Molecular Insights into Bromocriptine Binding to GPCRs Within Histamine-Linked Signaling Networks: Network Pharmacology, Pharmacophore Modeling, and Molecular Dynamics Simulation
Abstract
1. Introduction
2. Results
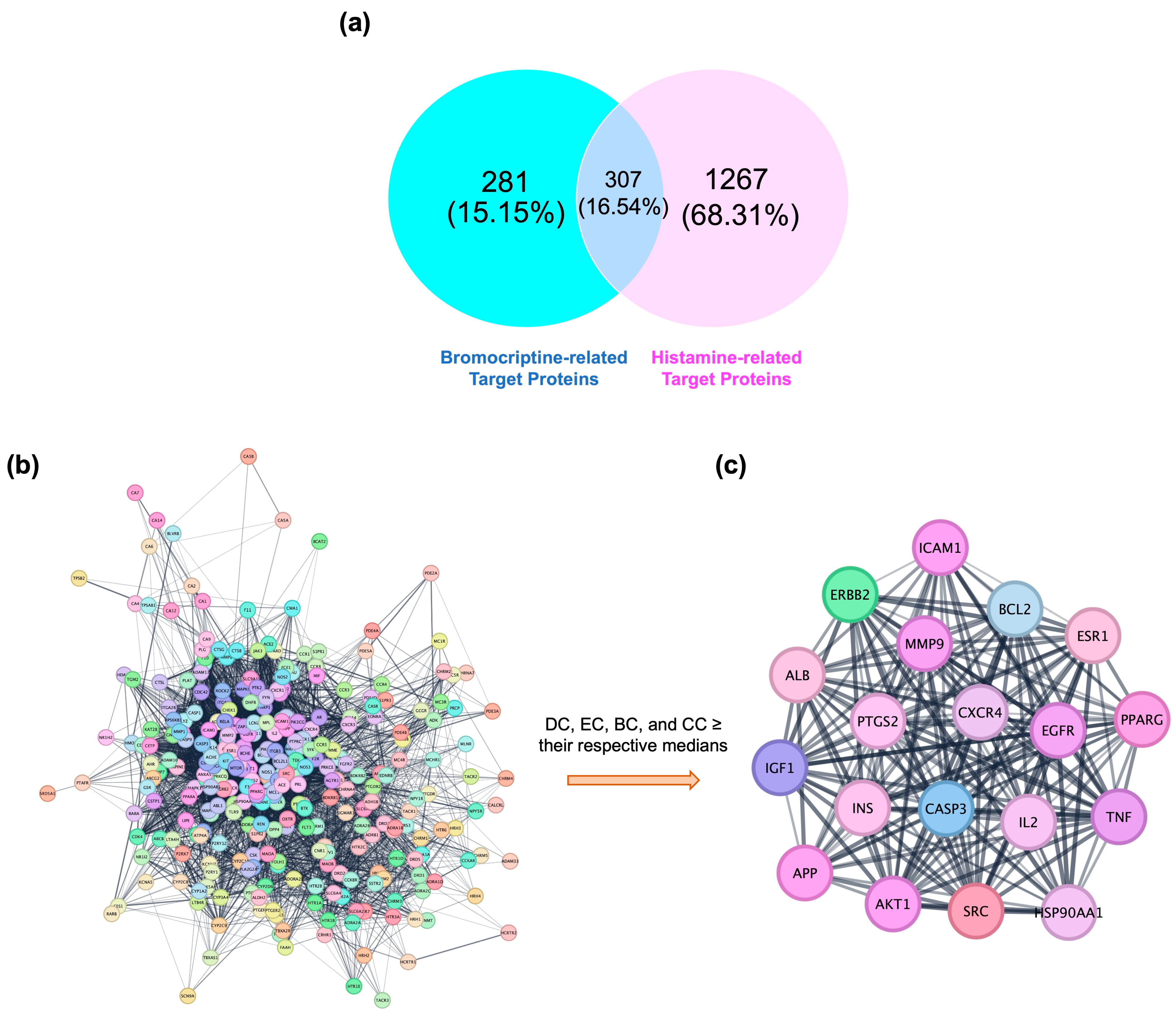
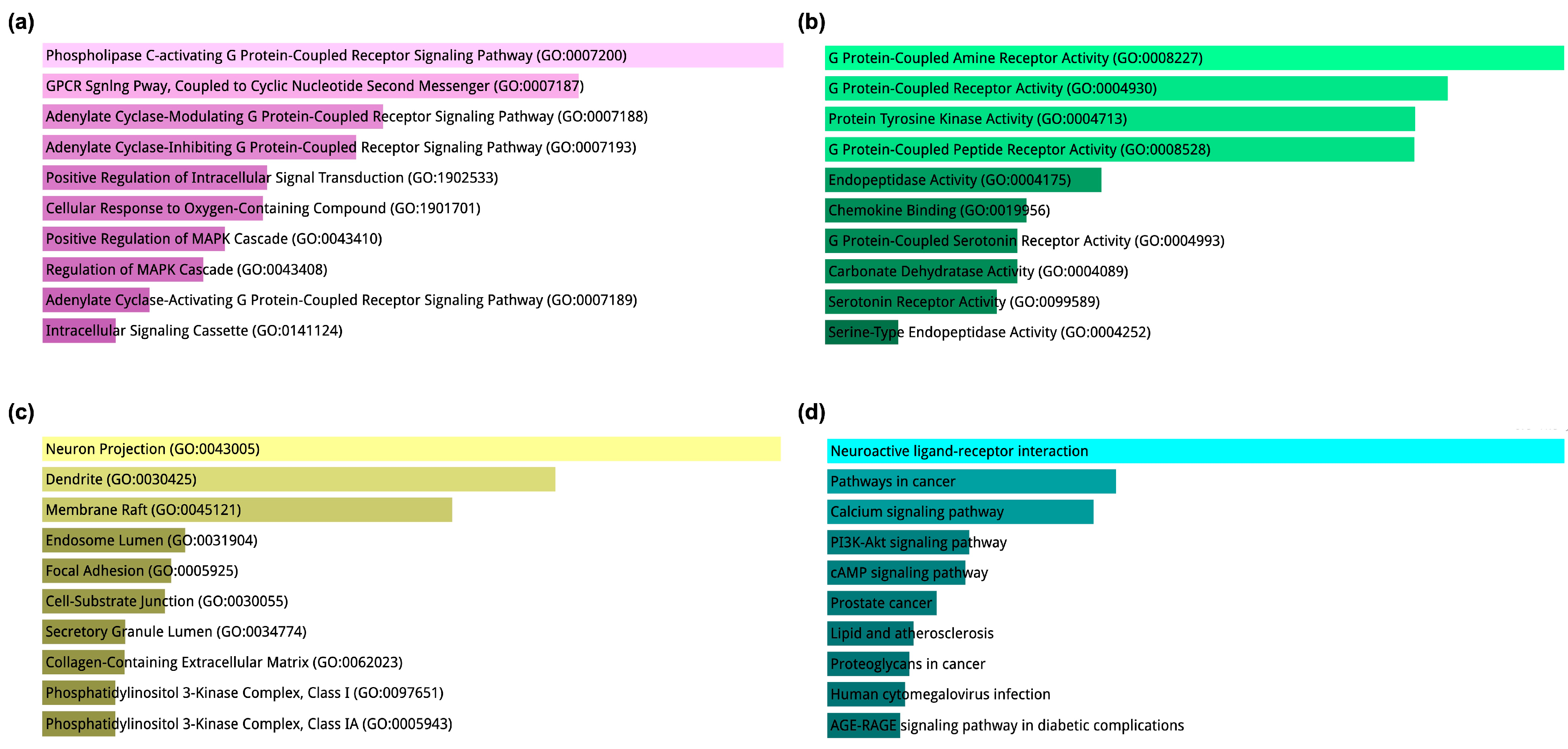
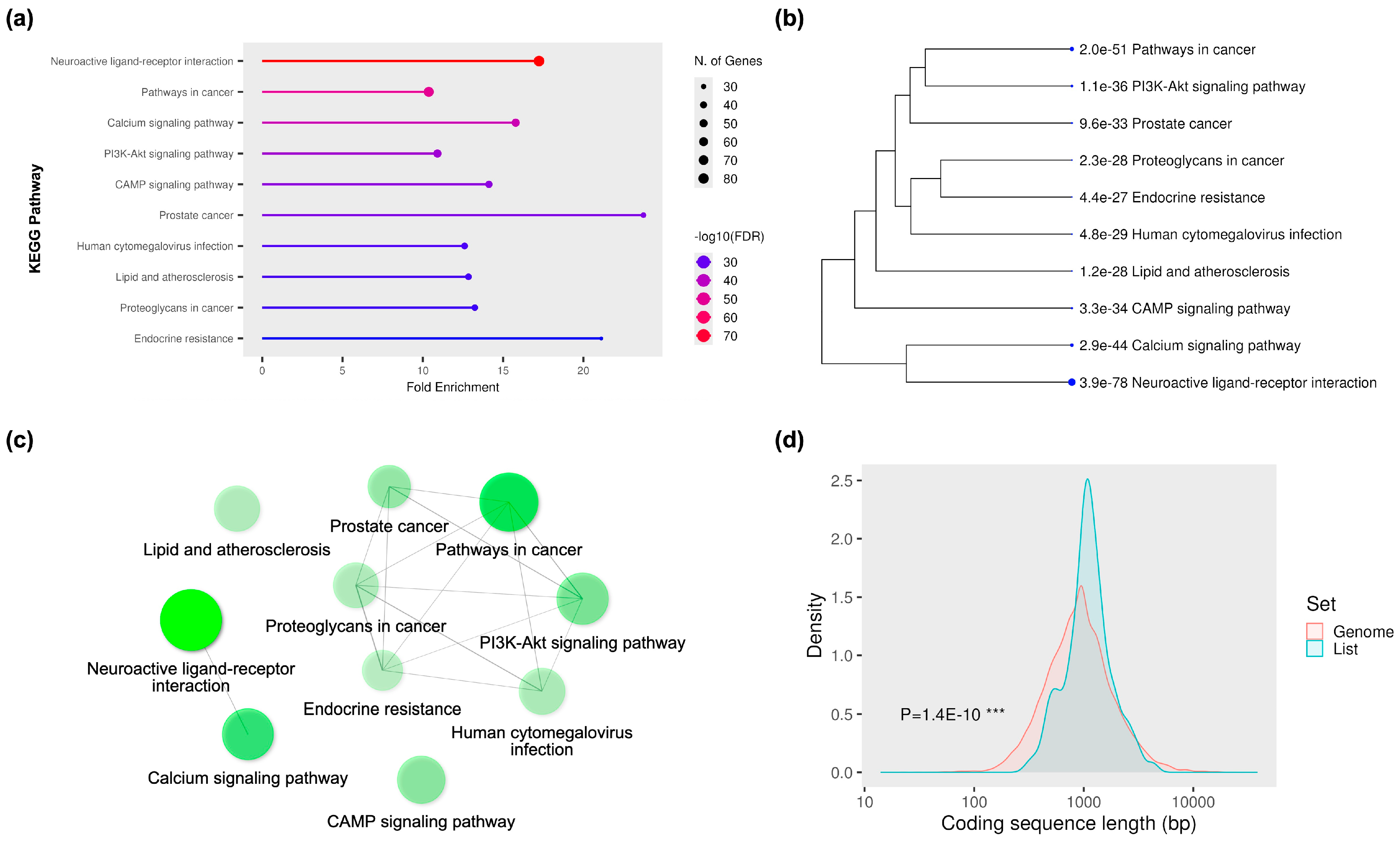
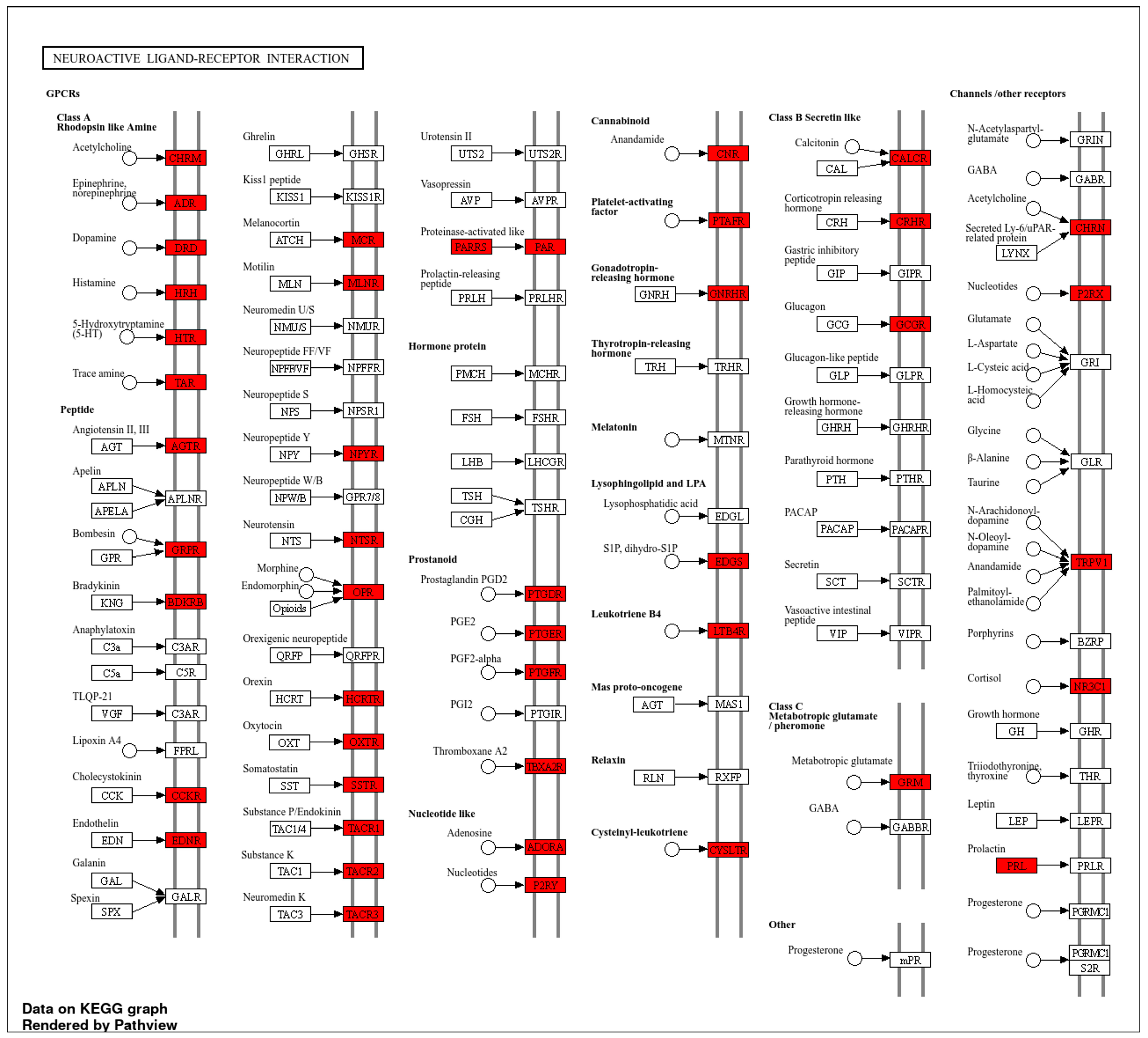
2.1. Network Pharmacology-Based Target Mapping Reveals Histamine-Related Polypharmacology of Bromocriptine
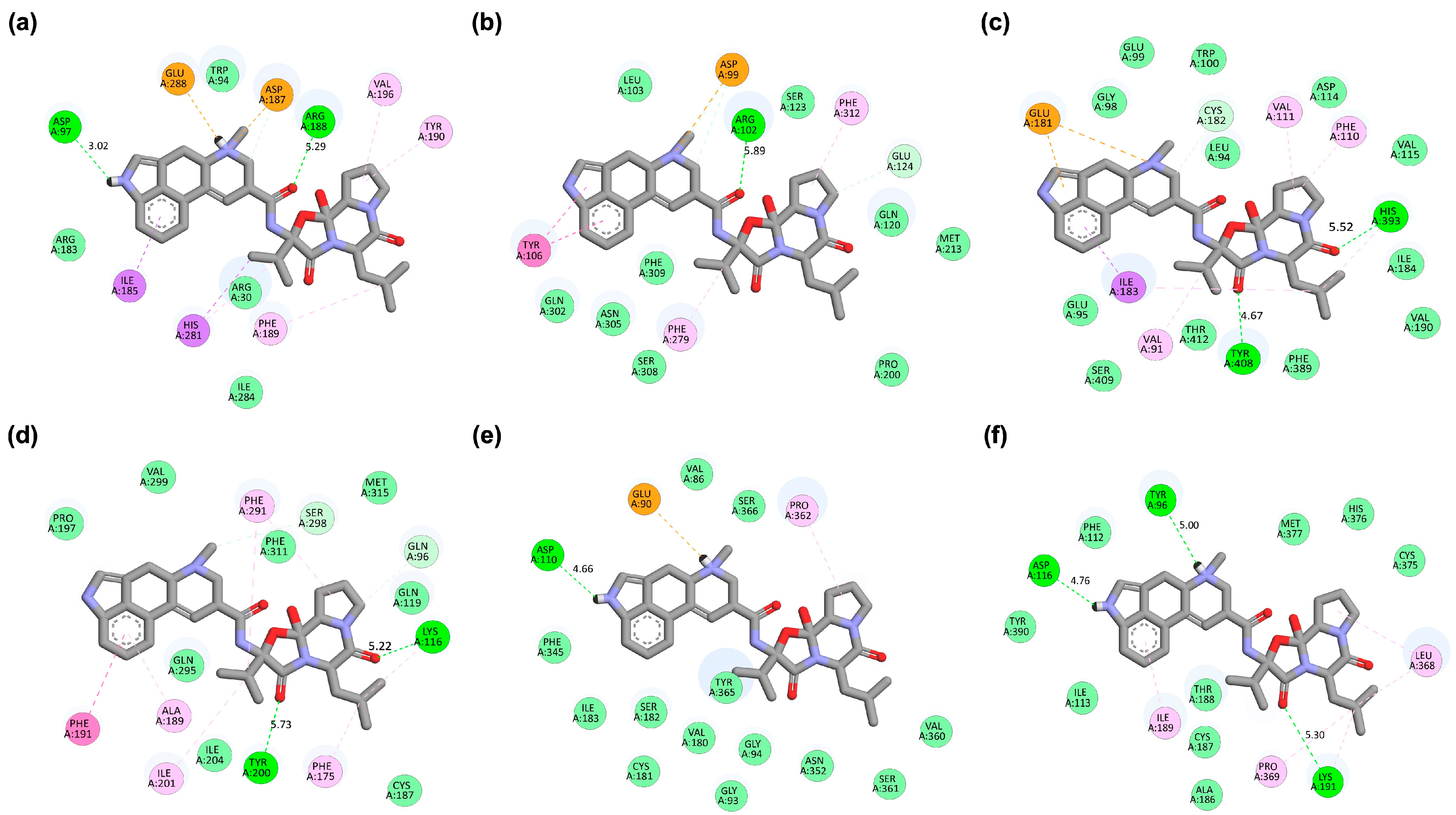
2.2. Binding Affinity and Interaction Profiles of Bromocriptine with Target Receptors Identified from Network Pharmacology
2.3. Pharmacophore Modeling Supports and Extends Molecular Docking Insights
2.4. MD Simulations Corroborate the Robustness and Binding Consistency of the Ligand–Receptor Complexes
3. Discussion
4. Materials and Methods
4.1. Database Collection
4.2. Protein–Protein Interaction (PPI) Network Construction
4.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analyses
4.4. Molecular Docking Simulations and Binding Affinity Analysis
4.5. Pharmacophore Modeling
4.6. Molecular Dynamics (MD) Simulation for Structural Stability and Interaction Analysis
4.7. MM/PBSA Binding Free Energy Calculation Using gmx_MMPBSA
5. Limitations, Clinical Implications, and Future Works
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AIRs | Ambiguous interaction restraints |
| AKT1 | AKT serine/threonine kinase 1 |
| APP | Amyloid precursor protein |
| BC | Betweenness centrality |
| BP | Biological process |
| CASP3 | Caspase-3 |
| CC | Closeness centrality |
| CDS | Coding sequence |
| CXCR4 | C-X-C chemokine receptor type 4 |
| DC | Degree centrality |
| DRD2 | D2 dopamine receptor |
| EC | Eigenvector centrality |
| EGFR | Epidermal growth factor receptor |
| FDA | Food and Drug Administration |
| FDR | False discovery rate |
| GABA | γ-aminobutyric acid |
| H1R | Histamine receptor H1 |
| HADDOCK | High ambiguity driven protein-protein docking |
| HBA | Hydrogen bond acceptor |
| HBD | Hydrogen bond donor |
| ICAM1 | Intercellular adhesion molecule 1 |
| IGF1 | Insulin-like growth factor 1 |
| IL2 | Interleukin-2 |
| iPSC | Induced pluripotent stem cell |
| LBD | Ligand-binding domain |
| MD | Molecular dynamics |
| MF | Molecular function |
| MM/PBSA | Molecular mechanics/Poisson-Boltzmann surface area |
| MMP9 | Matrix metalloproteinase-9 |
| NPT | Number of particles, pressure, and temperature |
| NVT | Number of particles, volume, and temperature |
| OMIM | Online Mendelian inheritance in man |
| PDB | Protein data bank |
| PI3K | Phosphatidylinositol 3-kinase |
| PK–PD | Pharmacokinetic–pharmacodynamic |
| PPI | Protein–protein interaction |
| PRODIGY | Protein binding energy prediction |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuation |
| RoG | Radius of gyration |
| SASA | Solvent-accessible surface area |
| SEA | Similarity ensemble approach |
| SMD | Steered molecular dynamics |
| SMILES | Simplified molecular input line entry system |
| SRC | Proto-oncogene tyrosine-protein kinase Src |
