Reduced Expression of Selected Exosomal MicroRNAs Is Associated with Poor Outcomes in Patients with Acute Stroke Receiving Reperfusion Therapy—Preliminary Study
Abstract
1. Introduction
2. Results
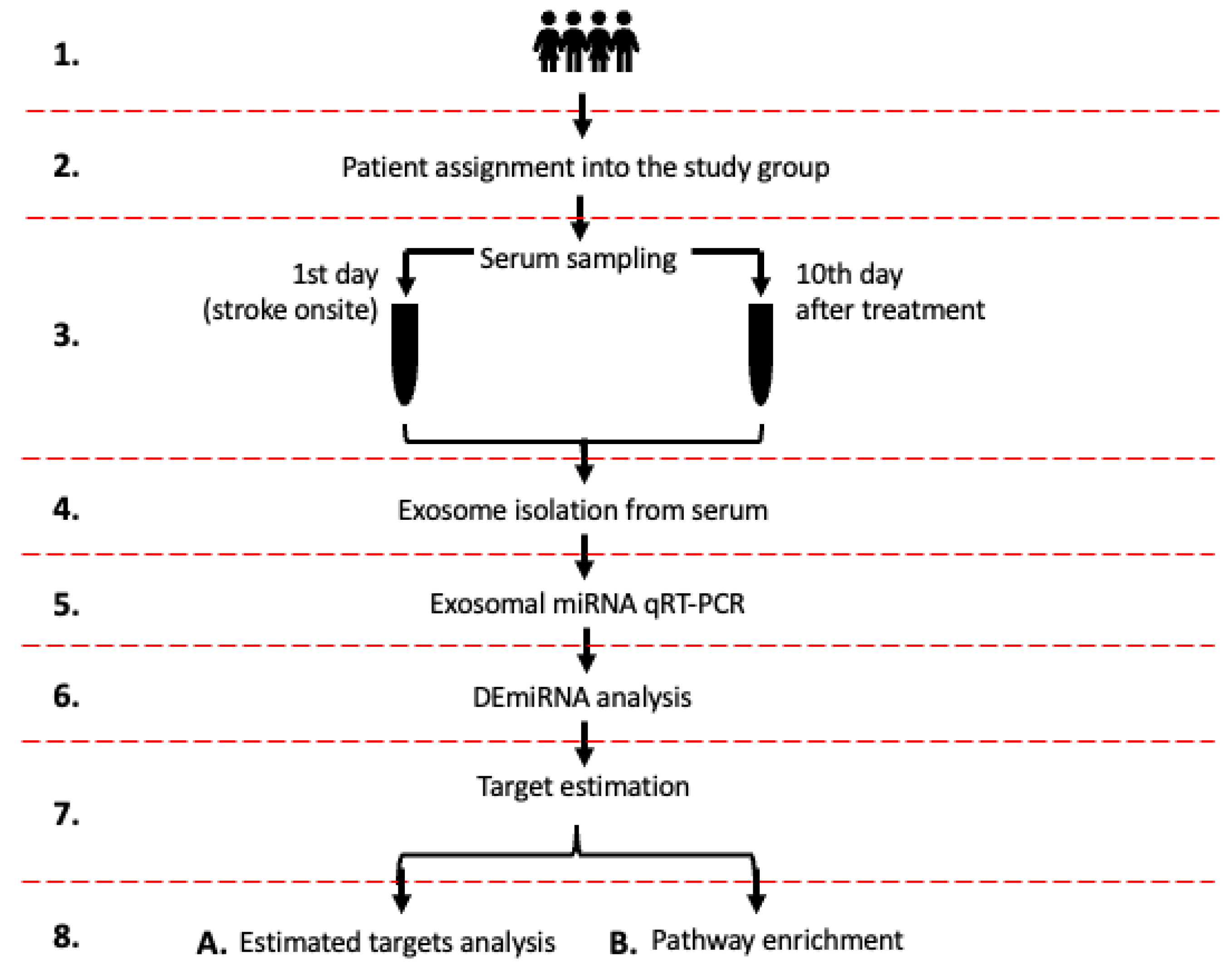
2.1. Study Design
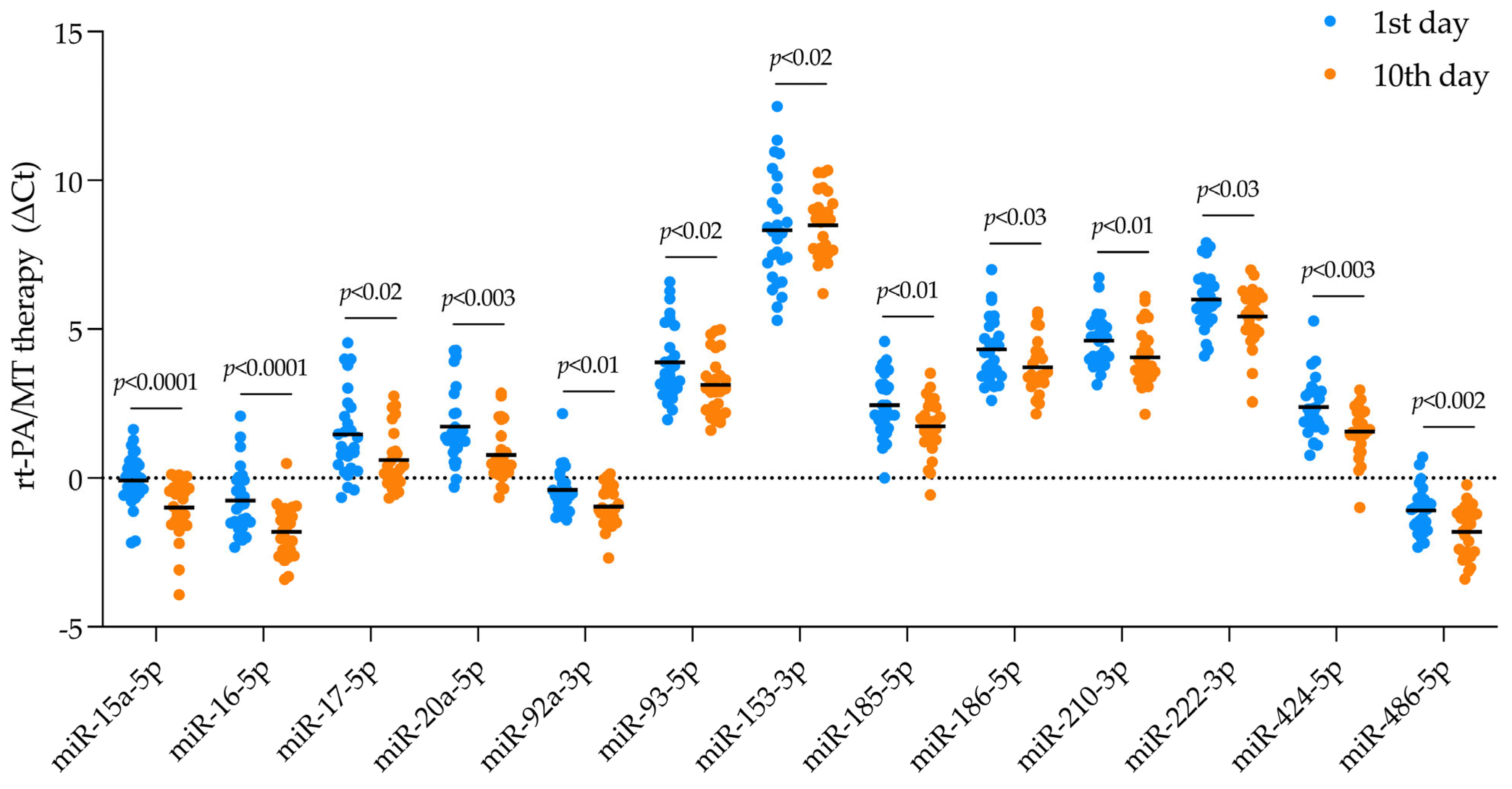
2.2. DEmiRNA Identification
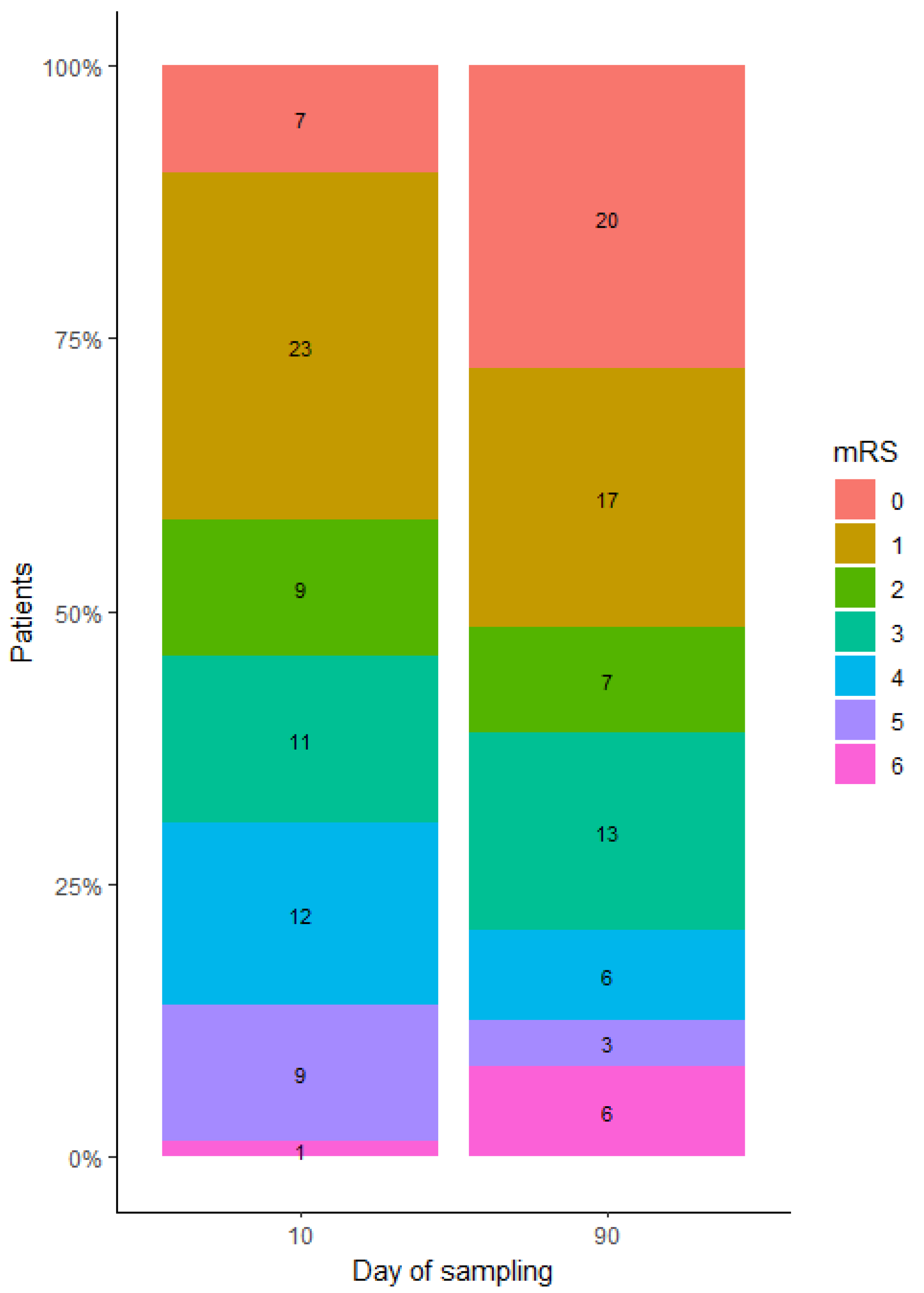
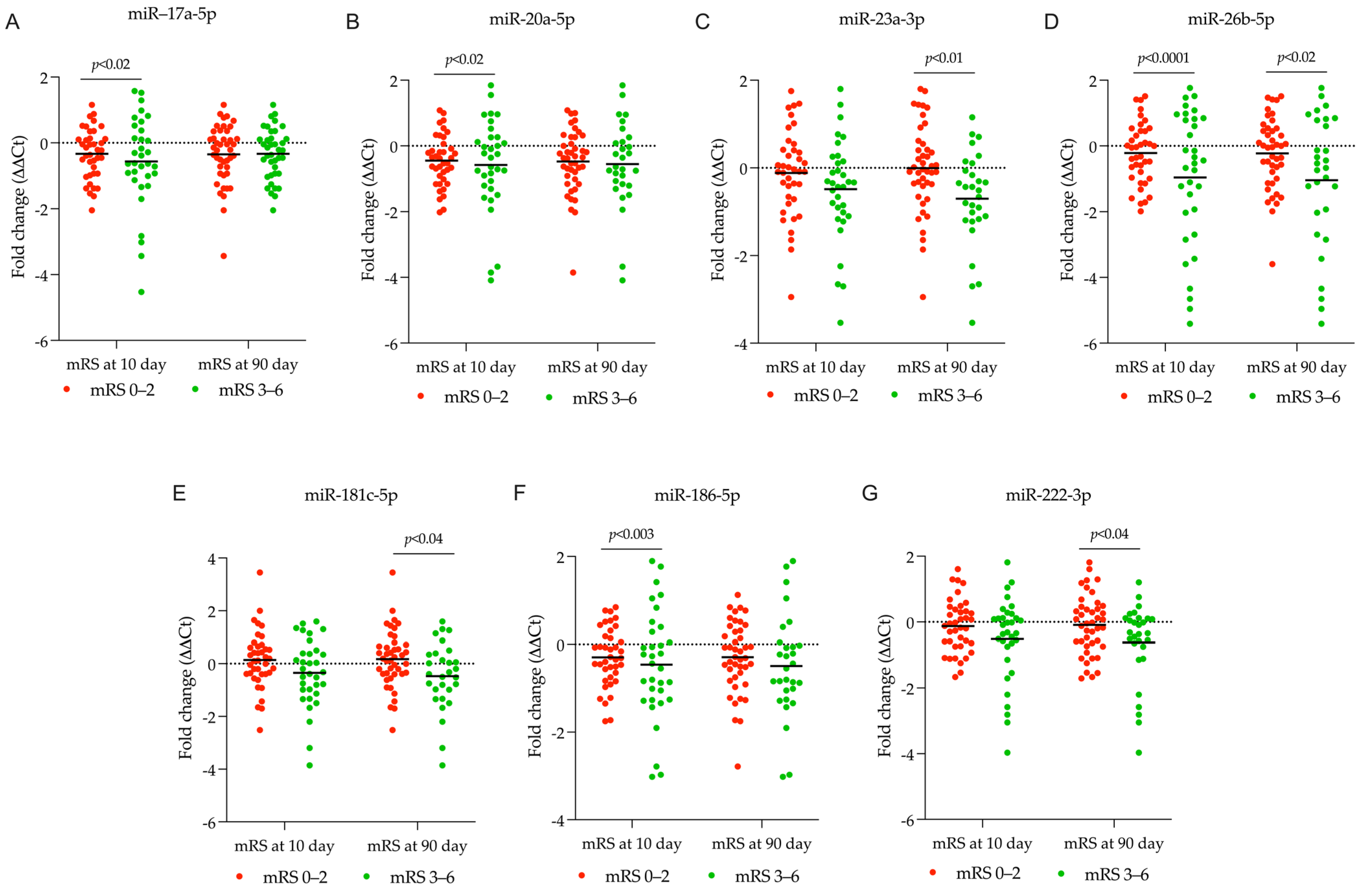
2.3. Identifying DEmiRNA Related to Stroke Patient’s Functional Outcome
2.4. Estimating Gene Targets for Differentially Expressed miRNAs and Their Enrichment Analysis
2.4.1. Estimating Gene Targets for DEmiRNA in Patients Receiving rt-PA/MT Treatment
2.4.2. Estimating Gene Targets for DEmiRNA in Patients Categorized Based on the 10-Day mRS Score
2.4.3. Estimating Gene Targets for DEmiRNA in Patients Categorized Based on the 90-Day mRS Score
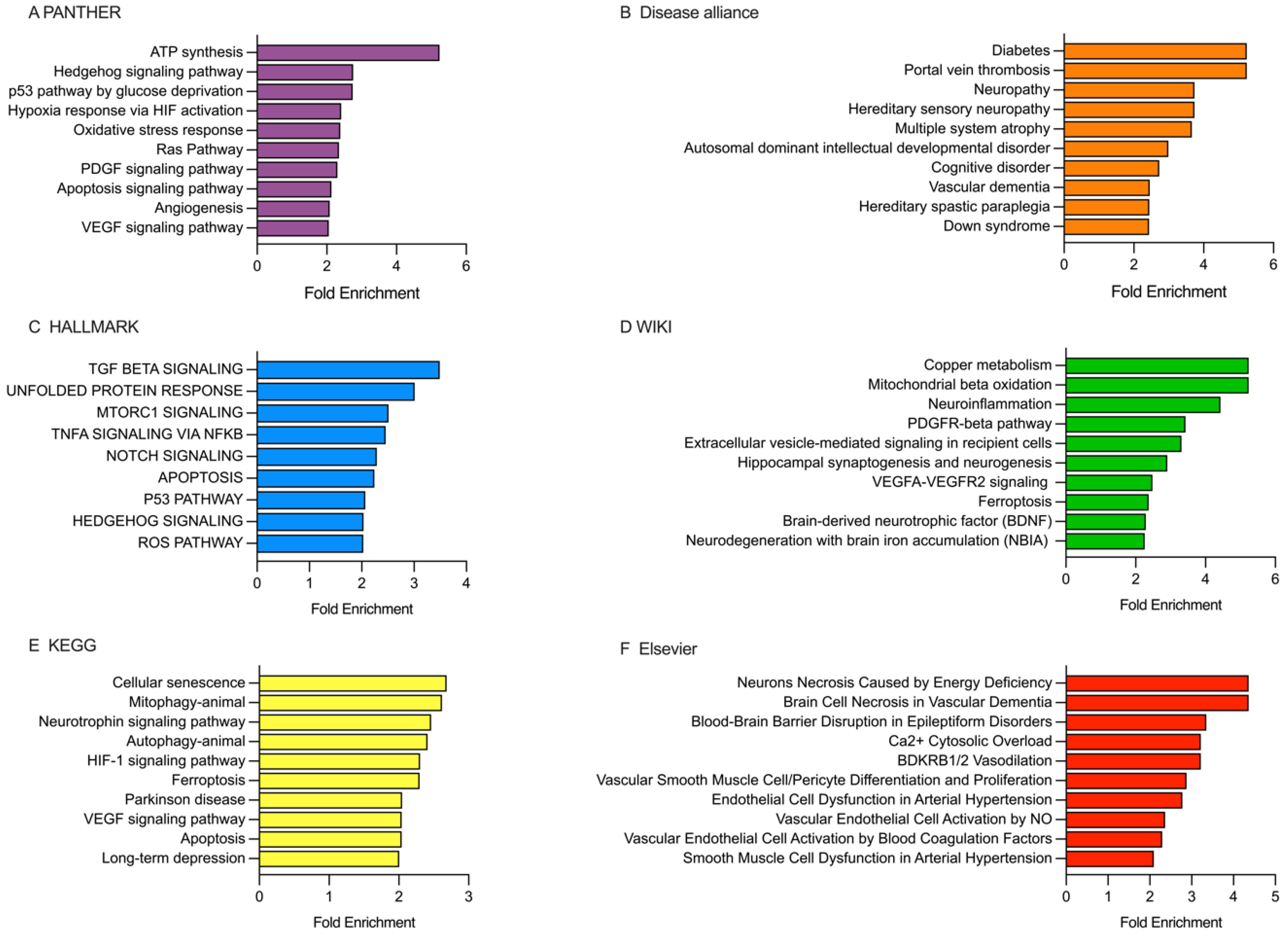
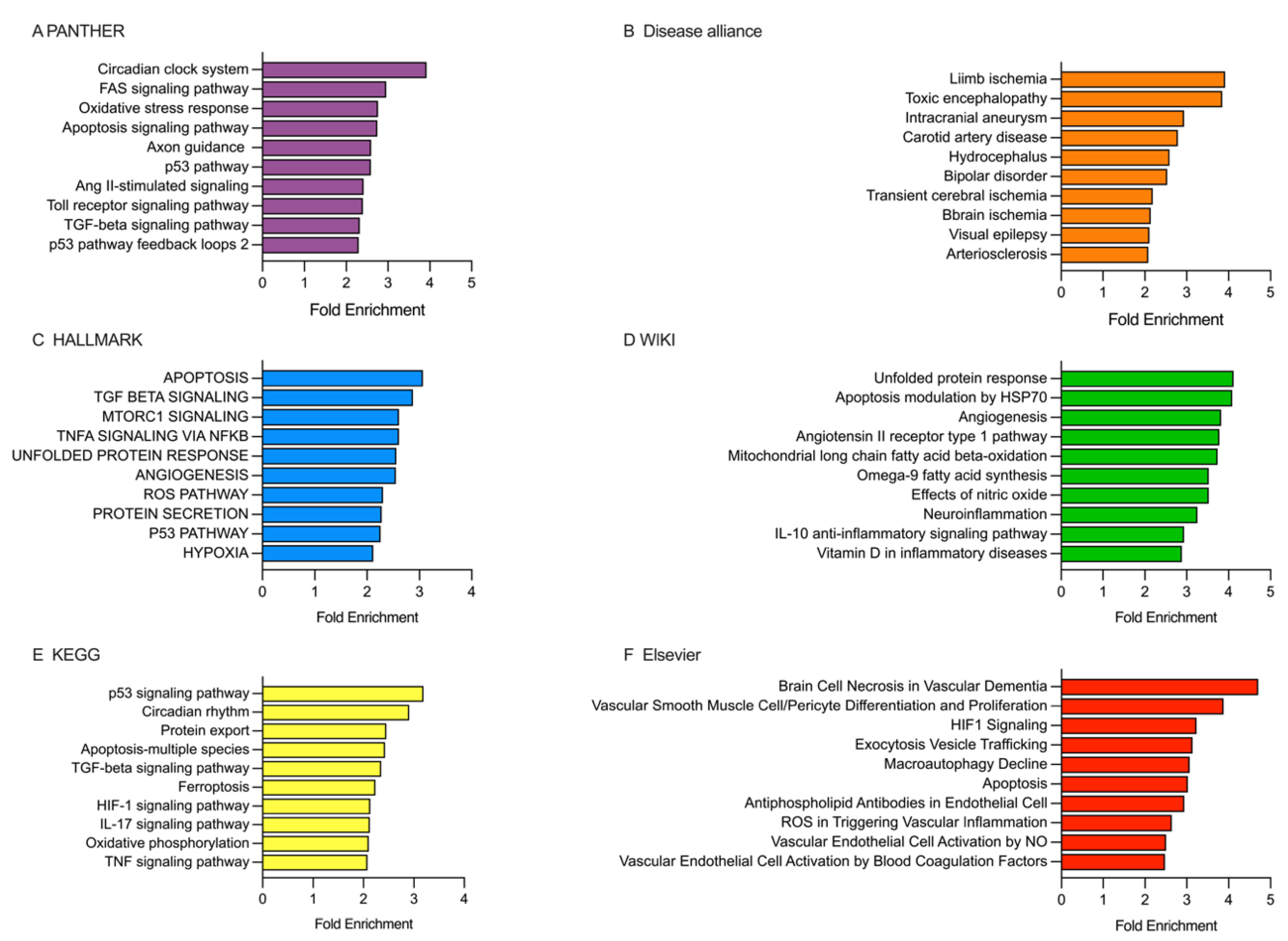
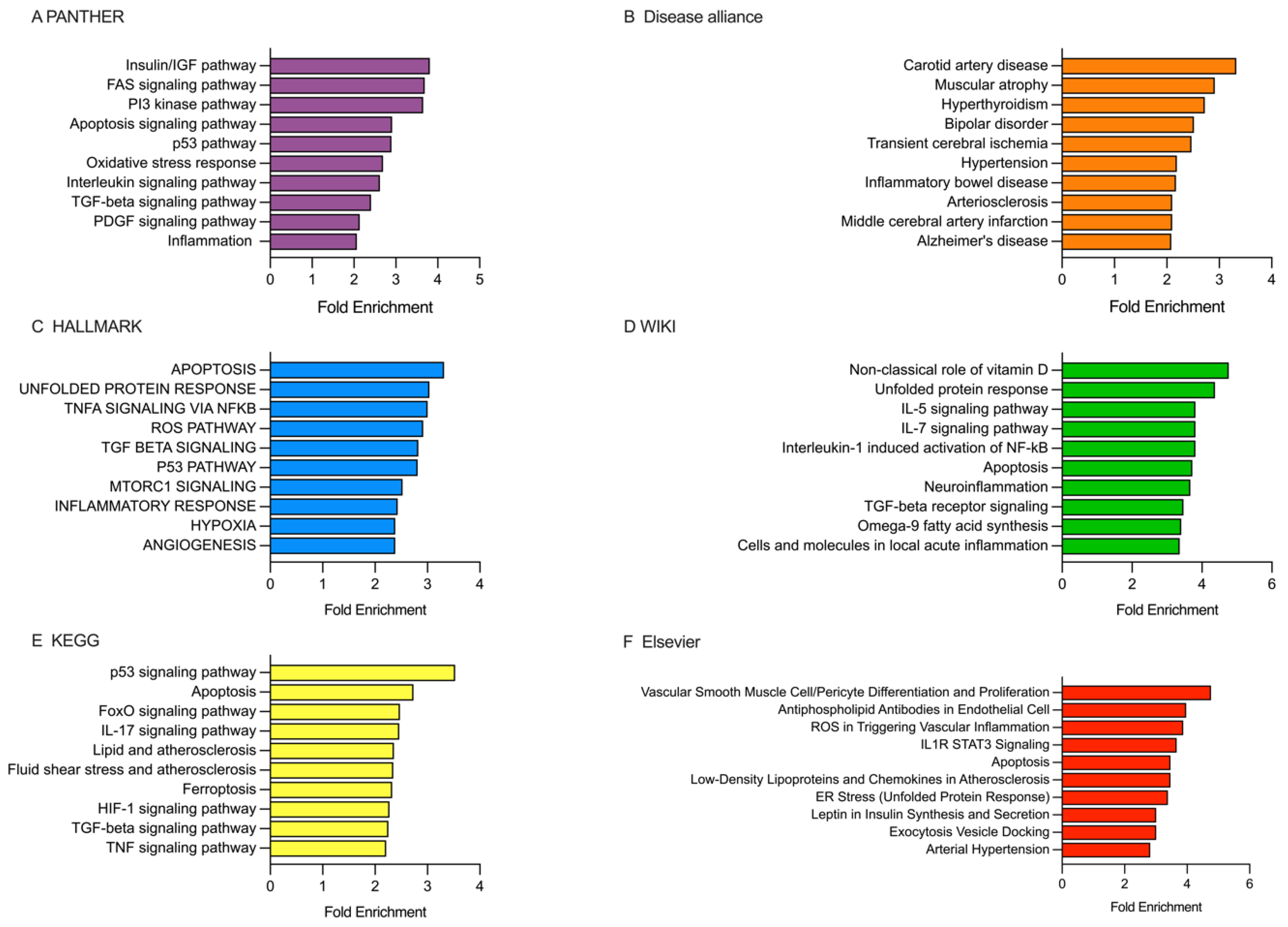
2.5. Analysis of the Enriched Pathways Targeted by DEmiRNAs Using Various Databases, Including PANTHER, Disease Alliance, HALLMARK, WIKI, KEGG, and Elsevier
2.5.1. Analysis of the Enriched Pathways Targeted by DEmiRNAs in the Patients Receiving rt-PA/MT Treatment
2.5.2. Analysis of the Enriched Pathways Targeted by DEmiRNAs in Patients Categorized Based on the 10-Day mRS Score
2.5.3. Analysis of the Enriched Pathways Targeted by DEmiRNAs in Patients Categorized Based on the 90-Day mRS Score
3. Discussion
Limitations
4. Materials and Methods
4.1. Inclusion Criteria
4.2. Serum Sampling
4.3. Exosome Isolation from Serum
4.4. Exosomal miRNA qPCR/RT
4.5. MiRNAs Quantification and Differential Expression Analysis
4.6. Estimation of DEmiRNAs Gene Targets and Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABL2 | Abelson Tyrosine-Protein Kinase 2 |
| ACTR2 | Actin-Related Protein 2 |
| AGO4 | Argonaute RISC Component 4 |
| APP | Amyloid Precursor Protein |
| ARHGAP12 | Rho GTPase-Activating Protein 12 |
| ATG14 | Autophagy-Related Protein 14 |
| BTG2 | BTG Anti-Proliferation Factor 2 |
| BTN3A3 | Butyrophilin Subfamily 3 Member A3 |
| BZW1 | Basic Leucine Zipper and W2 Domains 1 |
| CAPZA2 | Capping Actin Protein of Muscle Z-Line Subunit Alpha 2 |
| CCND1 | Cyclin D1 |
| CRIM1 | Cysteine-Rich Motor Neuron 1 |
| CRKL | CT10 Regulator of Kinase |
| DNAJC10 | DnaJ Heat Shock Protein Family Member C10 |
| EIF2B2 | Eukaryotic Translation Initiation Factor 2B Subunit Beta |
| EIF4G2 | Eukaryotic Translation Initiation Factor 4 Gamma 2 |
| FZD9 | Frizzled Class Receptor 9 |
| HSPA8 | Heat Shock Protein Family A (Hsp70) Member 8 |
| ITGA2 | Integrin Subunit Alpha 2 |
| KIF23 | Kinesin Family Member 23 |
| LAMC1 | Laminin Subunit Gamma 1 |
| MAP2K3 | Mitogen-Activated Protein Kinase 3 |
| MCL1 | Myeloid Cell Leukemia 1 |
| PMAIP1 | Phorbol-12-Myristate-13-Acetate-Induced Protein 1 |
| PTEN | Phosphatase and Tensin Homolog |
| RAB3IP | RAB3A Interacting Protein |
| RBM12B | RNA Binding Motif Protein 12B |
| RFK | Riboflavin Kinase |
| RPL14 | Ribosomal Protein L14 |
| RPRD2 | Regulation Of Nuclear Pre-MRNA Domain Containing 2 |
| SHOC2 | Leucine-Rich Repeat Protein SHOC2 |
| SIK1 | Salt-Inducible Kinase 1 |
| SKI | c-ski protooncogene |
| SLC28A1 | Solute Carrier Family 28 Member 1 |
| SMAD7 | SMAD Family Member 7 |
| SOCS5 | Suppressor Of Cytokine Signaling 5 |
| SPRED1 | Sprouty-Related, EVH1 Domain-Containing Protein 1 |
| TXNIP | Thioredoxin Interacting Protein |
| VEGFR2 | Vascular Endothelial Growth Factor Receptor 2 |
| WNK3 | WNK Lysine Deficient Protein Kinase 3 |
| YTHDC1 | YTH Domain Containing 1 |
| ZMAT3 | Zinc Finger Matrin-Type 3 |
References
- Murray, C.J.L.; Afshin, A.; Alam, T.; Ashbaugh, C.; Barthelemy, C.; Biehl, M.; Brauer, M.; Compton, K.; Cromwell, E.; Dandona, L.; et al. Global Burden of 369 Diseases and Injuries in 204 Countries and Territories, 1990–2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef] [PubMed]
- El Nawar, R.; Lapergue, B.; Piotin, M.; Gory, B.; Blanc, R.; Consoli, A.; Rodesch, G.; Mazighi, M.; Bourdain, F.; Kyheng, M.; et al. Higher Annual Operator Volume Is Associated with Better Reperfusion Rates in Stroke Patients Treated by Mechanical Thrombectomy: The ETIS Registry. JACC Cardiovasc. Interv. 2019, 12, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Froehler, M.T.; Saver, J.L.; Zaidat, O.O.; Jahan, R.; Aziz-Sultan, M.A.; Klucznik, R.P.; Haussen, D.C.; Hellinger, F.R.; Yavagal, D.R.; Yao, T.L.; et al. Interhospital Transfer Before Thrombectomy Is Associated with Delayed Treatment and Worse Outcome in the STRATIS Registry (Systematic Evaluation of Patients Treated with Neurothrombectomy Devices for Acute Ischemic Stroke). Circulation 2017, 136, 2311–2321. [Google Scholar] [CrossRef] [PubMed]
- Kaesmacher, J.; Gralla, J.; Mosimann, P.J.; Zibold, F.; Heldner, M.R.; Piechowiak, E.; Dobrocky, T.; Arnold, M.; Fischer, U.; Mordasini, P. Reasons for Reperfusion Failures in Stent-Retriever-Based Thrombectomy: Registry Analysis and Proposal of a Classification System. AJNR Am. J. Neuroradiol. 2018, 39, 1848–1853. [Google Scholar] [CrossRef]
- Coutinho, J.M.; Liebeskind, D.S.; Slater, L.A.; Nogueira, R.G.; Clark, W.; Dávalos, A.; Bonafé, A.; Jahan, R.; Fischer, U.; Gralla, J.; et al. Combined Intravenous Thrombolysis and Thrombectomy vs Thrombectomy Alone for Acute Ischemic Stroke: A Pooled Analysis of the SWIFT and STAR Studies. JAMA Neurol. 2017, 74, 268–274. [Google Scholar] [CrossRef]
