Integrative Transcriptomic and Network-Based Analysis of Neuromuscular Diseases
Abstract
1. Introduction
2. Results
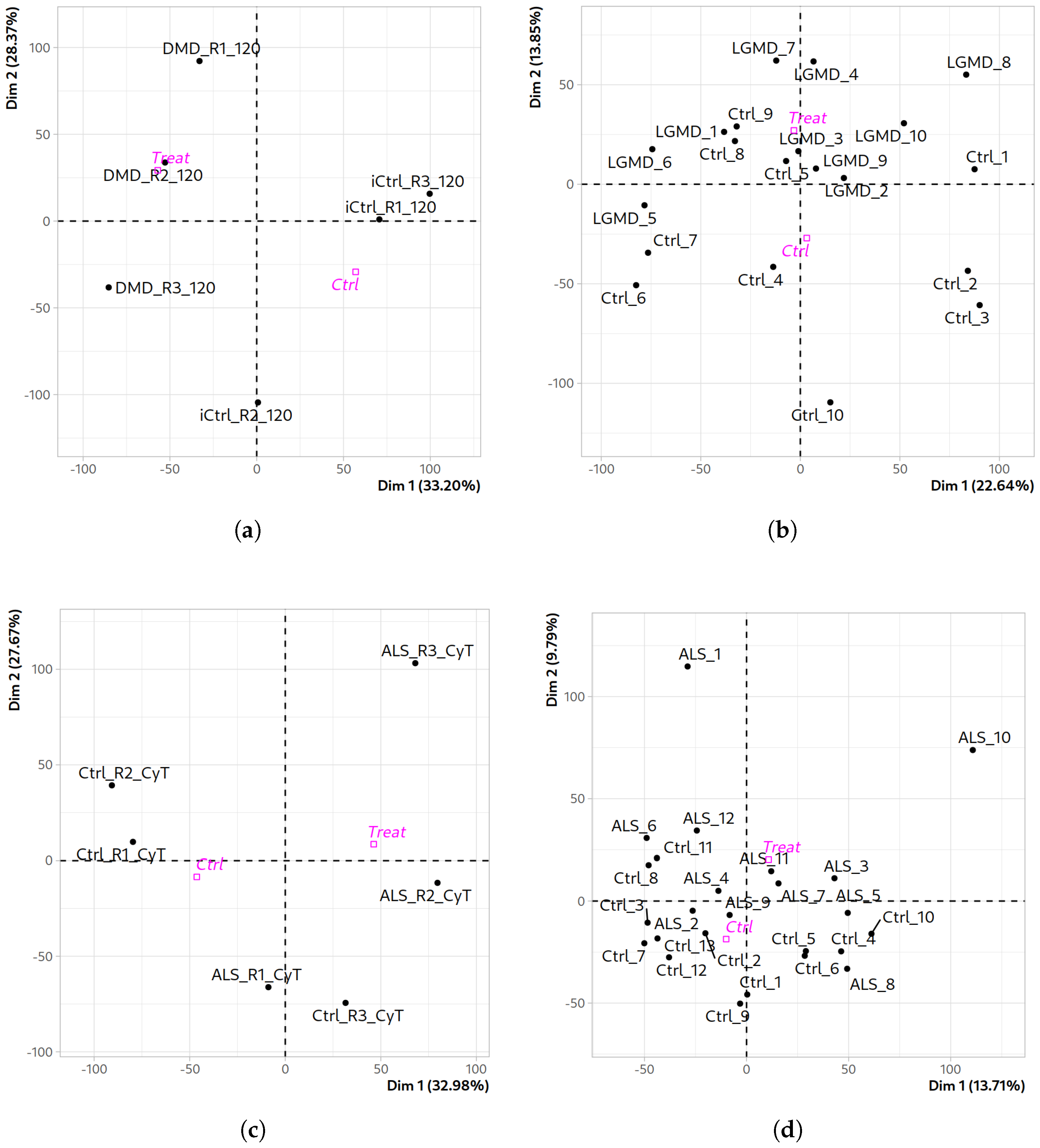
2.1. Quality Assessment and Differential Expression Analysis Across Datasets
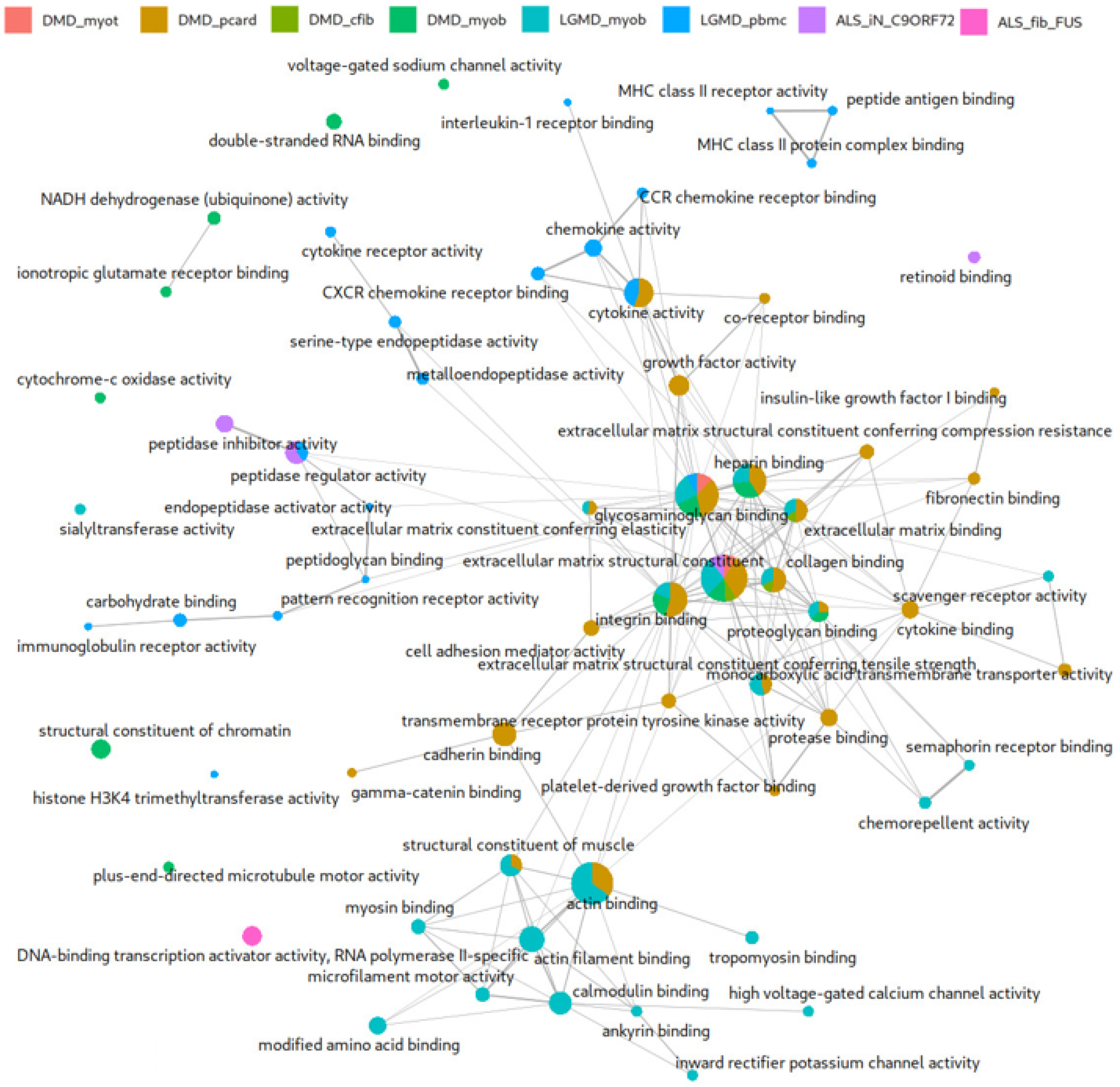
Comparative Functional Profiling Across Neuromuscular Disease Transcriptomes
2.2. Mapping Differential Expression Profiles onto the Human Interactome
Analysis of Unmapped Differentially Expressed Genes
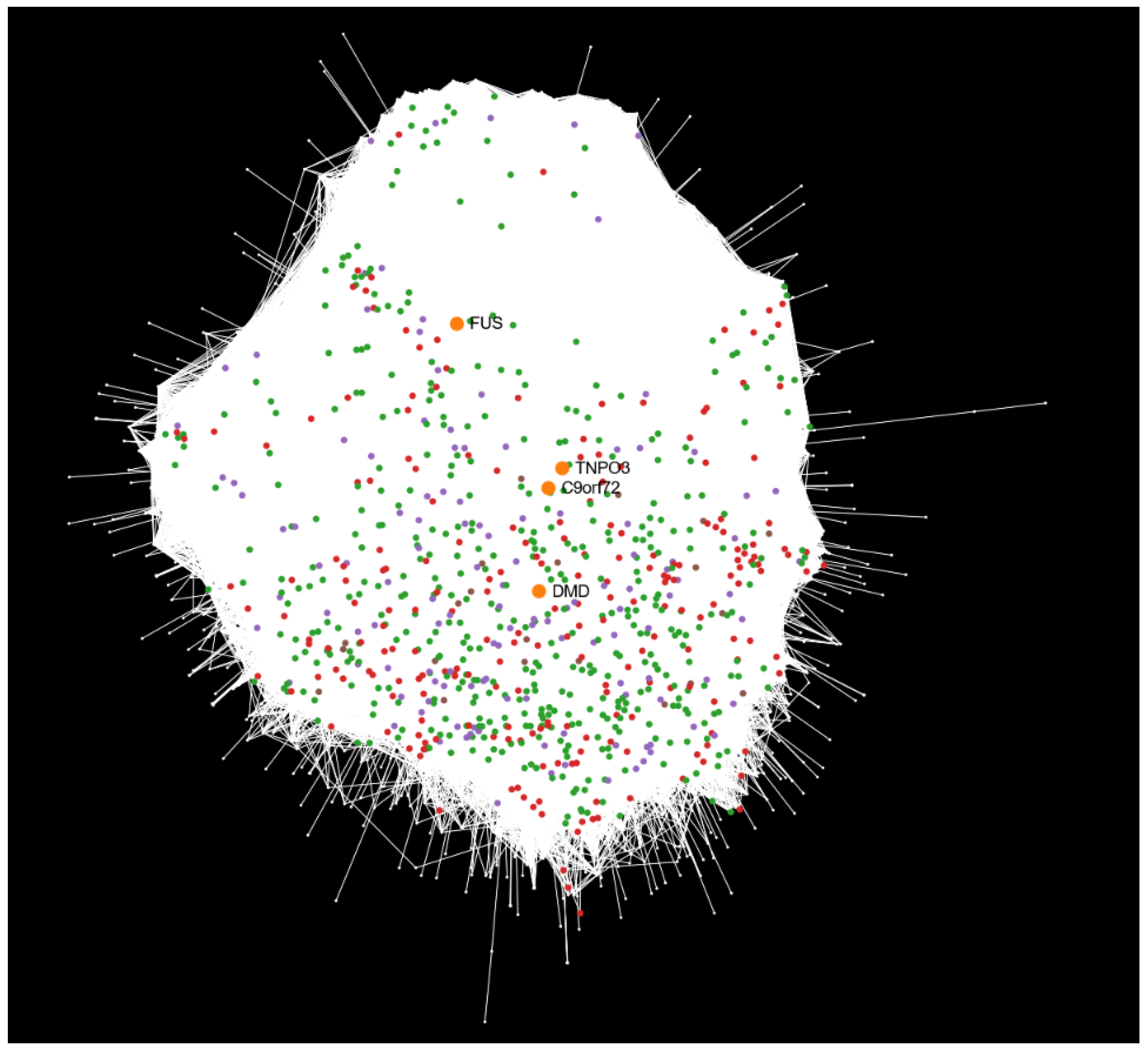
2.3. Mapping Disease-Associated Transcriptional Profiles onto the Human Interactome
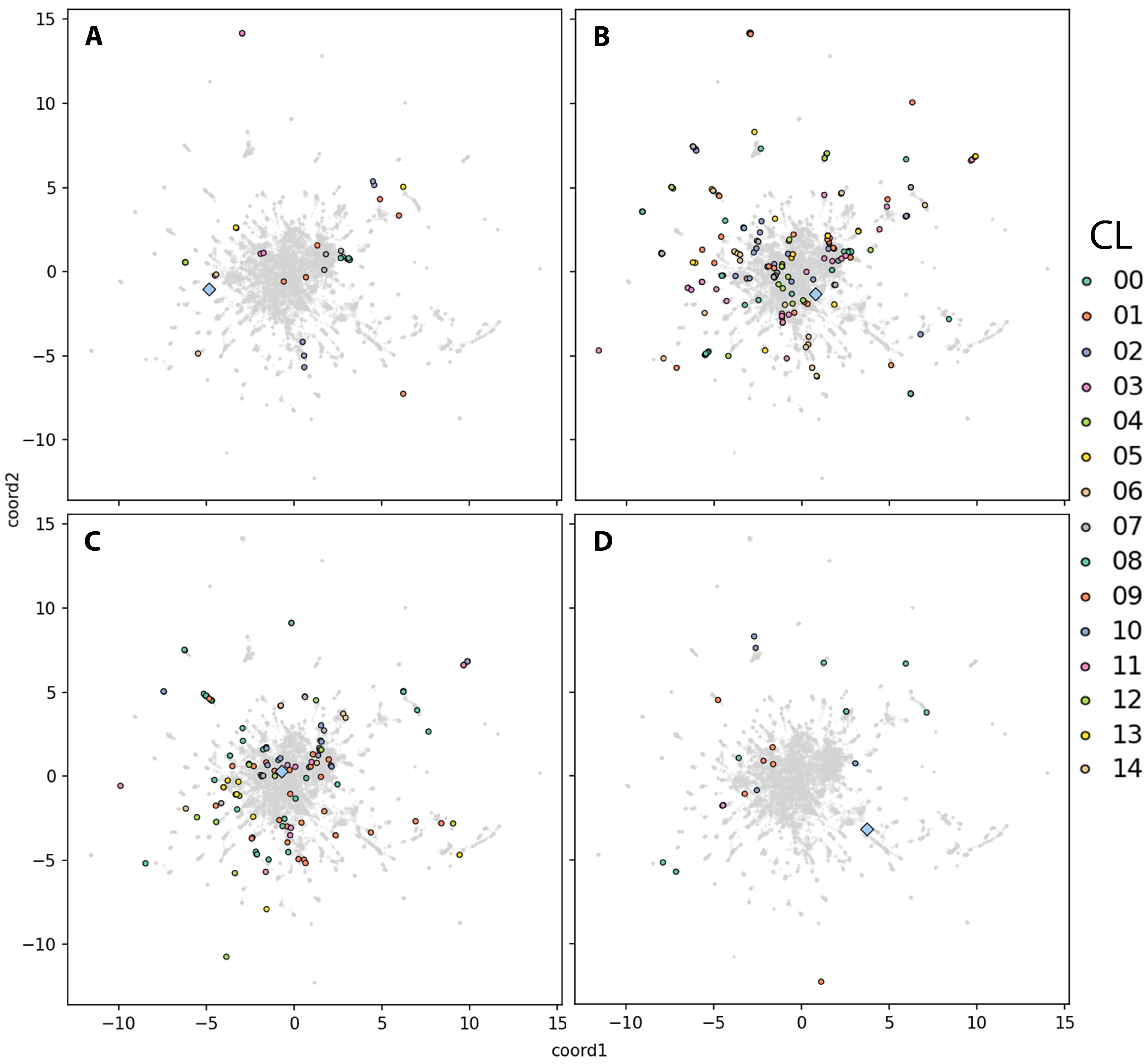
2.4. Embedding-Based Prioritization of DEG Clusters Relative to the Disease-Causal Gene
2.4.1. DEG Cluster Priorititazion and Functional Analysis
2.4.2. Prioritization of Isolated DEGs
3. Discussion
3.1. Differential Expression Analysis Reveals Novel Insights from NMD Dataset-Specific Comparisons
3.2. Comparative Functional Enrichment Extends Original DEG Findings
3.3. Insights from Mapping Differential Expression onto the Human Interactome
3.4. Biological Relevance of Unmapped DEGs
3.5. Integrating Differential Expression Profiles with the Human Interactome to Elucidate Disease Mechanisms
3.6. Biological Relevance and Interpretation of Isolated DEGs
3.7. Study Limitations
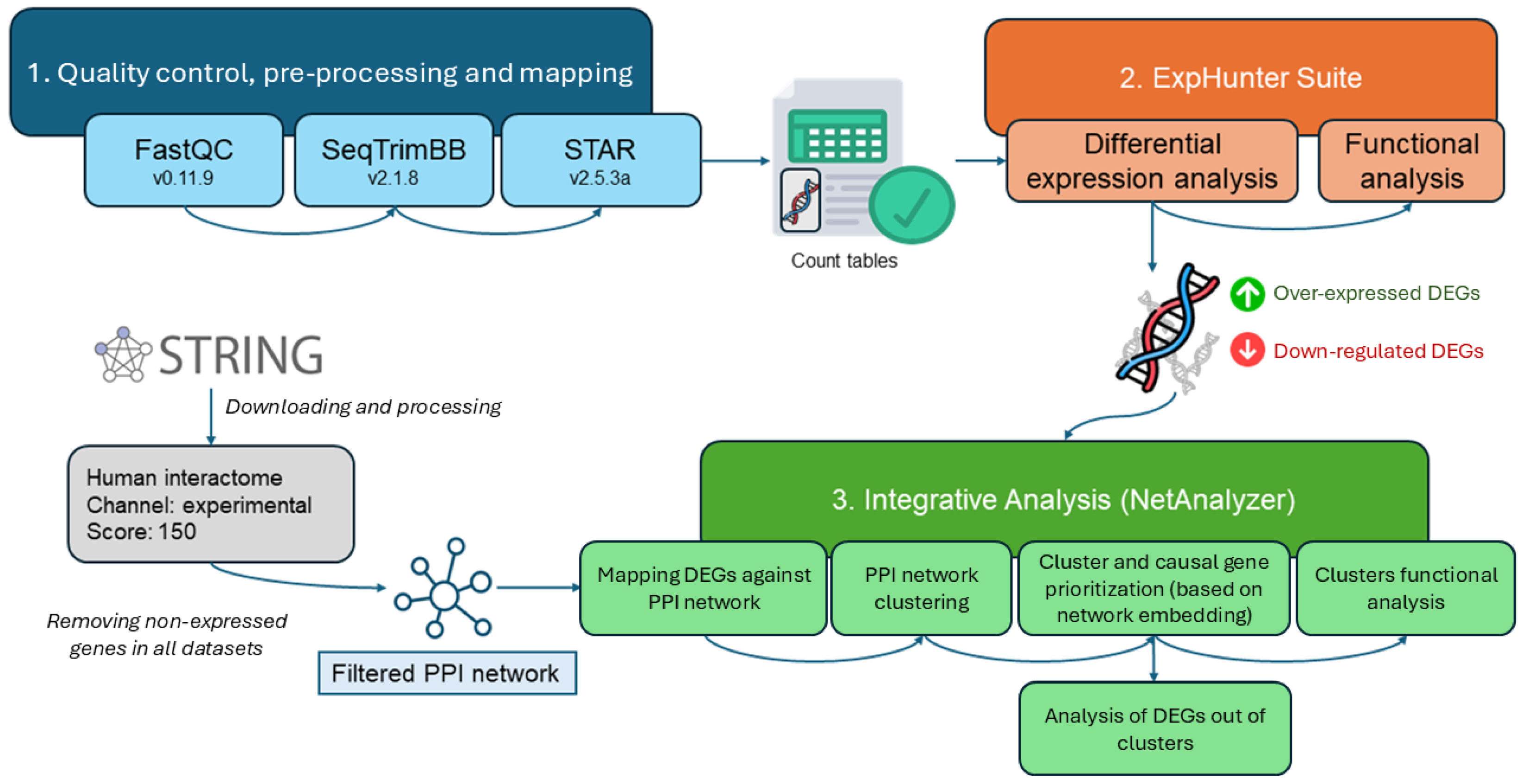
4. Materials and Methods
4.1. Dataset Description
4.2. Dataset Processing for Differential Expression Analysis
4.3. Integrative Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | Amyotrophic Lateral Sclerosis |
| ARS | Average Read Size |
| BH | Benjamini–Hochberg |
| CPM | Counts Per Million |
| CyT | Cytoplasmatic Transcriptome |
| DEG | Differentially Expressed Gene |
| DMD | Duchenne Muscular Dystrophy |
| ECM | Extracellular Matrix |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| HCPC | Hierarchical Clustering on Principal Components |
| hiPSC | Human-Induced Pluripotent Stem Cell |
| iPSCs | Induced Pluripotent Stem Cells |
| LGMD | Limb–Girdle Muscular Dystrophy |
| LGMDD2 | Limb–Girdle Muscular Dystrophy D2 |
| lncRNAs | Long Non-Coding RNAs |
| MLS | Minimum Libraries Selected |
| MRL | Minimum Read Length |
| NCBI | National Center for Biotechnology Information |
| NMDs | Neuromuscular Diseases |
| ORA | Over-Representation Analysis |
| PBMCs | Peripheral Blood Mononuclear Cells |
| PCA | Principal Component Analysis |
| PPI | Protein–Protein Interaction |
| RNA-seq | RNA Sequencing |
| SRA | Sequence Read Archive |
| UMAP | Uniform Manifold Approximation and Projection |
| WCT | Whole-Cell Transcriptome |
References
- Zambon, A.A.; Falzone, Y.M.; Bolino, A.; Previtali, S.C. Molecular mechanisms and therapeutic strategies for neuromuscular diseases. Cell. Mol. Life Sci. CMLS 2024, 81, 198, Correction in Cell. Mol. Life Sci. CMLS 2024, 81, 308. [Google Scholar] [CrossRef]
- Kelchtermans, J.; Mayer, O.H. Year in review 2021: Neuromuscular diseases. Pediatr. Pulmonol. 2023, 58, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, J.M. The Epidemiology of Neuromuscular Diseases. Neurol. Clin. 2016, 34, 999–1021. [Google Scholar] [CrossRef] [PubMed]