| STITCH | Search tool for interacting chemicals |
| STP | SwissTargetPrediction |
| TC | Tanimoto coefficient |
| TNF | Tumor necrosis factor |
References
- Parsons, M.E.; Ganellin, C.R. Histamine and its receptors. Br. J. Pharmacol. 2006, 147, S127–S135. [Google Scholar] [CrossRef] [PubMed]
- Panula, P.; Chazot, P.L.; Cowart, M.; Gutzmer, R.; Leurs, R.; Liu, W.L.; Stark, H.; Thurmond, R.L.; Haas, H.L. International Union of Basic and Clinical Pharmacology. XCVIII. Histamine Receptors. Pharmacol. Rev. 2015, 67, 601–655. [Google Scholar] [PubMed]
- Thurmond, R.L.; Venable, J.; Savall, B.; La, D.; Snook, S.; Dunford, P.J.; Edwards, J.P. Clinical Development of Histamine H4 Receptor Antagonists. Handb. Exp. Pharmacol. 2017, 241, 301–320. [Google Scholar] [PubMed]
- Thangam, E.B.; Jemima, E.A.; Singh, H.; Baig, M.S.; Khan, M.; Mathias, C.B.; Church, M.K.; Saluja, R. The Role of Histamine and Histamine Receptors in Mast Cell-Mediated Allergy and Inflammation: The Hunt for New Therapeutic Targets. Front. Immunol. 2018, 9, 1873. [Google Scholar] [CrossRef]
- Passani, M.B.; Lin, J.S.; Hancock, A.; Crochet, S.; Blandina, P. The histamine H3 receptor as a novel therapeutic target for cognitive and sleep disorders. Trends Pharmacol. Sci. 2004, 25, 618–625. [Google Scholar] [CrossRef]
- Esbenshade, T.A.; Fox, G.B.; Cowart, M.D. Histamine H3 receptor antagonists: Preclinical promise for treating obesity and cognitive disorders. Mol. Interv. 2006, 6, 77–88. [Google Scholar] [CrossRef]
- Salcedo, C.; Pontes, C.; Merlos, M. Is the H4 receptor a new drug target for allergies and asthma? Front. Biosci. (Elite Ed.) 2013, 5, 178–187. [Google Scholar] [CrossRef]
- Carthy, E.; Ellender, T. Histamine, Neuroinflammation and Neurodevelopment: A Review. Front. Neurosci. 2021, 15, 680214. [Google Scholar] [CrossRef]
- Szukiewicz, D. Histaminergic System Activity in the Central Nervous System: The Role in Neurodevelopmental and Neurodegenerative Disorders. Int. J. Mol. Sci. 2024, 25, 9859. [Google Scholar] [CrossRef]
- Zhu, H.X.; Lou, W.W.; Jiang, Y.M.; Ciobanu, A.; Fang, C.X.; Liu, C.Y.; Yang, Y.L.; Cao, J.Y.; Shan, L.; Zhuang, Q.X. Histamine Modulation of the Basal Ganglia Circuitry in the Motor Symptoms of Parkinson’s Disease. CNS Neurosci. Ther. 2025, 31, e70308. [Google Scholar] [CrossRef]
- Rocha, S.M.; Pires, J.; Esteves, M.; Graça, B.; Bernardino, L. Histamine: A new immunomodulatory player in the neuron-glia crosstalk. Front. Cell. Neurosci. 2014, 8, 120. [Google Scholar] [CrossRef]
- Rocha, S.M.; Saraiva, T.; Cristóvão, A.C.; Ferreira, R.; Santos, T.; Esteves, M.; Saraiva, C.; Je, G.; Cortes, L.; Valero, J.; et al. Histamine induces microglia activation and dopaminergic neuronal toxicity via H1 receptor activation. J. Neuroinflammation 2016, 13, 137. [Google Scholar] [CrossRef] [PubMed]