- Goyal, M.; Menon, B.K.; Van Zwam, W.H.; Dippel, D.W.J.; Mitchell, P.J.; Demchuk, A.M.; Dávalos, A.; Majoie, C.B.L.M.; Van Der Lugt, A.; De Miquel, M.A.; et al. Endovascular Thrombectomy After Large-Vessel Ischaemic Stroke: A Meta-Analysis of Individual Patient Data from Five Randomised Trials. Lancet 2016, 387, 1723–1731. [Google Scholar] [CrossRef]
- Mistry, E.A.; Mistry, A.M.; Nakawah, M.O.; Chitale, R.V.; James, R.F.; Volpi, J.J.; Fusco, M.R. Mechanical Thrombectomy Outcomes with and Without Intravenous Thrombolysis in Stroke Patients: A Meta-Analysis. Stroke 2017, 48, 2450–2456. [Google Scholar] [CrossRef]
- Kim, J.T.; Liebeskind, D.S.; Jahan, R.; Menon, B.K.; Goyal, M.; Nogueira, R.G.; Pereira, V.M.; Gralla, J.; Saver, J.L. Impact of Hyperglycemia According to the Collateral Status on Outcomes in Mechanical Thrombectomy. Stroke 2018, 49, 2706–2714. [Google Scholar] [CrossRef]
- Goyal, N.; Tsivgoulis, G.; Pandhi, A.; Dillard, K.; Katsanos, A.H.; Magoufis, G.; Chang, J.J.; Zand, R.; Hoit, D.; Safouris, A.; et al. Admission Hyperglycemia and Outcomes in Large Vessel Occlusion Strokes Treated with Mechanical Thrombectomy. J. Neurointerv. Surg. 2018, 10, 112–117. [Google Scholar] [CrossRef]
- Broocks, G.; Kemmling, A.; Aberle, J.; Kniep, H.; Bechstein, M.; Flottmann, F.; Leischner, H.; Faizy, T.D.; Nawabi, J.; Schön, G.; et al. Elevated Blood Glucose Is Associated with Aggravated Brain Edema in Acute Stroke. J. Neurol. 2020, 267, 440–448. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Shi, M.C.; Wang, Z.X.; Li, C.; Sun, M.Y.; Zhou, J.; Zhang, W.B.; Huo, L.W.; Wang, S.C. Factors Associated with Poor Outcomes in Patients Undergoing Endovascular Therapy for Acute Ischemic Stroke Due to Large-Vessel Occlusion in Acute Anterior Circulation: A Retrospective Study. World Neurosurg. 2021, 149, e128–e134. [Google Scholar] [CrossRef]
- Fischer, U.; Kaesmacher, J.; Strbian, D.; Eker, O.; Cognard, C.; Plattner, P.S.; Bütikofer, L.; Mordasini, P.; Deppeler, S.; Pereira, V.M.; et al. Thrombectomy Alone versus Intravenous Alteplase plus Thrombectomy in Patients with Stroke: An Open-Label, Blinded-Outcome, Randomised Non-Inferiority Trial. Lancet 2022, 400, 104–115. [Google Scholar] [CrossRef]
- Mitchell, P.J.; Yan, B.; Churilov, L.; Dowling, R.J.; Bush, S.J.; Bivard, A.; Huo, X.C.; Wang, G.; Zhang, S.Y.; Ton, M.D.; et al. Endovascular Thrombectomy versus Standard Bridging Thrombolytic with Endovascular Thrombectomy within 4·5 h of Stroke Onset: An Open-Label, Blinded-Endpoint, Randomised Non-Inferiority Trial. Lancet 2022, 400, 116–125. [Google Scholar] [CrossRef]
- Albers, G.W.; Marks, M.P.; Kemp, S.; Christensen, S.; Tsai, J.P.; Ortega-Gutierrez, S.; McTaggart, R.A.; Torbey, M.T.; Kim-Tenser, M.; Leslie-Mazwi, T.; et al. Thrombectomy for Stroke at 6 to 16 Hours with Selection by Perfusion Imaging. N. Engl. J. Med. 2018, 378, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Jovin, T.G.; Saver, J.L.; Ribo, M.; Pereira, V.; Furlan, A.; Bonafe, A.; Baxter, B.; Gupta, R.; Lopes, D.; Jansen, O.; et al. Diffusion-Weighted Imaging or Computerized Tomography Perfusion Assessment with Clinical Mismatch in the Triage of Wake up and Late Presenting Strokes Undergoing Neurointervention with Trevo (DAWN) Trial Methods. Int. J. Stroke 2017, 12, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Powers, W.J.; Rabinstein, A.A.; Ackerson, T.; Adeoye, O.M.; Bambakidis, N.C.; Becker, K.; Biller, J.; Brown, M.; Demaerschalk, B.M.; Hoh, B.; et al. Guidelines for the Early Management of Patients with Acute Ischemic Stroke: 2019 Update to the 2018 Guidelines for the Early Management of Acute Ischemic Stroke: A Guideline for Healthcare Professionals from the American Heart Association/American Stroke Association. Stroke 2019, 50, E344–E418. [Google Scholar] [CrossRef] [PubMed]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and Stoichiometric Analysis of the MicroRNA Content of Exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef]
- Krylova, S.V.; Feng, D. The Machinery of Exosomes: Biogenesis, Release, and Uptake. Int. J. Mol. Sci. 2023, 24, 1337. [Google Scholar] [CrossRef]
- Hong, S.B.; Yang, H.; Manaenko, A.; Lu, J.; Mei, Q.; Hu, Q. Potential of Exosomes for the Treatment of Stroke. Cell Transpl. 2019, 28, 662–670. [Google Scholar] [CrossRef]
- Sun, X.; Jung, J.H.; Arvola, O.; Santoso, M.R.; Giffard, R.G.; Yang, P.C.; Stary, C.M. Stem Cell-Derived Exosomes Protect Astrocyte Cultures from In Vitro Ischemia and Decrease Injury as Post-Stroke Intravenous Therapy. Front. Cell Neurosci. 2019, 13, 458548. [Google Scholar] [CrossRef]
- Sun, A.; Blecharz-Lang, K.G.; Małecki, A.; Meybohm, P.; Nowacka-Chmielewska, M.M.; Burek, M.; Decleces, X.; Vazquez Villasenor, I. Role of MicroRNAs in the Regulation of Blood-Brain Barrier Function in Ischemic Stroke and Under Hypoxic Conditions In Vitro. Front. Drug Deliv. 2022, 2, 1027098. [Google Scholar] [CrossRef]
- Vemuganti, R. All’s Well That Transcribes Well: Non-Coding RNAs and Post-Stroke Brain Damage. Neurochem. Int. 2013, 63, 438–449. [Google Scholar] [CrossRef]
- Long, G.; Wang, F.; Li, H.; Yin, Z.; Sandip, C.; Lou, Y.; Wang, Y.; Chen, C.; Wang, D.W. Circulating MiR-30a, MiR-126 and Let-7b as Biomarker for Ischemic Stroke in Humans. BMC Neurol. 2013, 13, 178. [Google Scholar] [CrossRef]
