Molecular Dynamics Simulation Reveals the Mechanism of Substrate Recognition by Lignin-Degrading Enzymes
Abstract
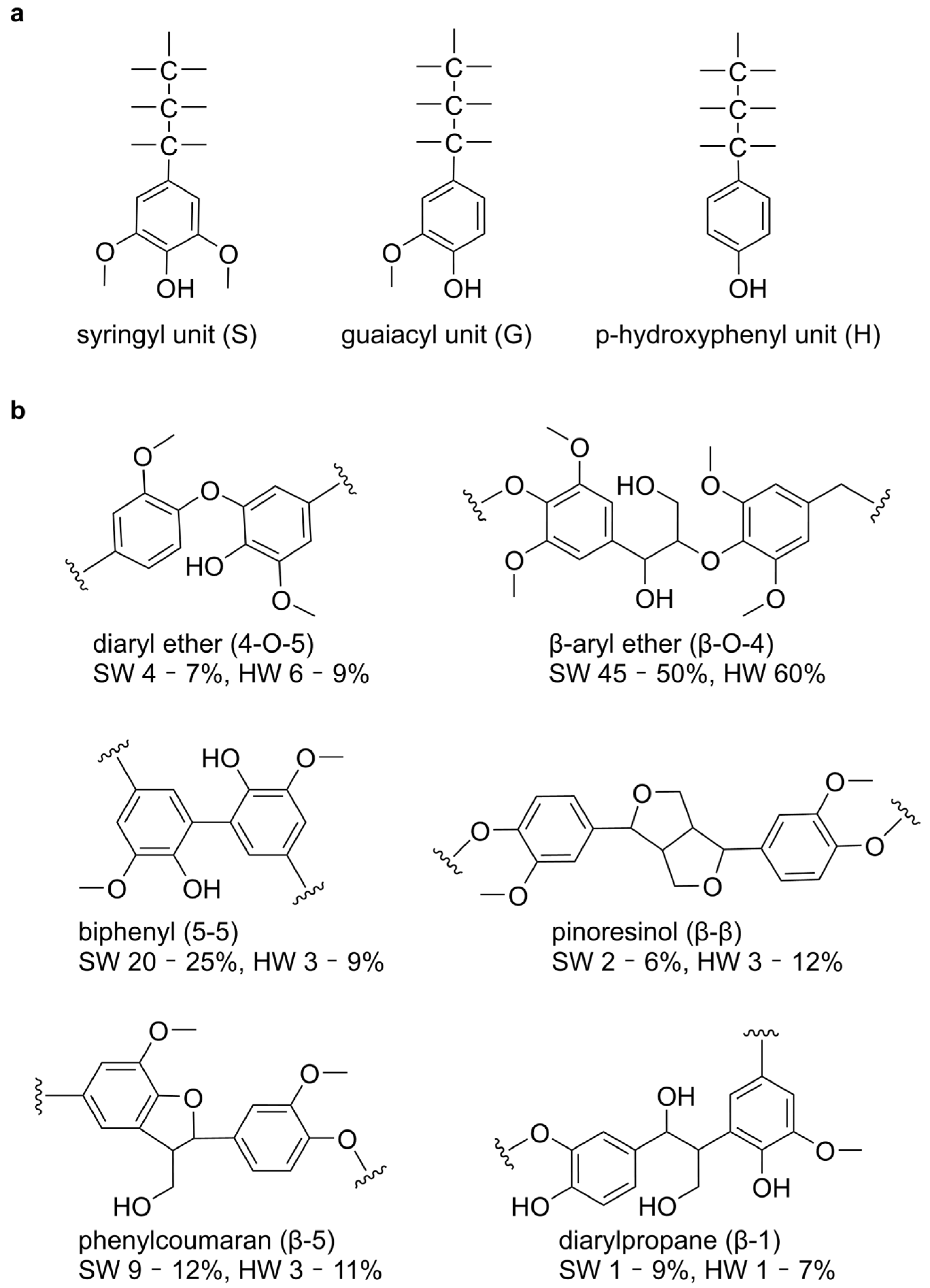
1. Introduction
2. Results
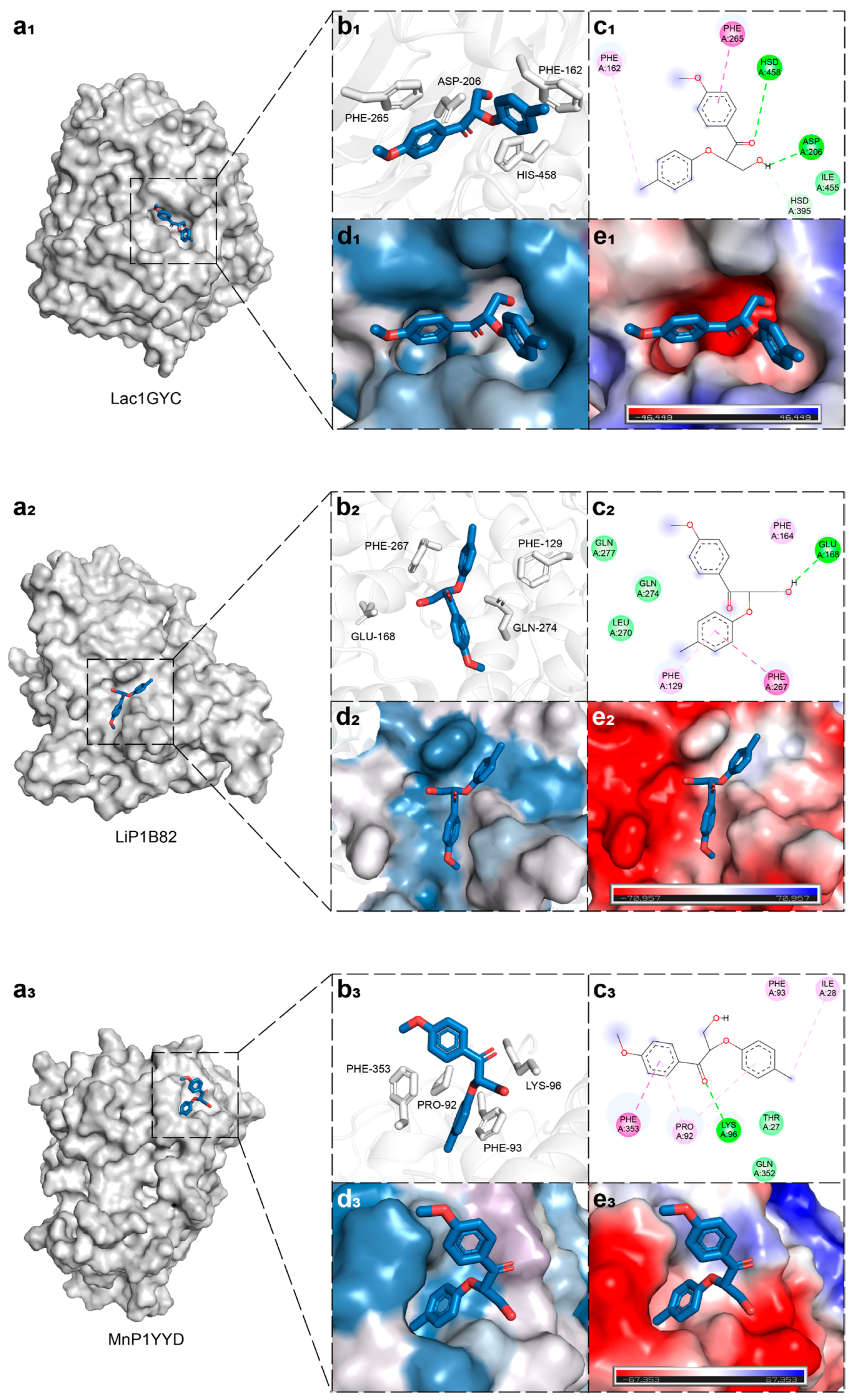
2.1. Structural Basis of Ligand Recognition by Lignin-Degrading Enzymes
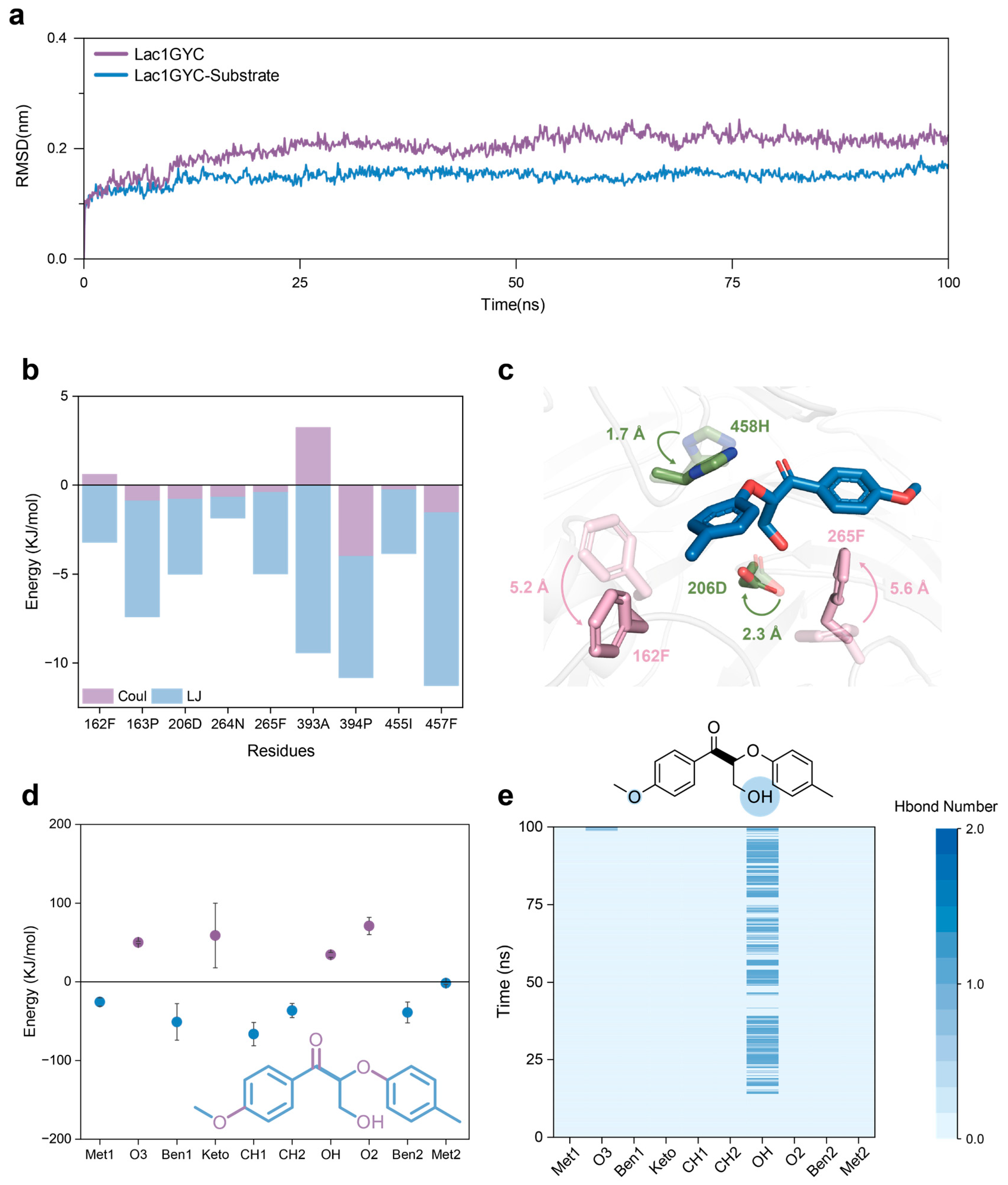
2.2. Dynamic Interaction Mechanisms in Specific Substrate Recognition
2.3. Functional Dissection of Hydrophobic and Hydrogen-Bonding Interactions via Site-Directed Mutagenesis
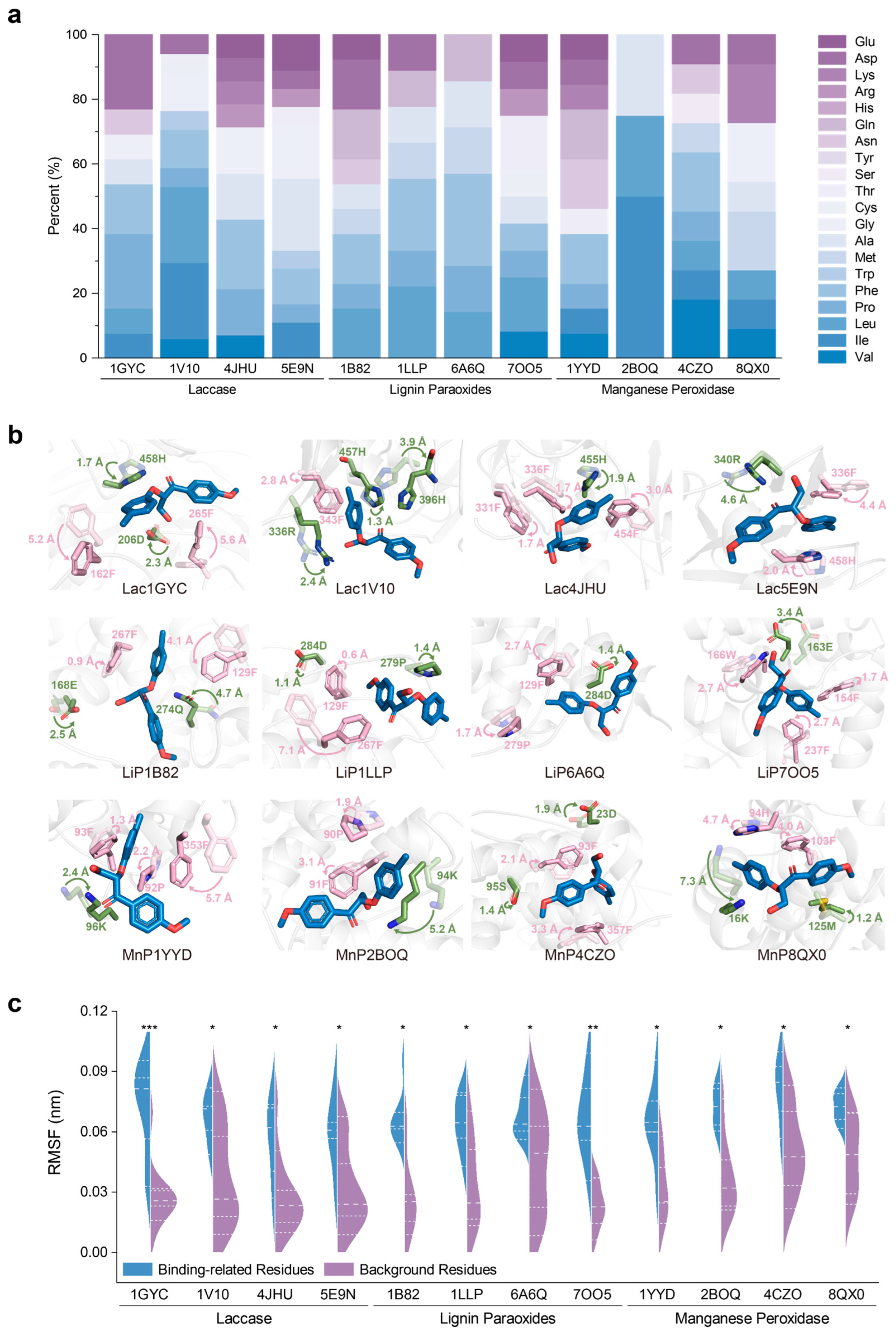
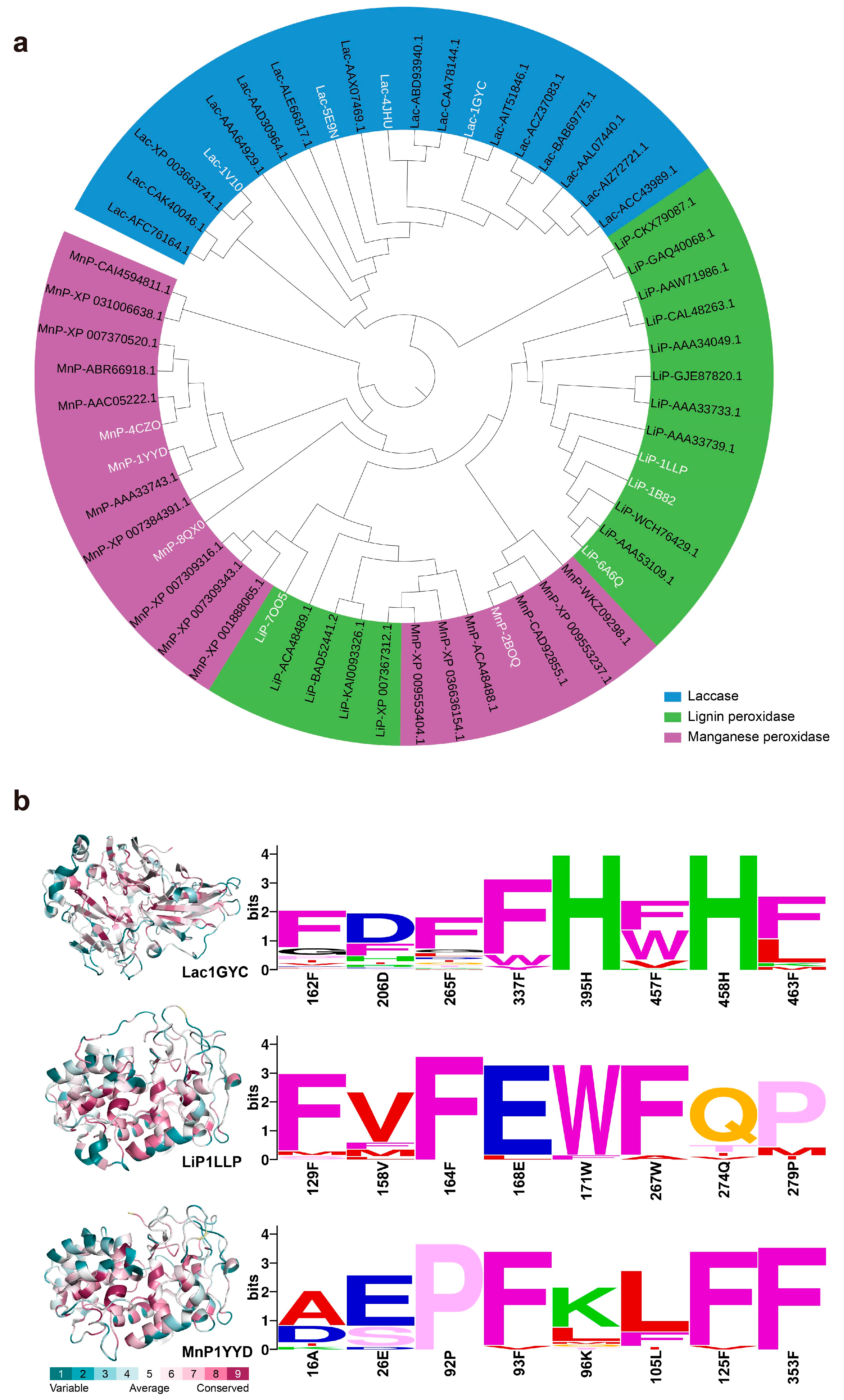
2.4. Universality of Catalytic Mechanisms Across Diverse Lignin-Degrading Enzyme
2.5. Conservation of Catalytic Motifs in Lignin-Degrading Enzyme
3. Discussion
4. Materials and Methods
4.1. Protein Preparation
4.2. Molecular Dynamics Simulations
4.3. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LAC | laccase |
| LiP | lignin peroxidase |
| MnP | manganese peroxidase |
References
- Jamaldheen, S.B.; Kurade, M.B.; Basak, B.; Yoo, C.G.; Oh, K.K.; Jeon, B.-H.; Kim, T.H. A review on physico-chemical delignification as a pretreatment of lignocellulosic biomass for enhanced bioconversion. Bioresour. Technol. 2022, 346, 126591. [Google Scholar] [CrossRef]
- Balat, M. Production of bioethanol from lignocellulosic materials via the biochemical pathway: A review. Energy Convers. Manag. 2011, 52, 858–875. [Google Scholar] [CrossRef]
- Liu, Z.-H.; Le, R.K.; Kosa, M.; Yang, B.; Yuan, J.; Ragauskas, A.J. Identifying and creating pathways to improve biological lignin valorization. Renew. Sustain. Energy Rev. 2019, 105, 349–362. [Google Scholar] [CrossRef]