- Panula, P.; Nuutinen, S. Histamine and H3 receptor in alcohol-related behaviors. J. Pharmacol. Exp. Ther. 2011, 336, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Passani, M.B.; Blandina, P. Histamine receptors in the CNS as targets for therapeutic intervention. Trends Pharmacol. Sci. 2011, 32, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Mamouni, K.; Li, X.; Chen, Y.; Kavuri, S.; Du, Y.; Fu, H.; Kucuk, O.; Wu, D. Repositioning Dopamine D2 Receptor Agonist Bromocriptine to Enhance Docetaxel Chemotherapy and Treat Bone Metastatic Prostate Cancer. Mol. Cancer Ther. 2018, 17, 1859–1870. [Google Scholar] [CrossRef]
- Holt, R.I.G.; Barnett, A.H.; Bailey, C.J. Bromocriptine: Old drug, new formulation and new indication. Diabetes Obes. Metab. 2010, 12, 1048–1057. [Google Scholar] [CrossRef]
- Ozery, M.; Wadhwa, R. Bromocriptine, in StatPearls [Internet]; StatPearls Publishing, Ed.; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Seol, I.-W.; Kuo, N.Y.; Kim, K.M. Effects of dopaminergic drugs on the mast cell degranulation and nitric oxide generation in RAW 264.7 cells. Arch. Pharm. Res. 2004, 27, 94–98. [Google Scholar] [CrossRef]
- Caldara, R.; Ferrari, C.; Romussi, M.; Paracchi, A. Effect of bromocriptine administration on gastric acid and gastrin secretion in man. J. Endocrinol. Investig. 1979, 2, 45–49. [Google Scholar] [CrossRef]
- Ghanbari, A.; Eliassi, A. The effect of bromocriptine on basal and histamine-stimulated gastric acid secretion in anesthetized rats. Physiol. Pharmacol. 2006, 10, 173–181. [Google Scholar]
- Dermawan, D.; Alotaiq, N. Unveiling Pharmacological Mechanisms of Bombyx mori (Abresham), a Traditional Arabic Unani Medicine for Ischemic Heart Disease: An Integrative Molecular Simulation Study. Pharmaceutics 2025, 17, 295. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, S.; Liu, K.; Hu, X.; Gu, X. Advances in drug discovery based on network pharmacology and omics technology. Curr. Pharm. Anal. 2024, 21, 33–43. [Google Scholar] [CrossRef]
- Alotaiq, N.; Dermawan, D. Advancements in Virtual Bioequivalence: A Systematic Review of Computational Methods and Regulatory Perspectives in the Pharmaceutical Industry. Pharmaceutics 2024, 16, 1414. [Google Scholar] [CrossRef] [PubMed]
- Noor, F.; Tahir Ul Qamar, M.; Ashfaq, U.A.; Albutti, A.; Alwashmi, A.S.S.; Aljasir, M.A. Network Pharmacology Approach for Medicinal Plants: Review and Assessment. Pharmaceuticals 2022, 15, 572. [Google Scholar] [CrossRef] [PubMed]
- Alotaiq, N.; Dermawan, D. Evaluation of Structure Prediction and Molecular Docking Tools for Therapeutic Peptides in Clinical Use and Trials Targeting Coronary Artery Disease. Int. J. Mol. Sci. 2025, 26, 462. [Google Scholar] [CrossRef]
- Alotaiq, N.; Dermawan, D.; Elwali, N.E. Leveraging Therapeutic Proteins and Peptides from Lumbricus Earthworms: Targeting SOCS2 E3 Ligase for Cardiovascular Therapy through Molecular Dynamics Simulations. Int. J. Mol. Sci. 2024, 25, 10818. [Google Scholar] [CrossRef]
- Dabrowska, S.; Andrzejewska, A.; Lukomska, B.; Janowski, M. Neuroinflammation as a target for treatment of stroke using mesenchymal stem cells and extracellular vesicles. J. Neuroinflammation 2019, 16, 178. [Google Scholar] [CrossRef]
- Bonham, L.W.; Karch, C.M.; Fan, C.C.; Tan, C.; Geier, E.G.; Wang, Y.; Wen, N.; Broce, I.J.; Li, Y.; Barkovich, M.J.; et al. CXCR4 involvement in neurodegenerative diseases. Transl. Psychiatry 2018, 8, 73. [Google Scholar] [CrossRef]