- Shah, J.S.; Soon, P.S.; Marsh, D.J. Comparison of Methodologies to Detect Low Levels of Hemolysis in Serum for Accurate Assessment of Serum MicroRNAs. PLoS ONE 2016, 11, e0153200. [Google Scholar] [CrossRef]
- Motameny, S.; Wolters, S.; Nürnberg, P.; Schumacher, B. Next Generation Sequencing of MiRNAs—Strategies, Resources and Methods. Genes 2010, 1, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Weisscher, N.; Vermeulen, M.; Roos, Y.B.; de Haan, R.J. What Should Be Defined as Good Outcome in Stroke Trials; a Modified Rankin Score of 0-1 or 0-2? J. Neurol. 2008, 255, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Fan, Y.; Tang, X.; Vitriol, E.; Chen, G.; Zheng, J.Q. Actin Capping Protein Is Required for Dendritic Spine Development and Synapse Formation. J. Neurosci. 2011, 31, 10228–10233. [Google Scholar] [CrossRef]
- Yu, W.; Cook, C.; Sauter, C.; Kuriyama, R.; Kaplan, P.L.; Baas, P.W. Depletion of a Microtubule-Associated Motor Protein Induces the Loss of Dendritic Identity. J Neurosci 2000, 20, 5782–5791. [Google Scholar] [CrossRef] [PubMed]
- Spence, E.F.; Kanak, D.J.; Carlson, B.R.; Soderling, S.H. The Arp2/3 Complex Is Essential for Distinct Stages of Spine Synapse Maturation, Including Synapse Unsilencing. J. Neurosci. 2016, 36, 9696. [Google Scholar] [CrossRef] [PubMed]
- Leon, G.; Sanchez-Ruiloba, L.; Perez-Rodriguez, A.; Gragera, T.; Martinez, N.; Hernandez, S.; Anta, B.; Calero, O.; Garcia-Dominguez, C.A.; Dura, L.M.; et al. Shoc2/Sur8 Protein Regulates Neurite Outgrowth. PLoS ONE 2014, 9, e114837. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, V.T.; Ramos-Fernández, E.; Henríquez, J.P.; Lorenzo, A.; Inestrosa, N.C. Wnt-5a/Frizzled9 Receptor Signaling through the Gαo-Gβγ Complex Regulates Dendritic Spine Formation. J. Biol. Chem. 2016, 291, 19092–19107. [Google Scholar] [CrossRef]
- Zedde, M.; Grisendi, I.; Assenza, F.; Napoli, M.; Moratti, C.; Di Cecco, G.; D’Aniello, S.; Valzania, F.; Pascarella, R. Stroke-Induced Secondary Neurodegeneration of the Corticospinal Tract—Time Course and Mechanisms Underlying Signal Changes in Conventional and Advanced Magnetic Resonance Imaging. J. Clin. Med. 2024, 13, 1969. [Google Scholar] [CrossRef]
- Sahni, V.; Itoh, Y.; Shnider, S.J.; Macklis, J.D. Crim1 and Kelch-like 14 Exert Complementary Dual-Directional Developmental Control over Segmentally Specific Corticospinal Axon Projection Targeting. Cell Rep. 2021, 37, 109842. [Google Scholar] [CrossRef]
- Wu, Q.; Yuan, K.; Yao, Y.; Yao, J.; Shao, J.; Meng, Y.; Wu, P.; Shi, H. LAMC1 Attenuates Neuronal Apoptosis via FAK/PI3K/AKT Signaling Pathway After Subarachnoid Hemorrhage. Exp. Neurol. 2024, 376, 114776. [Google Scholar] [CrossRef]
- Zhai, Y.; Ye, S.Y.; Wang, Q.S.; Xiong, R.P.; Fu, S.Y.; Du, H.; Xu, Y.W.; Peng, Y.; Huang, Z.Z.; Yang, N.; et al. Overexpressed Ski Efficiently Promotes Neurorestoration, Increases Neuronal Regeneration, and Reduces Astrogliosis After Traumatic Brain Injury. Gene Ther. 2022, 30, 75–87. [Google Scholar] [CrossRef]
- Hacisuleyman, E.; Hale, C.R.; Noble, N.; Luo, J.D.; Fak, J.J.; Saito, M.; Chen, J.; Weissman, J.S.; Darnell, R.B. Neuronal Activity Rapidly Reprograms Dendritic Translation via EIF4G2:UORF Binding. Nat. Neurosci. 2024, 27, 822–835. [Google Scholar] [CrossRef]
- Cheng, J.; Uchida, M.; Zhang, W.; Grafe, M.R.; Herson, P.S.; Hurn, P.D. Role of Salt-Induced Kinase 1 in Androgen Neuroprotection against Cerebral Ischemia. J. Cereb. Blood Flow. Metab. 2011, 31, 339–350. [Google Scholar] [CrossRef]
- Mohanan, A.; Deshpande, S.; Jamadarkhana, P.G.; Kumar, P.; Gupta, R.C.; Chauthaiwale, V.; Dutt, C. Delayed Intervention in Experimental Stroke with TRC051384—A Small Molecule HSP70 Inducer. Neuropharmacology 2011, 60, 991–999. [Google Scholar] [CrossRef]
- Zhan, X.; Kim, C.; Sharp, F.R. Very Brief Focal Ischemia Simulating Transient Ischemic Attacks (TIAs) Can Injure Brain and Induce Hsp70 Protein. Brain Res. 2008, 1234, 183–197. [Google Scholar] [CrossRef]
- Wang, P.; Shao, B.Z.; Deng, Z.; Chen, S.; Yue, Z.; Miao, C.Y. Autophagy in Ischemic Stroke. Prog. Neurobiol. 2018, 163–164, 98–117. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, S.S.; Malik, S.C.; Conforti, P.; di Lin, J.; Chu, Y.H.; Nath, S.; Greulich, F.; Dumbach, M.A.; Uhlenhaut, N.H.; Schachtrup, C. P75 Neurotrophin Receptor Controls Subventricular Zone Neural Stem Cell Migration after Stroke. Cell Tissue Res. 2022, 387, 415–431. [Google Scholar] [CrossRef] [PubMed]
- Xingyong, C.; Xicui, S.; Huanxing, S.; Jingsong, O.; Yi, H.; Xu, Z.; Ruxun, H.; Zhong, P. Upregulation of Myeloid Cell Leukemia-1 Potentially Modulates Beclin-1-Dependent Autophagy in Ischemic Stroke in Rats. BMC Neurosci. 2013, 14, 56. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Han, J.; Li, X.; Zhang, Z.; Wang, D. Identification of the Biological Function of MiR-9 in Spinal Cord Ischemia-Reperfusion Injury in Rats. PeerJ 2021, 9, e11440. [Google Scholar] [CrossRef]
- Yan, C.; Yu, H.; Liu, Y.; Wu, P.; Wang, C.; Zhao, H.; Yang, K.; Shao, Q.; Zhong, Y.; Zhao, W.; et al. C-Abl Tyrosine Kinase-Mediated Neuronal Apoptosis in Subarachnoid Hemorrhage by Modulating the LRP-1-Dependent Akt/GSK3β Survival Pathway. J. Mol. Neurosci. 2021, 71, 2514–2525. [Google Scholar] [CrossRef]
- Motaln, H.; Rogelj, B. The Role of C-Abl Tyrosine Kinase in Brain and Its Pathologies. Cells 2023, 12, 2041. [Google Scholar] [CrossRef]