- Akiyama, T.; Goto, H.; Nawawi, D.S.; Syafii, W.; Matsumoto, Y.; Meshitsuka, G. Erythro/threo ratio of β-O-4-5 structures as an important structural characteristic of lignin. Part 4: Variation in the erythro/threo ratio in softwood and hardwood lignins and its relation to syringyl/guaiacyl ratio. Holzforschung 2005, 59, 276–281. [Google Scholar] [CrossRef]
- Palazzolo, M.A.; Kurina-Sanz, M. Microbial utilization of lignin: Available biotechnologies for its degradation and valorization. World J. Microbiol. Biotechnol. 2016, 32, 173. [Google Scholar] [CrossRef]
- Wang, X.; Wang, G.; Yu, X.; Chen, H.; Sun, Y.; Chen, G. Pretreatment of corn stover by solid acid for d-lactic acid fermentation. Bioresour. Technol. 2017, 239, 490–495. [Google Scholar] [CrossRef]
- Iandolo, D.; Amore, A.; Birolo, L.; Leo, G.; Olivieri, G.; Faraco, V. Fungal solid state fermentation on agro-industrial wastes for acid wastewater decolorization in a continuous flow packed-bed bioreactor. Bioresour. Technol. 2011, 102, 7603–7607. [Google Scholar] [CrossRef]
- Bilal, M.; Iqbal, H.M.N. Lignin peroxidase immobilization on Ca-alginate beads and its dye degradation performance in a packed bed reactor system. Biocatal. Agric. Biotechnol. 2019, 20, 101205. [Google Scholar] [CrossRef]
- Zhu, D.; Liang, N.; Zhang, R.; Ahmad, F.; Zhang, W.; Yang, B.; Wu, J.; Geng, A.; Gabriel, M.; Sun, J. Insight into Depolymerization Mechanism of Bacterial Laccase for Lignin. ACS Sustain. Chem. Eng. 2020, 8, 12920–12933. [Google Scholar] [CrossRef]
