Candidate Transcript Panel in Semen Extracellular Vesicles Can Improve Prediction of Aggressiveness of Prostate Cancer
Abstract
1. Introduction
2. Results
2.1. Selection of Candidate RNA Transcripts to Study in Semen sEVs
2.2. Clinical Evaluation of Individuals Included in the Study
2.3. PSA and Citric Acid in Semen Compared with PSA in Blood
2.4. Altered Expression of mRNAs Contained in Semen sEVs
2.5. Evaluation of the Potential of Semen Variables as PCa Diagnostic Biomarkers
2.6. Evaluation of the Potential of Semen Variables as PCa Prognostic Biomarkers
3. Discussion
4. Materials and Methods
4.1. Subjects of Study
4.2. PSA Determination by Electrochemiluminescent Immunoassay in Semen Samples
4.3. Citric Acid Quantification by Photometry in Semen Samples
4.4. Semen Sample Processing and sEV Isolation
4.5. Total RNA Isolation
4.6. Transcript Detection by RT-qPCR
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am. J. Surg. Pathol. 2016, 40, 244–252. [Google Scholar] [CrossRef]
- Hayes, J.H.; Barry, M.J. Screening for prostate cancer with the prostate-specific antigen test: A review of current evidence. JAMA 2014, 311, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Oesterling, J.E. Prostate specific antigen: A critical assessment of the most useful tumor marker for adenocarcinoma of the prostate. J. Urol. 1991, 145, 907–923. [Google Scholar] [CrossRef] [PubMed]
- Hessels, D.; Klein Gunnewiek, J.M.; van Oort, I.; Karthaus, H.F.; van Leenders, G.J.; van Balken, B.; Kiemeney, L.A.; Witjes, J.A.; Schalken, J.A. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 2003, 44, 8–15, discussion 15–16. [Google Scholar] [CrossRef] [PubMed]
- Tutrone, R.; Donovan, M.J.; Torkler, P.; Tadigotla, V.; McLain, T.; Noerholm, M.; Skog, J.; McKiernan, J. Clinical utility of the exosome based ExoDx Prostate(IntelliScore) EPI test in men presenting for initial Biopsy with a PSA 2-10 ng/mL. Prostate Cancer Prostatic Dis. 2020, 23, 607–614. [Google Scholar] [CrossRef]
