Selecting Optimal Housekeeping Genes for RT-qPCR in Endometrial Cancer Studies: A Narrative Review
Abstract
1. Introduction
2. A Critique of Glyceraldehyde-3-Phosphate Dehydrogenase as a Housekeeping Gene
3. Housekeeping Gene(s) for Studies of Normal Endometrium
4. Housekeeping Gene(s) for Studies of Endometrial Cancer
5. Our Experience
6. Discrepancies in RT-qPCR Studies on Sex Hormone Receptors in Human Endometrial Cancer Tissue
7. Necessity of Validation of Housekeeping Genes Before Experimentation
8. Emerging Strategies and Innovative Tools in HKG Selection
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Hopkins, N.H.; Roberts, J.W.; Steitz, J.A.; Weiner, A.M. The functioning of higher eucaryotic genes. In Molecular Biology of the Gene, 1st ed.; Benjamin/Cummings: Menlo Park, CA, USA, 1965; Volume 1, Chapter 21; p. 704. [Google Scholar]
- Warrington, J.A.; Nair, A.; Mahadevappa, M.; Tsyganskaya, M. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol. Genom. 2000, 2, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Butte, A.J.; Dzau, V.J.; Glueck, S.B. Further defining housekeeping, or “maintenance,” genes. Focus on “A compendium of gene expression in normal human tissues”. Physiol. Genom. 2001, 7, 95–96. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; He, F.; Song, S.; Wang, J.; Yu, J. How many human genes can be defined as housekeeping with current expression data? BMC Genom. 2008, 9, 172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, W.-H. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol. Biol. Evol. 2004, 21, 236–239. [Google Scholar] [CrossRef]
- Lercher, M.J.; Urrutia, A.O.; Hurst, L.D. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 2002, 31, 180–183. [Google Scholar] [CrossRef]
- Zhu, J.; He, F.; Hu, S.; Yu, J. On the nature of human housekeeping genes. Trends Genet. 2008, 24, 481–484. [Google Scholar] [CrossRef]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes are compact. Trends Genet. 2003, 19, 362–365. [Google Scholar] [CrossRef]
- Hsiao, L.-L.; Dangond, F.; Yoshida, T.; Hong, R.; Jensen, R.V.; Misra, J.; Dillon, W.; Lee, K.F.; Clark, K.E.; Haverty, P.; et al. A compendium of gene expression in normal human tissues. Physiol. Genom. 2001, 7, 97–104. [Google Scholar] [CrossRef]
- Karge, W.H., 3rd; Schaefer, E.J.; Ordovas, J.M. Quantification of mRNA by polymerase chain reaction (PCR) using an internal standard and a nonradioactive detection method. Methods Mol. Biol. 1998, 110, 43–61. [Google Scholar] [CrossRef]
- de Kok, J.B.; Roelofs, R.W.; Giesendorf, B.A.; Pennings, J.L.; Waas, E.T.; Feuth, T.; Swinkels, D.W.; Span, P.N. Normalization of gene expression measurements in tumor tissues: Comparison of 13 endogenous control genes. Mod. Pathol. 2005, 85, 154–159. [Google Scholar] [CrossRef]
- Feroze-Merzoug, F.; Berquin, I.; Dey, J.; Chen, Y. Peptidylprolyl Isomerase A (PPIA) as a Preferred Internal Control Over GAPDH and β-Actin in Quantitative RNA Analyses. BioTechniques 2002, 32, 776–782. [Google Scholar] [CrossRef]
- Sadek, K.H.; Cagampang, F.R.; Bruce, K.D.; Shreeve, N.; Macklon, N.; Cheong, Y. Variation in stability of housekeeping genes in endometrium of healthy and polycystic ovarian syndrome women. Hum. Reprod. 2011, 27, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Sirover, M.A. New insights into an old protein: The functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1999, 1432, 159–184. [Google Scholar] [CrossRef] [PubMed]
- Sirover, M.A. Pleiotropic effects of moonlighting glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in cancer progression, invasiveness, and metastases. Cancer Metastasis Rev. 2018, 37, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Sirover, M.A. The role of posttranslational modification in moonlighting glyceraldehyde-3-phosphate dehydrogenase structure and function. Amino Acids 2021, 53, 507–515. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Zhang, F.; Hong, C.-Q.; Giuliano, A.E.; Cui, X.-J.; Zhou, G.-J.; Zhang, G.-J.; Cui, Y.-K. Critical protein GAPDH and its regulatory mechanisms in cancer cells. Cancer Biol. Med. 2015, 12, 10–22. [Google Scholar] [CrossRef]
- Tristan, C.; Shahani, N.; Sedlak, T.W.; Sawa, A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell. Signal. 2011, 23, 317–323. [Google Scholar] [CrossRef]
- Barber, R.D.; Harmer, D.W.; Coleman, R.A.; Clark, B.J. GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genom. 2005, 21, 389–395. [Google Scholar] [CrossRef]
- Ke, L.; Chen, Z.; Yung, W. A reliability test of standard-based quantitative PCR: Exogenous vs endogenous standards. Mol. Cell. Probes 2000, 14, 127–135. [Google Scholar] [CrossRef]
- Blanquicett, C.; Johnson, M.R.; Heslin, M.; Diasio, R.B. Housekeeping gene variability in normal and carcinomatous colorectal and liver tissues: Applications in pharmacogenomic gene expression studies. Anal. Biochem. 2002, 303, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Zakrajsek, B.A. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 2000, 46, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Simons, J.W. Direct comparison of GAPDH, β-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem. Biophys. Res. Commun. 1999, 259, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Caradec, J.; Sirab, N.; Keumeugni, C.; Moutereau, S.; Chimingqi, M.; Matar, C.; Revaud, D.; Bah, M.; Manivet, P.; Conti, M.; et al. ‘Desperate house genes’: The dramatic example of hypoxia. Br. J. Cancer 2010, 102, 1037–1043. [Google Scholar] [CrossRef]