- Kim, S.; Chmely, S.C.; Nimlos, M.R.; Bomble, Y.J.; Foust, T.D.; Paton, R.S.; Beckham, G.T. Computational Study of Bond Dissociation Enthalpies for a Large Range of Native and Modified Lignins. J. Phys. Chem. Lett. 2011, 2, 2846–2852. [Google Scholar] [CrossRef]
- Jiang, X.; Lu, Q.; Hu, B.; Liu, J.; Dong, C.; Yang, Y. A Comprehensive Study on Pyrolysis Mechanism of Substituted beta-O-4 Type Lignin Dimers. Int. J. Mol. Sci. 2017, 18, 2364. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Romson, J.; Elder, T.; Emmer, Å.; Lawoko, M. Lignin Structure and Reactivity in the Organosolv Process Studied by NMR Spectroscopy, Mass Spectrometry, and Density Functional Theory. Biomacromolecules 2023, 24, 2314–2326. [Google Scholar] [CrossRef] [PubMed]
- Park, G.W.; Gong, G.; Joo, J.C.; Song, J.; Lee, J.; Lee, J.P.; Kim, H.T.; Ryu, M.H.; Sirohi, R.; Zhuang, X.; et al. Recent progress and challenges in biological degradation and biotechnological valorization of lignin as an emerging source of bioenergy: A state-of-the-art review. Renew. Sustain. Energy Rev. 2022, 157, 112025. [Google Scholar] [CrossRef]
- Kishimoto, T.; Uraki, Y.; Ubukata, M. Easy synthesis of β-O-4 type lignin related polymers. Org. Biomol. Chem. 2005, 3, 1067–1073. [Google Scholar] [CrossRef]
- Ding, Y.; Cui, K.; Guo, Z.; Cui, M.; Chen, Y. Manganese peroxidase mediated oxidation of sulfamethoxazole: Integrating the computational analysis to reveal the reaction kinetics, mechanistic insights, and oxidation pathway. J. Hazard. Mater. 2021, 415, 125719. [Google Scholar] [CrossRef]
- Kamimura, N.; Sakamoto, S.; Mitsuda, N.; Masai, E.; Kajita, S. Advances in microbial lignin degradation and its applications. Curr. Opin. Biotechnol. 2019, 56, 179–186. [Google Scholar] [CrossRef]