- Lilja, H. A kallikrein-like serine protease in prostatic fluid cleaves the predominant seminal vesicle protein. J. Clin. Investig. 1985, 76, 1899–1903. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B.; Narayan, P. Citrate in the diagnosis of prostate cancer. Prostate 1999, 38, 237–245. [Google Scholar] [CrossRef]
- Gregorio, E.P.; Alexandrino, A.P.; Schuquel, I.T.A.; da Costa, W.F.; Rodrigues, M.A.F. Seminal citrate is superior to PSA for detecting clinically significant prostate cancer. Int. Braz J Urol Off. J. Braz. Soc. Urol. 2019, 45, 1113–1121. [Google Scholar] [CrossRef]
- Kline, E.E.; Treat, E.G.; Averna, T.A.; Davis, M.S.; Smith, A.Y.; Sillerud, L.O. Citrate concentrations in human seminal fluid and expressed prostatic fluid determined via 1H nuclear magnetic resonance spectroscopy outperform prostate specific antigen in prostate cancer detection. J. Urol. 2006, 176, 2274–2279. [Google Scholar] [CrossRef]
- Zheng, H.; Zhu, Y.; Shao, X.; Cai, A.; Dong, B.; Xue, W.; Gao, H. Distinct Metabolic Signatures of Hormone-Sensitive and Castration-Resistant Prostate Cancer Revealed by a 1H NMR-Based Metabolomics of Biopsy Tissue. J. Proteome Res. 2020, 19, 3741–3749. [Google Scholar] [CrossRef]
- Barceló, M.; Castells, M.; Bassas, L.; Vigués, F.; Larriba, S. Semen miRNAs Contained in Exosomes as Non-Invasive Biomarkers for Prostate Cancer Diagnosis. Sci. Rep. 2019, 9, 13772. [Google Scholar] [CrossRef] [PubMed]