- Waxman, S.; Wurmbach, E. De-regulation of common housekeeping genes in hepatocellular carcinoma. BMC Genom. 2007, 8, 243. [Google Scholar] [CrossRef]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control selection for RNA quantitation. BioTechniques 2000, 29, 332–337. [Google Scholar] [CrossRef]
- Ohkubo, M.; Sawada, K.; Sakiyama, S. Enhanced expression of a 37,000-dalton protein in human lung cancers. Jpn. J. Cancer Res. 1986, 77, 546–553. [Google Scholar] [CrossRef]
- Ohkubo, M.; Nakamura, Y.; Tokunaga, K.; Sakiyama, S. Similarity between glyceraldehyde-3-phosphate dehydrogenase and a 37,000-dalton protein which is abundantly expressed in human lung cancers. Jpn. J. Cancer Res. 1986, 77, 554–559. [Google Scholar] [CrossRef]
- Tokunaga, K.; Nakamura, Y.; Sakata, K.; Fujimori, K.; Ohkubo, M.; Sawada, K.; Sakiyama, S. Enhanced expression of a glyceraldehyde-3-phosphate dehydrogenase gene in human lung cancers. Cancer Res. 1987, 47, 5616–5619. [Google Scholar]
- Schek, N.; Hall, B.L.; Finn, O.J. Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in human pancreatic adenocarcinoma. Cancer Res. 1988, 48, 6354–6359. [Google Scholar]
- Persons, D.A.; Schek, N.; Hall, B.L.; Finn, O.J. Increased expression of glycolysis-associated genes in oncogene-transformed and growth-accelerated states. Mol. Carcinog. 1989, 2, 88–94. [Google Scholar] [CrossRef]
- Perfetti, V.; Manenti, G.; Dragani, T.A. Expression of housekeeping genes in Hodgkin’s disease lymph nodes. Leukemia 1991, 5, 1110–1112. [Google Scholar]
- Ripple, M.O.; Wilding, G. Alteration of glyceraldehyde-3-phosphate dehydrogenase activity and messenger RNA content by androgen in human prostate carcinoma cells. Cancer Res. 1995, 55, 4234–4236. [Google Scholar] [PubMed]
- Gong, Y.; Cui, L.; Minuk, G.Y. Comparison of glyceraldehyde-3-phosphate dehydrogenase and 28S-ribosomal RNA gene expression in human hepatocellular carcinoma. Hepatology 1996, 23, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Rondinelli, R.; Epner, D.; Tricoli, J. Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in late pathological stage human prostate cancer. Prostate Cancer Prostatic Dis. 1997, 1, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Kim, S.J.; Han, S.M.; Paik, S.Y.; Hur, S.Y.; Kim, Y.W.; Lee, J.M.; Namkoong, S.E. Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in human cervical cancers. Gynecol. Oncol. 1998, 71, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Gerard, C.J.; Andrejka, L.M.; Macina, R.A. Mitochondrial ATP synthase 6 as an endogenous control in the quantitative RT-PCR analysis of clinical cancer samples. Mol. Diagn. 2000, 5, 39–46. [Google Scholar] [CrossRef]
- Révillion, F.; Pawlowski, V.; Hornez, L.; Peyrat, J.-P. Glyceraldehyde-3-phosphate dehydrogenase gene expression in human breast cancer. Eur. J. Cancer 2000, 36, 1038–1042. [Google Scholar] [CrossRef]
- Vilà, M.R.; Nicolás, A.; Morote, J.; de Torres, I.; Meseguer, A. Increased glyceraldehyde-3-phosphate dehydrogenase expression in renal cell carcinoma identified by RNA-based, arbitrarily primed polymerase chain reaction. Cancer 2000, 89, 152–164. [Google Scholar] [CrossRef]
- Goidin, D.; Mamessier, A.; Staquet, M.-J.; Schmitt, D.; Berthier-Vergnes, O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and β-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 2001, 295, 17–21. [Google Scholar] [CrossRef]
- Aerts, J.L.; Gonzales, M.I.; Topalian, S.L. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. BioTechniques 2004, 36, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Altenberg, B.; Greulich, K. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 2004, 84, 1014–1020. [Google Scholar] [CrossRef]
- Rubie, C.; Kempf, K.; Hans, J.; Su, T.; Tilton, B.; Georg, T.; Brittner, B.; Ludwig, B.; Schilling, M. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol. Cell. Probes 2005, 19, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Ohl, F.; Jung, M.; Radonić, A.; Sachs, M.; Loening, S.A.; Jung, K. Identification and validation of suitable endogenous reference genes for gene expression studies of human bladder cancer. J. Urol. 2006, 175, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Yuan, S.; Hu, Y.; Zhang, H.; Wu, W.; Zeng, Z.; Yang, J.; Yun, J.; Xu, R.; Huang, P. Over-expression of GAPDH in human colorectal carcinoma as a preferred target of 3-bromopyruvate propyl ester. J. Bioenerg. Biomembr. 2012, 44, 117–125. [Google Scholar] [CrossRef]
- Wang, D.; Moothart, D.R.; Lowy, D.R.; Qian, X.; Coles, J.A. The expression of glyceraldehyde-3-phosphate dehydrogenase associated cell cycle (GACC) genes correlates with cancer stage and poor survival in patients with solid tumors. PLoS ONE 2013, 8, e61262. [Google Scholar] [CrossRef]
- Ramos, D.; Pellín-Carcelén, A.; Agustí, J.; Murgui, A.; Jordá, E.; Pellín, A.; Monteagudo, C. Deregulation of glyceraldehyde-3-phosphate dehydrogenase expression during tumor progression of human cutaneous melanoma. Anticancer Res. 2015, 35, 439–444. [Google Scholar]
- Shen, C.; Li, W.; Wang, Y. Research on the oncogenic role of the house-keeping gene GAPDH in human tumors. Transl. Cancer Res. 2023, 12, 525–535. [Google Scholar] [CrossRef]
- Wang, J.; Yu, X.; Cao, X.; Tan, L.; Jia, B.; Chen, R.; Li, J. GAPDH: A common housekeeping gene with an oncogenic role in pan-cancer. Comput. Struct. Biotechnol. J. 2023, 21, 4056–4069. [Google Scholar] [CrossRef]
- Gresner, P.; Gromadzinska, J.; Wasowicz, W. Reference genes for gene expression studies on non-small cell lung cancer. Acta Biochim. Pol. 2009, 56, 307–316. [Google Scholar] [CrossRef]