- Zhang, W.; Xiao, D.; Mao, Q.; Xia, H. Role of neuroinflammation in neurodegeneration development. Signal Transduct. Target. Ther. 2023, 8, 267. [Google Scholar] [CrossRef]
- Singh, R.; Letai, A.; Sarosiek, K. Regulation of apoptosis in health and disease: The balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019, 20, 175–193. [Google Scholar] [CrossRef]
- Khan, S.; Ahmad, K.; Alshammari, E.M.; Adnan, M.; Baig, M.H.; Lohani, M.; Somvanshi, P.; Haque, S. Implication of Caspase-3 as a Common Therapeutic Target for Multineurodegenerative Disorders and Its Inhibition Using Nonpeptidyl Natural Compounds. Biomed. Res. Int. 2015, 2015, 379817. [Google Scholar] [CrossRef]
- Cevenini, A.; Orrù, S.; Mancini, A.; Alfieri, A.; Buono, P.; Imperlini, E. Molecular Signatures of the Insulin-like Growth Factor 1-mediated Epithelial-Mesenchymal Transition in Breast, Lung and Gastric Cancers. Int. J. Mol. Sci. 2018, 19, 2411. [Google Scholar] [CrossRef]
- Levenga, J.; Wong, H.; Milstead, R.A.; Keller, B.N.; LaPlante, L.E.; Hoeffer, C.A. AKT isoforms have distinct hippocampal expression and roles in synaptic plasticity. Elife 2017, 6, e30640. [Google Scholar] [CrossRef]
- Du, Y.; Karatekin, F.; Wang, W.K.; Hong, W.; Boopathy, G.T.K. Cracking the EGFR code: Cancer biology, resistance mechanisms, and future therapeutic frontiers. Pharmacol. Rev. 2025, 77, 100076. [Google Scholar] [CrossRef]
- Schultz, N.; Nielsen, H.M.; Minthon, L.; Wennström, M. Involvement of matrix metalloproteinase-9 in amyloid-β 1-42-induced shedding of the pericyte proteoglycan NG2. J. Neuropathol. Exp. Neurol. 2014, 73, 684–692. [Google Scholar] [CrossRef]
- Radosinska, D.; Radosinska, J. The Link Between Matrix Metalloproteinases and Alzheimer’s Disease Pathophysiology. Mol. Neurobiol. 2025, 62, 885–899. [Google Scholar] [CrossRef]
- Guo, N.; Wang, X.; Xu, M.; Bai, J.; Yu, H.; Le, Z. PI3K/AKT signaling pathway: Molecular mechanisms and therapeutic potential in depression. Pharmacol. Res. 2024, 206, 107300. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Sun, M. Phytochemical regulation of CaMKII in Alzheimer’s disease: A review of molecular mechanisms and therapeutic potential. Pharmacol. Res. 2025, 216, 107790. [Google Scholar] [CrossRef]
- Yao, Y.; Baronio, D.; Chen, Y.C.; Jin, C.; Panula, P. The Roles of Histamine Receptor 1 (hrh1) in Neurotransmitter System Regulation, Behavior, and Neurogenesis in Zebrafish. Mol. Neurobiol. 2023, 60, 6660–6675. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.; Stark, H. Histamine receptor subtypes: A century of rational drug design. Front. Biosci. (Schol. Ed.) 2012, 4, 461–488. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Liu, J.; Chen, Z. The Histaminergic System in Neuropsychiatric Disorders. Biomolecules 2021, 11, 1345. [Google Scholar] [CrossRef]
