Four Large Indels in Barley Chloroplast Mutator (cpm) Seedlings Reinforce the Hypothesis of a Malfunction in the MMR System
Abstract
1. Introduction
2. Results
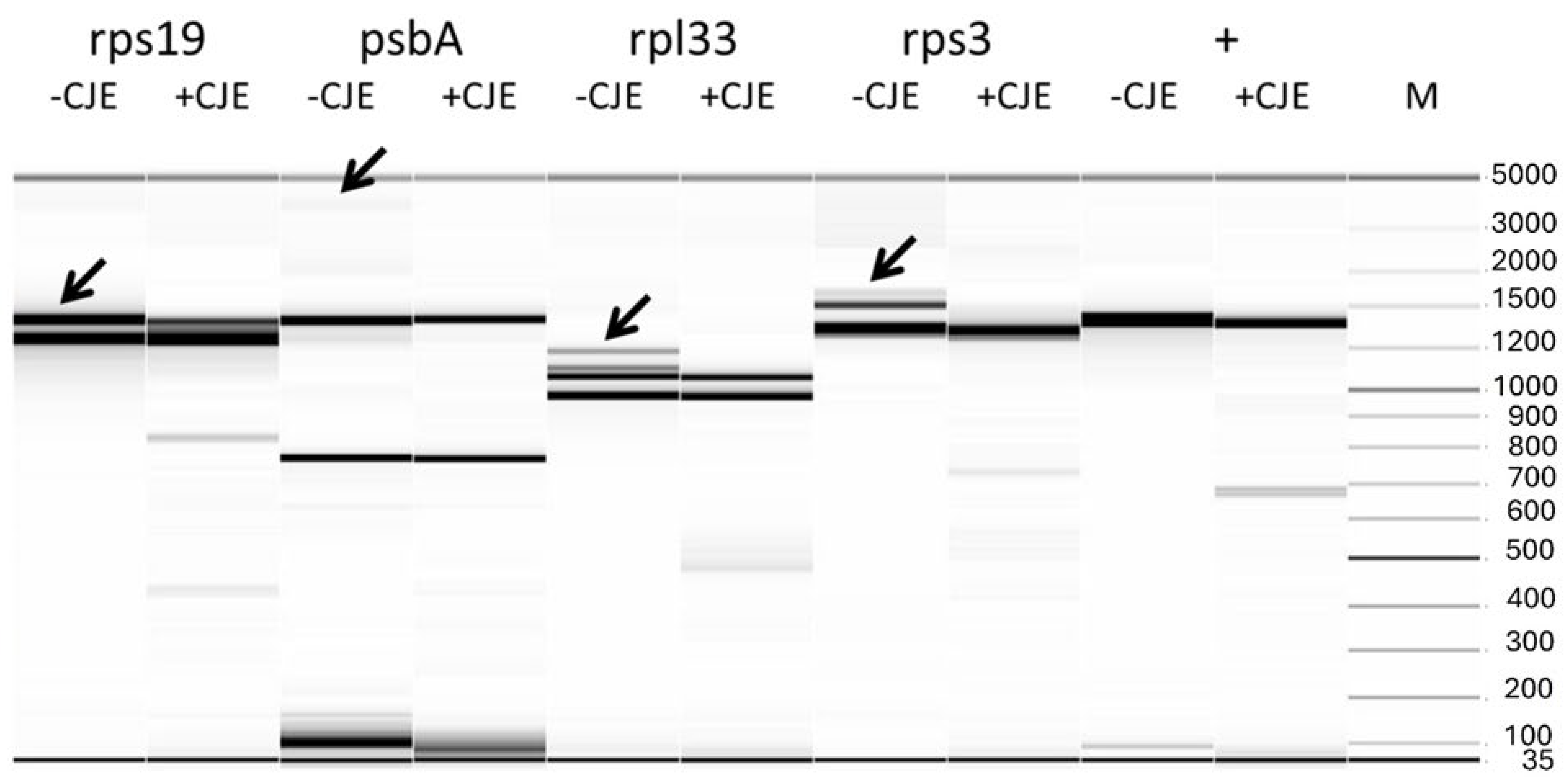
2.1. Four Large Indels Detected by CJE Digestion
2.2. Presence of Perfect or Imperfect Direct Repeats Flanking the Large Indels
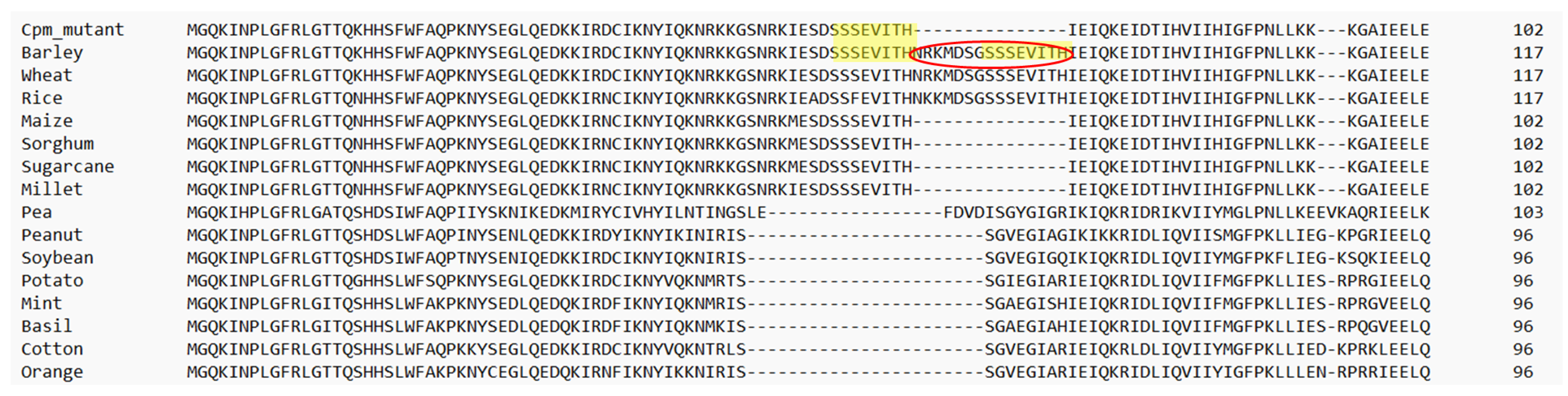
2.3. The RPS3 Protein Encoded by the Deleted rps3 Gene in Barley Is the Same Size as the WT RPS3 Protein in Other C4 Plants
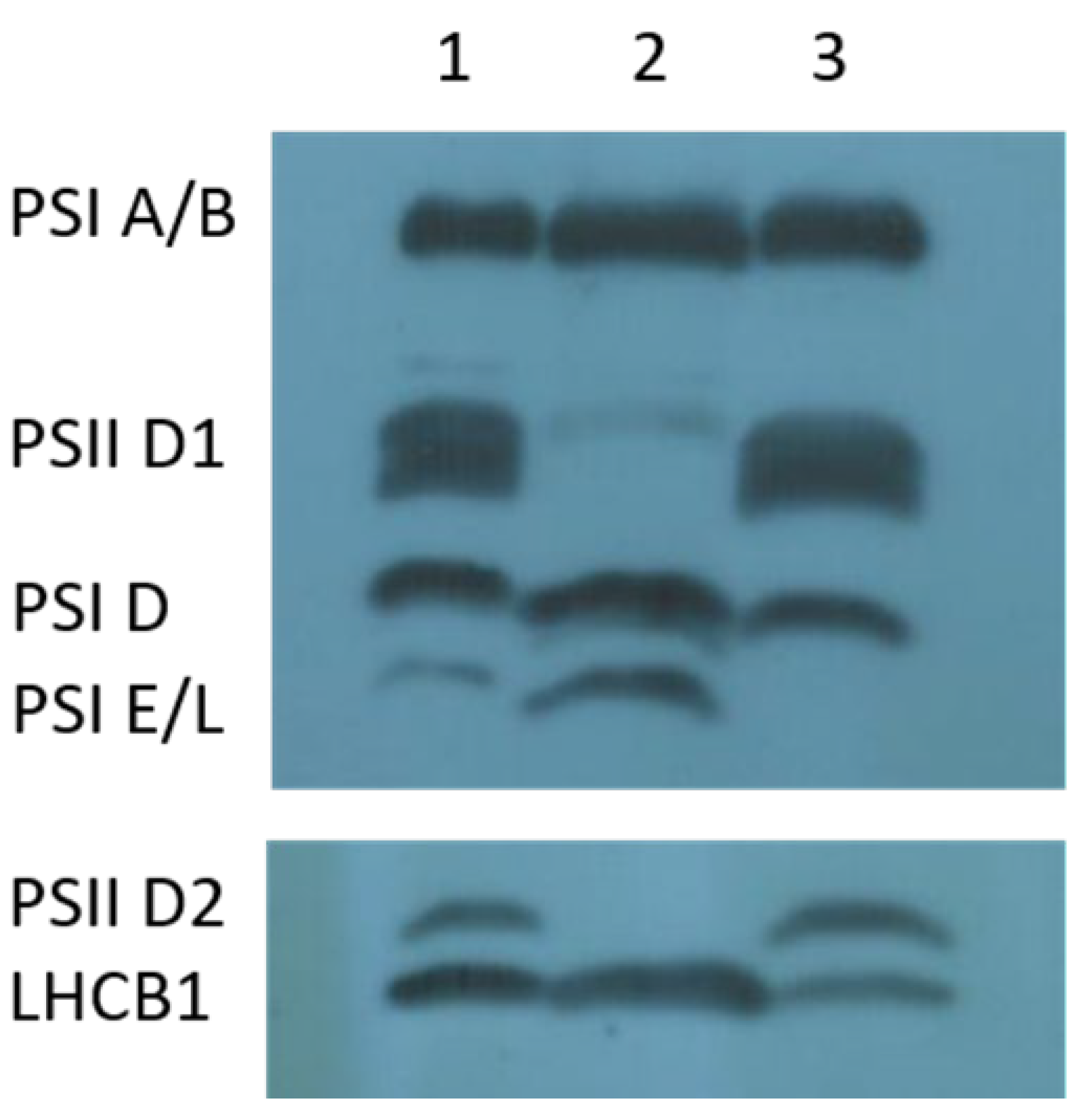
2.4. The Deletion in the psbA Gene Disrupts PSII Function and Causes Seedling Lethality
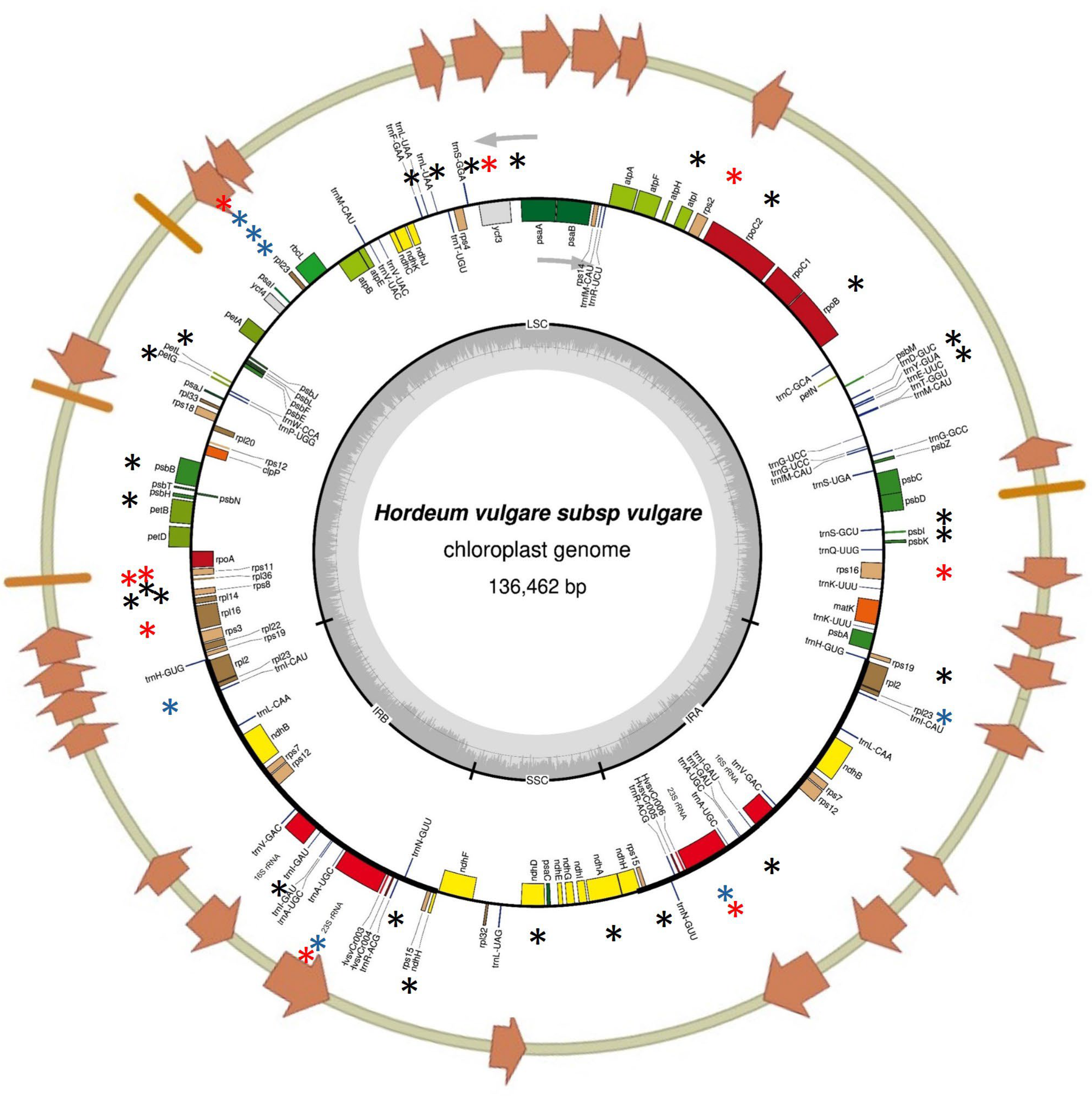
2.5. Complete Plastome Sequencing Confirms Large Indels Identified by cpTILLING and Reveals Additional Polymorphisms
2.5.1. Plastome of the rps19 Amplicon Insertion Mutant
2.5.2. Plastome of the psbA Gene Deletion Mutant
2.5.3. Plastome of the rpl33 Amplicon Deletion Mutant
2.5.4. Plastome of the rps3 Gene Deletion Mutant
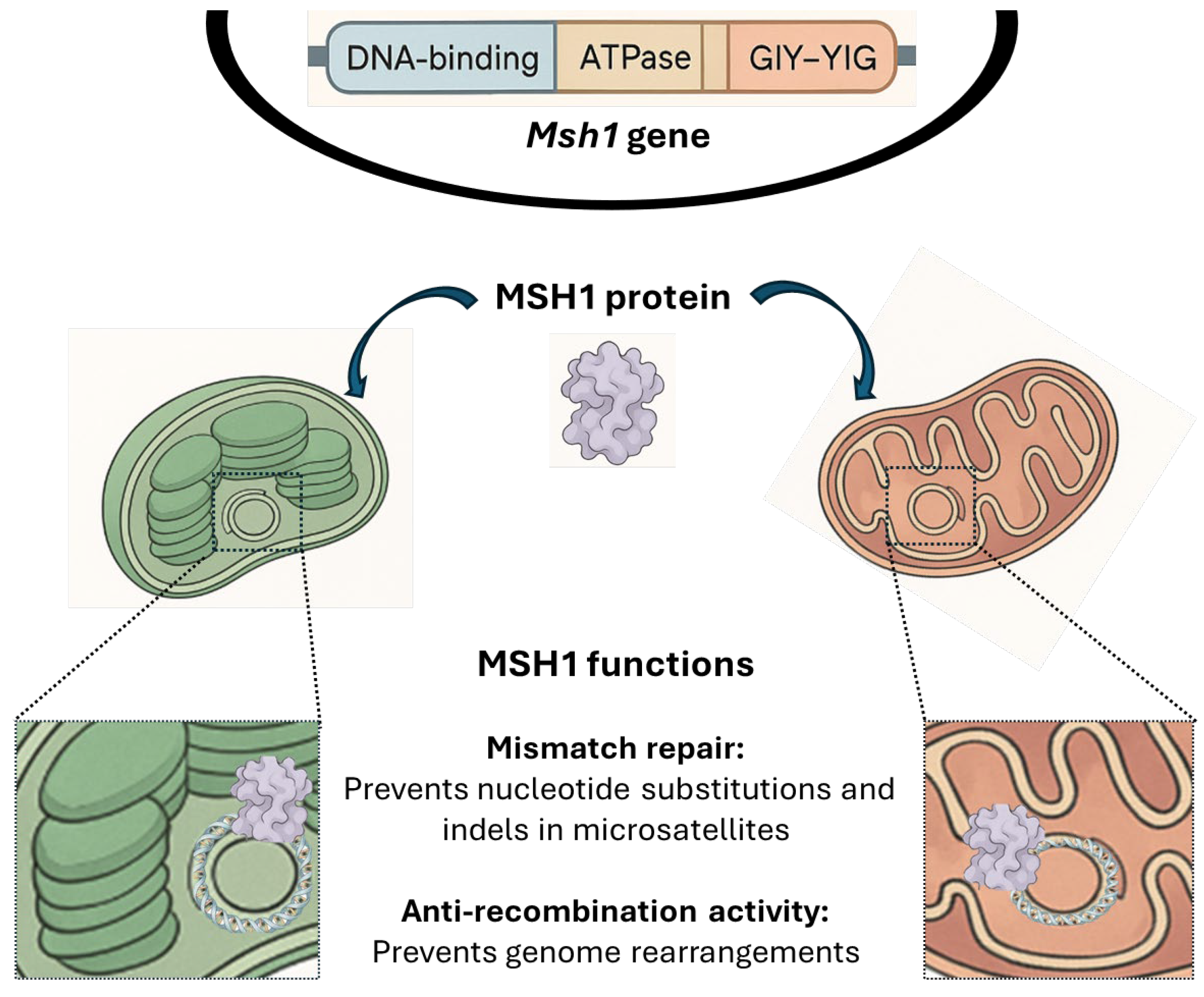
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. DNA Isolation
4.3. CJE Digestion and Electrophoresis of Amplicons
4.4. Sequencing of the Msh1 Gene Region Carrying the Premature Stop Codon Mutation in the Seedlings with Large Indels
4.5. Plastome Sequencing and Bioinformatic Analysis
4.6. Thylakoid Membrane Isolation and Immunoblotting
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clegg, M.T.; Gaut, B.S.; Learn, G.H.; Morton, B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6795–6801. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, Y.; Terachi, T.; Sasakuma, T. Structural analysis of length mutations in a hot-spot region of wheat chloroplast DNAs. Curr. Genet. 1992, 22, 251–258. [Google Scholar] [CrossRef]