- Tasker, R.C.; Darras, B.T. Neuromuscular disorders: From diagnosis to translational research, drug development and clinical trials. Curr. Opin. Pediatr. 2013, 25, 674–675. [Google Scholar] [CrossRef]
- Audag, N.; Goubau, C.; Toussaint, M.; Reychler, G. Screening and evaluation tools of dysphagia in adults with neuromuscular diseases: A systematic review. Ther. Adv. Chronic Dis. 2019, 10, 2040622318821622. [Google Scholar] [CrossRef]
- Holm, A.; Hansen, S.N.; Klitgaard, H.; Kauppinen, S. Clinical advances of RNA therapeutics for treatment of neurological and neuromuscular diseases. RNA Biol. 2022, 19, 594–608. [Google Scholar] [CrossRef]
- Duan, D.; Goemans, N.; Takeda, S.; Mercuri, E.; Aartsma-Rus, A. Duchenne muscular dystrophy. Nat. Rev. Dis. Prim. 2021, 7, 13. [Google Scholar] [CrossRef]
- Feldman, E.L.; Goutman, S.A.; Petri, S.; Mazzini, L.; Savelieff, M.G.; Shaw, P.J.; Sobue, G. Amyotrophic lateral sclerosis. Lancet 2022, 400, 1363–1380. [Google Scholar] [CrossRef]
- Angelini, C. LGMD. Identification, description and classification. Acta Myol. 2020, 39, 207–217. [Google Scholar] [CrossRef]
- Mathur, P.; Kaur, A.; Vijay, U.; Gupta, A.; Agarwal, K.; Agrawal, L. Limb-Girdle Muscular Dystrophies (LGMD): Clinical features, diagnosis and genetic variability through next generation sequencing. Glob. Med Genet. 2025, 12, 100035. [Google Scholar] [CrossRef]
- Dobrescu, M.; Chelu, G.; Tache, D.; Purcaru, S.; Petrescu, I. Differential Diagnosis between Duchenne Muscular Dystrophy and Limb Girdle Muscular Dystrophy 2a. Curr. Health Sci. J. 2015, 41, 385–389. [Google Scholar] [CrossRef]
- Narayanaswami, P.; Weiss, M.; Selcen, D.; David, W.; Raynor, E.; Carter, G.; Wicklund, M.; Barohn, R.J.; Ensrud, E.; Griggs, R.C.; et al. Evidence-based guideline summary: Diagnosis and treatment of limb-girdle and distal dystrophies. Neurology 2014, 83, 1453–1463. [Google Scholar] [CrossRef] [PubMed]
- Doherty, L.; Chaudhry, V. Inpatient Diagnosis and Management of Neuromuscular Disorders. Semin. Neurol. 2021, 41, 493–510. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Santiago, E.; Claros, M.G.; Yahyaoui, R.; de Diego-Otero, Y.; Calvo, R.; Hoenicka, J.; Palau, F.; Ranea, J.A.; Perkins, J.R. Decoding Neuromuscular Disorders Using Phenotypic Clusters Obtained from Co-Occurrence Networks. Front. Mol. Biosci. 2021, 8, 635074. [Google Scholar] [CrossRef]
- Vo, A.H.; McNally, E.M. Modifier Genes and their effect on Duchenne Muscular Dystrophy. Curr. Opin. Neurol. 2015, 28, 528–534. [Google Scholar] [CrossRef]
- Nijs, M.; Van Damme, P. The genetics of amyotrophic lateral sclerosis. Curr. Opin. Neurol. 2024, 37, 560–569. [Google Scholar] [CrossRef]
- Wagner, K.N.; Nagaraja, H.N.; Allain, D.C.; Quick, A.; Kolb, S.J.; Roggenbuck, J. Patients with sporadic and familial amyotrophic lateral sclerosis found value in genetic testing. Mol. Genet. Genom. Med. 2017, 6, 224–229. [Google Scholar] [CrossRef]
- Sturmey, E.; Malaspina, A. Blood biomarkers in ALS: Challenges, applications and novel frontiers. Acta Neurol. Scand. 2022, 146, 375–388. [Google Scholar] [CrossRef]
- Cho, A. Neuromuscular diseases: Genomics-driven advances. Genom. Inform. 2024, 22, 24. [Google Scholar] [CrossRef]