- Ferre-Giraldo, A.; Castells, M.; Sánchez-Herrero, J.F.; López-Rodrigo, O.; de Rocco-Ponce, M.; Bassas, L.; Vigués, F.; Sumoy, L.; Larriba, S. Semen sEV tRF-Based Models Increase Non-Invasive Prediction Accuracy of Clinically Significant Prostate Cancer among Patients with Moderately Altered PSA Levels. Int. J. Mol. Sci. 2024, 25, 10122. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.B.; Palumbo, A.; de Mello, K.D.; Sternberg, C.; Caetano, M.S.; de Oliveira, F.L.; Neves, A.F.; Nasciutti, L.E.; Goulart, L.R.; Gimba, E.R. PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling. BMC Cancer 2012, 12, 507. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Coetzee, G.A. Prostate specific antigen gene regulation by androgen receptor. J. Cell. Biochem. 2004, 93, 233–241. [Google Scholar] [CrossRef]
- Kim, T.H.; Park, J.M.; Kim, M.Y.; Ahn, Y.H. The role of CREB3L4 in the proliferation of prostate cancer cells. Sci. Rep. 2017, 7, srep45300. [Google Scholar] [CrossRef]
- Keum, S.; Lee, Y.J.; Kim, J.W.; Rhee, S. Dual-specificity phosphatase 23 functions as a promising prognostic biomarker in non-small cell lung cancer. Genes Genom. 2025, 47, 321–329. [Google Scholar] [CrossRef]
- Li, H.; Huang, S.; Guo, C.; Guan, H.; Xiong, C. Cell-free seminal mRNA and microRNA exist in different forms. PLoS ONE 2012, 7, e34566. [Google Scholar] [CrossRef]
- Plata-Peña, L.; López-Rodrigo, O.; Bassas, L.; Larriba, S. Experimental validation of seminal miR-31-5p as biomarker for azoospermia and evaluation of the effect of preanalytical variables. Andrology 2023, 11, 668–676. [Google Scholar] [CrossRef]
- Hu, L.; Chen, X.; Narwade, N.; Lim, M.G.L.; Chen, Z.; Tennakoon, C.; Guan, P.; Chan, U.I.; Zhao, Z.; Deng, M.; et al. Single-cell analysis reveals androgen receptor regulates the ER-to-Golgi trafficking pathway with CREB3L2 to drive prostate cancer progression. Oncogene 2021, 40, 6479–6493. [Google Scholar] [CrossRef]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol 2016, 2, 882–889. [Google Scholar] [CrossRef]