- Chen, L.; Jin, Y.; Wang, L.; Sun, F.; Yang, X.; Shi, M.; Zhan, C.; Shi, Y.; Wang, Q. Identification of reference genes and miRNAs for qRT-PCR in human esophageal squamous cell carcinoma. Med. Oncol. 2016, 34, 2. [Google Scholar] [CrossRef]
- Janssens, N.; Janicot, M.; Perera, T.; Bakker, A. Housekeeping genes as internal standards in cancer research. Mol. Diagn. 2004, 8, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Cairns, R.A.; Harris, I.; McCracken, S.; Mak, T.W. Cancer cell metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Cairns, R.A. Drivers of the Warburg phenotype. Cancer J. 2015, 21, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Cale, J.M.; Millican, D.S.; Itoh, H.; Magness, R.R.; Bird, I.M. Pregnancy induces an increase in the expression of glyceraldehyde-3-phosphate dehydrogenase in uterine artery endothelial cells. J. Soc. Gynecol. Investig. 1997, 4, 284–292. [Google Scholar] [CrossRef]
- Xu, H.; Bionaz, M.; Sloboda, D.M.; Ehrlich, L.; Li, S.; Newnham, J.P.; Dudenhausen, J.W.; Henrich, W.; Plagemann, A.; Challis, J.R.; et al. The dilution effect and the importance of selecting the right internal control genes for RT-qPCR: A paradigmatic approach in fetal sheep. BMC Res. Notes 2015, 8, 58. [Google Scholar] [CrossRef]
- Zainuddin, A.; Chua, K.H.; Rahim, N.A.; Makpol, S. Effect of experimental treatment on GAPDH mRNA expression as a housekeeping gene in human diploid fibroblasts. BMC Mol. Biol. 2010, 11, 59. [Google Scholar] [CrossRef]
- Finnegan, M.C.; Goepel, J.R.; Hancock, B.W.; Goyns, M.H. Investigation of the expression of housekeeping genes in non-Hodgkin’s lymphoma. Leuk. Lymphoma 1993, 10, 387–393. [Google Scholar] [CrossRef]
- Seykora, J.T.; Jih, D.; Elenitsas, R.; Horng, W.-H.; Elder, D.E. Gene expression profiling of melanocytic lesions. Am. J. Dermatopathol. 2003, 25, 6–11. [Google Scholar] [CrossRef]
- Jóźwik, M.; Okungbowa, O.E.; Lipska, A.; Jóźwik, M.; Smoktunowicz, M.; Semczuk, A.; Jóźwik, M.; Radziwon, P. Surface antigen expression on peripheral blood monocytes in women with gynecologic malignancies. BMC Cancer 2015, 15, 129. [Google Scholar] [CrossRef]
- Carson, D.D.; Lagow, E.; Thathiah, A.; Al-Shami, R.; Farach-Carson, M.C.; Vernon, M.; Yuan, L.; Fritz, M.A.; Lessey, B. Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening. Mol. Hum. Reprod. 2002, 8, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Kao, L.C.; Tulac, S.; Lobo, S.; Imani, B.; Yang, J.P.; Germeyer, A.; Osteen, K.; Taylor, R.N.; Lessey, B.A.; Giudice, L.C. Global gene profiling in human endometrium during the window of implantation. Endocrinology 2002, 143, 2119–2138. [Google Scholar] [CrossRef] [PubMed]
- Gebeh, A.K.; Marczylo, E.L.; Amoako, A.A.; Willets, J.M.; Konje, J.C. Variation in stability of endogenous reference genes in fallopian tubes and endometrium from healthy and ectopic pregnant women. Int. J. Mol. Sci. 2012, 13, 2810–2826. [Google Scholar] [CrossRef] [PubMed]
- Caracausi, M.; Piovesan, A.; Antonaros, F.; Strippoli, P.; Vitale, L.; Pelleri, M.C. Systematic identification of human housekeeping genes possibly useful as references in gene expression studies. Mol. Med. Rep. 2017, 16, 2397–2410. [Google Scholar] [CrossRef]
- Dirnhofer, S.; Berger, C.; Untergasser, G.; Geley, S.; Berger, P. Human β-actin retropseudogenes interfere with RT-PCR. Trends Genet. 1995, 11, 380–381. [Google Scholar] [CrossRef]
- Tonner, P.; Srinivasasainagendra, V.; Zhang, S.; Zhi, D. Detecting transcription of ribosomal protein pseudogenes in diverse human tissues from RNA-seq data. BMC Genom. 2012, 13, 412. [Google Scholar] [CrossRef]
- Krasnov, G.S.; Kudryavtseva, A.V.; Snezhkina, A.V.; Lakunina, V.A.; Beniaminov, A.D.; Melnikova, N.V.; Dmitriev, A.A. Pan-cancer analysis of TCGA data revealed promising reference genes for qPCR normalization. Front. Genet. 2019, 10, 97. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Luo, D.; Liao, D.J.; Rishi, A. Pseudogenes as weaknesses of ACTB (Actb) and GAPDH (Gapdh) used as reference genes in reverse transcription and polymerase chain reactions. PLoS ONE 2012, 7, e41659. [Google Scholar] [CrossRef]
- Bhuva, D.D.; Cursons, J.; Davis, M.J. Stable gene expression for normalisation and single-sample scoring. Nucleic Acids Res. 2020, 48, e113. [Google Scholar] [CrossRef]
- Gu, W.; Wang, Y.; Xu, R.; Li, J.; Jin, J.; Zhao, J.; Chen, Y.; Lu, Y.; Zhang, G. Experimental assessment of robust reference genes for qRT-PCR in lung cancer studies. Front. Oncol. 2023, 13, 1178629. [Google Scholar] [CrossRef]
- Sharan, R.N.; Vaiphei, S.T.; Nongrum, S.; Keppen, J.; Ksoo, M. Consensus reference gene(s) for gene expression studies in human cancers: End of the tunnel visible? Cell. Oncol. (Dordr.) 2015, 38, 419–431. [Google Scholar] [CrossRef]
- Kheirelseid, E.A.; Chang, K.H.; Newell, J.; Kerin, M.J.; Miller, N. Identification of endogenous control genes for normalisation of real-time quantitative PCR data in colorectal cancer. BMC Mol. Biol. 2010, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Guertler, R.; Naim, S.; Nixdorf, S.; Fedier, A.; Hacker, N.F.; Heinzelmann-Schwarz, V.; Huang, S. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS ONE 2013, 8, e59180. [Google Scholar] [CrossRef]
- Rácz, G.A.; Nagy, N.; Tóvári, J.; Apáti, Á.; Vértessy, B.G. Identification of new reference genes with stable expression patterns for gene expression studies using human cancer and normal cell lines. Sci. Rep. 2021, 11, 19459. [Google Scholar] [CrossRef] [PubMed]