- Yao, X.; Wang, Y.; Zhang, D. MicroRNA-21 Confers Neuroprotection Against Cerebral Ischemia-Reperfusion Injury and Alleviates Blood-Brain Barrier Disruption in Rats via the MAPK Signaling Pathway. J. Mol. Neurosci. 2018, 65, 43–53. [Google Scholar] [CrossRef]
- Liu, Y.; Shang, G.; Zhang, X.; Liu, F.; Zhang, C.; Li, Z.; Jia, J.; Xu, Y.; Zhang, Z.; Yang, S.; et al. CAMTA1 Gene Affects the Ischemia-Reperfusion Injury by Regulating CCND1. Front. Cell Neurosci. 2022, 16, 868291. [Google Scholar] [CrossRef]
- Tomasevic, G.; Shamloo, M.; Israeli, D.; Wieloch, T. Activation of P53 and Its Target Genes P21WAF1/Cip1 and PAG608/Wig-1 in Ischemic Preconditioning. Mol. Brain Res. 1999, 70, 304–313. [Google Scholar] [CrossRef]
- Shibue, T.; Suzuki, S.; Okamoto, H.; Yoshida, H.; Ohba, Y.; Takaoka, A.; Taniguchi, T. Differential Contribution of Puma and Noxa in Dual Regulation of P53-Mediated Apoptotic Pathways. EMBO J. 2006, 25, 4952–4962. [Google Scholar] [CrossRef]
- Li, X.; An, P.; Han, F.; Yu, M.; Yu, Z.; Li, Y. Silencing of YTHDF1 Attenuates Cerebral Stroke by Inducing PTEN Degradation and Activating the PTEN/AKT/MTOR Pathway. Mol. Biotechnol. 2023, 65, 822–832. [Google Scholar] [CrossRef]
- Schädlich, I.S.; Winzer, R.; Stabernack, J.; Tolosa, E.; Magnus, T.; Rissiek, B. The Role of the ATP-Adenosine Axis in Ischemic Stroke. Semin. Immunopathol. 2023, 45, 347. [Google Scholar] [CrossRef] [PubMed]
- Begum, G.; Yuan, H.; Kahle, K.T.; Li, L.; Wang, S.; Shi, Y.; Shmukler, B.E.; Yang, S.S.; Lin, S.H.; Alper, S.L.; et al. Inhibition of WNK3 Kinase Signaling Reduces Brain Damage and Accelerates Neurological Recovery after Stroke. Stroke 2015, 46, 1956–1965. [Google Scholar] [CrossRef] [PubMed]
- Koester, S.K.; Dougherty, J.D. A Proposed Role for Interactions between Argonautes, MiRISC, and RNA Binding Proteins in the Regulation of Local Translation in Neurons and Glia. J. Neurosci. 2022, 42, 3291–3301. [Google Scholar] [CrossRef] [PubMed]
- Trollmann, R.; Rehrauer, H.; Schneider, C.; Krischke, G.; Huemmler, N.; Keller, S.; Rascher, W.; Gassmann, M. Late-Gestational Systemic Hypoxia Leads to a Similar Early Gene Response in Mouse Placenta and Developing Brain. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 299, 1489–1499. [Google Scholar] [CrossRef]
- Johnson, E.C.B.; Dammer, E.B.; Duong, D.M.; Ping, L.; Zhou, M.; Yin, L.; Higginbotham, L.A.; Guajardo, A.; White, B.; Troncoso, J.C.; et al. Large-Scale Proteomic Analysis of Alzheimer’s Disease Brain and Cerebrospinal Fluid Reveals Early Changes in Energy Metabolism Associated with Microglia and Astrocyte Activation. Nat. Med. 2020, 26, 769–780. [Google Scholar] [CrossRef]
- Hu, Y.; Huang, S.; Shen, T.; Wang, R.; Geng, M.; Wang, Y.; Zheng, Y.; Luo, Y.; Li, S. Prognostic Significance of Plasma VEGFA and VEGFR2 in Acute Ischemic Stroke-a Prospective Cohort Study. Mol. Neurobiol. 2024, 61, 6341–6353. [Google Scholar] [CrossRef]
- Wei, L.; Li, X.; Wei, Q.; Chen, L.; Xu, L.; Zhou, P. Oxidative Stress-Mediated Sprouty-Related Protein with an EVH1 Domain 1 Down-Regulation Contributes to Resisting Oxidative Injury in Microglia. Neuroscience 2023, 526, 13–20. [Google Scholar] [CrossRef]
- Cevik, O.; Baykal, A.T.; Sener, A. Platelets Proteomic Profiles of Acute Ischemic Stroke Patients. PLoS ONE 2016, 11, 158287. [Google Scholar] [CrossRef]
- Al-Qaisi, R.M.; Al-Ani, L.; Al-Halbosiy, M.M.F. Genetic Polymorphism of ITGA2 Gene and the Risk of Heart Attack and Stroke in Al-Anbar Population/Iraq. J. Pharm. Sci. Res. 2018, 10, 2305–2308. [Google Scholar] [CrossRef]
- Lu, J.X.; Lu, Z.Q.; Zhang, S.L.; Zhi, J.; Chen, Z.P.; Wang, W.X. Polymorphism in Integrin ITGA2 Is Associated with Ischemic Stroke and Altered Serum Cholesterol in Chinese Individuals. Balk. Med. J. 2014, 31, 55. [Google Scholar] [CrossRef] [PubMed]
- Mendes, F.R.; Leclerc, J.L.; Liu, L.; Kamat, P.K.; Naziripour, A.; Hernandez, D.; Li, C.; Ahmad, A.S.; Doré, S. Effect of Experimental Ischemic Stroke and PGE2 EP1 Selective Antagonism in Alzheimer’s Disease Mouse Models. J. Alzheimer’s Dis. 2020, 74, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Goulay, R.; Mena Romo, L.; Hol, E.M.; Dijkhuizen, R.M. From Stroke to Dementia: A Comprehensive Review Exposing Tight Interactions Between Stroke and Amyloid-β Formation. Transl. Stroke Res. 2020, 11, 601. [Google Scholar] [CrossRef]
- Kashima, R.; Hata, A. The Role of TGF-β Superfamily Signaling in Neurological Disorders. Acta Biochim. Biophys. Sin. 2018, 50, 106–120. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, Y.; Ma, L.; Wei, Y.; Li, H. Possible Mechanisms of Tau Spread and Toxicity in Alzheimer’s Disease. Front. Cell Dev. Biol. 2021, 9, 707268. [Google Scholar] [CrossRef]
- Montibeller, L.; Tan, L.Y.; Kim, J.K.; Paul, P.; de Belleroche, J. Tissue-Selective Regulation of Protein Homeostasis and Unfolded Protein Response Signalling in Sporadic ALS. J. Cell Mol. Med. 2020, 24, 6055–6069. [Google Scholar] [CrossRef]
- Suzuki, K.; Shinohara, M.; Uno, Y.; Tashiro, Y.; Gheni, G.; Yamamoto, M.; Fukumori, A.; Shindo, A.; Mashimo, T.; Tomimoto, H.; et al. Deletion of B-Cell Translocation Gene 2 (BTG2) Alters the Responses of Glial Cells in White Matter to Chronic Cerebral Hypoperfusion. J. Neuroinflammation 2021, 18, 86. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, T.; Jiang, Z.; Gai, C.; Yu, S.; Xin, D.; Li, T.; Liu, D.; Wang, Z. The MiR-9-5p/CXCL11 Pathway Is a Key Target of Hydrogen Sulfide-Mediated Inhibition of Neuroinflammation in Hypoxic Ischemic Brain Injury. Neural Regen. Res. 2024, 19, 1084. [Google Scholar] [CrossRef]