- Bonache, S.; Mata, A.; Ramos, M.D.; Bassas, L.; Larriba, S. Sperm gene expression profile is related to pregnancy rate after insemination and is predictive of low fecundity in normozoospermic men. Hum. Reprod. 2012, 27, 1556–1567. [Google Scholar] [CrossRef]
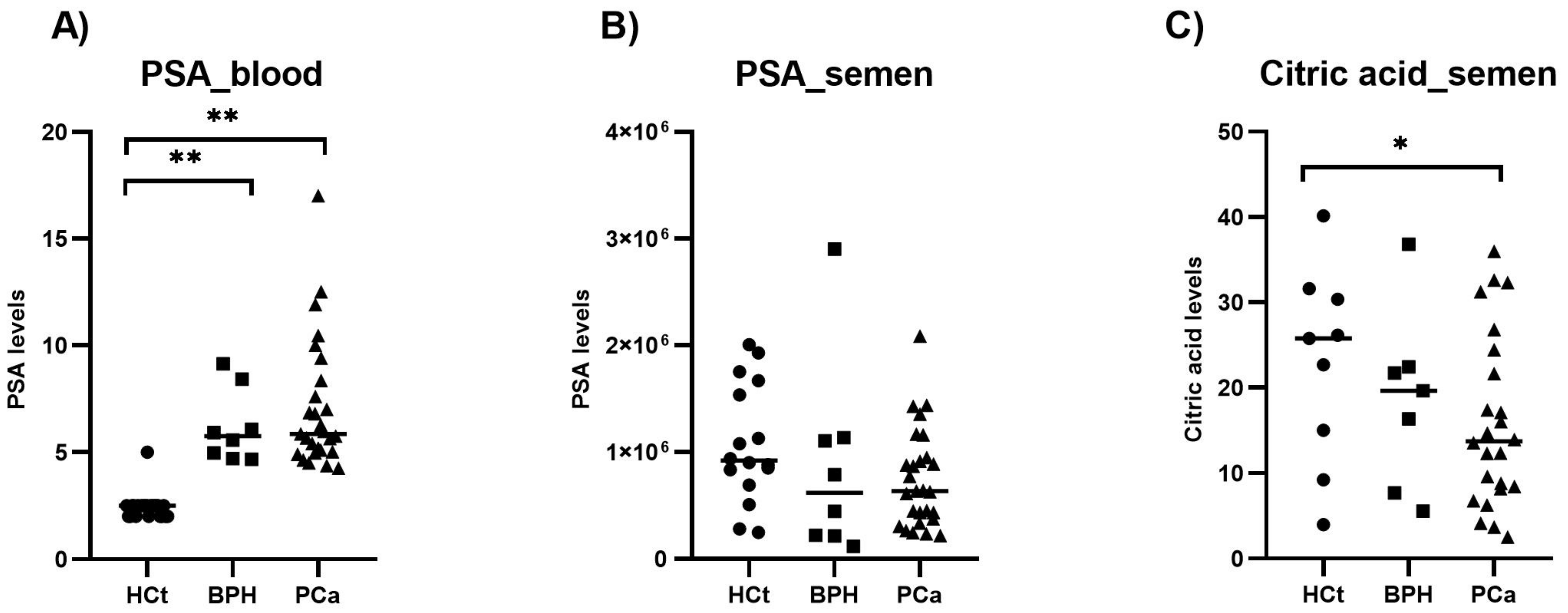
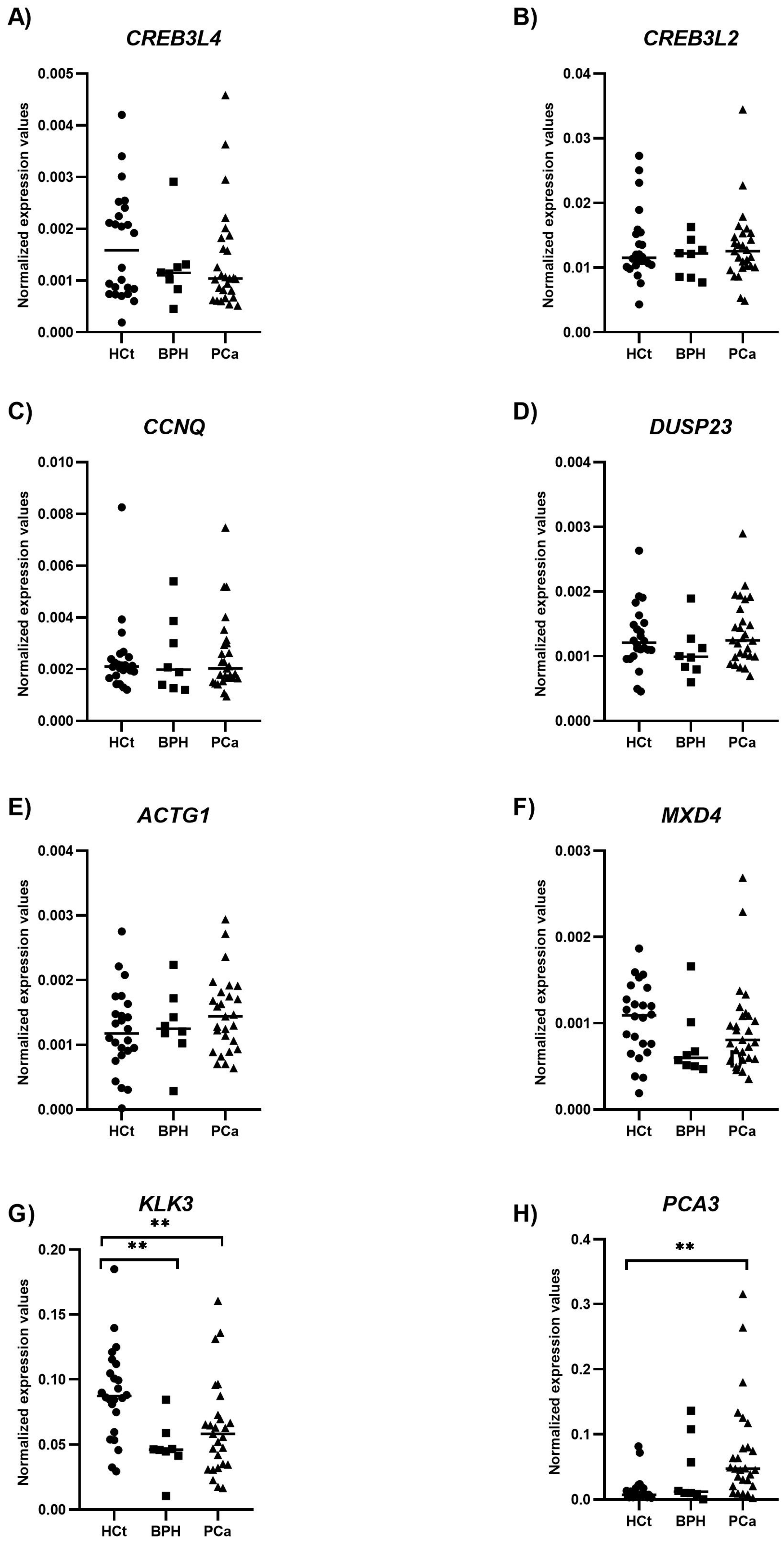
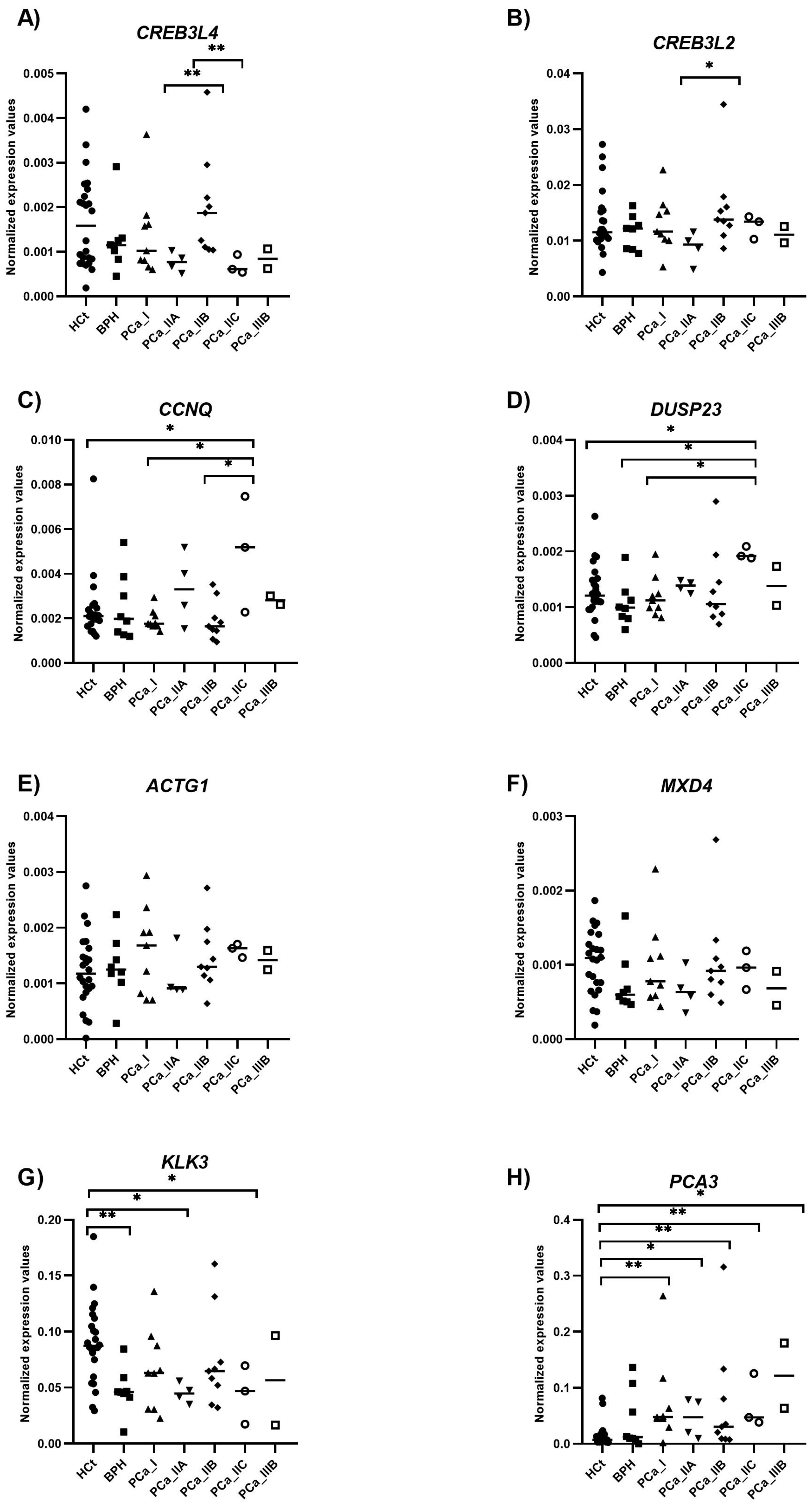
| Variable | HCt_noV | HCt_V | BPH | PCa_noV | PCa_V |
|---|---|---|---|---|---|
| Total (n) | 7 | 17 | 8 | 20 | 7 |
| Age, range (years) | 37–44 | 32–48 | 52–64 | 50–72 | 43–68 |
| Age, mean ±SD (years) | 40.57 ± 2.94 | 40 ± 3.95 | 58.63 ± 4.50 | 59.45 ± 6.48 | 58 ± 9.70 |
| Pre-biopsy PSA | |||||
| ≤10 ng/mL (n) | nd | nd | 8 | 19 | 4 |
| >10 ng/mL (n) | nd | nd | 0 | 1 | 3 |
| Pre-biopsy PSA, mean ±SD (ng/mL) | nd | nd | 6.19 | 6.43 | 8.97 |
| Gleason score | |||||
| Gleason score biopsy 6(3+3) (n) | nd | nd | nd | 10 | 3 |
| Gleason score biopsy 7(3+4) (n) | nd | nd | nd | 7 | 4 |
| Gleason score biopsy 7(4+3) (n) | nd | nd | nd | 2 | 0 |
| Gleason score biopsy 8(4+4) (n) | nd | nd | nd | 1 | 0 |
| Clinical stage | |||||
| T1c (n) | nd | nd | nd | 14 | 2 |
| T2a (n) | nd | nd | nd | 5 | 1 |
| T2c (n) | nd | nd | nd | 0 | 3 |
| T3a (n) | nd | nd | nd | 1 | 1 |
| Prognostic group * | |||||
| I (n) | nd | nd | nd | 7 | 2 |
| IIA (n) | nd | nd | nd | 3 | 1 |
| IIB (n) | nd | nd | nd | 6 | 3 |
| IIC (n) | nd | nd | nd | 3 | 0 |
| IIIB (n) | nd | nd | nd | 1 | 1 |
| Markers | AUC (p-Value) | 95% CI | Sn % | Sp % | PPV % | NPV % |
|---|---|---|---|---|---|---|
| A. (HCt + BPH) vs. PCa | ||||||
| CREB3L4 | 0.576 (0.319) | 0.428–0.724 | 11.1 | 90.6 | 50 | 54.71 |
| CREB3L2 | 0.522 (0.773) | 0.372–0.672 | 3.7 | 100 | 100 | 55.17 |
| CCNQ | 0.504 (0.958) | 0.351–0.657 | 3.7 | 96.9 | 50 | 54.38 |
| DUSP23 | 0.578 (0.308) | 0.430–0.725 | 25.9 | 81.3 | 53.84 | 56.52 |
| ACTG1 | 0.593 (0.221) | 0.447–0.740 | 40.7 | 75 | 57.89 | 60 |