- Romani, C.; Calza, S.; Todeschini, P.; Tassi, R.A.; Zanotti, L.; Bandiera, E.; Sartori, E.; Pecorelli, S.; Ravaggi, A.; Santin, A.D.; et al. Identification of optimal reference genes for gene expression normalization in a wide cohort of endometrioid endometrial carcinoma tissues. PLoS ONE 2014, 9, e113781. [Google Scholar] [CrossRef] [PubMed]
- Ayakannu, T.; Taylor, A.H.; Willets, J.M.; Brown, L.; Lambert, D.G.; McDonald, J.; Davies, Q.; Moss, E.L.; Konje, J.C. Validation of endogenous control reference genes for normalizing gene expression studies in endometrial carcinoma. Mol. Hum. Reprod. 2015, 21, 723–735. [Google Scholar] [CrossRef]
- Ayakannu, T.; Taylor, A.H.; Konje, J.C. Cannabinoid receptor expression in estrogen-dependent and estrogen-independent endometrial cancer. J. Recept. Signal Transduct. 2018, 38, 385–392. [Google Scholar] [CrossRef]
- Ayakannu, T.; Taylor, A.H.; Konje, J.C. Selection of endogenous control reference genes for studies on Type 1 or Type 2 endometrial cancer. Sci. Rep. 2020, 10, 8468. [Google Scholar] [CrossRef]
- Bokhman, J.V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol. 1983, 15, 10–17. [Google Scholar] [CrossRef]
- Voss, M.A.; Ganesan, R.; Ludeman, L.; McCarthy, K.; Gornall, R.; Schaller, G.; Wei, W.; Sundar, S. Should grade 3 endometrioid endometrial carcinoma be considered a type 2 cancer—A clinical and pathological evaluation. Gynecol. Oncol. 2012, 124, 15–20. [Google Scholar] [CrossRef]
- Setiawan, V.W.; Yang, H.P.; Pike, M.C.; McCann, S.E.; Yu, H.; Xiang, Y.-B.; Wolk, A.; Wentzensen, N.; Weiss, N.S.; Webb, P.M.; et al. Type I and II endometrial cancers: Have they different risk factors? J. Clin. Oncol. 2013, 31, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Kowalewska, M.; Danska-Bidzinska, A.; Bakula-Zalewska, E.; Bidzinski, M. Identification of suitable reference genes for gene expression measurement in uterine sarcoma and carcinosarcoma tumors. Clin. Biochem. 2012, 45, 368–371. [Google Scholar] [CrossRef] [PubMed]
- Steg, A.; Wang, W.; Blanquicett, C.; Grunda, J.M.; Eltoum, I.A.; Wang, K.; Buchsbaum, D.J.; Vickers, S.M.; Russo, S.; Diasio, R.B.; et al. Multiple gene expression analyses in paraffin-embedded tissues by TaqMan low-density array: Application to Hedgehog and Wnt pathway analysis in ovarian endometrioid adenocarcinoma. J. Mol. Diagn. 2006, 8, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Asiabi, P.; Ambroise, J.; Giachini, C.; Coccia, M.E.; Bearzatto, B.; Chiti, M.C.; Dolmans, M.M.; Amorim, C.A. Assessing and validating housekeeping genes in normal, cancerous, and polycystic human ovaries. J. Assist. Reprod. Genet. 2020, 37, 2545–2553. [Google Scholar] [CrossRef]
- Casas, A.I.; Hassan, A.A.; Manz, Q.; Wiwie, C.; Kleikers, P.; Egea, J.; López, M.G.; List, M.; Baumbach, J.; Schmidt, H.H.H.W. Un-biased housekeeping gene panel selection for high-validity gene expression analysis. Sci. Rep. 2022, 12, 12324. [Google Scholar] [CrossRef]
- Sidorkiewicz, I.; Jóźwik, M.; Buczyńska, A.; Erol, A.; Jóźwik, M.; Moniuszko, M.; Jarząbek, K.; Niemira, M.; Krętowski, A. Identification and subsequent validation of transcriptomic signature associated with metabolic status in endometrial cancer. Sci. Rep. 2023, 13, 13763. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Muñoz, J.J.; Anauate, A.C.; Amaral, A.G.; Ferreira, F.M.; Watanabe, E.H.; Meca, R.; Ormanji, M.S.; Boim, M.A.; Onuchic, L.F.; Heilberg, I.P. Ppia is the most stable housekeeping gene for qRT-PCR normalization in kidneys of three Pkd1-deficient mouse models. Sci. Rep. 2021, 11, 19798. [Google Scholar] [CrossRef]
- Sørby, L.A.; Andersen, S.N.; Bukholm, I.R.; Jacobsen, M.B. Evaluation of suitable reference genes for normalization of real-time reverse transcription PCR analysis in colon cancer. J. Exp. Clin. Cancer Res. 2010, 29, 144. [Google Scholar] [CrossRef]
- Nigro, P.; Pompilio, G.; Capogrossi, M.C. Cyclophilin A: A key player for human disease. Cell Death Dis. 2013, 4, e888. [Google Scholar] [CrossRef]
- Chang, T.J.; Juan, C.C.; Yin, P.H.; Chi, C.W.; Tsay, H.J. Up-regulation of beta-actin, cyclophilin and GAPDH in N1S1 rat hepatoma. Oncol. Rep. 1998, 5, 469–540. [Google Scholar] [CrossRef]
- Guo, C.; Liu, S.; Wang, J.; Sun, M.-Z.; Greenaway, F.T. ACTB in cancer. Clin. Chim. Acta 2013, 417, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Santarius, T.; Shipley, J.; Brewer, D.; Stratton, M.R.; Cooper, C.S. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer 2010, 10, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Nyholm, H.C.J. Estrogen and progesterone receptors in endometrial cancer. Clinicopathological correlations and prognostic significance. J. Pathol. Microbiol. Immunol. 1996, 104 (Suppl. 65), 5–33. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, K.B.; Alexander, P.S. In situ photolinked nuclear progesterone receptors of human breast cancer cells: Subunit molecular weights after transformation and translocation. Endocrinology 1983, 113, 2195–2201. [Google Scholar] [CrossRef]
- Feil, P.D.; Clarke, C.L.; Satyaswaroop, P.G. Progestin-mediated changes in progesterone receptor forms in the normal human endometrium. Endocrinology 1988, 123, 2506–2513. [Google Scholar] [CrossRef]
- Krett, N.L.; Wei, L.L.; Francis, M.D.; Nordeen, S.K.; Gordon, D.F.; Wood, W.M.; Horwitz, K.B. Human progesterone A-receptors can be synthesized intracellularly and are biologically functional. Biochem. Biophys. Res. Commun. 1988, 157, 278–285. [Google Scholar] [CrossRef]