- Wang, L.; Yao, C.; Chen, J.; Ge, Y.; Wang, C.; Wang, Y.; Wang, F.; Sun, Y.; Dai, M.; Lin, Y.; et al. Γδ T Cell in Cerebral Ischemic Stroke: Characteristic, Immunity-Inflammatory Role, and Therapy. Front. Neurol. 2022, 13, 842212. [Google Scholar] [CrossRef]
- Zou, Y.X.; Zhang, X.H.; Su, F.Y.; Liu, X. Importance of Riboflavin Kinase in the Pathogenesis of Stroke. CNS Neurosci. Ther. 2012, 18, 834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, H.; Zhang, W.; Liu, Y.; Ding, L.; Gong, J.; Ma, R.; Zheng, S.; Zhang, Y. Biomimetic Remodeling of Microglial Riboflavin Metabolism Ameliorates Cognitive Impairment by Modulating Neuroinflammation. Adv. Sci. 2023, 10, 2300180. [Google Scholar] [CrossRef] [PubMed]
- Nasoohi, S.; Ismael, S.; Ishrat, T. Thioredoxin-Interacting Protein (TXNIP) in Cerebrovascular and Neurodegenerative Diseases: Regulation and Implication. Mol. Neurobiol. 2018, 55, 7900–7920. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Cheng, P.; Feng, T.; Xie, Z.; Yang, M.; Chen, Z.; Hu, S.; Han, D.; Chen, W. Nrf2/HO-1 Blocks TXNIP/NLRP3 Interaction via Elimination of ROS in Oxygen-Glucose Deprivation-Induced Neuronal Necroptosis. Brain Res. 2023, 1817, 148482. [Google Scholar] [CrossRef]
- Mao, L.L.; Hao, D.L.; Mao, X.W.; Xu, Y.F.; Huang, T.T.; Wu, B.N.; Wang, L.H. Neuroprotective Effects of Bisperoxovanadium on Cerebral Ischemia by Inflammation Inhibition. Neurosci. Lett. 2015, 602, 120–125. [Google Scholar] [CrossRef]
- Deng, Y.; Huang, P.; Zhang, F.; Chen, T. Association of MicroRNAs With Risk of Stroke: A Meta-Analysis. Front. Neurol. 2022, 13, 865265. [Google Scholar] [CrossRef]
- Eyileten, C.; Wicik, Z.; De Rosa, S.; Mirowska-Guzel, D.; Soplinska, A.; Indolfi, C.; Jastrzebska-Kurkowska, I.; Czlonkowska, A.; Postula, M. Cells MicroRNAs as Diagnostic and Prognostic Biomarkers in Ischemic Stroke-A Comprehensive Review and Bioinformatic Analysis. Cells 2018, 7, 249. [Google Scholar] [CrossRef]
- Otero-Ortega, L.; Alonso-López, E.; Pérez-Mato, M.; Laso-García, F.; Gómez-De Frutos, M.C.; Diekhorst, L.; García-Bermejo, M.L.; Conde-Moreno, E.; Fuentes, B.; de Leciñana, M.A.; et al. Circulating Extracellular Vesicle Proteins and MicroRNA Profiles in Subcortical and Cortical-Subcortical Ischaemic Stroke. Biomedicines 2021, 9, 786. [Google Scholar] [CrossRef]
- Song, P.; Sun, H.; Chen, H.; Wang, Y.; Zhang, Q. Decreased Serum Exosomal MiR-152-3p Contributes to the Progression of Acute Ischemic Stroke. Clin. Lab. 2020, 66, 1615–1622. [Google Scholar] [CrossRef]
- Wang, W.; Sun, G.; Zhang, L.; Shi, L.; Zeng, Y. Circulating MicroRNAs as Novel Potential Biomarkers for Early Diagnosis of Acute Stroke in Humans. J. Stroke Cerebrovasc. Dis. 2014, 23, 2607–2613. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wu, J.; Geng, Y.; Zhang, Y.; Wang, Y.; Wu, J.; Lu, C.; Luo, G.; Zan, J.; Zhang, Y. Identification of Differentially Expressed MicroRNAs Associated with Ischemic Stroke by Integrated Bioinformatics Approaches. Int. J. Genom. 2022, 2022, 9264555. [Google Scholar] [CrossRef] [PubMed]
- van Kralingen, J.C.; McFall, A.; Ord, E.N.J.; Coyle, T.F.; Bissett, M.; McClure, J.D.; McCabe, C.; Macrae, I.M.; Dawson, J.; Work, L.M. Altered Extracellular Vesicle MicroRNA Expression in Ischemic Stroke and Small Vessel Disease. Transl. Stroke Res. 2019, 10, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xu, C.; Chao, D.; Chen, Z.; Li, S.; Shi, M.; Pei, Y.; Dai, Y.; Ji, J.; Ji, Y.; et al. Acute Cerebral Ischemia Increases a Set of Brain-Specific MiRNAs in Serum Small Extracellular Vesicles. Front. Mol. Neurosci. 2022, 15, 874903. [Google Scholar] [CrossRef]
- Eyileten, C.; Jakubik, D.; Shahzadi, A.; Gasecka, A.; van der Pol, E.; De Rosa, S.; Siwik, D.; Gajewska, M.; Mirowska-Guzel, D.; Kurkowska-Jastrzebska, I.; et al. Diagnostic Performance of Circulating MiRNAs and Extracellular Vesicles in Acute Ischemic Stroke. Int. J. Mol. Sci. 2022, 23, 4530. [Google Scholar] [CrossRef]
- Gendosz de Carrillo, D.; Kocikowska, O.; Rak, M.; Krzan, A.; Student, S.; Jędrzejowska-Szypułka, H.; Pawletko, K.; Lasek-Bal, A. The Relevance of Reperfusion Stroke Therapy for MiR-9-3p and MiR-9-5p Expression in Acute Stroke—A Preliminary Study. Int. J. Mol. Sci. 2024, 25, 2766. [Google Scholar] [CrossRef]
- Curtaz, C.J.; Reifschläger, L.; Strähle, L.; Feldheim, J.; Feldheim, J.J.; Schmitt, C.; Kiesel, M.; Herbert, S.L.; Wöckel, A.; Meybohm, P.; et al. Analysis of MicroRNAs in Exosomes of Breast Cancer Patients in Search of Molecular Prognostic Factors in Brain Metastases. Int. J. Mol. Sci. 2022, 23, 3683. [Google Scholar] [CrossRef]
- Wu, J.; Du, K.; Lu, X. Elevated Expressions of Serum MiR-15a, MiR-16, and MiR-17-5p Are Associated with Acute Ischemic Stroke. Int. J. Clin. Exp. Med. 2015, 8, 21071, ISSN:1940-5901/IJCEM0015102. [Google Scholar]
- Zhao, H.; Wang, J.; Gao, L.; Wang, R.; Liu, X.; Gao, Z.; Tao, Z.; Xu, C.; Song, J.; Ji, X.; et al. MiRNA-424 Protects against Permanent Focal Cerebral Ischemia Injury in Mice Involving Suppressing Microglia Activation. Stroke 2013, 44, 1706–1713. [Google Scholar] [CrossRef]
- Guo, C.; Yao, Y.; Li, Q.; Gao, Y.; Cao, H. Expression and Clinical Value of MiR-185 and MiR-424 in Patients with Acute Ischemic Stroke. Int. J. Gen. Med. 2022, 15, 71. [Google Scholar] [CrossRef]