| MXD4 | 0.549 (0.523) | 0.400–0.697 | 0 | 100 | 0 | 54.23 |
| KLK3 | 0.634 (0.078) | 0.489–0.779 | 48.1 | 65.6 | 54.16 | 60 |
| PCA3 | 0.796 (0.000 * | 0.677–0.915 | 59.3 | 84.4 | 76.19 | 71.05 |
| PCA3/KLK3 | 0.808 (0.000 *) | 0.688–0.927 | 44.4 | 90.6 | 80 | 65.9 |
| PSA_blood | 0.884 (0.000 *) | 0.797–0.970 | 77.8 | 84.4 | 84 | 81.81 |
| PSA_semen | 0.550 (0.513) | 0.399–0.701 | 0 | 100 | 0 | 54.23 |
| Citric acid_semen | 0.633 (0.080) | 0.487–0.779 | 25.9 | 68.8 | 41.17 | 52.38 |
| B. BPH vs. PCa | ||||||
| CREB3L4 | 0.472 (0.814) | 0.252–0.692 | 100 | 0 | 77.14 | 0 |
| CREB3L2 | 0.593 (0.432) | 0.365–0.820 | 100 | 0 | 77.14 | 0 |
| CCNQ | 0.525 (0.829) | 0.268–0.783 | 100 | 0 | 77.14 | 0 |
| DUSP23 | 0.708 (0.077) | 0.502–0.915 | 100 | 0 | 77.14 | 0 |
| ACTG1 | 0.569 (0.556) | 0.352–0.787 | 100 | 0 | 77.14 | 0 |
| MXD4 | 0.648 (0.209) | 0.425–0.871 | 100 | 0 | 77.14 | 0 |
| KLK3 | 0.634 (0.255) | 0.435–0.833 | 100 | 0 | 77.14 | 0 |
| PCA3 | 0.644 (0.223) | 0.401–0.886 | 100 | 0 | 77.14 | 0 |
| PCA3/KLK3 | 0.634 (0.255) | 0.392–0.877 | 100 | 0 | 77.14 | 0 |
| PSA_blood | 0.563 (0.596) | 0.349–0.776 | 100 | 0 | 77.14 | 0 |
| PSA_semen | 0.560 (0.610) | 0.288–0.832 | 100 | 0 | 77.14 | 0 |
| Citric acid_semen | 0.433 (0.569) | 0.192–0.674 | 100 | 0 | 77.14 | 0 |
| Combined RNA model (CCNQ+DUSP23) | 0.722 (0.059) | 0.521–0.923 | 100 | 25 | 81.81 | 100 |
| Combined PSA_blood+RNA model (PSA_blood+PCA3+CREB3L4+CCNQ+DUSP23) | 0.806 (0.010) | 0.647–0.964 | 96.3 | 25 | 66.66 | 81.25 |
| Combined PSA+citric+RNA model (PSA_blood+citric acid+PCA3+CREB3L4+CCNQ+DUSP23) | 0.866 (0.002) | 0.742–0.989 | 96.3 | 50 | 86.66 | 80 |
| C. (HCt + BPH+PCa_GS6) vs. (PCa GS7+GS8) | ||||||
| CREB3L4 | 0.532 (0.722) | 0.359–0.704 | 0 | 100 | 0 | 76.27 |
| CREB3L2 | 0.616 (0.193) | 0.455–0.777 | 7.1 | 100 | 100 | 77.58 |
| CCNQ | 0.520 (0.824) | 0.324–0.716 | 0 | 100 | 0 | 76.27 |