- Zhang, S.; Dong, Z.; Shi, J.; Yang, C.; Fang, Y.; Chen, G.; Chen, H.; Tian, C. Enzymatic hydrolysis of corn stover lignin by laccase, lignin peroxidase, and manganese peroxidase. Bioresour. Technol. 2022, 361, 127699. [Google Scholar] [CrossRef]
- Piontek, K.; Antorini, M.; Choinowski, T. Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-A resolution containing a full complement of coppers. J. Biol. Chem. 2002, 277, 37663–37669. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Ruiz, M.I.; Ayuso-Fernández, I.; Rencoret, J.; González-Ramírez, A.M.; Linde, D.; Davó-Siguero, I.; Romero, A.; Gutiérrez, A.; Martínez, A.T.; Ruiz-Dueñas, F.J. Agaricales Mushroom Lignin Peroxidase: From Structure-Function to Degradative Capabilities. Antioxidants 2021, 10, 1446. [Google Scholar] [CrossRef]
- Blodig, W.; Smith, A.T.; Doyle, W.A.; Piontek, K. Crystal structures of pristine and oxidatively processed lignin peroxidase expressed in Escherichia coli and of the W171F variant that eliminates the redox active tryptophan 171. Implications for the reaction mechanism. J. Mol. Biol. 2001, 305, 851–861. [Google Scholar] [CrossRef]
- Zeng, J.; Helms, G.L.; Gao, X.; Chen, S. Quantification of Wheat Straw Lignin Structure by Comprehensive NMR Analysis. J. Agric. Food Chem. 2013, 61, 10848–10857. [Google Scholar] [CrossRef] [PubMed]
- Pham, L.T.M.; Seo, H.; Kim, K.J.; Kim, Y.H. In silico-designed lignin peroxidase from Phanerochaete chrysosporium shows enhanced acid stability for depolymerization of lignin. Biotechnol. Biofuels 2018, 11, 325. [Google Scholar] [CrossRef] [PubMed]