- Tian, C.; Li, Z.; Yang, Z.; Huang, Q.; Liu, J.; Hong, B. Plasma MicroRNA-16 Is a Biomarker for Diagnosis, Stratification, and Prognosis of Hyperacute Cerebral Infarction. PLoS ONE 2016, 11, 166688. [Google Scholar] [CrossRef]
- Wang, Y.; Su, X.; Leung, G.H.D.; Ren, B.; Zhang, Q.; Xiong, Z.; Zhou, J.; Yang, L.; Lu, G.; Chan, W.Y.; et al. Circulating MicroRNAs as Diagnostic Biomarkers for Ischemic Stroke: Evidence from Comprehensive Analysis and Real-World Validation. Int. J. Med. Sci. 2023, 20, 1009. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Wei, G.; Wang, C.; Lu, Y.; Liu, C.; Wang, R.; Shi, X.; Yang, J.; Wei, Y. A Functional Polymorphism in the Promoter of MiR-17-92 Cluster Is Associated with Decreased Risk of Ischemic Stroke. BMC Med. Genom. 2019, 12, 159. [Google Scholar] [CrossRef]
- Ma, Q.; Li, G.; Tao, Z.; Wang, J.; Wang, R.; Liu, P.; Luo, Y.; Zhao, H. Blood MicroRNA-93 as an Indicator for Diagnosis and Prediction of Functional Recovery of Acute Stroke Patients. J. Clin. Neurosci. 2019, 62, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Bao, H.; Zhang, S.; Li, R.; Chen, L.; Zhu, Y. MiR-186-5p Promotes Apoptosis by Targeting IGF-1 in SH-SY5Y OGD/R Model. Int. J. Biol. Sci. 2018, 14, 1791. [Google Scholar] [CrossRef]
- You, J.; Qian, F.; Huang, Y.; Guo, Y.; Lv, Y.; Yang, Y.; Lu, X.; Guo, T.; Wang, J.; Gu, B. LncRNA WT1-AS Attenuates Hypoxia/Ischemia-Induced Neuronal Injury During Cerebral Ischemic Stroke via MiR-186-5p/XIAP Axis. Open Med. 2022, 17, 1338–1349. [Google Scholar] [CrossRef]
- Tiedt, S.; Prestel, M.; Malik, R.; Schieferdecker, N.; Duering, M.; Kautzky, V.; Stoycheva, I.; Böck, J.; Northoff, B.H.; Klein, M.; et al. RNA-Seq Identifies Circulating MIR-125a-5p, MIR-125b-5p, and MIR-143-3p as Potential Biomarkers for Acute Ischemic Stroke. Circ. Res. 2017, 121, 970–980. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, K.; Huang, J.; Zheng, G.; Lv, Y.; Luo, N.; Liang, M.; Huang, L. Upregulated Serum MiR-146b Serves as a Biomarker for Acute Ischemic Stroke. Cell. Physiol. Biochem. 2018, 45, 397–405. [Google Scholar] [CrossRef]
- Salman, A.T.; Shaker, O.; Elshaer, S.S.; Elshafei, A. The Expression Profiling of Serum MiR-92a, MiR-134 and MiR-375 in Acute Ischemic Stroke. Future Sci. OA 2022, 8, FSO829. [Google Scholar] [CrossRef]
- Aldous, E.K.; Toor, S.M.; Parray, A.; Al-Sarraj, Y.; Diboun, I.; Abdelalim, E.M.; Arredouani, A.; El-Agnaf, O.; Thornalley, P.J.; Akhtar, N.; et al. Identification of Novel Circulating MiRNAs in Patients with Acute Ischemic Stroke. Int. J. Mol. Sci. 2022, 23, 3387. [Google Scholar] [CrossRef]
- Ma, Q.; Zhao, H.; Tao, Z.; Wang, R.; Liu, P.; Han, Z.; Ma, S.; Luo, Y.; Jia, J. MicroRNA-181c Exacerbates Brain Injury in Acute Ischemic Stroke. Aging Dis. 2016, 7, 705. [Google Scholar] [CrossRef]
- Jin, F.; Xing, J. Circulating Pro-Angiogenic and Anti-Angiogenic MicroRNA Expressions in Patients with Acute Ischemic Stroke and Their Association with Disease Severity. Neurol. Sci. 2017, 38, 2015–2023. [Google Scholar] [CrossRef] [PubMed]
- Rahmati, M.; Ferns, G.A.; Mobarra, N. The Lower Expression of Circulating MiR-210 and Elevated Serum Levels of HIF-1α in Ischemic Stroke; Possible Markers for Diagnosis and Disease Prediction. J. Clin. Lab. Anal. 2021, 35, e24073. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.L.; He, X.S.; Liu, J.R.; Zheng, C.B.; Wang, Y.T.; Yang, G.Y. Lentivirus-Mediated Overexpression of MicroRNA-210 Improves Long-Term Outcomes After Focal Cerebral Ischemia in Mice. CNS Neurosci. Ther. 2016, 22, 961. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Liu, J.; Wang, Y.; Wang, L.; Weng, S.; Tang, Y.; Zheng, C.; Cheng, Q.; Chen, S.; Yang, G.Y. MicroRNA-210 as a Novel Blood Biomarker in Acute Cerebral Ischemia. Front. Biosci. 2011, 3, 1265–1272. [Google Scholar] [CrossRef]
- Kong, Y.; Li, S.; Cheng, X.; Ren, H.; Zhang, B.; Ma, H.; Li, M.; Zhang, X.A. Brain Ischemia Significantly Alters MicroRNA Expression in Human Peripheral Blood Natural Killer Cells. Front. Immunol. 2020, 11, 502798. [Google Scholar] [CrossRef]
- van Wijk, N.; Zohar, K.; Linial, M. Challenging Cellular Homeostasis: Spatial and Temporal Regulation of MiRNAs. Int. J. Mol. Sci. 2022, 23, 16152. [Google Scholar] [CrossRef]
- Burek, M.; König, A.; Lang, M.; Fiedler, J.; Oerter, S.; Roewer, N.; Bohnert, M.; Thal, S.C.; Blecharz-Lang, K.G.; Woitzik, J.; et al. Hypoxia-Induced MicroRNA-212/132 Alter Blood-Brain Barrier Integrity Through Inhibition of Tight Junction-Associated Proteins in Human and Mouse Brain Microvascular Endothelial Cells. Transl. Stroke Res. 2019, 10, 672–683. [Google Scholar] [CrossRef]
- Teles, R.H.G.; Engelmayr, D.; Meybohm, P.; Burek, M. Isolation of Extracellular Vesicles Using Formulas to Adapt Centrifugation to Different Centrifuges. Methods Mol. Biol. 2024, 2761, 39–48. [Google Scholar] [CrossRef]
- Exosomal miRNAs, A.; Dugandži, A.; Maria Ciaccio, A.; Tuttolomondo, A. Exosomal MiRNAs as Biomarkers of Ischemic Stroke. Brain Sci. 2023, 13, 1647. [Google Scholar] [CrossRef]
- Eskandar, S. Exploring Common Distance Measures for Machine Learning and Data Science: A Comparative | Medium [Internet]. Available online: https://medium.com/@eskandar.sahel/exploring-common-distance-measures-for-machine-learning-and-data-science-a-comparative-analysis-ea0216c93ba3 (accessed on 9 September 2025).