- Conneely, O.M.; Kettelberger, D.M.; Tsai, M.J.; Schrader, W.T.; O’Malley, B.W. The chicken progesterone receptor A and B isoforms are products of an alternate translation initiation event. J. Biol. Chem. 1989, 264, 14062–14064. [Google Scholar] [CrossRef]
- Vegeto, E.; Shahbaz, M.M.; Wen, D.X.; Goldman, M.E.; O’Malley, B.W.; McDonnell, D.P. Human progesterone receptor A form is a cell- and promoter-specific repressor of human progesterone receptor B function. Mol. Endocrinol. 1993, 7, 1244–1255. [Google Scholar] [CrossRef]
- Wen, D.X.; Xu, Y.-F.; Mais, D.E.; Goldman, M.E.; Mcdonnell, D.P. The A and B Isoforms of the human progesterone receptor operate through distinct signaling pathways within target cells. Mol. Cell. Biol. 1994, 14, 8356–8364. [Google Scholar] [CrossRef]
- Green, S.; Walter, P.; Kumar, V.; Krust, A.; Bornert, J.-M.; Argos, P.; Chambon, P. Human oestrogen receptor cDNA: Sequence, expression and homology to v-erb-A. Nature 1986, 320, 134–139. [Google Scholar] [CrossRef]
- Greene, G.L.; Gilna, P.; Waterfield, M.; Baker, A.; Hort, Y.; Shine, J. Sequence and expression of human estrogen receptor complementary DNA. Science 1986, 231, 1150–1154. [Google Scholar] [CrossRef]
- Koike, S.; Sakai, M.; Muramatsu, M. Molecular cloning and characterization of rat estrogen receptor cDNA. Nucleic Acids Res. 1987, 15, 2499–2513. [Google Scholar] [CrossRef] [PubMed]
- White, R.; Lees, J.A.; Needham, M.; Ham, J.; Parker, M. Structural organization and expression of the mouse estrogen receptor. Mol. Endocrinol. 1987, 1, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, G.G.J.M.; Enmark, E.; Pelto-Huikko, M.; Nilsson, S.; Gustafsson, J.-Å. Cloning of a novel receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. USA 1996, 93, 5925–5930. [Google Scholar] [CrossRef] [PubMed]
- Mosselman, S.; Polman, J.; Dijkema, R. ERβ: Identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996, 392, 49–53. [Google Scholar] [CrossRef]
- Saegusa, M.; Okayasu, I. Changes in expression of estrogen receptors α and β in relation to progesterone receptor and pS2 status in normal and malignant endometrium. Jpn. J. Cancer Res. 2000, 91, 510–518. [Google Scholar] [CrossRef]
- Utsunomiya, H.; Suzuki, T.; Harada, N.; Ito, K.; Matsuzaki, S.; Konno, R.; Sato, S.; Yajima, A.; Sasano, H. Analysis of estrogen receptor α and β in endometrial carcinomas: Correlation with ERβ and clinicopathologic findings in 45 cases. Int. J. Gynecol. Pathol. 2000, 19, 335–341. [Google Scholar] [CrossRef]
- Jazaeri, A.; Nunes, K.J.; Dalton, M.S.; Xu, M.; Shupnik, M.A.; Rice, L.W. Well-differentiated endometrial adenocarcinomas and poorly differentiated mixed mullerian tumors have altered ER and PR isoform expression. Oncogene 2001, 20, 6965–6969. [Google Scholar] [CrossRef]
- Kershah, S.M.; Desouki, M.M.; Koterba, K.L.; Rowan, B.G. Expression of estrogen receptor coregulators in normal and malignant human endometrium. Gynecol. Oncol. 2004, 92, 304–313. [Google Scholar] [CrossRef]
- Skrzypczak, M.; Bieche, I.; Szymczak, S.; Tozlu, S.; Lewandowski, S.; Girault, I.; Radwanska, K.; Szczylik, C.; Jakowicki, J.A.; Lidereau, R.; et al. Evaluation of mRNA expression of estrogen receptor β and its isoforms in human normal and neoplastic endometrium. Int. J. Cancer 2004, 110, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Pathirage, N.; Di Nezza, L.A.; Salmonsen, L.A.; Jobling, T.; Simpson, E.R.; Clyne, C.D. Expression of aromatase, estrogen receptors, and their coactivators in patients with endometrial cancer. Fertil. Steril. 2006, 86, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Srinivasan, R.; Ghosh, S.; Gopalan, S.; Rajwanshi, A.; Majumdar, S. Estrogen receptor β1 and the β2/βcx isoforms in nonneoplastic endometrium and in endometrioid carcinoma. Int. J. Gynecol. Cancer 2007, 17, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Šmuc, T.; Lanišnik Rižner, T. Aberrant pre-receptor regulation of estrogen and progesterone action in endometrial cancer. Mol. Cell. Endocrinol. 2009, 301, 74–82. [Google Scholar] [CrossRef]
- Treeck, O.; Häring, J.; Skrzypczak, M.; Stegerer, A.; Lattrich, C.; Weber, F.; Görse, R.; Ortmann, O. Estrogen receptor β transcript variants associate with oncogene expression in endometrial cancer. Int. J. Mol. Med. 2012, 29, 1127–1136. [Google Scholar] [CrossRef]
- Jarzabek, K.; Koda, M.; Walentowicz-Sadlecka, M.; Grabiec, M.; Laudanski, P.; Wolczynski, S. Altered expression of ERs, aromatase, and COX2 connected to estrogen action in type 1 endometrial cancer biology. Tumour Biol. 2013, 34, 4007–4016. [Google Scholar] [CrossRef]
- Wik, E.; Ræder, M.B.; Krakstad, C.; Trovik, J.; Birkeland, E.; Hoivik, E.A.; Mjos, S.; Werner, H.M.; Mannelqvist, M.; Stefansson, I.M.; et al. Lack of estrogen receptor-α is associated with epithelial–mesenchymal transition and PI3K alterations in endometrial carcinoma. Clin. Cancer Res. 2013, 19, 1094–1105. [Google Scholar] [CrossRef]
- Kamal, A.M.; Bulmer, J.N.; DeCruze, S.B.; Stringfellow, H.F.; Martin-Hirsch, P.; Hapangama, D.K. Androgen receptors are acquired by healthy postmenopausal endometrial epithelium and their subsequent loss in endometrial cancer is associated with poor survival. Br. J. Cancer 2016, 114, 688–696. [Google Scholar] [CrossRef]
- Kasoha, M.; Dernektsi, C.; Seibold, A.; Bohle, R.M.; Takacs, Z.; Ioan-Iulian, I.; Solomayer, E.-F.; Juhasz-Böss, I. Crosstalk of estrogen receptors and Wnt/β-catenin signaling in endometrial cancer. J. Cancer Res. Clin. Oncol. 2019, 146, 315–327. [Google Scholar] [CrossRef]