- Marchant, R.G.; Bryen, S.J.; Bahlo, M.; Cairns, A.; Chao, K.R.; Corbett, A.; Davis, M.R.; Ganesh, V.S.; Ghaoui, R.; Jones, K.J.; et al. Genome and RNA sequencing boost neuromuscular diagnoses to 62% from 34% with exome sequencing alone. Ann. Clin. Transl. Neurol. 2024, 11, 1250–1266. [Google Scholar] [CrossRef]
- Lamar, K.M.; McNally, E.M. Genetic Modifiers for Neuromuscular Diseases. J. Neuromuscul. Dis. 2014, 1, 3–13. [Google Scholar] [CrossRef]
- Capobianco, E. RNA-Seq Data: A Complexity Journey. Comput. Struct. Biotechnol. J. 2014, 11, 123–130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, P.; Itan, Y. Biological Network Approaches and Applications in Rare Disease Studies. Genes 2019, 10, 797. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Guan, B.; Dai, L.; Liu, J.x.; Li, F.; Shang, J. Network embedding framework for driver gene discovery by combining functional and structural information. BMC Genom. 2023, 24, 426. [Google Scholar] [CrossRef]
- Kojaku, S.; Radicchi, F.; Ahn, Y.Y.; Fortunato, S. Network community detection via neural embeddings. Nat. Commun. 2024, 15, 9446. [Google Scholar] [CrossRef]
- Kovács, B.; Kojaku, S.; Palla, G.; Fortunato, S. Iterative embedding and reweighting of complex networks reveals community structure. Sci. Rep. 2024, 14, 17184. [Google Scholar] [CrossRef] [PubMed]
- García-Criado, F.; Seoane, P.; Rojano, E.; Ranea, J.A.G.; Perkins, J.R. Advancing edge-based clustering and graph embedding for biological network analysis: A case study in RASopathies. Brief. Bioinform. 2025, 26, bbaf320. [Google Scholar] [CrossRef]
- Costa, R.; Rodia, M.T.; Pacilio, S.; Angelini, C.; Cenacchi, G. LGMD D2 TNPO3-Related: From Clinical Spectrum to Pathogenetic Mechanism. Front. Neurol. 2022, 13, 840683. [Google Scholar] [CrossRef]
- Prudencio, M.; Belzil, V.V.; Batra, R.; Ross, C.A.; Gendron, T.F.; Pregent, L.J.; Murray, M.E.; Overstreet, K.K.; Piazza-Johnston, A.E.; Desaro, P.; et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 2015, 18, 1175–1182. [Google Scholar] [CrossRef]
- Vahsen, B.F.; Nalluru, S.; Morgan, G.R.; Farrimond, L.; Carroll, E.; Xu, Y.; Cramb, K.M.L.; Amein, B.; Scaber, J.; Katsikoudi, A.; et al. C9orf72-ALS human iPSC microglia are pro-inflammatory and toxic to co-cultured motor neurons via MMP9. Nat. Commun. 2023, 14, 5898. [Google Scholar] [CrossRef]
- Kumbier, K.; Roth, M.; Li, Z.; Lazzari-Dean, J.; Waters, C.; Hammerlindl, S.; Rinaldi, C.; Huang, P.; Korobeynikov, V.A.; Phatnani, H.; et al. Identifying FUS amyotrophic lateral sclerosis disease signatures in patient dermal fibroblasts. Dev. Cell 2024, 59, 2134–2142.e6. [Google Scholar] [CrossRef]
- Mariani, D.; Setti, A.; Castagnetti, F.; Vitiello, E.; Stufera Mecarelli, L.; Di Timoteo, G.; Giuliani, A.; D’Angelo, A.; Santini, T.; Perego, E.; et al. ALS-associated FUS mutation reshapes the RNA and protein composition of stress granules. Nucleic Acids Res. 2024, 52, 13269–13289. [Google Scholar] [CrossRef] [PubMed]
- Akçimen, F.; Lopez, E.R.; Landers, J.E.; Nath, A.; Chiò, A.; Chia, R.; Traynor, B.J. Amyotrophic lateral sclerosis: Translating genetic discoveries into therapies. Nat. Rev. Genet. 2023, 24, 642–658. [Google Scholar] [CrossRef]