- Kirk, J.T.O.; Tilney-Bassett, R.A.E. The Plastids, Their Chemistry, Structure, Growth, and Inheritance; W.H. Freeman and Company: London, UK, 1967. [Google Scholar]
- Börner, T.; Sears, B. Plastome mutants. Plant Mol. Biol. Rep. 1986, 4, 69–72. [Google Scholar] [CrossRef]
- Prina, A.R.; Pacheco, M.G.; Landau, A.M. Mutation Induction in Cytoplasmic Genomes. In Plant Mutation Breeding and Biotechnology; Shu, Q.S., Forster, B.P., Nakagawa, H., Eds.; CABI: Wallingford, UK, 2012; pp. 201–206. [Google Scholar]
- Greiner, S. Plastome Mutants of Higher Plants. In Genomics of Chloroplasts and Mitochondria; Bock, R., Knoop, V., Eds.; Springer: Dordrecht, The Netherlands, 2012; Volume 35, pp. 237–266. [Google Scholar]
- Prina, A.R. A mutator nuclear gene inducing a wide spectrum of cytoplasmically inherited chlorophyll deficiencies in barley. Theor. Appl. Genet. 1992, 85, 245–251. [Google Scholar] [CrossRef]
- Prina, A.R. Mutator-Induced Cytoplasmic Mutants in Barley: Genetic Evidence of Activation of a Putative Chloroplast Transposon. J. Hered. 1996, 87, 385–389. [Google Scholar] [CrossRef][Green Version]
- Landau, A.; Brizuela, V.; Lencina, F.; Martínez, A.; Tcach, M.; Díaz, D.; Pacheco, M.G.; Prina, A.R. Induced mutations in crop plants: Mutants of scientific and/or agronomic interest at the Institute of Genetics “Ewald A. Favret”. ACI Av. Cienc. Ing. 2021, 12, 14. [Google Scholar] [CrossRef]
- Landau, A.; Lencina, F.; Pacheco, M.G.; Prina, A.R. Plastome mutations and recombination events in barley chloroplast mutator seedlings. J. Hered. 2016, 107, 266–273. [Google Scholar] [CrossRef][Green Version]