- Hojnik, M.; Sinreih, M.; Anko, M.; Hevir-Kene, N.; Knific, T.; Pirš, B.; Frković Grazio, S.; Lanišnik Rižner, T. The co-expression of estrogen receptors ERα, ERβ, and GPER in endometrial cancer. Int. J. Mol. Sci. 2023, 24, 3009. [Google Scholar] [CrossRef] [PubMed]
- Kunth, K.; Höfler, H.; Atkinson, M.J. Quantifizierung der messenger RNA-Expression in Tumoren: Welcher Standard eignet sich am besten für den RNA-Abgleich? [Quantification of messenger RNA expression in tumors: Which standard should be used for best RNA normalization?]. Verh. Dtsch. Ges. Pathol. 1994, 78, 226–230. (In German) [Google Scholar] [PubMed]
- Jo, J.; Choi, S.; Oh, J.; Lee, S.-G.; Choi, S.Y.; Kim, K.K.; Park, C. Conventionally used reference genes are not outstanding for normalization of gene expression in human cancer research. BMC Bioinform. 2019, 20, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Tricarico, C.; Pinzani, P.; Bianchi, S.; Paglierani, M.; Distante, V.; Pazzagli, M.; Bustin, S.A.; Orlando, C. Quantitative real-time reverse transcription polymerase chain reaction: Normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal. Biochem. 2002, 309, 293–300. [Google Scholar] [CrossRef]
- Li, Y.-L.; Ye, F.; Hu, Y.; Lu, W.-G.; Xie, X. Identification of suitable reference genes for gene expression studies of human serous ovarian cancer by real-time polymerase chain reaction. Anal. Biochem. 2009, 394, 110–116. [Google Scholar] [CrossRef]
- Chapman, J.R.; Waldenström, J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef]
- Wang, C.; Taciroglu, A.; Maetschke, S.R.; Nelson, C.C.; Ragan, M.A.; Davis, M.J. mCOPA: Analysis of heterogeneous features in cancer expression data. J. Clin. Bioinforma. 2012, 2, 22. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genomics 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, E.B.; Khayat, A.S.; Assumpção, P.P.; Casseb, S.M.; Moreira-Nunes, C.A.; Moreira, F.C. EndoGeneAnalyzer: A tool for selection and validation of reference genes. PLoS ONE 2024, 19, e0299993. [Google Scholar] [CrossRef]
- Montagnana, M.; Benati, M.; Danese, E.; Giudici, S.; Perfranceschi, M.; Ruzzenenete, O.; Salvagno, G.L.; Bassi, A.; Gelati, M.; Paviati, E.; et al. Aberrant microRNA expression in patients with endometrial cancer. Int. J. Gynecol. Cancer 2017, 27, 459–466. [Google Scholar] [CrossRef]
- Yanokura, M.; Banno, K.; Aoki, D. MicroRNA-34b expression enhances chemosensitivity of endometrial cancer cells to paclitaxel. Int. J. Oncol. 2020, 57, 1145–1156. [Google Scholar] [CrossRef]
- Veryaskina, Y.A.; Titov, S.E.; Zhimulev, I.F. Reference genes for qPCR-based miRNA expression profiling in 14 human tissues. Med. Princ. Pract. 2022, 31, 322–332. [Google Scholar] [CrossRef]
- Hermyt, E.; Zmarzły, N.; Grabarek, B.; Kruszniewska-Rajs, C.; Gola, J.; Jęda-Golonka, A.; Szczepanek, K.; Mazurek, U.; Witek, A. Interplay between miRNAs and genes associated with cell proliferation in endometrial cancer. Int J Mol Sci 2019, 20, 6011. [Google Scholar] [CrossRef]
- Sidorkiewicz, I.; Jóźwik, M.; Niemira, M.; Krętowski, A. Insulin resistance and endometrial cancer: Emerging role for microRNA. Cancers 2020, 12, 2559. [Google Scholar] [CrossRef]
- Bogaczyk, A.; Zawlik, I.; Zuzak, T.; Kluz, M.; Potocka, N.; Kluz, T. The role of miRNAs in the development, proliferation, and progression of endometrial cancer. Int. J. Mol. Sci. 2023, 24, 11489. [Google Scholar] [CrossRef]
- Torres, A.; Torres, K.; Wdowiak, P.; Paszkowski, T.; Maciejewski, R. Selection and validation of endogenous controls for microRNA expression studies in endometrioid endometrial cancer tissues. Gynecol. Oncol. 2013, 130, 588–594. [Google Scholar] [CrossRef]
- Bogaczyk, A.; Potocka, N.; Paszek, S.; Skrzypa, M.; Zuchowska, A.; Kośny, M.; Kluz, M.; Zawlik, I.; Kluz, T. Absolute quantification of selected microRNAs expression in endometrial cancer by digital PCR. Int. J. Mol. Sci. 2024, 25, 3286. [Google Scholar] [CrossRef]
- Tsukamoto, O.; Miura, K.; Mishima, H.; Abe, S.; Kaneuchi, M.; Higashijima, A.; Miura, S.; Kinoshita, A.; Yoshiura, K.-I.; Masuzaki, H. Identification of endometrioid endometrial carcinoma-associated microRNAs in tissue and plasma. Gynecol. Oncol. 2014, 132, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Li, B.-L.; Wan, X.-P. The role of lncRNAs in the development of endometrial carcinoma (Review). Oncol. Lett. 2018, 16, 3424–3429. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-E.; Li, Y.; Cai, B.; He, Q.; Chen, G.; Wang, M.; Wang, K.; Wan, X.; Yan, Q. Phenotyping of immune and endometrial epithelial cells in endometrial carcinomas revealed by single-cell RNA sequencing. Aging (Albany N. Y.) 2021, 13, 6565–6591. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhang, J.; Zhang, Q.; Wei, S.; Shi, R.; Zhao, R.; An, L.; Grose, R.; Feng, D.; Wang, H. Single-cell sequencing reveals the heterogeneity and intratumoral crosstalk in human endometrial cancer. Cell Prolif. 2022, 55, e13249. [Google Scholar] [CrossRef]
- Boldu-Fernández, S.; Lliberos, C.; Simon, C.; Mas, A. Mapping human uterine disorders through single-cell transcriptomics. Cells 2025, 14, 156. [Google Scholar] [CrossRef]
- Dong, C.; Zhao, L.; Liu, X.; Dang, L.; Zhang, X. Single-cell analysis reveals landscape of endometrial cancer response to estrogen and identification of early diagnostic markers. PLoS ONE 2024, 19, e0301128. [Google Scholar] [CrossRef]
- Jin, Y.; Zuo, Y.; Li, G.; Liu, W.; Pan, Y.; Fan, T.; Fu, X.; Yao, X.; Peng, Y. Advances in spatial transcriptomics and its applications in cancer research. Mol. Cancer 2024, 23, 129. [Google Scholar] [CrossRef]
