipopolysaccharide assembly at the outer membrane of Gram-negative bacteria. J. Biol. Chem. 2017, 3, 17981–17990. [Google Scholar] [CrossRef]
- Sperandeo, P.; Martorana, A.M.; Zaccaria, M.; Polissi, A. Targeting the LPS export pathway for the development of novel therapeutics. Biochim. Biophys. Acta. Mol. Cell. Res. 2023, 1870, 119406. [Google Scholar] [CrossRef]
- Güntert, P. Automated NMR protein structure calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378.2526. [Google Scholar]
- Berjanskii, M.V.; Neal, S.; Wishart, D.S. PREDITOR: A web server for predicting protein torsion angle restraints. Nucleic Acids. Res. 2006, 34, W63–W69. [Google Scholar]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 4, 477–486. [Google Scholar] [CrossRef]
| MIC (µM) | |||||
|---|---|---|---|---|---|
| Gram-Negative | Gram-Positive | ||||
| EC | KP | SE | SP | EF | |
| Ana-thanatin | 2 | 1–2 | 1 | 1 | >16 |
| Free Solution | Complex of the LPS Micelle |
|---|---|
| Y4δHs-L16H | I3δHs-R14ƐHs |
| C6αH-R14H | I3δHs-R15ƐHs |
| C6βHs-R14H | Y4δHs-R15H |
| C13αH-N7H | Y4δHs-L16H |
| R15βHs-Y4δHs | C6αH-R14H |
| R15αH-Y5H | C6βHs-R14H |
| C13αH-N7H | |
| R14δHs-Y5δHs | |
| R14δHs-Y5ƐHs | |
| R14γHs-Y5δHs | |
| R14γHs-Y5ƐHs | |
| R15αH-Y4δHs | |
| R15αH-Y4ƐHs | |
| R15βHs-Y4δHs | |
| R15βHs-Y4ƐH3 | |
| R15δHs-Y5δHs | |
| L16γHs-Y4δHs | |
| L16γHs-Y5δHs | |
| L16δHs-C6H |
| Free | LPS | |
|---|---|---|
| Distance Constraints | ||
| Intra residue [|i-j| = 0] | 65 | 82 |
| Sequential [|i-j|= 1] | 29 | 46 |
| Medium range [1 < |i-j| < 4] | 5 | 10 |
| Long range [|i-j| ≥ 4] | 6 | 19 |
| Total NOE | 105 | 158 |
| Dihedral—angle constraints | 24 | 24 |
| Deviation from mean structure | ||
| Backbone atoms (Å) | 1.22 | 0.57 |
| All heavy atoms (Å) | 2.11 | 1.06 |
| Ramachandran plot for the mean structure | ||
| % of residues in most favored region and additional allowed region | 100 | 100 |
| % of residues in generously allowed region | 0 | 0 |
| % of residues in disallowed region | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah, S.J.; Guan, J.S.; Mu, Y.; Bhattacharjya, S. Structures, Interactions, and Antimicrobial Activity of the Shortest Thanatin Peptide from Anasa tristis. Int. J. Mol. Sci. 2025, 26, 9571. https://doi.org/10.3390/ijms26199571
Abdullah SJ, Guan JS, Mu Y, Bhattacharjya S. Structures, Interactions, and Antimicrobial Activity of the Shortest Thanatin Peptide from Anasa tristis. International Journal of Molecular Sciences. 2025; 26(19):9571. https://doi.org/10.3390/ijms26199571
Chicago/Turabian StyleAbdullah, Swaleeha Jaan, Jia Sheng Guan, Yuguang Mu, and Surajit Bhattacharjya. 2025. "Structures, Interactions, and Antimicrobial Activity of the Shortest Thanatin Peptide from Anasa tristis" International Journal of Molecular Sciences 26, no. 19: 9571. https://doi.org/10.3390/ijms26199571
APA StyleAbdullah, S. J., Guan, J. S., Mu, Y., & Bhattacharjya, S. (2025). Structures, Interactions, and Antimicrobial Activity of the Shortest Thanatin Peptide from Anasa tristis. International Journal of Molecular Sciences, 26(19), 9571. https://doi.org/10.3390/ijms26199571