- Lencina, F.; Landau, A.; Petterson, M.E.; Pacheco, M.G.; Prina, A.R. The rpl23 gene and pseudogene are hotspots of illegitimate recombination in barley chloroplast mutator seedlings. Sci. Rep. 2019, 9, 9960. [Google Scholar] [CrossRef] [PubMed]
- Spampinato, C.P.; Gomez, R.L.; Galles, C.; Lario, L.D. From Bacteria to Plants: A Compendium of Mismatch Repair Assays. Mutat. Res. 2009, 682, 110–128. [Google Scholar] [CrossRef] [PubMed]
- Lencina, F.; Landau, A.; Prina, A.R. The Barley Chloroplast Mutator (cpm) Mutant: All Roads Lead to the Msh1 Gene. Int. J. Mol. Sci. 2022, 23, 1814. [Google Scholar] [CrossRef]
- Ogihara, Y.; Ohsawa, T. Molecular analysis of the complete set of length mutations found in the plastomes of Triticum-Aegilops species. Genome 2002, 45, 956–962. [Google Scholar] [CrossRef]
- Stoike, L.L.; Sears, B.B. Plastome mutator-induced alterations arise in Oenothera chloroplast DNA through template slippage. Genetics 1998, 149, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, Y.; Terachi, T.; Sasakuma, T. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc. Natl. Acad. Sci. USA 1988, 85, 8573–8577. [Google Scholar] [CrossRef]
- Ogihara, Y.; Terachi, T.; Sasakuma, T. Molecular analysis of the hot spot region related to length mutations in wheat chloroplast DNAs. I. Nucleotide divergence of genes and intergenic spacer regions located in the hot spot region. Genetics 1991, 129, 873–884. [Google Scholar] [CrossRef]