- Hosseinzadeh, P.; Mirts, E.N.; Pfister, T.D.; Gao, Y.-G.; Mayne, C.; Robinson, H.; Tajkhorshid, E.; Lu, Y. Enhancing Mn(II)-Binding and Manganese Peroxidase Activity in a Designed Cytochrome c Peroxidase through Fine-Tuning Secondary-Sphere Interactions. Biochemistry 2016, 55, 1494–1502. [Google Scholar] [CrossRef]
- Johjima, T.; Itoh, N.; Kabuto, M.; Tokimura, F.; Nakagawa, T.; Wariishi, H.; Tanaka, H. Direct interaction of lignin and lignin peroxidase from Phanerochaete chrysosporium. Proc. Natl. Acad. Sci. USA 1999, 96, 1989–1994. [Google Scholar] [CrossRef]
- Tanaka, T.; Hinohara, R.; Duron, O.A.C.; Aso, Y.; Kobayashi, N.; Saito, K.; Watanabe, T. Exploration of lignin-binding synthetic polymers with pendant hydrophobic amino acids. RSC Sustain. 2025, 3, 875–880. [Google Scholar] [CrossRef]
- Singh, A.K.; Katari, S.K.; Umamaheswari, A.; Raj, A. In silico exploration of lignin peroxidase for unraveling the degradation mechanism employing lignin model compounds. RSC Adv. 2021, 11, 14632–14653. [Google Scholar] [CrossRef]
- Awasthi, M.; Jaiswal, N.; Singh, S.; Pandey, V.P.; Dwivedi, U.N. Molecular docking and dynamics simulation analyses unraveling the differential enzymatic catalysis by plant and fungal laccases with respect to lignin biosynthesis and degradation. J. Biomol. Struct. Dyn. 2014, 33, 1835–1849. [Google Scholar] [CrossRef]
- Fu, N.; Li, J.; Wang, M.; Ren, L.; Luo, Y. Genes Identification, Molecular Docking and Dynamics Simulation Analysis of Laccases from Amylostereum areolatum Provides Molecular Basis of Laccase Bound to Lignin. Int. J. Mol. Sci. 2020, 21, 8845. [Google Scholar] [CrossRef]