- Lemos, J.P.; Tenório, L.P.G.; Mouly, V.; Butler-Browne, G.; Mendes-da Cruz, D.A.; Savino, W.; Smeriglio, P. T cell biology in neuromuscular disorders: A focus on Duchenne Muscular Dystrophy and Amyotrophic Lateral Sclerosis. Front. Immunol. 2023, 14, 1202834. [Google Scholar] [CrossRef] [PubMed]
- Toupenet Marchesi, L.; Stockholm, D.; Esteves, T.; Leblanc, M.; Auger, N.; Branchu, J.; El Hachimi, K.H.; Stevanin, G. Transcriptomic analysis reinforces the implication of spatacsin in neuroinflammation and neurodevelopment. Sci. Rep. 2025, 15, 2370. [Google Scholar] [CrossRef] [PubMed]
- Ferrandi, P.J.; Khan, M.M.; Paez, H.G.; Pitzer, C.R.; Alway, S.E.; Mohamed, J.S. Transcriptome Analysis of Skeletal Muscle Reveals Altered Proteolytic and Neuromuscular Junction Associated Gene Expressions in a Mouse Model of Cerebral Ischemic Stroke. Genes 2020, 11, 726. [Google Scholar] [CrossRef]
- Gonorazky, H.; Liang, M.; Cummings, B.; Lek, M.; Micallef, J.; Hawkins, C.; Basran, R.; Cohn, R.; Wilson, M.D.; MacArthur, D.; et al. RNAseq analysis for the diagnosis of muscular dystrophy. Ann. Clin. Transl. Neurol. 2015, 3, 55–60. [Google Scholar] [CrossRef]
- Suárez-Calvet, X.; Fernández-Simón, E.; Natera, D.; Jou, C.; Pinol-Jurado, P.; Villalobos, E.; Ortez, C.; Monceau, A.; Schiava, M.; Codina, A.; et al. Decoding the transcriptome of Duchenne muscular dystrophy to the single nuclei level reveals clinical-genetic correlations. Cell Death Dis. 2023, 14, 596. [Google Scholar] [CrossRef]
- Gittings, L.M.; Alsop, E.B.; Antone, J.; Singer, M.; Whitsett, T.G.; Sattler, R.; Van Keuren-Jensen, K. Cryptic exon detection and transcriptomic changes revealed in single-nuclei RNA sequencing of C9ORF72 patients spanning the ALS-FTD spectrum. Acta Neuropathol. 2023, 146, 433–450. [Google Scholar] [CrossRef]
- Verhaert, D.; Richards, K.; Rafael-Fortney, J.A.; Raman, S.V. Cardiac Involvement in Patients with Muscular Dystrophies: Magnetic Resonance Imaging Phenotype and Genotypic Considerations. Circ. Cardiovasc. Imaging 2011, 4, 67–76. [Google Scholar] [CrossRef]
- Myszczynska, M.; Ferraiuolo, L. New In Vitro Models to Study Amyotrophic Lateral Sclerosis. Brain Pathol. 2016, 26, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.S.; Davis, J.; Lee, G.; Mack, D.L.; Kim, D.H. Muscular dystrophy in a dish: Engineered human skeletal muscle mimetics for disease modeling and drug discovery. Drug Discov. Today 2016, 21, 1387–1398. [Google Scholar] [CrossRef] [PubMed]
- Diez-Fuertes, F.; López-Huertas, M.R.; García-Pérez, J.; Calonge, E.; Bermejo, M.; Mateos, E.; Martí, P.; Muelas, N.; Vílchez, J.J.; Coiras, M.; et al. Transcriptomic Evidence of the Immune Response Activation in Individuals with Limb Girdle Muscular Dystrophy Dominant 2 (LGMDD2) Contributes to Resistance to HIV-1 Infection. Front. Cell Dev. Biol. 2022, 10, 839813. [Google Scholar] [CrossRef] [PubMed]
- Atmanli, A.; Chai, A.C.; Cui, M.; Wang, Z.; Nishiyama, T.; Bassel-Duby, R.; Olson, E.N. Cardiac Myoediting Attenuates Cardiac Abnormalities in Human and Mouse Models of Duchenne Muscular Dystrophy. Circ. Res. 2021, 129, 602–616. [Google Scholar] [CrossRef]