| DUSP23 | 0.600 (0.262) | 0.409–0.791 | 7.1 | 97.8 | 50 | 77.19 |
| ACTG1 | 0.613 (0.203) | 0.463–0.764 | 0 | 100 | 0 | 76.27 |
| MXD4 | 0.508 (0.929) | 0.348–0.668 | 0 | 100 | 0 | 76.27 |
| KLK3 | 0.568 (0.444) | 0.391–0.746 | 0 | 100 | 0 | 76.27 |
| PCA3 | 0.717 (0.015) | 0.572–0.863 | 14.3 | 97.8 | 66.66 | 78.57 |
| PCA3/KLK3 | 0.716 (0.015) | 0.552–0.880 | 7.1 | 97.8 | 50 | 77.19 |
| PSA_blood | 0.832 (0.000 *) | 0.724–0.939 | 28.6 | 88.9 | 44.44 | 80 |
| PSA_semen | 0.537 (0.682) | 0.373–0.700 | 0 | 100 | 0 | 76.27 |
| Citric acid_semen | 0.538 (0.669) | 0.384–0.693 | 0 | 100 | 0 | 76.27 |
| Combined RNA model (PCA3+CREB3L4+KLK3+CCNQ+DUSP23) | 0.746 (0.006) | 0.573–0.919 | 35.7 | 97.8 | 83.33 | 83.01 |
| Combined PSA_blood+RNA model (PSA_blood+PCA3+CREB3L4+KLK3+DUSP23) | 0.906 (0.000 *) | 0.831–0.982 | 64.3 | 91.1 | 69.23 | 89.1 |
| Combined PSA+citric+RNA model (PSA_blood+citric acid+PCA3+DUSP23) | 0.881 (0.000 *) | 0.777–0.984 | 64.3 | 93.3 | 75 | 89.36 |
| D. (BPH + PCa_GS6) vs. (PCa_GS7+GS8) | ||||||
| CREB3L4 | 0.604 (0.304) | 0.406–0.801 | 14.3 | 90.5 | 50 | 61.29 |
| CREB3L2 | 0.658 (0.117) | 0.476–0.841 | 21.4 | 85.7 | 50 | 62.06 |
| CCNQ | 0.522 (0.827) | 0.316–0.728 | 7.1 | 100 | 100 | 61.76 |
| DUSP23 | 0.639 (0.167) | 0.441–0.838 | 42.9 | 90.5 | 75 | 70.37 |
| ACTG1 | 0.578 (0.439) | 0.385–0.772 | 7.1 | 95.2 | 50 | 60.6 |
| MXD4 | 0.612 (0.266) | 0.420–0.804 | 7.1 | 90.5 | 33.33 | 59.37 |
| KLK3 | 0.592 (0.363) | 0.388–0.796 | 14.3 | 95.2 | 66.66 | 62.5 |
| PCA3 | 0.558 (0.567) | 0.357–0.758 | 14.3 | 95.2 | 66.66 | 62.5 |
| PCA3/KLK3 | 0.575 (0.459) | 0.372–0.778 | 7.1 | 95.2 | 50 | 60.6 |
| PSA_blood | 0.646 (0.148) | 0.451–0.842 | 28.6 | 85.7 | 57.14 | 64.28 |
| PSA_semen | 0.514 (0.893) | 0.311–0.717 | 0 | 100 | 0 | 60 |
| Citric acid_semen | 0.612 (0.266) | 0.422–0.803 | 21.4 | 90.5 | 60 | 63.33 |