- Zhuang, Y.; Xu, P.; Mao, C.; Wang, L.; Krumm, B.; Zhou, X.E.; Huang, S.; Liu, H.; Cheng, X.; Huang, X.-P.; et al. Structural insights into the human D1 and D2 dopamine receptor signaling complexes. Cell 2021, 184, 931–942.e18. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, B.; Sun, F.; Xu, Y.; Luan, H.; Yang, M.; Wang, C.; Zhang, T.; Zhou, Z.; Yan, H. Histamine H3R antagonist counteracts the impaired hippocampal neurogenesis in Lipopolysaccharide-induced neuroinflammation. Int. Immunopharmacol. 2022, 110, 109045. [Google Scholar] [CrossRef] [PubMed]
- Kononoff Vanhanen, J.; Nuutinen, S.; Tuominen, M.; Panula, P. Histamine H3 Receptor Regulates Sensorimotor Gating and Dopaminergic Signaling in the Striatum. J. Pharmacol. Exp. Ther. 2016, 357, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Chien, E.Y.; Mol, C.D.; Fenalti, G.; Liu, W.; Katritch, V.; Abagyan, R.; Brooun, A.; Wells, P.; Bi, F.C.; et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 2010, 330, 1066–1071. [Google Scholar] [CrossRef]
- Congreve, M.; Langmead, C.; Marshall, F.H. Chapter 1—The Use of GPCR Structures in Drug Design. In Advances in Pharmacology; Neubig, R.R., Ed.; Academic Press: Cambridge, MA, USA, 2011; pp. 1–36. [Google Scholar]
- Giorgioni, G.; Del Bello, F.; Quaglia, W.; Botticelli, L.; Cifani, C.; Micioni Di Bonaventura, E.; Micioni Di Bonaventura, M.V.; Piergentili, A. Advances in the Development of Nonpeptide Small Molecules Targeting Ghrelin Receptor. J. Med. Chem. 2022, 65, 3098–3118. [Google Scholar] [CrossRef]
- Moulin, A.; Brunel, L.; Boeglin, D.; Demange, L.; Ryan, J.; M’Kadmi, C.; Denoyelle, S.; Martinez, J.; Fehrentz, J.A. The 1,2,4-triazole as a scaffold for the design of ghrelin receptor ligands: Development of JMV 2959, a potent antagonist. Amino Acids 2013, 44, 301–314. [Google Scholar] [CrossRef]
- Breton, C.; Chellil, H.; Kabbaj-Benmansour, M.; Carnazzi, E.; Seyer, R.; Phalipou, S.; Morin, D.; Durroux, T.; Zingg, H.; Barberis, C.; et al. Direct identification of human oxytocin receptor-binding domains using a photoactivatable cyclic peptide antagonist: Comparison with the human V1a vasopressin receptor. J. Biol. Chem. 2001, 276, 26931–26941. [Google Scholar] [CrossRef]
- Hauser, A.S.; Kooistra, A.J.; Munk, C.; Heydenreich, F.M.; Veprintsev, D.B.; Bouvier, M.; Babu, M.M.; Gloriam, D.E. GPCR activation mechanisms across classes and macro/microscales. Nat. Struct. Mol. Biol. 2021, 28, 879–888. [Google Scholar] [CrossRef]
- Fleetwood, O.; Matricon, P.; Carlsson, J.; Delemotte, L. Energy Landscapes Reveal Agonist Control of G Protein-Coupled Receptor Activation via Microswitches. Biochemistry 2020, 59, 880–891. [Google Scholar] [CrossRef]
- Uy, G.L.; Rettig, M.P.; Cashen, A.F. Plerixafor, a CXCR4 antagonist for the mobilization of hematopoietic stem cells. Expert Opin. Biol. Ther. 2008, 8, 1797–1804. [Google Scholar] [CrossRef]
- Jørgensen, A.S.; Daugvilaite, V.; De Filippo, K.; Berg, C.; Mavri, M.; Benned-Jensen, T.; Juzenaite, G.; Hjortø, G.; Rankin, S.; Våbenø, J.; et al. Biased action of the CXCR4-targeting drug plerixafor is essential for its superior hematopoietic stem cell mobilization. Commun. Biol. 2021, 4, 569. [Google Scholar] [CrossRef]