- Hirschberger, S.; Hübner, M.; Strauß, G.; Effinger, D.; Bauer, M.; Weis, S.; Giamarellos-Bourboulis, E.J.; Kreth, S. Identification of Suitable Controls for MiRNA Quantification in T-Cells and Whole Blood Cells in Sepsis. Sci. Rep. 2019, 9, 15735. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the Evolution of Gene Function, and Other Gene Attributes, in the Context of Phylogenetic Trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.; Mi, H. PANTHER: Making Genome-scale Phylogenetics Accessible to All. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Grissa, D.; Junge, A.; Oprea, T.I.; Jensen, L.J. Diseases 2.0: A Weekly Updated Database of Disease-Gene Associations from Text Mining and Data Integration. Database 2022, 2022, baac019. [Google Scholar] [CrossRef]
- Agrawal, A.; Balcı, H.; Hanspers, K.; Coort, S.L.; Martens, M.; Slenter, D.N.; Ehrhart, F.; Digles, D.; Waagmeester, A.; Wassink, I.; et al. WikiPathways 2024: Next Generation Pathway Database. Nucleic Acids Res. 2024, 52, D679–D689. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for Inferring Cellular Functions from Protein Sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG Mapping Tools for Uncovering Hidden Features in Biological Data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef]
- Nesterova, A.P.; Klimov, E.A.; Zharkova, M.; Sozin, S.; Sobolev, V.; Ivanikova, N.V.; Shkrob, M.; Yuryev, A. Disease Pathways: An Atlas of Human Disease Signaling Pathways. In Disease Pathways: An Atlas of Human Disease Signaling Pathways; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–704. [Google Scholar] [CrossRef]
- Wiebe, D.S.; Omelyanchuk, N.A.; Mukhin, A.M.; Grosse, I.; Lashin, S.A.; Zemlyanskaya, E.V.; Mironova, V.V. Fold-Change-Specific Enrichment Analysis (FSEA): Quantification of Transcriptional Response Magnitude for Functional Gene Groups. Genes 2020, 11, 434. [Google Scholar] [CrossRef]
| Baseline Demographic and Clinical Characteristics | |
|---|---|
| Cohort, n | 72 |
| Age, mean., med. [ref.] | 70, 72.5 [35–93] |
| Gender M/F | 37/35 |
| BMI (kg/m2), med. [ref.] | 27.5 [17–36] |
| Medical history | |
| Atrial Fibrillation | 28 (39%) |
| Arterial Hypertension | 65 (90%) |
| Diabetes Mellitus | 25 (35%) |
| Coronary Artery Disease | 21 (29%) |
| Peripheral Artery Disease | 25 (35%) |
| Lipid Disorders | 31 (43%) |
| Smoking | 18 (25%) |
| Stroke Characteristics | |
| Occluded artery | |
| Left Middle cerebral artery | 23 (32%) |
| Right Middle cerebral artery | 14 (19%) |
| Left Internal carotid artery | 4 (6%) |
| Basilar artery | 1 (1%) |
| No thrombus | 30 (42%) |
| Circulation territory (Oxfordshire community stroke project) | |
| Total anterior cerebral artery | 20 (28%) |
| Partial anterior cerebral artery | 23 (32%) |
| Lacunar infarct | 23 (32%) |
| Posterior circulation infarct | 6 (8%) |
| Stroke etiology (TOAST-trial of ORG 10172 in acute stroke treatment) | |
| Atherosclerosis | 23 (32%) |
| Cardioembolism | 29 (40%) |
| Small vessel occlusion | 14 (19%) |
| Unknown/others origin of stroke | 6 (8%) |
| Cohort Treatment | |
| Antithrombotic therapy (aspirin) | 19 (26%) |
| Reperfusion therapy | |
| rt-Pa | 17 (24%) |
| MT | 9 (12.5%) |
| rt-Pa + MT | 27 (27.5%) |
| Mechanical Thrombectomy | |
| MT (n = 36) Stent retriever Aspiration | 25 (69%) 11 (31%) |
| Stroke onset-groin puncture, mean [ref.] min. | 262 [140–360] |
| TICI (Treatment in cerebral infarction) | |
| 0 | 4 (11%) |
| 1 | 0 (0%) |
| 2b | 1 (3%) |
| 2c | 2 (5.5%) |
| 3 | 29 (80.5%) |
| Successful recanalization (TICI 2b/2c/3) | 32 (89%) |
| Blood Tests | |
| Day 1 [normal range] | |
| RBC_1 | 4.33 × 106/µL [4.00–5.00] |
| WBC_1 | 9.68 × 103/µL [4.00–10.00] |
| Lymphocyte_1 | 1.77 × 103/µL [1.00–4.50] |
| Neutrophile_1 | 6.11 × 103/µL [2.00–6.14] |
| Basophile_1 | 0.03 × 103/µL [0.00–0.10] |
| Eosinophile_1 | 0.07 × 103/µL [0.05–0.50] |
| PLT_1 | 200 × 103/µL [135–350] |
| HCT_1 | 37.93% [36.00–47.00] |
| Hb_1 | 13.19 g/dL [12.00–16.00] |
| creatinine | 2.39 mg/dL [0.51–0.95] |
| eGFR | 73 mL/min/1.73 m2 [>60] |
| CRP | 13 mg/L [<5.0] |
| Day 10 [normal range] | |
| RBC_10 | 4.36 × 106/µL [4.00–5.00] |
| WBC_10 | 7.94 × 106/µL [4.00–10.00] |
| PLT_10 | 272 × 103/µL [135–350] |
| HCT_10 | 38.17% [36.00–47.00] |
| Hb_10 | 13.12 g/dL [12.00–16.00] |
| Functional Outcome | |
| NIHSS day 1, med. [ref.] | 9 [0–28] |
| NIHSS day 2 | 4 [0–28] |
| NIHSS day 10 | 2 [0–24] |
| 10-day mRS, med. [ref.] | 2 [0–6] |
| 90-day mRS | 1 [0–6] |
| TREATMENT (Mean Ct level) | ||||||||
|---|---|---|---|---|---|---|---|---|
| miRNA | Aspirin | rt-PA | MT | rt-PA + MT | ||||
| Day | Day | Day | Day | |||||
| 1st | 10th | 1st | 10th | 1st | 10th | 1st | 10th | |
| let-7g-5p | 4.90 | 4.83 | 5.08 | 4.80 | 5.02 | 4.45 | 5.21 | 4.60 |
| miR-15a-5p | −0.07 | −0.17 | −0.17 | −0.29 | −0.43 | −0.71 | −0.08 | −0.99 * |
| miR-16-5p | −1.02 | −1.30 | −1.15 | −1.29 | −1.23 | −1.38 | −0.76 | −1.80 * |
| miR-17-5p | 1.15 | 1.17 | 1.25 | 1.10 | 1.40 | 0.67 | 1.46 | 0.61 * |
| miR-20a-5p | 1.22 | 1.10 | 1.37 | 1.07 | 1.21 | 0.82 | 1.73 | 0.78 * |
| miR-21-5p | 0.64 | 0.78 | 0.87 | 0.68 | 0.27 | −0.16 | 0.38 | −0.10 |
| miR-23a-3p | 0.99 | 0.90 | 0.72 | 0.53 | 0.66 | −0.14 | 0.53 | 0.22 |
| miR-26b-5p | −0.09 | 0.00 | 0.32 | −0.54 | 0.11 | −0.78 | 0.17 | −0.54 |
| miR-30b-5p | 1.37 | 1.13 | 1.41 | 1.06 | 1.58 | 0.97 | 1.78 | 1.05 |
| miR-92a-3p | −0.51 | −0.65 | −0.58 | −0.80 | −0.70 | −0.71 | −0.40 | −0.96 * |
| miR-93-5p | 3.87 | 3.52 | 3.98 | 3.67 | 3.75 | 3.46 | 3.89 | 3.12 * |
| miR-103a-3p | 3.29 | 2.77 | 3.06 | 3.01 | 3.23 | 2.68 | 3.11 | 2.49 |
| miR-107 | 3.66 | 3.21 | 4.09 | 3.47 | 3.93 | 3.18 | 4.17 | 3.62 |
| miR-125b-5p | 5.84 | 5.86 | 6.11 | 5.59 | 5.46 | 4.90 | 5.24 | 4.77 |
| miR-126-3p | 0.25 | 0.48 | 0.16 | 0.11 | −0.24 | −1.06 | 0.17 | 0.03 |
| miR-130a-3p | 3.17 | 3.69 | 3.25 | 3.33 | 3.59 | 2.93 | 2.86 | 2.57 |
| miR-142-3p | 2.11 | 2.27 | 1.62 | 2.21 | 1.86 | 1.38 | 2.53 | 1.76 |
| miR-143-3p | 3.45 | 3.55 | 3.99 | 3.17 | 3.44 | 2.15 | 2.96 | 2.65 |
| miR-148a-3p | 3.61 | 3.73 | 3.77 | 3.74 | 3.31 | 3.05 | 3.09 | 2.80 |
| miR-150-5p | 2.25 | 2.20 | 2.22 | 1.88 | 3.81 | 1.97 | 2.37 | 2.03 |
| miR-152-3p | 4.31 | 4.64 | 4.25 | 5.21 * | 4.94 | 3.67 | 4.24 | 3.80 |
| miR-153-3p | 8.46 | 8.06 | 8.60 | 8.48 | 7.72 | 7.40 | 8.32 | 8.49 * |
| miR-181c-5p | 6.81 | 6.99 | 6.72 | 6.73 | 6.94 | 6.10 | 7.00 | 6.92 |
| miR-185-5p | 2.07 | 2.03 | 2.02 | 2.05 | 1.97 | 1.99 | 2.45 | 1.74 * |
| miR-186-5p | 4.14 | 4.06 | 4.08 | 3.79 | 4.11 | 3.64 | 4.32 | 3.72 * |
| miR-193b-3p | 6.82 | 6.44 | 6.64 | 6.43 | 5.63 | 5.82 | 5.87 | 5.96 |
| miR-193a-5p | 4.88 | 5.24 | 5.27 | 4.62 | 4.28 | 2.82 | 3.86 | 4.20 |
| miR-199a-3p | 2.69 | 3.02 | 2.89 | 2.62 | 3.04 | 1.78 | 2.53 | 2.37 |
| miR-205-5p | 2.13 | 1.96 | 0.39 | 0.52 | 2.27 | 1.34 | 1.43 | 1.07 |
| miR-210-3p | 4.22 | 4.68 | 4.92 | 4.85 | 4.21 | 4.28 | 4.62 | 4.05 * |