- Monaco, A.; Pantaleo, E.; Amoroso, N.; Lacalamita, A.; Lo Giudice, C.; Fonzino, A.; Fosso, B.; Picardi, E.; Tangaro, S.; Pesole, G.; et al. A primer on machine learning techniques for genomic applications. Comput. Struct. Biotechnol. J. 2021, 19, 4345–4359. [Google Scholar] [CrossRef]
- Chen, T.; Yang, Y.; Huang, Z.; Pan, F.; Xiao, Z.; Gong, K.; Huang, W.; Xu, L.; Liu, X.; Fang, C. Prognostic risk modeling of endometrial cancer using programmed cell death-related genes: A comprehensive machine learning approach. Discov. Oncol. 2025, 16, 280. [Google Scholar] [CrossRef]
- Li, L.; Sun, M.; Wang, J.; Wan, S. Multi-omics based artificial intelligence for cancer research. Adv. Cancer Res. 2024, 163, 303–356. [Google Scholar] [CrossRef]
| Gene Name | Full Gene Name |
|---|---|
| ACTG1 | actin gamma 1 |
| RPS18 | ribosomal protein S18 |
| POM121C | POM121 transmembrane nucleoporin C |
| MRPL18 | mitochondrial ribosomal protein L18 |
| TOMM5 | translocase of outer mitochondrial membrane 5 |
| YTHDF1 | YTH N6-methyladenosine RNA binding protein F1 |
| TPT1 | tumor protein, translationally-controlled 1 |
| RPS27 | ribosomal protein S27 |
| Selected Tissue Type | ||
|---|---|---|
| Number of HKGs in Combination | Endometrium | Uterus |
| 2 | RPS3 | EXOSC4 |
| RPL19 | MIDN | |
| 3 | EXOSC4 | RPS9 |
| MIDN | RPS18 (ENSG00000223367) | |
| NANS | RPS18 (ENSG00000226225) | |
| 5 | RPLP0 | RPS9 |
| RPS18 (ENSG00000223367) | RPS18 (ENSG00000223367) | |
| RPS18 (ENSG00000226225) | RPS18 (ENSG00000226225) | |
| TSPO | RPL31 | |
| HINT1 | RPS5 | |
| 10 | RPS9 | RPS9 |
| RPS18 (ENSG00000223367) | RPS18 (ENSG00000223367) | |
| RPS18 (ENSG00000226225) | RPS18 (ENSG00000226225) | |
| RPS5 | RPS5 | |
| RPL31 | RPL31 | |
| CD63 | CD63 | |
| ATP5PD | ATP5PD | |
| DAD1 | DAD1 | |
| RPS18 (ENSG00000227794) | RPS18 (ENSG00000227794) | |
| EEF1G | RPLP0 | |
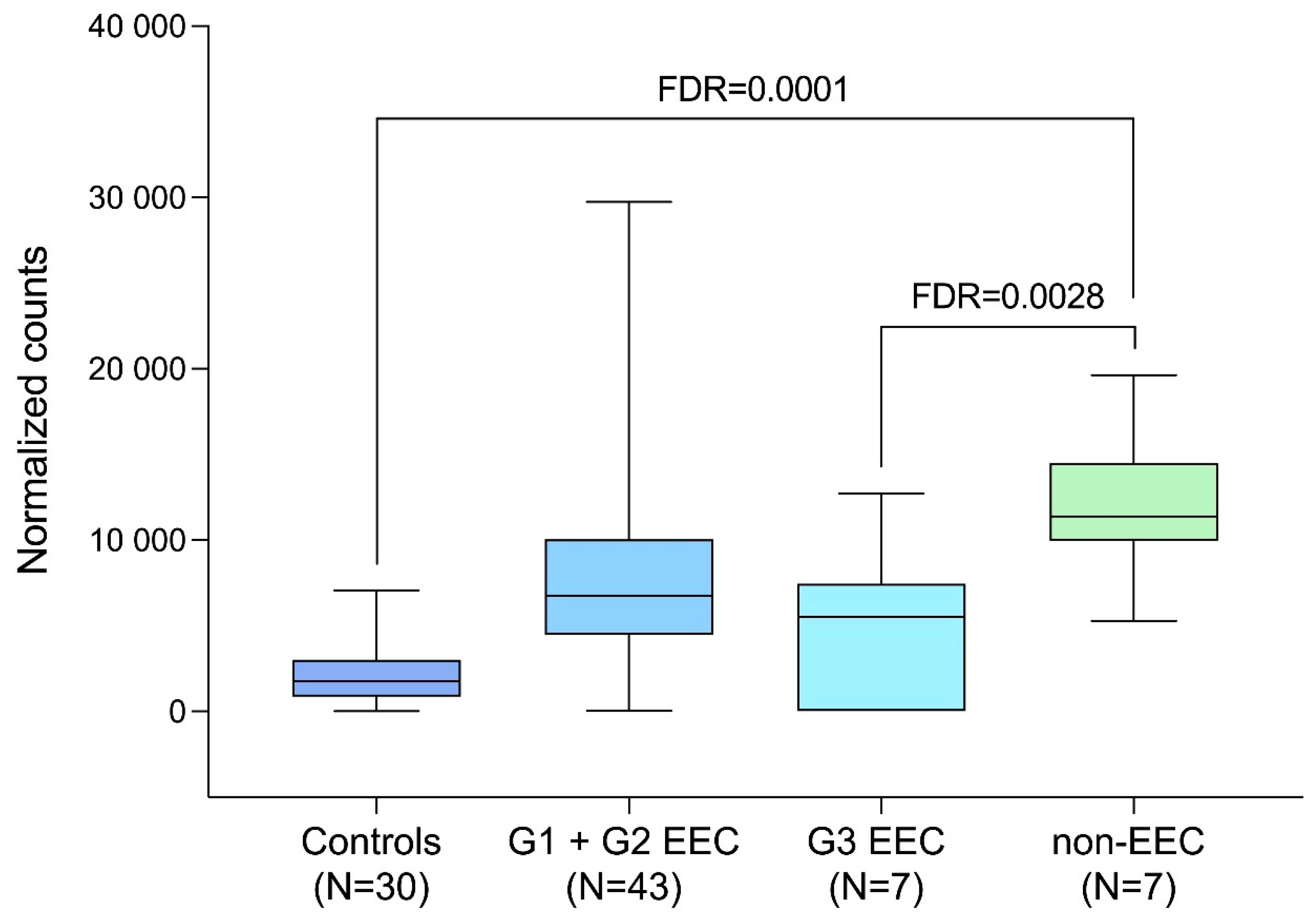
| GAPDH Expression | FC | p-Value | FDR-Adjusted p-Value |
|---|---|---|---|
| EC (N = 57) vs. Control (N = 30) | 4.15 | 0.000456 | 0.0700 |
| EEC (N = 50) vs. Control (N = 30) | 3.41 | 0.002482 | 0.2700 |
| EEC G1 + EEC G2 (N = 43) vs. Control (N = 30) | 4.00 | 0.000508 | 0.0700 |
| non-EEC (N = 7) vs. Control (N = 30) | 10.73 | 0.000000 | 0.0001 |
| non-EEC (N = 7) vs. EEC (N = 50) | 3.15 | 0.000393 | 0.0028 |
| EEC G3 (N = 7) vs. EEC G1 + EEC G2 (N = 43) | 4.16 | 0.249911 | 1.00 |
| non-EEC (N = 7) vs. EEC G1 + EEC G2 (N = 43) | 2.02 | 0.003000 | 0.4910 |
| EEC G3 + non-EEC (N = 14) vs. EEC G1 + EEC G2 (N = 43) | 1.72 | 0.239000 | 1.00 |
| EEC G3 (N = 7) vs. non-EEC (N = 7) | 12.13 | 0.077883 | 1.00 |
| Authors, Publication Year | RT-PCR HKG(s) | Evaluation Method(s) | ERα | ERß | PRA | PRB |
|---|---|---|---|---|---|---|
| Saegusa & Okayasu, 2000 [110] | β-actin | RT-PCR and IHC | Wild type transcripts detected in 41/48 (85.4%) of tumors; ↓ ERα with increasing grade; varied immunointensity and distribution of immuno-positive cells by IHC | Wild type expression observed in 22/34 (64.7%) of tumors; weak immune-reactivity sporadically observed in a few cases of tumor epithelial cells; no relation with histologic grade by IHC | No distinction between PRA and PRB made; PR mRNA positive in 47/48 (97.9%) of tumors, ↓ PR with increasing grade; by IHC, varied immunoreactivity; immunoreactivity scores for PR higher than for ERα | |
| Utsunomiya et al., 2000 [111] | β-actin | mRNA in situ hybridization, RT-PCR and IHC | 45 tumors were studied with no controls; ERα mRNA detected in 36/45 (80.0%) of cases | ERβ mRNA detected in 16/45 (35.6%) of cases; among the 16 ERβ positive cases, 15 were also ERα positive; thus, ERβ is coexpressed with ERα | Solely PR labeling index was given | |
| Jazaeri et al., 2001 [112] | β2-microglobulin | RT-PCR and Western blot | ↓ ERα mRNA expression in EC (N = 7) | ERα mRNA expression exceeds that of ERβ | PRA mRNA levels inferred by subtracting PRB mRNA values from total PR (N = 8) | A relative abundance of PRB mRN |
| Kershah et al., 2004 [113] | β-actin | RT-PCR and Western blot | ↑ in the expression of ERα mRNA in EC, yet not at the protein level; no difference between endometrioid and non-endometrioid tumors in mRNA expression | ERβ NS | No distinction between PRA and PRB made; no significant changes in expression levels between normal premenopausal endometrium (N = 26) and EC (N = 30 of varied histology) | |
| Skrzypczak et al., 2004 [114] | GAPDH | RT-PCR | ↓ ERα in EC (N = 19) compared to normal endometrium (N = 21) | Expression of total ERβ, ERβ1, ERβ2, ERβ2Δ5, ERβ3, ERβ4, and ERβ5 were studied; ↓ of ERβ2Δ5, no expression of ERβ3, very low expression of ERβ4, and ↑ of ERβ5 in EC were reported | No distinction between PRA and PRB made; no significant changes in expression levels between normal endometrium (N = 21) and EC (N = 19) | |
| Pathirage et al., 2006 [115] | 18S rRNA | RT-PCR | ↑ ERα in postmenopausal G1 EC (N = 7) compared to higher grade tumors (N = 10) and normal premenopausal endometrium (N = 20) | A trend for G1 tumors to express higher ERβ levels than G2 and -3 tumors | NS | NS |
| Chakravarty et al., 2007 [116] | β-actin | RT-PCR, Western blotting, and IHC | Studied yet data not reported | Low levels of expression of ERβ1 in EC (N = 26); ↓ ERβ2/βcx at the protein level when compared to normal proliferative endometrium (N = 22), yet no statistical difference at the transcript level; a significant ↓ ERβ2/βcx in G2 tumors compared to G1 tumors | Studied yet data not reported; PR expression correlated with ERα expression, no correlation of PR with ERβ1 or -β2/βcx expression | |