- Richardson, R.S.; Sulima, A.; Rice, K.C.; Kucharczk, J.A.; Janda, K.D.; Nisbett, K.E.; Koob, G.F.; Vendruscolo, L.F.; Leggio, L. Pharmacological GHSR (ghrelin receptor) blockade reduces alcohol binge-like drinking in male and female mice. Neuropharmacology 2023, 238, 109643. [Google Scholar] [CrossRef] [PubMed]
- Park, S.B.; King, S.; MacDonald, D.; Wilson, A.; MacKay, H.; Woodside, B.; Abizaid, A. Contribution of growth hormone secretagogue receptor (GHSR) signaling in the ventral tegmental area (VTA) to the regulation of social motivation in male mice. Transl. Psychiatry 2021, 11, 230. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, Y.; Onaka, T. Roles of Oxytocin in Stress Responses, Allostasis and Resilience. Int. J. Mol. Sci. 2021, 23, 150. [Google Scholar] [CrossRef]
- Ellenbroek, B.A. Histamine H3 receptors, the complex interaction with dopamine and its implications for addiction. Br. J. Pharmacol. 2013, 170, 46–57. [Google Scholar] [CrossRef]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.L.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef]
- Kuhn, M.; von Mering, C.; Campillos, M.; Jensen, L.J.; Bork, P. STITCH: Interaction networks of chemicals and proteins. Nucleic Acids Res. 2008, 36, D684–D688. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef]
- Yao, Z.J.; Dong, J.; Che, Y.J.; Zhu, M.F.; Wen, M.; Wang, N.N.; Wang, S.; Lu, A.P.; Cao, D.S. TargetNet: A web service for predicting potential drug-target interaction profiling via multi-target SAR models. J. Comput. Aided Mol. Des. 2016, 30, 413–424. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2025 update. Nucleic Acids Res. 2025, 53, D1516–D1525. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Vilar, S.; Quezada, E.; Uriarte, E.; Costanzi, S.; Borges, F.; Viña, D.; Hripcsak, G. Computational Drug Target Screening through Protein Interaction Profiles. Sci. Rep. 2016, 6, 36969. [Google Scholar] [CrossRef]
- The UniProt Consortium, UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Hamosh, A.; Scott, A.F.; Amberger, J.; Valle, D.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM). Hum. Mutat. 2000, 15, 57–61. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.-X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.-P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Soo, G.C.; Cartledge, F.K.; Unwalla, R.J.; Profeta, S. Development of a molecular mechanics (MM2) force field for α-chlorosilanes. Tetrahedron 1990, 46, 8005–8018. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein−Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- van Zundert, G.C.P.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Vangone, A.; Bonvin, A. PRODIGY: A Contact-based Predictor of Binding Affinity in Protein-protein Complexes. Bio-Protocol 2017, 7, e2124. [Google Scholar] [CrossRef]
- Wolber, G.; Langer, T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Mishra, S.K.; Shaheen, M.M.; Sultana, S.; Al-Dies, A.-A.M.; Tayyeb, J.Z.; Alqahtani, T.; Tiruneh, Y.K.; de Farias Morais, G.C.; Oliveira, J.I.N.; Zaki, M.E.A. Computational analysis of lupenone derivatives as potential inhibitor of human papillomavirus oncoprotein E6 associated cervical cancer. Sci. Rep. 2025, 15, 15402. [Google Scholar] [CrossRef]