- Castelli, L.M.; Cutillo, L.; Souza, C.D.S.; Sanchez-Martinez, A.; Granata, I.; Lin, Y.H.; Myszczynska, M.A.; Heath, P.R.; Livesey, M.R.; Ning, K.; et al. SRSF1-dependent inhibition of C9ORF72-repeat RNA nuclear export: Genome-wide mechanisms for neuroprotection in amyotrophic lateral sclerosis. Mol. Neurodegener. 2021, 16, 53. [Google Scholar] [CrossRef]
- Lemoine, J.; Dubois, A.; Dorval, A.; Jaber, A.; Warthi, G.; Mamchaoui, K.; Wang, T.; Corre, G.; Bovolenta, M.; Richard, I. Correction of exon 2, exon 2–9 and exons 8–9 duplications in DMD patient myogenic cells by a single CRISPR/Cas9 system. Sci. Rep. 2024, 14, 21238. [Google Scholar] [CrossRef]
- Poyatos-García, J.; Blázquez-Bernal, A.; Selva-Giménez, M.; Bargiela, A.; Espinosa-Espinosa, J.; Vázquez-Manrique, R.P.; Bigot, A.; Artero, R.; Vilchez, J.J. CRISPR-Cas9 editing of a TNPO3 mutation in a muscle cell model of limb-girdle muscular dystrophy type D2. Mol. Ther. Nucleic Acids 2023, 31, 324–338. [Google Scholar] [CrossRef]
- Paredes-Redondo, A.; Harley, P.; Maniati, E.; Ryan, D.; Louzada, S.; Meng, J.; Kowala, A.; Fu, B.; Yang, F.; Liu, P.; et al. Optogenetic modeling of human neuromuscular circuits in Duchenne muscular dystrophy with CRISPR and pharmacological corrections. Sci. Adv. 2021, 7, eabi8787. [Google Scholar] [CrossRef]
- Soussi, S.; Savchenko, L.; Rovina, D.; Iacovoni, J.S.; Gottinger, A.; Vialettes, M.; Pioner, J.M.; Farini, A.; Mallia, S.; Rabino, M.; et al. IPSC derived cardiac fibroblasts of DMD patients show compromised actin microfilaments, metabolic shift and pro-fibrotic phenotype. Biol. Direct 2023, 18, 41. [Google Scholar] [CrossRef]
- Evans, A.D.; Pournoori, N.; Saksala, E.; Oommen, O.P. Glycosaminoglycans’ for brain health: Harnessing glycosaminoglycan based biomaterials for treating central nervous system diseases. Biomaterials 2024, 309, 122629. [Google Scholar] [CrossRef]
- Wang, Q.; Chi, L. The Alterations and Roles of Glycosaminoglycans in Human Diseases. Polymers 2022, 14, 5014. [Google Scholar] [CrossRef]
- Shi, D.; Sheng, A.; Chi, L. Glycosaminoglycan-Protein Interactions and Their Roles in Human Disease. Front. Mol. Biosci. 2021, 8, 639666. [Google Scholar] [CrossRef]
- Negroni, E.; Henault, E.; Chevalier, F.; Gilbert-Sirieix, M.; Van Kuppevelt, T.H.; Papy-Garcia, D.; Uzan, G.; Albanese, P. Glycosaminoglycan Modifications in Duchenne Muscular Dystrophy: Specific Remodeling of Chondroitin Sulfate/Dermatan Sulfate. J. Neuropathol. Exp. Neurol. 2014, 73, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Carmen, L.; Maria, V.; Morales-Medina, J.C.; Vallelunga, A.; Palmieri, B.; Iannitti, T. Role of proteoglycans and glycosaminoglycans in Duchenne muscular dystrophy. Glycobiology 2019, 29, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Sahadevan, S.; Hembach, K.M.; Tantardini, E.; Pérez-Berlanga, M.; Hruska-Plochan, M.; Megat, S.; Weber, J.; Schwarz, P.; Dupuis, L.; Robinson, M.D.; et al. Synaptic FUS accumulation triggers early misregulation of synaptic RNAs in a mouse model of ALS. Nat. Commun. 2021, 12, 3027. [Google Scholar] [CrossRef] [PubMed]
- Nowak, K.; McCullagh, K.; Poon, E.; Davies, K.E. Muscular dystrophies related to the cytoskeleton/nuclear envelope. Novartis Found. Symp. 2005, 264, 98–111; discussion 112–117, 227–230. [Google Scholar]