| miR-221-3p | −0.01 | 0.21 | −0.24 | −0.41 | −0.33 | −0.91 | −0.34 | −0.47 |
| miR-222-3p | 6.27 | 6.43 | 6.48 | 6.33 | 5.98 | 5.23 | 6.00 | 5.24 * |
| miR-223-3p | −0.87 | −0.85 | −0.61 | −1.09 | −0.65 | −1.84 | −0.62 | −1.16 |
| miR-224-5p | 9.50 | 8.90 | 9.32 | 8.60 | 8.74 | 7.21 | 8.72 | 8.08 |
| miR-326 | 2.34 | 2.86 | 2.18 | 2.04 | 2.40 | 1.93 | 2.12 | 2.09 |
| miR-339-5p | 3.60 | 3.74 | 3.86 | 3.54 | 3.70 | 3.02 | 3.53 | 3.07 |
| miR-342-3p | 5.20 | 5.26 | 5.51 | 5.45 | 5.77 | 4.76 | 5.56 | 5.06 |
| miR-361-5p | 5.79 | 5.71 | 5.80 | 5.50 | 5.63 | 4.57 | 5.67 | 5.16 |
| miR-376a-3p | 5.98 | 5.60 | 5.58 | 5.28 | 5.24 | 4.85 | 5.63 | 6.23 |
| miR-423-5p | 1.60 | 1.54 | 1.77 | 1.66 | 1.62 | 1.81 | 1.53 | 1.43 |
| miR-424-5p | 2.62 | 2.22 | 2.80 | 2.49 | 2.68 | 1.83 | 2.38 | 1.56 * |
| miR-484 | 1.47 | 1.50 | 1.55 | 1.62 | 1.67 | 1.25 | 1.48 | 1.20 |
| miR-486-5p | −1.46 | −1.62 | −1.42 | −1.29 | −1.60 | −1.28 | −1.08 | −1.80 * |
| miR-505-3p | 6.71 | 6.77 | 6.17 | 6.70 | 4.79 | 5.66 | 6.07 | 5.25 |
| miR-576-5p | 2.97 | 2.78 | 2.82 | 2.35 | 2.76 | 4.25 | 3.34 | 2.81 |
| miR-652-3p | 3.70 | 3.68 | 3.79 | 3.48 | 3.64 | 3.21 | 3.73 | 3.46 |
| miR-744-5p | 2.36 | 2.76 | 2.66 | 2.13 | 2.14 | 1.45 | 2.70 | 2.64 |
| TREATMENT (deltaCt) | ||||
|---|---|---|---|---|
| miRNA | Aspirin | rt-PA | MT | rt-PA + MT |
| let-7g-5p | −0.0694 | −0.2738 | −0.5620 | −0.6083 |
| miR-15a-5p | −0.1049 | −0.1199 | −0.2750 | −0.9136 A*,B* |
| miR-16-5p | −0.2776 | −0.1423 | −0.1535 | −1.0421 B* |
| miR-17-5p | 0.0247 | −0.1487 | −0.7379 | −0.8489 A* |
| miR-20a-5p | −0.1165 | −0.2951 | −0.3943 | −0.9500 |
| miR-21-5p | 0.1431 | −0.1907 | −0.4275 | −0.4769 |
| miR-23a-3p | −0.0880 | −0.1931 | −0.8014 | −0.3126 |
| miR-26b-5p | 0.0941 | −0.8565 | −0.8884 | −0.7090 |
| miR-30b-5p | −0.2360 | −0.2725 | −0.6944 | −0.7352 |
| miR-92a-3p | −0.1393 | −0.2153 | −0.0081 | −0.5579 |
| miR-93-5p | −0.3415 | −0.3073 | −0.2982 | −0.7645 |
| miR-103a-3p | −0.5179 | −0.0470 | −0.5547 | −0.6271 |
| miR-107 | −0.4478 | −0.6237 | −0.7437 | −0.5458 |
| miR-125b-5p | 0.0212 | −0.5198 | −0.5544 | −0.4658 |
| miR-126-3p | 0.2287 | −0.0438 | −0.8228 | −0.1455 |
| miR-130a-3p | 0.5158 | 0.0752 | −0.4787 | −0.8639 |
| miR-142-3p | 0.1614 | 0.5838 | −0.4787 | −0.8639 B* |
| miR-143-3p | 0.0969 | −0.8183 | −1.2904 | −0.3064 |
| miR-148a-3p | 0.1146 | −0.0216 | −0.2655 | −0.2943 |
| miR-150-5p | −0.0463 | −0.3419 | −1.8395 | −0.3348 |
| miR-152-3p | 0.3355 | 0.9672 | −1.2705 D* | −0.4368 B* |
| miR-153-3p | −0.3953 | −0.1152 | −0.3229 | 0.1719 |
| miR-181c-5p | 0.1744 | 0.0159 | −0.8334 | −0.0790 |
| miR-185-5p | −0.0397 | 0.0226 | 0.0298 | −0.7151 |
| miR-186-5p | −0.0813 | −0.2855 | −0.4653 | −0.6009 |
| miR-193b-3p | 0.2956 | −0.0798 | −0.7962 | 0.4360 |
| miR-193a-5p | −0.3834 | −0.2044 | 0.1922 | 0.0928 |
| cmiR-199a-3p | 0.3269 | −0.2650 | −1.2600 | −0.1582 |
| miR-205-5p | 0.4140 | −0.7130 | −0.1341 | −0.5216 |
| miR-210-3p | 0.4585 | −0.0682 | 0.0717 | −0.5635 |
| miR-221-3p | 0.2189 | −0.1704 | −0.5812 | −0.1273 |
| miR-222-3p | 0.1647 | −0.1553 | −0.7543 | −0.5779 |
| miR-223-3p | 0.0165 | −0.4776 | −1.1839 | −0.5409 |
| miR-224-5p | −0.6883 | −0.3592 | −1.5274 | −0.6447 |
| miR-326 | 0.5234 | −0.1400 | −0.4710 | −0.0317 |
| miR-339-5p | 0.1477 | −0.3231 | −0.6830 | −0.4544 |
| miR-342-3p | 0.0605 | −0.0595 | −1.0074 | −0.5013 |
| miR-361-5p | −0.0882 | −0.2986 | −1.0626 | −0.5139 |
| miR-376a-3p | −0.3842 | −0.3047 | −0.3927 | 0.6082 |
| miR-423-5p | −0.0533 | −0.1089 | 0.1881 | −0.0986 |
| miR-424-5p | −0.3941 | −0.3157 | −0.8417 | −0.8297 |
| miR-484 | 0.0297 | 0.0716 | −0.4219 | −0.2730 |
| miR-486-5p | −0.1620 | 0.1333 | 0.3238 | −0.7204 B* |
| miR-505-3p | 0.0578 | 0.5247 | 0.8710 | −0.8124 B*,C* |
| miR-576-5p | −0.2973 | −0.4956 | −0.3756 | −0.7959 |
| miR-652-3p | −0.0238 | −0.3095 | −0.4288 | −0.2717 |
| miR-744-5p | 0.4028 | −0.5303 F* | −0.6964 D* | 0.0582 |
| DEmiRNA Effect via Estimated Targets in Stroke | ||
|---|---|---|
| General biological effect | Potential positive effect of targets on stroke-cell survival | Potential negative effect of targets on stroke-cell death |
| Neurogenesis, neurite outgrowth, neuronal re-networking, synaptic plasticity | CAPZA2 [30], KIF23 [31], ACTR2 [32], SHOC2 [33], FZD9 [34], CRIM1 [35,36], LAMC1 [37], SKI [38], EIFG2 [39] | |
| Oxidative stress response | YTHDC1 79, EIF2B2 80, BZW1 83, MCL1 138 | SIK1 [40] |
| Autophagy | HSPA8 [41,42], ATG14 [43] | |
| Neurite/synapse rejuvenation | ARHGAP12 [44] | |
| Acute/delayed cell death, apoptosis regulation | MCL1 [45], | RAB3IP [46], ABL2 [47,48], MAP2K3 [49], CCDN1 [50], ZMAT3 [51], PMAIP1 [52], PTEN [53] |
| Tissue ions and nucleotide homoeostasis | SLC28A1 [54] | WNK3 [55] |
| Translation regulation | AGO4 [56] | |
| Protein synthesis | RBM12B [57] | RPL14 [58] |
| Angiogenesis | VEGFR2 [59], SPRED1 [60] | |
| Platelets aggregation | CRKL [61], ITGA2 [62,63] | |
| Protein aggregation—dementia | APP [64,65], SMAD7 [66], DNAJC [67,68], | |
| Neuroinflammation | BTG2 [69] 142 | SOC5 [70], BTN3A3 [71], RFK [72,73], TXNIP [74,75], PTEN [76] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gendosz de Carrillo, D.; Kocikowska, O.; Krzan, A.; Student, S.; Rak, M.; Nowak-Andraka, M.; Mi, J.; Burek, M.; Lasek-Bal, A.; Jędrzejowska-Szypułka, H. Reduced Expression of Selected Exosomal MicroRNAs Is Associated with Poor Outcomes in Patients with Acute Stroke Receiving Reperfusion Therapy—Preliminary Study. Int. J. Mol. Sci. 2025, 26, 9533. https://doi.org/10.3390/ijms26199533
Gendosz de Carrillo D, Kocikowska O, Krzan A, Student S, Rak M, Nowak-Andraka M, Mi J, Burek M, Lasek-Bal A, Jędrzejowska-Szypułka H. Reduced Expression of Selected Exosomal MicroRNAs Is Associated with Poor Outcomes in Patients with Acute Stroke Receiving Reperfusion Therapy—Preliminary Study. International Journal of Molecular Sciences. 2025; 26(19):9533. https://doi.org/10.3390/ijms26199533
Chicago/Turabian StyleGendosz de Carrillo, Daria, Olga Kocikowska, Aleksandra Krzan, Sebastian Student, Małgorzata Rak, Magdalena Nowak-Andraka, Junqiao Mi, Małgorzata Burek, Anetta Lasek-Bal, and Halina Jędrzejowska-Szypułka. 2025. "Reduced Expression of Selected Exosomal MicroRNAs Is Associated with Poor Outcomes in Patients with Acute Stroke Receiving Reperfusion Therapy—Preliminary Study" International Journal of Molecular Sciences 26, no. 19: 9533. https://doi.org/10.3390/ijms26199533
APA StyleGendosz de Carrillo, D., Kocikowska, O., Krzan, A., Student, S., Rak, M., Nowak-Andraka, M., Mi, J., Burek, M., Lasek-Bal, A., & Jędrzejowska-Szypułka, H. (2025). Reduced Expression of Selected Exosomal MicroRNAs Is Associated with Poor Outcomes in Patients with Acute Stroke Receiving Reperfusion Therapy—Preliminary Study. International Journal of Molecular Sciences, 26(19), 9533. https://doi.org/10.3390/ijms26199533