- Morton, B.R.; Clegg, M.T. A chloroplast DNA mutational hotspot and gene conversion in a noncoding region near rbcL in the grass family (Poaceae). Curr. Genet. 1993, 24, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Xia, Y.; Wang, G. An improved heteroduplex analysis for rapid genotyping of SNPs and single base pair indels. Biotechniques 2019, 67, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Kakui, H.; Yamazaki, M.; Shimizu, K. PRIMA: A rapid and cost effective genotyping method to detect single nucleotide differences using probe induced heteroduplexes. Sci. Rep. 2021, 11, 20741. [Google Scholar] [CrossRef]
- Tsai, H.; Howell, T.; Nitcher, R.; Missirian, V.; Watson, B.; Ngo, K.J.; Lieberman, M.; Fass, J.; Uauy, C.; Tran, R.K.; et al. Discovery of Rare Mutations in Populations: TILLING by Sequencing. Plant Physiol. 2011, 156, 1257–1268. [Google Scholar] [CrossRef]
- Lovett, S.T. Encoded errors: Mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 2004, 52, 1243–1253. [Google Scholar] [CrossRef]
- Maréchal, A.; Brisson, N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010, 186, 299–317. [Google Scholar] [CrossRef]
- Fischer, N.; Stampacchia, O.; Redding, K.; Rochaix, J.D. Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 1996, 251, 373–380. [Google Scholar] [CrossRef]
- Bi, X.; Liu, L. recA-independent and recA-dependent intramolecular plasmid recombination. Differential homology requirement and distance effect. J. Mol. Biol. 1994, 235, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Dauvillee, D.; Hilbig, L.; Preiss, S.; Johanningmeier, U. Minimal extent of sequence homology required for homologous recombination at the psbA locus in Chlamydomonas reinhardtii chloroplasts using PCR-generated DNA fragments. Photosynth. Res. 2004, 79, 219–224. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, P.; Chang, W.; Yu, K.; Lin, C. Plastid Transformation: How Does it Work? Can it Be Applied to Crops? What Can it Offer? Int. J. Mol. Sci. 2020, 21, 4854. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, U.; Alani, E. Understanding how mismatch repair proteins participate in the repair/antirecombination decision. FEMS Yeast Res. 2016, 16, fow071. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, Y.Z.; Arrieta-Montiel, M.P.; Virdi, K.S.; de Paula, W.B.; Widhalm, J.R.; Basset, G.J.; Davila, J.I.; Elthon, T.E.; Elowsky, C.G.; Sato, S.J. MSH1 is a nucleoid protein that alters mitochondrial and plastid properties and plant response to high light. Plant Cell 2011, 23, 3428–3441. [Google Scholar] [CrossRef]
- Wu, Z.; Waneka, G.; Broz, A.K.; King, C.R.; Sloan, D.B. MSH1 Is Required for Maintenance of the Low Mutation Rates in Plant Mitochondrial and Plastid Genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 16448–16455. [Google Scholar] [CrossRef]
- Zou, Y.; Zhu, W.; Sloan, D.B.; Wu, Z. Long-read sequencing characterizes mitochondrial and plastid genome variants in Arabidopsis msh1 mutants. Plant J. 2022, 112, 738–755. [Google Scholar] [CrossRef]
- Wang, J.; Zou, Y.; Mower, J.P.; Reeve, W.; Wu, Z. Rethinking the mutation hypotheses of plant organellar DNA. Genom. Commun. 2024, 1, e003. [Google Scholar] [CrossRef]
- de Vitry, C.; Olive, J.; Drapier, D.; Recouvreur, M.; Wollman, F.A. Posttranslational events leading to the assembly of photosystem II protein complex: A study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol. 1989, 109, 991–1006. [Google Scholar] [CrossRef]