| Combined RNA model (PCA3+DUSP23) | 0.670 (0.092) | 0.469–0.871 | 50 | 85.7 | 70 | 72 |
| Combined PSA_blood+RNA model (PSA_blood+DUSP23) | 0.721 (0.029) | 0.534–0.908 | 71.4 | 85.7 | 76.92 | 81.81 |
| E. [BPH + PCa_I + PCa_IIA+PCa_IIB] vs. [PCa_IIC + PCa_IIIB] | ||||||
| CREB3L4 | 0.844 (0.052) | 0.680–1.000 | 0 | 100 | 0 | 91.42 |
| CREB3L2 | 0.552 (0.768) | 0.317–0.787 | 0 | 100 | 0 | 91.42 |
| CCNQ | 0.865 (0.039) | 0.667–1.000 | 33.3 | 100 | 100 | 94.11 |
| DUSP23 | 0.917 (0.018) | 0.820–1.000 | 0 | 96.9 | 0 | 91.17 |
| ACTG1 | 0.656 (0.377) | 0.493–0.820 | 0 | 100 | 0 | 91.42 |
| MXD4 | 0.635 (0.444) | 0.391–0.880 | 0 | 100 | 0 | 91.42 |
| KLK3 | 0.583 (0.637) | 0.244–0.922 | 0 | 100 | 0 | 91.42 |
| PCA3 | 0.615 (0.517) | 0.380–0.849 | 0 | 100 | 0 | 91.42 |
| PCA3/KLK3 | 0.719 (0.216) | 0.497–0.940 | 0 | 100 | 0 | 91.42 |
| PSA_blood | 0.604 (0.556) | 0.423–0.786 | 0 | 100 | 0 | 91.42 |
| PSA_semen | 0.688 (0.289) | 0.301–1.000 | 0 | 100 | 0 | 91.42 |
| Citric acid_semen | 0.594 (0.596) | 0.198–0.989 | 0 | 100 | 0 | 91.42 |
| Combined RNA model (PCA3+CREB3L4+DUSP23) | 1.000 (0.005) | 1.000–1.000 | 100 | 100 | 100 | 100 |
| Combined PSA_blood+RNA model (PSA_blood+PCA3+CREB3L4+DUSP23+KLK3) | 1.000 (0.005) | 1.000–1.000 | 100 | 100 | 100 | 100 |
| Gene | NCBI Transcript Name | Primer Forward | Primer Reverse | Amplicon (nt) |
|---|---|---|---|---|
| CREB3L4 | NM_001255978.2 | AAACCCTGTTCCTGACCGAT | ACCAAGGAGATGTTGTGCCT | 268 |
| CREB3L2 | NM_194071.4 | CACTGGGGTTGATTCCTCGTG * | AATGCAGGTGGTCCACTGGG * | 116 |
| CCNQ | NM_152274.5 | ATCATGGAGGCAGGTGTCAA | GTAAGGGTCATAGGCGTCCA | 111 |
| DUSP23 | NM_001319658.2 | GGAGAGGCTGTGGGAGTG | CGTAGTCGTCGGATTTCAGC | 125 |
| ACTG1 | NM_001199954.3 | GGAACAAAAGGCGGGGTC | ATGGGGTACTTCAGGGTCAG | 233 |
| MXD4 | NM_006454.3 | GAAAAGCACAGACGAGCCAA | GTCCTGCTCCTCCAGTTTCT | 144 |