- Schultz, T.I.; Raucci, F.J.; Salloum, F.N. Cardiovascular Disease in Duchenne Muscular Dystrophy. JACC Basic Transl. Sci. 2022, 7, 608–625. [Google Scholar] [CrossRef]
- Yao, Y.; Yan, C.; Huang, H.; Wang, S.; Li, J.; Chen, Y.; Qu, X.; Bao, Q.; Xu, L.; Zhang, Y.; et al. LncRNA-MEG3 Regulates Muscle Mass and Metabolic Homeostasis by Facilitating SUZ12 Liquid–Liquid Phase Separation. Adv. Sci. 2025, 12, 2417715. [Google Scholar] [CrossRef]
- Piccoli, M.T.; Gupta, S.K.; Viereck, J.; Foinquinos, A.; Samolovac, S.; Kramer, F.L.; Garg, A.; Remke, J.; Zimmer, K.; Batkai, S.; et al. Inhibition of the Cardiac Fibroblast–Enriched lncRNA Meg3 Prevents Cardiac Fibrosis and Diastolic Dysfunction. Circ. Res. 2017, 121, 575–583. [Google Scholar] [CrossRef]
- Zha, W.; Li, X.; Tie, X.; Xing, Y.; Li, H.; Gao, F.; Ye, T.; Du, W.; Chen, R.; Liu, Y. The molecular mechanisms of the long noncoding RNA SBF2-AS1 in regulating the proliferation of oesophageal squamous cell carcinoma. Sci. Rep. 2021, 11, 805. [Google Scholar] [CrossRef]
- Long, A.M.; Lee, G.; Demonbreun, A.R.; McNally, E.M. Extracellular matrix contribution to disease progression and dysfunction in myopathy. Am. J. Physiol.-Cell Physiol. 2023, 325, C1244–C1251. [Google Scholar] [CrossRef]
- Taiana, E.; Ronchetti, D.; Todoerti, K.; Nobili, L.; Tassone, P.; Amodio, N.; Neri, A. LncRNA NEAT1 in Paraspeckles: A Structural Scaffold for Cellular DNA Damage Response Systems? Non-Coding RNA 2020, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Shelkovnikova, T.A.; Kukharsky, M.S.; An, H.; Dimasi, P.; Alexeeva, S.; Shabir, O.; Heath, P.R.; Buchman, V.L. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.G.; Whitehead, N.P.; Froehner, S.C. Absence of Dystrophin Disrupts Skeletal Muscle Signaling: Roles of Ca2+, Reactive Oxygen Species, and Nitric Oxide in the Development of Muscular Dystrophy. Physiol. Rev. 2016, 96, 253–305. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, S.; Öztürk, A.; Hicks, G.G. FUS-regulated RNA metabolism and DNA damage repair. Rare Dis. 2014, 2, e29515. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, X.; Zhang, Q. Cdh11: Roles in different diseases and potential value in disease diagnosis and treatment. Biochem. Biophys. Rep. 2023, 36, 101576. [Google Scholar] [CrossRef]
- Kim, J.W.; Bae, J.H.; Go, G.Y.; Lee, J.R.; Jeong, Y.; Kim, J.Y.; Kim, T.H.; Kim, Y.K.; Han, J.W.; Oh, J.E.; et al. Epsti1 Regulates the Inflammatory Stage of Early Muscle Regeneration through STAT1-VCP Interaction. Int. J. Biol. Sci. 2024, 20, 3530–3543. [Google Scholar] [CrossRef]
- Desai, M.S.; Eblimit, Z.; Thevananther, S.; Kosters, A.; Moore, D.D.; Penny, D.J.; Karpen, S.J. Cardiomyopathy reverses with recovery of liver injury, cholestasis and cholanemia in mouse model of biliary fibrosis. Liver Int. Off. J. Int. Assoc. Study Liver 2015, 35, 1464–1477. [Google Scholar] [CrossRef]
- Chua, J.P.; De Calbiac, H.; Kabashi, E.; Barmada, S.J. Autophagy and ALS: Mechanistic insights and therapeutic implications. Autophagy 2022, 18, 254–282. [Google Scholar] [CrossRef]