- Järvi, S.; Suorsa, M.; Aro, E. Photosystem II repair in plant chloroplasts. Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 2015, 1847, 900–909. [Google Scholar] [CrossRef]
- Saski, C.; Lee, S.B.; Fjellheim, S.; Guda, C.; Jansen, R.K.; Luo, H.; Tomkins, J.; Rognli, O.A.; Daniell, H.; Clarke, J.L. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor. Appl. Genet. 2007, 115, 571–590. [Google Scholar] [CrossRef]
- Chevigny, N.; Schatz-Daas, D.; Lotfi, F.; Gualberto, J.M. DNA Repair and the Stability of the Plant Mitochondrial Genome. Int. J. Mol. Sci. 2020, 21, 328. [Google Scholar] [CrossRef]
- Dellaporta, S. Plant DNA miniprep and microprep: Versions 2.1–2.3. In The Maize Handbook; Freeling, M., Walbot, V., Eds.; Springer: New York, NY, USA, 1994; pp. 522–525. [Google Scholar]
- Till, B.J.; Zerr, T.; Comai, L.; Henikoff, S. A protocol for TILLING and Ecotilling in plants and animals. Nat. Protoc. 2006, 1, 2465–2477. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses. Nucleic Acids Res. 2018, 46, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. VISTA: Visualizing Global DNA Sequence Alignments of Arbitrary Length. Bioinformatics 2000, 16, 1046. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Guiamet, J.J.; Tyystjärvi, E.; Tyystjärvi, T.; John, I.; Kairavuo, M.; Pichersky, E.; Noodén, L.D. Photoinhibition and loss of photosystem II reaction centre proteins during senescence of soybean leaves. Enhancement of photoinhibition by the ‘stay-green’ mutation cytG. Physiol. Plant. 2002, 115, 468–478. [Google Scholar] [CrossRef]
- ClickUp. ClickUp Brain [Artificial Intelligence Tool]. 2025. Available online: https://clickup.com/ (accessed on 14 August 2025).