- Alkhatabi, H.A.; Alatyb, H.N. Pharmacophore-Based Study: An In Silico Perspective for the Identification of Potential New Delhi Metallo-β-lactamase-1 (NDM-1) Inhibitors. Pharmaceuticals 2024, 17, 1183. [Google Scholar] [CrossRef]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef]
- Alotaiq, N.; Dermawan, D. Computational Investigation of Montelukast and Its Structural Derivatives for Binding Affinity to Dopaminergic and Serotonergic Receptors: Insights from a Comprehensive Molecular Simulation. Pharmaceuticals 2025, 18, 559. [Google Scholar] [CrossRef]
- Kim, M.; Kim, E.; Lee, S.; Kim, J.S.; Lee, S. New Method for Constant-NPT Molecular Dynamics. J. Phys. Chem. A 2019, 123, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Kleinerman, D.S.; Czaplewski, C.; Liwo, A.; Scheraga, H.A. Implementations of Nosé-Hoover and Nosé-Poincaré thermostats in mesoscopic dynamic simulations with the united-residue model of a polypeptide chain. J. Chem. Phys. 2008, 128, 245103. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Sousa da Silva, A.W.; Vranken, W.F. ACPYPE-AnteChamber PYthon Parser interfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]




| Complex | HADDOCK Score (a.u.) | Binding Energy (kcal/mol) | Van der Waals Energy | Electrostatic Energy | Desolvation Energy | RMSD |
|---|---|---|---|---|---|---|
| CXCR4_Bromocriptine | −56.1 ± 0.7 | −10.67 | −36.9 ± 0.6 | −124.0 ± 9.1 | −7.2 ± 0.4 | 0.2 ± 0.1 |
| GHSR_Bromocriptine | −62.9 ± 1.6 | −10.36 | −39.4 ± 1.5 | −54.3 ± 7.5 | −18.4 ± 0.7 | 0.3 ± 0.2 |
| DRD2_Bromocriptine | −57.4 ± 0.5 | −10.29 | −35.5 ± 1.4 | −71.5 ± 7.3 | −14.9 ± 0.6 | 0.2 ± 0.2 |
| OXTR_Bromocriptine | −56.5 ± 0.9 | −9.93 | −32.0 ± 0.4 | −21.3 ± 7.3 | −22.8 ± 1.3 | 0.4 ± 0.2 |
| DRD3_Bromocriptine | −52.8 ± 0.3 | −9.77 | −30.3 ± 0.3 | −71.8 ± 5.0 | −15.4 ± 0.5 | 0.3 ± 0.2 |
| HTR1A_Bromocriptine | −52.6 ± 0.1 | −9.50 | −33.0 ± 0.6 | −53.3 ± 1.9 | −14.4 ± 0.4 | 0.3 ± 0.2 |
| ERBB2_Bromocriptine | −42.5 ± 1.7 | −9.37 | −32.5 ± 0.8 | −79.5 ± 8.4 | −3.3 ± 0.4 | 0.6 ± 0.0 |
| HRH3_Bromocriptine | −56.7 ± 0.2 | −9.15 | −37.3 ± 0.4 | −84.0 ± 3.6 | −11.1 ± 0.4 | 0.3 ± 0.2 |
| TSHR_Bromocriptine | −40.9 ± 2.9 | −9.08 | −31.0 ± 1.1 | −76.0 ± 6.6 | −6.7 ± 0.2 | 0.3 ± 0.0 |
| EGFR_Bromocriptine | −43.8 ± 1.0 | −9.02 | −35.7 ± 1.6 | −36.7 ± 5.8 | −5.1 ± 1.2 | 0.5 ± 0.0 |
| Complex | CC | CO | CN | CX | OO | OX | NO | NN | NX | XX |
|---|---|---|---|---|---|---|---|---|---|---|
| CXCR4_Bromocriptine | 3179 | 1357 | 1366 | 35 | 133 | 2 | 267 | 132 | 4 | 0 |
| GHSR_Bromocriptine | 3658 | 1545 | 1414 | 47 | 162 | 9 | 283 | 126 | 8 | 0 |
| DRD2_Bromocriptine | 3298 | 1504 | 1212 | 58 | 149 | 10 | 270 | 100 | 9 | 0 |
| OXTR_Bromocriptine | 3376 | 1234 | 1164 | 43 | 111 | 12 | 193 | 90 | 7 | 0 |
| DRD3_Bromocriptine | 2949 | 1302 | 1150 | 32 | 131 | 1 | 239 | 112 | 5 | 0 |
| HTR1A_Bromocriptine | 3015 | 1216 | 1189 | 62 | 115 | 10 | 242 | 120 | 14 | 0 |
| ERBB2_Bromocriptine | 2719 | 1172 | 1183 | 44 | 101 | 5 | 218 | 121 | 6 | 0 |
| HRH3_Bromocriptine | 2468 | 936 | 1091 | 27 | 83 | 0 | 196 | 108 | 4 | 0 |