| KLK3 | NM_001648.2 | TTGTCTTCCTCACCCTGTCC | ACGCTTTTGTTCCTGATGCA | 202 |
| PCA3 | NR_015342.2 | GCACATTTCCAGCCCCTTTA # | GGCATTTCTCCCAGGGATCT # | 78 |
| Normalizer | NCBI Transcript Name | Primer Forward | Primer Reverse | Amplicon (nt) |
| RPL19 | NM_000981.4 | GCACATGGGCATAGGTAAGC | CAGGCTGTGATACATGTGGC | 148 |
| RPS17 | NM_001021.6 | CGCCATTATCCCCAGCAAAA | CTGCCGAAGTCCAAAAGCTT | 225 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferre-Giraldo, A.; Castells, M.; Madurga, A.; Arbiol-Roca, A.; de Rocco-Ponce, M.; Bassas, L.; Vigués, F.; Larriba, S. Candidate Transcript Panel in Semen Extracellular Vesicles Can Improve Prediction of Aggressiveness of Prostate Cancer. Int. J. Mol. Sci. 2025, 26, 9562. https://doi.org/10.3390/ijms26199562
Ferre-Giraldo A, Castells M, Madurga A, Arbiol-Roca A, de Rocco-Ponce M, Bassas L, Vigués F, Larriba S. Candidate Transcript Panel in Semen Extracellular Vesicles Can Improve Prediction of Aggressiveness of Prostate Cancer. International Journal of Molecular Sciences. 2025; 26(19):9562. https://doi.org/10.3390/ijms26199562
Chicago/Turabian StyleFerre-Giraldo, Adriana, Manel Castells, Alicia Madurga, Ariadna Arbiol-Roca, Maurizio de Rocco-Ponce, Lluís Bassas, Francesc Vigués, and Sara Larriba. 2025. "Candidate Transcript Panel in Semen Extracellular Vesicles Can Improve Prediction of Aggressiveness of Prostate Cancer" International Journal of Molecular Sciences 26, no. 19: 9562. https://doi.org/10.3390/ijms26199562
APA StyleFerre-Giraldo, A., Castells, M., Madurga, A., Arbiol-Roca, A., de Rocco-Ponce, M., Bassas, L., Vigués, F., & Larriba, S. (2025). Candidate Transcript Panel in Semen Extracellular Vesicles Can Improve Prediction of Aggressiveness of Prostate Cancer. International Journal of Molecular Sciences, 26(19), 9562. https://doi.org/10.3390/ijms26199562