- Bischoff, M.E.; Zang, Y.; Chu, J.; Price, A.D.; Ehmer, B.; Talbot, N.J.; Newbold, M.J.; Paul, A.; Guan, J.L.; Plas, D.R.; et al. Selective MAP1LC3C (LC3C) autophagy requires noncanonical regulators and the C-terminal peptide. J. Cell Biol. 2021, 220, e202004182. [Google Scholar] [CrossRef]
- Davis, K.; Azarcon, P.; Hickenlooper, S.; Bia, R.; Horiuchi, E.; Szulik, M.W.; Franklin, S. The role of demethylases in cardiac development and disease. J. Mol. Cell. Cardiol. 2021, 158, 89–100. [Google Scholar] [CrossRef]
- Oliveira, D.; Assoni, A.F.; Alves, L.M.; Sakugawa, A.; Melo, U.S.; Teles e Silva, A.L.; Sertie, A.L.; Caires, L.C.; Goulart, E.; Ghirotto, B.; et al. ALS-associated VRK1 R321C mutation causes proteostatic imbalance and mitochondrial defects in iPSC-derived motor neurons. Neurobiol. Dis. 2024, 198, 106540. [Google Scholar] [CrossRef] [PubMed]
- Crisafulli, S.; Sultana, J.; Fontana, A.; Salvo, F.; Messina, S.; Trifirò, G. Global epidemiology of Duchenne muscular dystrophy: An updated systematic review and meta-analysis. Orphanet J. Rare Dis. 2020, 15, 141. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.L.; Bassel-Duby, R.; Olson, E.N. CRISPR Correction of Duchenne Muscular Dystrophy. Annu. Rev. Med. 2019, 70, 239–255. [Google Scholar] [CrossRef]
- Cordoba-Caballero, J.; Perkins, J.R.; García-Criado, F.; Gallego, D.; Navarro-Sánchez, A.; Moreno-Estellés, M.; Garcés, C.; Bonet, F.; Romá-Mateo, C.; Toro, R.; et al. Exploring miRNA–target gene pair detection in disease with coRmiT. Brief. Bioinform. 2024, 25, bbae060. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap. 2015. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 17 September 2025).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15. [Google Scholar] [CrossRef]
- Jabato, F.M.; Córdoba-Caballero, J.; Rojano, E.; Romá-Mateo, C.; Sanz, P.; Pérez, B.; Gallego, D.; Seoane, P.; Ranea, J.A.; Perkins, J.R. Gene expression analysis method integration and co-expression module detection applied to rare glucide metabolism disorders using ExpHunterSuite. Sci. Rep. 2021, 11, 15062. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Jabato, F.M.; Rojano, E.; Perkins, J.R.; Ranea, J.A.G.; Seoane-Zonjic, P. Kernel Based Approaches to Identify Hidden Connections in Gene Networks Using NetAnalyzer. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2020; pp. 763–774. [Google Scholar] [CrossRef]
- Blondel, V.D.; Guillaume, J.L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Núñez-Carpintero, I.; Rigau, M.; Bosio, M.; O’Connor, E.; Spendiff, S.; Azuma, Y.; Topf, A.; Thompson, R.; ’t Hoen, P.A.C.; Chamova, T.; et al. Rare disease research workflow using multilayer networks elucidates the molecular determinants of severity in Congenital Myasthenic Syndromes. Nat. Commun. 2024, 15, 1227. [Google Scholar] [CrossRef]
- Rojano, E.; Seoane, P.; Bueno-Amoros, A.; Perkins, J.R.; Garcia-Ranea, J.A. Revealing the Relationship Between Human Genome Regions and Pathological Phenotypes Through Network Analysis. In Proceedings of the Bioinformatics and Biomedical Engineering: 5th International Work-Conference, IWBBIO 2017, Granada, Spain, 26–28 April 2017; Springer International Publishing: Cham, Switzerland, 2017. Proceedings, Part I. Volume 10208 LNCS, pp. 197–207, ISBN 9783319561479. [Google Scholar] [CrossRef]