| TSHR_Bromocriptine | 2371 | 1252 | 892 | 14 | 145 | 5 | 233 | 84 | 3 | 0 |
| EGFR_Bromocriptine | 2919 | 1260 | 1321 | 55 | 113 | 4 | 263 | 142 | 9 | 0 |
| Complex | HADDOCK Score (a.u.) | Binding Energy (kcal/mol) | Van der Waals Energy | Electrostatic Energy | Desolvation Energy | RMSD |
|---|---|---|---|---|---|---|
| CXCR4_Bromocriptine | −56.1 ± 0.7 | −10.67 | −36.9 ± 0.6 | −124.0 ± 9.1 | −7.2 ± 0.4 | 0.2 ± 0.1 |
| CXCR4_CXCL-12 (agonist) | −21.0 ± 2.6 | −7.06 | −17.3 ± 2.2 | −62.2 ± 21.1 | 1.2 ± 0.5 | 0.5 ± 0.0 |
| CXCR4_Plerixafor (antagonist) | −79.7 ± 4.2 | −14.21 | −26.1 ± 1.8 | −530.0 ± 19.2 | −3.3 ± 0.4 | 0.5 ± 0.1 |
| GHSR_Bromocriptine | −62.9 ± 1.6 | −10.36 | −39.4 ± 1.5 | −54.3 ± 7.5 | −18.4 ± 0.7 | 0.3 ± 0.2 |
| GHSR_GHRP-6 (agonist) | −48.6 ± 0.4 | −8.58 | −37.9 ± 1.2 | −19.9 ± 20.4 | −12.2 ± 1.5 | 0.7 ± 0.1 |
| GHSR_JMV-2959 (antagonist) | −62.5 ± 1.3 | −10.49 | −36.3 ± 0.8 | −89.8 ± 9.2 | −17.5 ± 1.3 | 0.3 ± 0.2 |
| OXTR_Bromocriptine | −56.5 ± 0.9 | −9.93 | −32.0 ± 0.4 | −21.3 ± 7.3 | −22.8 ± 1.3 | 0.4 ± 0.2 |
| OXTR_Oxytocin (agonist) | −58.6 ± 1.3 | −8.93 | −35.9 ± 2.4 | −29.7 ± 4.4 | −21.6 ± 1.5 | 0.2 ± 0.1 |
| OXTR_Atosiban (antagonist) | −56.5 ± 1.1 | −8.99 | −33.9 ± 0.7 | −36.5 ± 9.8 | −19.4 ± 0.7 | 0.6 ± 0.1 |
| Complex | MM/PBSA Free Binding Energy ΔG_Binding (kcal/mol) |
|---|---|
| CXCR4_Bromocriptine | −22.67 ± 3.70 |
| CXCR4_CXCL-12 (agonist) | −13.33 ± 4.62 |
| CXCR4_Plerixafor (antagonist) | −28.43 ± 2.11 |
| GHSR_Bromocriptine | −22.11 ± 3.55 |
| GHSR_GHRP-6 (agonist) | −14.96 ± 4.78 |
| GHSR_JMV-2959 (antagonist) | −24.98 ± 3.12 |
| OXTR_Bromocriptine | −21.43 ± 2.41 |
| OXTR_Oxytocin (agonist) | −16.56 ± 4.72 |
| OXTR_Atosiban (antagonist) | −19.87 ± 3.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dermawan, D.; Elbouamri, L.; Chtita, S.; Alotaiq, N. Molecular Insights into Bromocriptine Binding to GPCRs Within Histamine-Linked Signaling Networks: Network Pharmacology, Pharmacophore Modeling, and Molecular Dynamics Simulation. Int. J. Mol. Sci. 2025, 26, 8717. https://doi.org/10.3390/ijms26178717
Dermawan D, Elbouamri L, Chtita S, Alotaiq N. Molecular Insights into Bromocriptine Binding to GPCRs Within Histamine-Linked Signaling Networks: Network Pharmacology, Pharmacophore Modeling, and Molecular Dynamics Simulation. International Journal of Molecular Sciences. 2025; 26(17):8717. https://doi.org/10.3390/ijms26178717
Chicago/Turabian StyleDermawan, Doni, Lamiae Elbouamri, Samir Chtita, and Nasser Alotaiq. 2025. "Molecular Insights into Bromocriptine Binding to GPCRs Within Histamine-Linked Signaling Networks: Network Pharmacology, Pharmacophore Modeling, and Molecular Dynamics Simulation" International Journal of Molecular Sciences 26, no. 17: 8717. https://doi.org/10.3390/ijms26178717
APA StyleDermawan, D., Elbouamri, L., Chtita, S., & Alotaiq, N. (2025). Molecular Insights into Bromocriptine Binding to GPCRs Within Histamine-Linked Signaling Networks: Network Pharmacology, Pharmacophore Modeling, and Molecular Dynamics Simulation. International Journal of Molecular Sciences, 26(17), 8717. https://doi.org/10.3390/ijms26178717