- Poulos, T.; Edwards, S.; Wariishi, H.; Gold, M.H. Crystallographic refinement of lignin peroxidase at 2 A. J. Biol. Chem. 1993, 268, 4429–4440. [Google Scholar] [CrossRef]
- Bhatt, P.; Bhatt, K.; Chen, W.-J.; Huang, Y.; Xiao, Y.; Wu, S.; Lei, Q.; Zhong, J.; Zhu, X.; Chen, S. Bioremediation potential of laccase for catalysis of glyphosate, isoproturon, lignin, and parathion: Molecular docking, dynamics, and simulation. J. Hazard. Mater. 2022, 443, 130319. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, M.; Zheng, Y.; Zhao, X.; Li, B.; Huan, W. Atomic-scale investigation of the interaction between coniferyl alcohol and laccase for lignin degradation using molecular dynamics simulations and spectroscopy. J. Dispers. Sci. Technol. 2018, 40, 686–694. [Google Scholar] [CrossRef]
- Lanzarotti, E.; Defelipe, L.A.; Marti, M.A.; Turjanski, A.G. Aromatic clusters in protein-protein and protein-drug complexes. J. Cheminform. 2020, 12, 30. [Google Scholar] [CrossRef]
- Fernández-Fueyo, E.; Acebes, S.; Ruiz-Dueñas, F.J.; Martínez, M.J.; Romero, A.; Medrano, F.J.; Guallar, V.; Martínez, A.T. Structural implications of the C-terminal tail in the catalytic and stability properties of manganese peroxidases from ligninolytic fungi. Acta Crystallogr. D Biol. Crystallogr. 2014, 70 Pt 12, 3253–3265. [Google Scholar] [CrossRef]