- Grover, A.; Leskovec, J. node2vec: Scalable Feature Learning for Networks. In Proceedings of the KDD’16: The 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 855–864, ISBN 9781450342322. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2020, arXiv:1802.03426. [Google Scholar] [CrossRef]

| Dataset | Expressed Genes | DEGs | Over-Expressed | Down-Regulated |
|---|---|---|---|---|
| DMD_myot | 13,085 | 176 | 137 | 39 |
| DMD_pCard | 12,832 | 437 | 38 | 399 |
| DMD_cfib | 13,157 | 80 | 30 | 50 |
| DMD_myob | 14,466 | 580 | 356 | 224 |
| LGMD_myob | 13,838 | 548 | 199 | 349 |
| LGMD_pbmc | 13,598 | 160 | 108 | 52 |
| ALS_iN_C9ORF72 | 13,453 | 325 | 284 | 41 |
| ALS_fib_FUS | 13,004 | 171 | 96 | 75 |
| Dataset | Mapped DEGs | uDEGs | Unmap. Coding | ncRNA | Pseudo | Clusters |
|---|---|---|---|---|---|---|
| DMD_myot | 34 | 17 | 2 | 13 | 2 | 8 |
| DMD_pCard | 247 | 31 | 6 | 22 | 3 | 17 |
| DMD_cfib | 13 | 14 | 2 | 9 | 3 | 4 |
| DMD_myob | 239 | 164 | 14 | 114 | 30 | 13 |
| LGMD_myob | 206 | 120 | 11 | 85 | 20 | 15 |
| LGMD_pbmc | 19 | 53 | 12 | 19 | 19 | 4 |
| ALS_iN_C9ORF72 | 127 | 50 | 3 | 40 | 2 | 10 |
| ALS_fib_FUS | 21 | 36 | 5 | 25 | 3 | 4 |
| Dataset | Samples | Causal Gene | ARS | MRL | MLS | log2FC | Ref. |
|---|---|---|---|---|---|---|---|
| DMD_myot | 6 (3 isogenic controls + 3 DMD) | DMD | 75 | 60 | 2 | 1.5 | [48] |
| DMD_pCard | 6 (3 CRISPR-Cas9 corrected + 3 DMD) | DMD | 74 | 60 | 2 | 1.5 | [44] |
| DMD_cfib | 8 (4 healthy controls + 4 DMD) | DMD | 100 | 85 | 2 | 1 | [49] |
| DMD_myob | 12 (3 controls + 9 patients) | DMD | 150 | 135 | 2 | 1.5 | [46] |
| LGMD_myob | 6 (3 controls + 3 patients) | TNPO3 | 150 | 135 | 2 | 2.5 | [47] |
| LGMD_pbmc | 20 (10 controls and 10 patients) | TNPO3 | 76 | 61 | 2 | 1 | [43] |
| ALS_iN_C9ORF72 | 12 (6 controls + 6 C9ORF72-ALS) | C9ORF72 | 150 | 135 | 2 | 0.6 | [45] |
| ALS_fib_FUS | 25 (13 controls + 12 FUS-ALS) | FUS | 150 | 135 | 8 | 0.6 | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Criado, F.; Hurtado-García, L.; Rojano, E.; Esteban-Martos, Á.; Pérez-García, J.; Seoane, P.; Ranea, J.A.G. Integrative Transcriptomic and Network-Based Analysis of Neuromuscular Diseases. Int. J. Mol. Sci. 2025, 26, 9376. https://doi.org/10.3390/ijms26199376
García-Criado F, Hurtado-García L, Rojano E, Esteban-Martos Á, Pérez-García J, Seoane P, Ranea JAG. Integrative Transcriptomic and Network-Based Analysis of Neuromuscular Diseases. International Journal of Molecular Sciences. 2025; 26(19):9376. https://doi.org/10.3390/ijms26199376
Chicago/Turabian StyleGarcía-Criado, Federico, Lucia Hurtado-García, Elena Rojano, Álvaro Esteban-Martos, Jesús Pérez-García, Pedro Seoane, and Juan A. G. Ranea. 2025. "Integrative Transcriptomic and Network-Based Analysis of Neuromuscular Diseases" International Journal of Molecular Sciences 26, no. 19: 9376. https://doi.org/10.3390/ijms26199376
APA StyleGarcía-Criado, F., Hurtado-García, L., Rojano, E., Esteban-Martos, Á., Pérez-García, J., Seoane, P., & Ranea, J. A. G. (2025). Integrative Transcriptomic and Network-Based Analysis of Neuromuscular Diseases. International Journal of Molecular Sciences, 26(19), 9376. https://doi.org/10.3390/ijms26199376