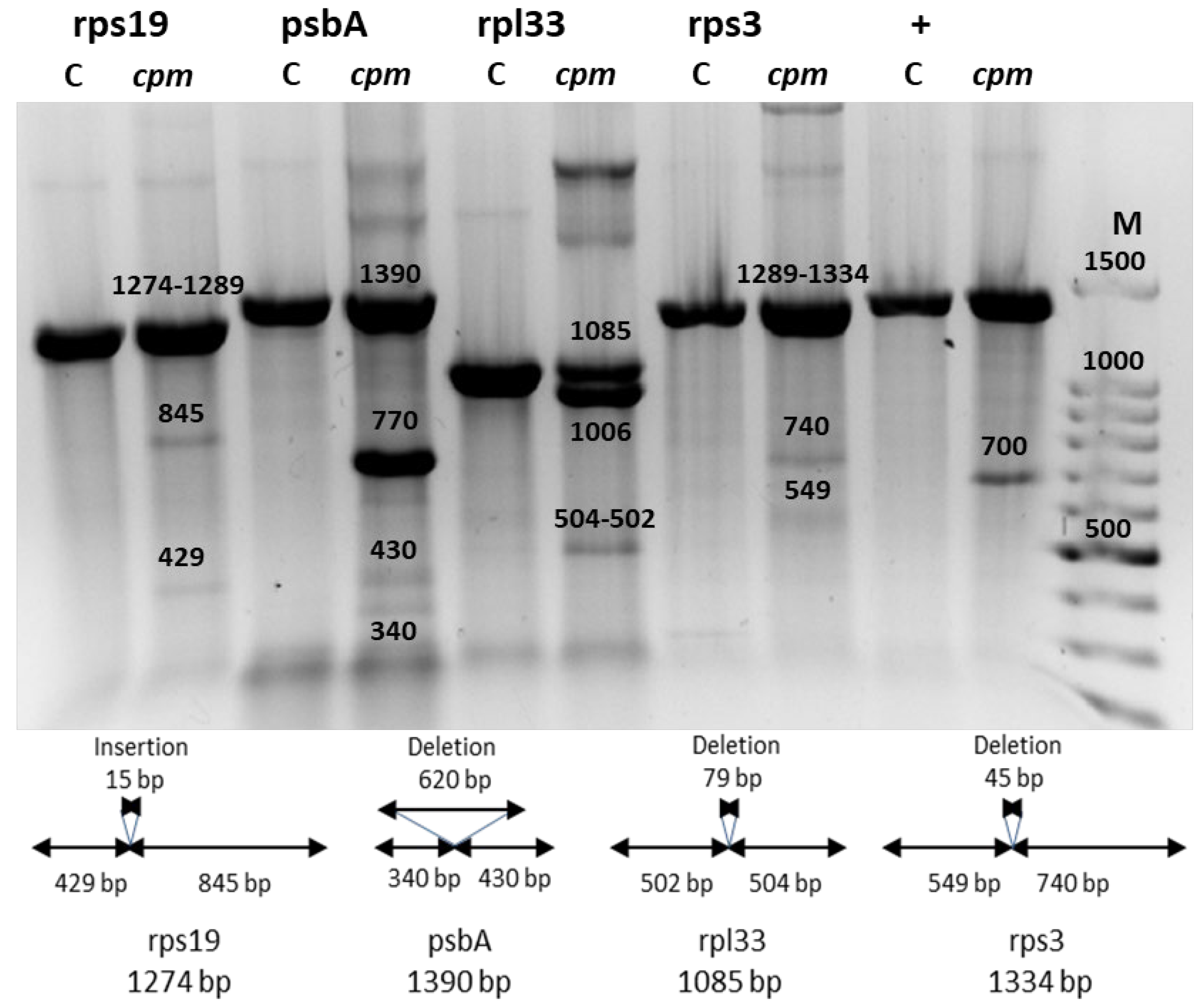
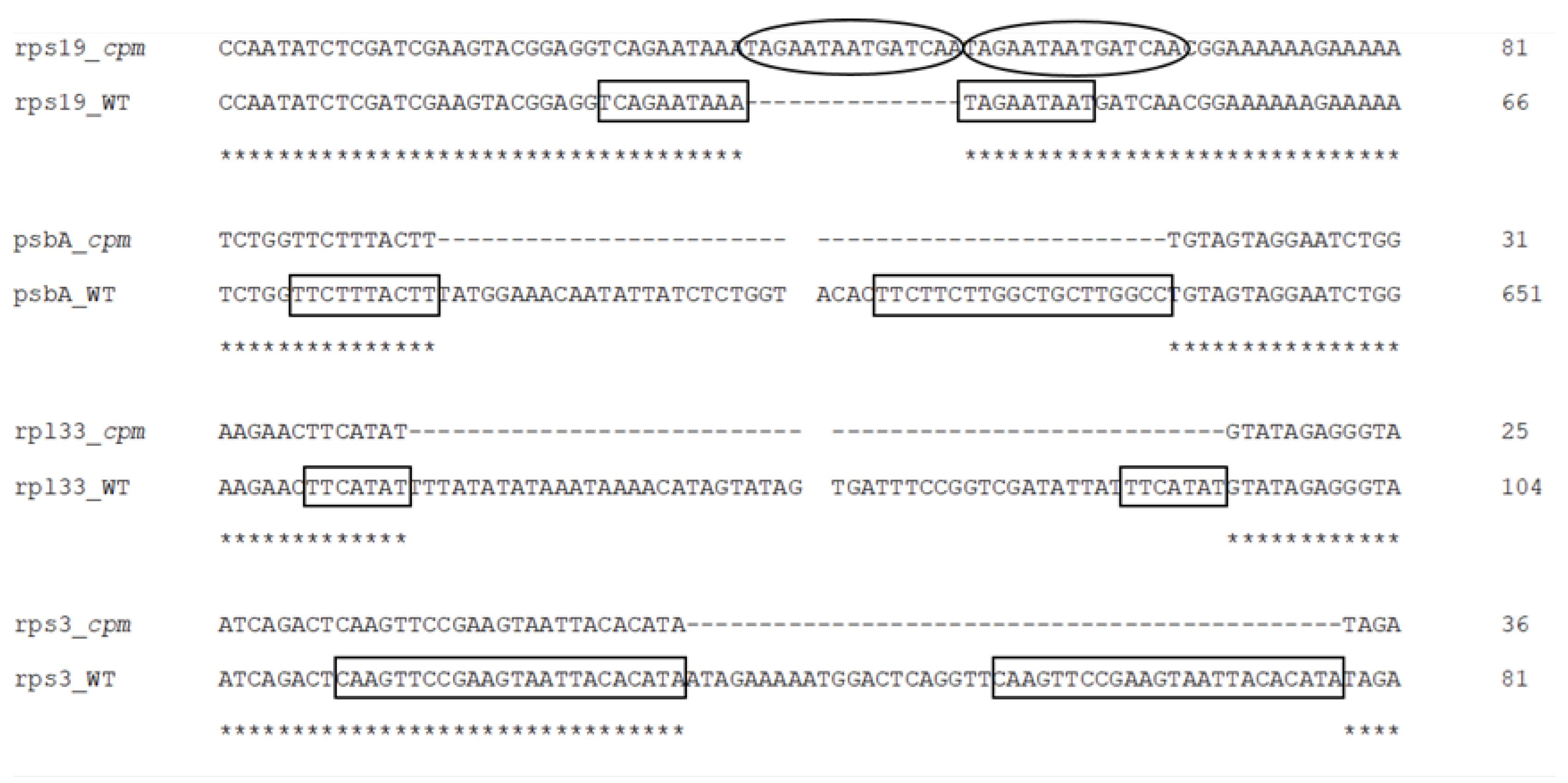

| Large Indel | Reference Genome Position | Plastome Region | Indel Length (bp) | Alignment of Direct Repeats |
|---|---|---|---|---|
| Insertion in tRNAHis–rps19 | 81544 | rps19 intron IRa | 15 | 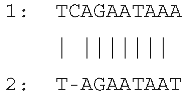 |
| Deletion in psbA | 845 | psbA gene LSC | 620 (217–836) |  |
| Deletion in rpl33–rps18 | 66047 | Intergenic region LSC | 79 | 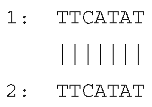 |
| Deletion in rps3 | 80340 | rps3 gene LSC | 45 (188–232) |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lencina, F.; Prina, A.R.; Pacheco, M.G.; Kobayashi, K.; Landau, A.M. Four Large Indels in Barley Chloroplast Mutator (cpm) Seedlings Reinforce the Hypothesis of a Malfunction in the MMR System. Int. J. Mol. Sci. 2025, 26, 8644. https://doi.org/10.3390/ijms26178644
Lencina F, Prina AR, Pacheco MG, Kobayashi K, Landau AM. Four Large Indels in Barley Chloroplast Mutator (cpm) Seedlings Reinforce the Hypothesis of a Malfunction in the MMR System. International Journal of Molecular Sciences. 2025; 26(17):8644. https://doi.org/10.3390/ijms26178644
Chicago/Turabian StyleLencina, Franco, Alberto R. Prina, María G. Pacheco, Ken Kobayashi, and Alejandra M. Landau. 2025. "Four Large Indels in Barley Chloroplast Mutator (cpm) Seedlings Reinforce the Hypothesis of a Malfunction in the MMR System" International Journal of Molecular Sciences 26, no. 17: 8644. https://doi.org/10.3390/ijms26178644
APA StyleLencina, F., Prina, A. R., Pacheco, M. G., Kobayashi, K., & Landau, A. M. (2025). Four Large Indels in Barley Chloroplast Mutator (cpm) Seedlings Reinforce the Hypothesis of a Malfunction in the MMR System. International Journal of Molecular Sciences, 26(17), 8644. https://doi.org/10.3390/ijms26178644