- Nemec, T. Nucleation parameters of SPC/E and TIP4P/2005 water vapor measured in NPT molecular dynamics simulations. J. Mol. Model. 2022, 28, 174. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, Y.; Cheng, H.; Zeng, X.; Zhang, X.; Sang, P.; Chen, B.; Yang, L. Deciphering gp120 sequence variation and structural dynamics in HIV neutralization phenotype by molecular dynamics simulations and graph machine learning. Proteins 2022, 90, 1413–1424. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, F.; Niu, H.; Yuan, L.; Tian, J.; Cai, S.; Bi, X.; Zhou, L. Structural studies and molecular dynamic simulations of polyphenol oxidase treated by high pressure processing. Food Chem. 2022, 372, 131243. [Google Scholar] [CrossRef]
- Braun, E.; Moosavi, S.M.; Smit, B. Anomalous Effects of Velocity Rescaling Algorithms: The Flying Ice Cube Effect Revisited. J. Chem. Theory Comput. 2018, 14, 5262–5272. [Google Scholar] [CrossRef]
- Martonak, R.; Laio, A.; Parrinello, M. Predicting crystal structures: The Parrinello-Rahman method revisited. Phys. Rev. Lett. 2003, 90, 075503. [Google Scholar] [CrossRef]
- Wang, S.; Xu, Y.; Yu, X.W. A phenylalanine dynamic switch controls the interfacial activation of Rhizopus chinensis lipase. Int. J. Biol. Macromol. 2021, 173, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 2004, 13, 952–962. [Google Scholar] [CrossRef]
- Simmonett, A.C.; Brooks, B.R. Analytical Hessians for Ewald and particle mesh Ewald electrostatics. J. Chem. Phys. 2021, 154, 104101. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Wu, X.; Xiu, Z.; Wang, J.; Yin, L.; Li, G. Understanding the Molecular Mechanism of Binding Modes of Aurora a Inhibitors by Long Time Scale Gpu Dynamics. J. Theor. Comput. Chem. 2013, 12, 1341003. [Google Scholar] [CrossRef]