| Šmuc & Lanišnik Rižner, 2009 [117] | PPIA for RT-PCR, β-actin for Western blotting | RT-PCR, Western blot and IHC | ↓ ERα in EC (N = 16) compared to adjacent normal endometrium | ↓ ERβ in EC (N = 16) compared to adjacent normal endometrium | PRA: NS; PR-AB studied instead (N = 16) and found ↓ in EC | ↑ expression of PRB at the protein level in EC (N = 16) in Western blot |
| Häring et al., 2012 [118] | β-actin | RT-PCR, cell culture, and Western blot | ↓ ERα in G3 EC (N = 15) compared to normal pre- and postmenopausal endometrium (N = 28) or to G1 (N = 15) or G2 (N = 16) tumors | Compared to normal endometrium (N = 28), no difference in expression for ERβ1 and -2; ↑ ERβ5, ↑ ERβ∆1, ↑ ERβ∆2/3 and ↓ ERβ∆4 in cancer (N = 46); expression of ERβ1 and ERβ2 strongly correlated with ERα expression | No distinction between PRA and PRB; data only reported as a strong correlation of ERα transcript levels with PR expression | |
| Jarzabek et al., 2013 [119] | 18S rRNA | RT-PCR and IHC | Significantly decreased mRNA and protein expression levels in EC (N = 48) as compared to normal endometrium (N = 15); a positive correlation between ERα and ERβ | Significantly decreased mRNA and protein expression levels in EC (N = 48) as compared to normal endometrium (N = 15); negative correlations between levels of ERα and ERβ transcripts and depth of myoinvasion; a negative correlation of ERβ mRNA expression with FIGO staging | NS | NS |
| Wik et al., 2013 [120] | GAPDH | RT-PCR, IHC, single-nucleotide polymorphism array, and Sanger sequencing | ERα negativity: in 19/76 (25%) of primary investigation cases, in 35/155 (22.6%) of prospective validation cohort cases, and in 68/286 (23.8%) of retrospective validation cohort cases; over 50% of ERα-negative tumors were G3; low ERα was strongly associated with poor patient survival | NS | NS | NS |
| Kamal et al., 2016 [121] | YWHAZ | RT-PCR and IHC | A reduction in stromal expression of ERα in EC when compared with healthy premenopausal controls (N = 28) | ERβ was the predominant steroid receptor expressed in both low-grade- (N = 37) and high-grade (N = 48) EC; a general reduction in the expression of steroid receptors in high-grade EC compared with healthy premenopausal tissue | No distinction between PRA and PRB; a reduction in stromal expression of PR in EC when compared with healthy premenopausal controls | |
| Kasoha et al., 2020 [122] | β-actin | RT-PCR on prospective samples and IHC on retrospective samples | No expression difference between EC (N = 16) and normal endometrium (N = 6) | No expression difference between EC (N = 17) and normal endometrium (N = 6); no immunostaining differences for ERβ1 and ERβ5 either; significantly lower immunopositivity for ERβ2 in EC | NS | NS |
| Hojnik et al. 2023 [123] | HPRT1 and POLR2A for RT-PCR, GAPDH for Western blot | RT-PCR, Western blot and IHC | Decreased mRNA expression in 44 tissue pairs of EC and adjacent normal endometrium; a significant correlation of ERα and ERβ expression at the mRNA level | Decreased mRNA expression in EC in 34 tissue pairs; no significant changes in the ERα/ERβ expression ratio in EC | NS | NS |
| Tool | Methodological Principle(s) | Advantage(s) | Limitations | Reference |
|---|---|---|---|---|
| geNorm™ | Pairwise variation of gene expression (M value); optimal gene number (V value) | Simple; indicates required number of reference genes | Assumes no co-regulation; favors similarly expressed genes | [127] |
| NormFinder | Model-based estimation of intra- and inter-group variation | Accounts for experimental groups; robust to co-regulation | Requires grouping information; less intuitive | [132] |
| BestKeeper | Ct-based SD, CV, and correlation analysis | Easy to use; works directly with raw data | Assumes normality; sensitive to outliers | [133] |
| RefFinder | Integrates geNorm™, NormFinder, BestKeeper, and ΔCt method | Combines results for consensus ranking; user-friendly | Limited customization; dependent on included algorithms’ assumptions | [134] |
| EndoGeneAnalyzer | Combines multiple statistical approaches (NormFinder, SD, correlation analysis); integrates stability assessment with normalization | Raw qRT-PCR Ct data | Stability ranking of HKGs; outlier detection; impact assessment on target gene normalization | [135] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jóźwik, M.; Sidorkiewicz, I.; Wojtkiewicz, J.; Sulkowski, S.; Semczuk, A.; Jóźwik, M. Selecting Optimal Housekeeping Genes for RT-qPCR in Endometrial Cancer Studies: A Narrative Review. Int. J. Mol. Sci. 2025, 26, 8610. https://doi.org/10.3390/ijms26178610
Jóźwik M, Sidorkiewicz I, Wojtkiewicz J, Sulkowski S, Semczuk A, Jóźwik M. Selecting Optimal Housekeeping Genes for RT-qPCR in Endometrial Cancer Studies: A Narrative Review. International Journal of Molecular Sciences. 2025; 26(17):8610. https://doi.org/10.3390/ijms26178610
Chicago/Turabian StyleJóźwik, Maciej, Iwona Sidorkiewicz, Joanna Wojtkiewicz, Stanisław Sulkowski, Andrzej Semczuk, and Marcin Jóźwik. 2025. "Selecting Optimal Housekeeping Genes for RT-qPCR in Endometrial Cancer Studies: A Narrative Review" International Journal of Molecular Sciences 26, no. 17: 8610. https://doi.org/10.3390/ijms26178610
APA StyleJóźwik, M., Sidorkiewicz, I., Wojtkiewicz, J., Sulkowski, S., Semczuk, A., & Jóźwik, M. (2025). Selecting Optimal Housekeeping Genes for RT-qPCR in Endometrial Cancer Studies: A Narrative Review. International Journal of Molecular Sciences, 26(17), 8610. https://doi.org/10.3390/ijms26178610