- Yadav, S.; Kardam, V.; Tripathi, A.; G, S.T.; Dubey, K.D. The Performance of Different Water Models on the Structure and Function of Cytochrome P450 Enzymes. J. Chem. Inf. Model. 2022, 62, 6679–6690. [Google Scholar] [CrossRef]
- Li, Z.L.; Buck, M. Modified Potential Functions Result in Enhanced Predictions of a Protein Complex by All-Atom Molecular Dynamics Simulations, Confirming a Stepwise Association Process for Native Protein-Protein Interactions. J. Chem. Theory Comput. 2019, 15, 4318–4331. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Cao, X.; Ma, Z.; Zhu, J.; Yang, L.; Xiao, M.; Jiang, X. Molecular Dynamics Simulation Reveals the Mechanism of Substrate Recognition by Lignin-Degrading Enzymes. Int. J. Mol. Sci. 2025, 26, 9378. https://doi.org/10.3390/ijms26199378
Ma X, Cao X, Ma Z, Zhu J, Yang L, Xiao M, Jiang X. Molecular Dynamics Simulation Reveals the Mechanism of Substrate Recognition by Lignin-Degrading Enzymes. International Journal of Molecular Sciences. 2025; 26(19):9378. https://doi.org/10.3390/ijms26199378
Chicago/Turabian StyleMa, Xue, Xueting Cao, Zhenyu Ma, Jingyi Zhu, Letian Yang, Min Xiao, and Xukai Jiang. 2025. "Molecular Dynamics Simulation Reveals the Mechanism of Substrate Recognition by Lignin-Degrading Enzymes" International Journal of Molecular Sciences 26, no. 19: 9378. https://doi.org/10.3390/ijms26199378
APA StyleMa, X., Cao, X., Ma, Z., Zhu, J., Yang, L., Xiao, M., & Jiang, X. (2025). Molecular Dynamics Simulation Reveals the Mechanism of Substrate Recognition by Lignin-Degrading Enzymes. International Journal of Molecular Sciences, 26(19), 9378. https://doi.org/10.3390/ijms26199378




