Endogenous Ethanol Metabolism and Development of MASLD-MASH
Abstract
1. Introduction
2. The Role of the Gut Microbiome in MASLD-MASH
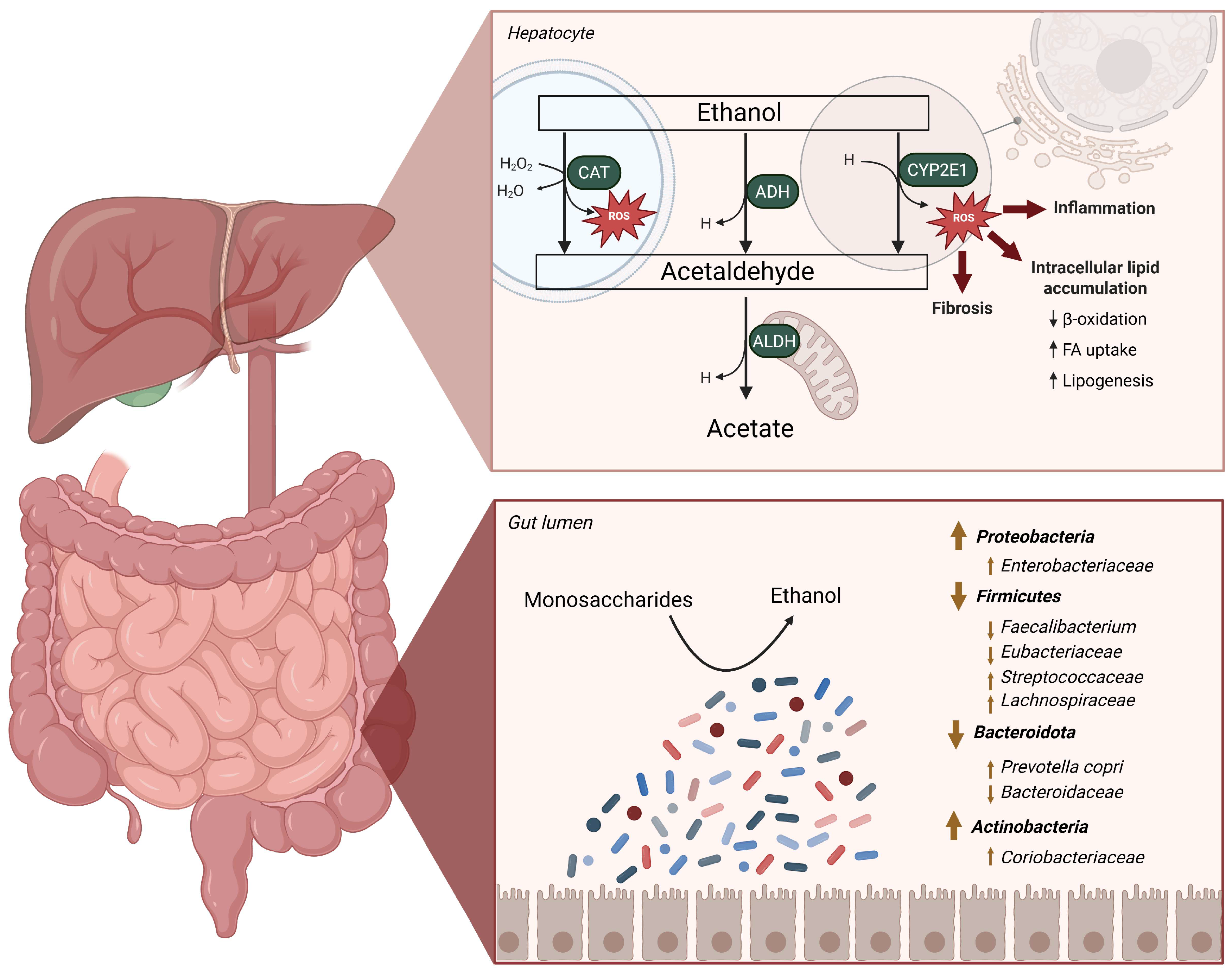
2.1. Implications of the Metabolization of Ethanol in the Liver
2.2. Alterations in the Gut Microbiome in MASLD-MASH
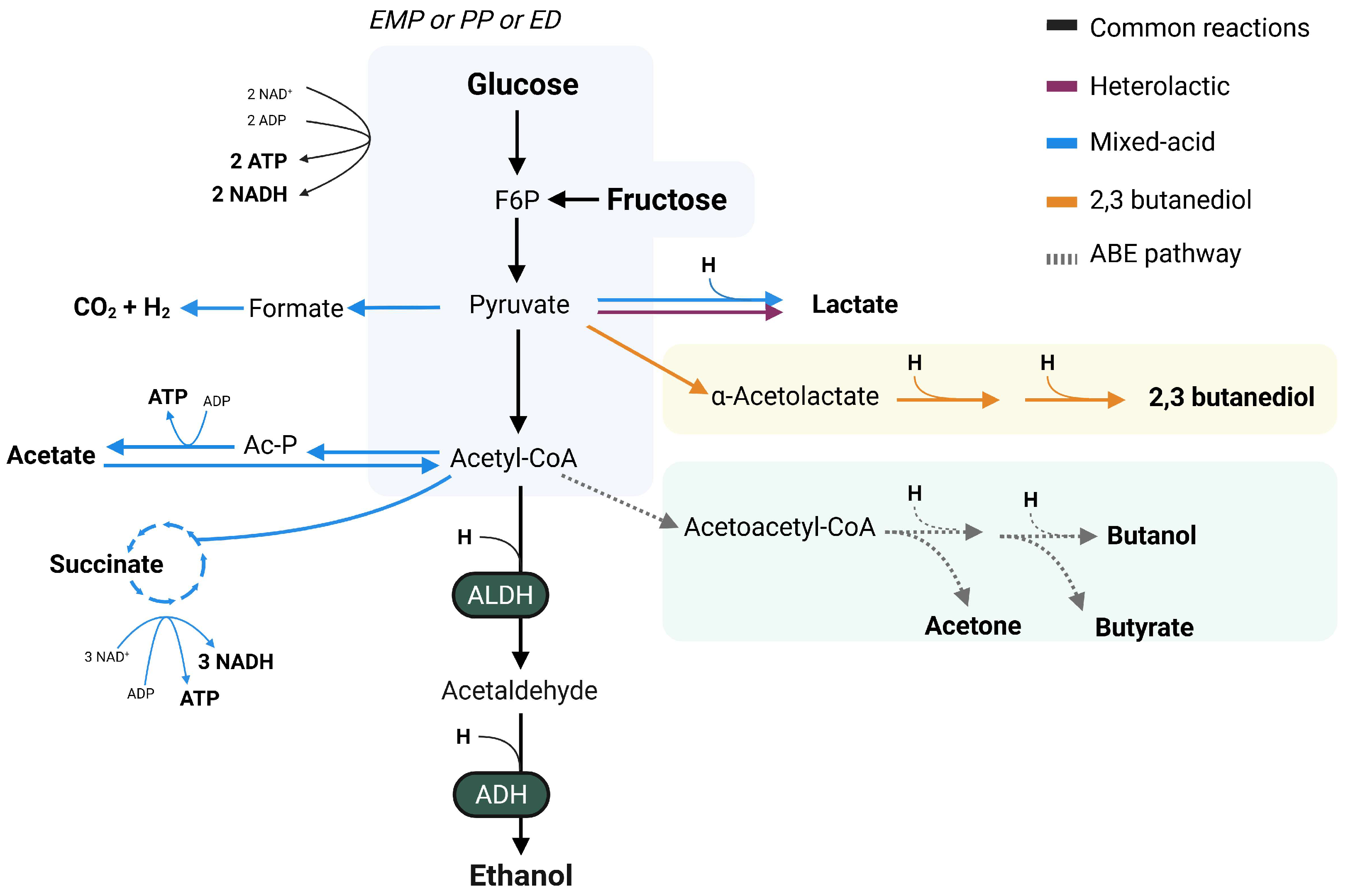
3. Pathways for Endogenous Ethanol Production
| Bacterial Strain | Original Isolation Source | Substrates | Pathway | References |
|---|---|---|---|---|
| Bifidobacterium longum BB536 | Infant fecal sample, healthy | Fructose | Heterolactic | [55,56] |
| Enterocloster bolteae | Fecal sample, MASLD | Glucose, arabinose, fructose, sucrose, glycerol, maltose, mannose, melezitose, sorbitol, trehalose and xylose | Mixed acid | [57] |
| Enterocloster bolteae s28/42 | Fecal sample, hepB | Glucose | Mixed acid | [58] |
| Klebsiella pneumoniae TH1 | Fecal sample, MASH | Glucose and fructose | 2,3-butanediol | [59] |
| Klebsiella pneumoniae W14 | Fecal sample, MASH | Glucose and fructose | 2,3-butanediol | [54,59] |
| Lactobacillus fermentum 92294 | Food, residing in gut | Glucose | Heterolactic | [48] |
| Lactobacillus reuteri ATCC 6475 | Breast milk | Glucose | Heterolactic | [60] |
| Limosilactobacillus fermentum | Fecal sample, MASH | Glucose | Heterolactic | [57] |
| Streptococcus mutans | Fecal sample, MASH | Glucose | n.s. | [57] |
| Weissella confusa | Fecal sample, healthy | Glucose, fructose | Heterolactic | [48,61] |
4. Ethanol as a Substrate for Microbial Metabolism
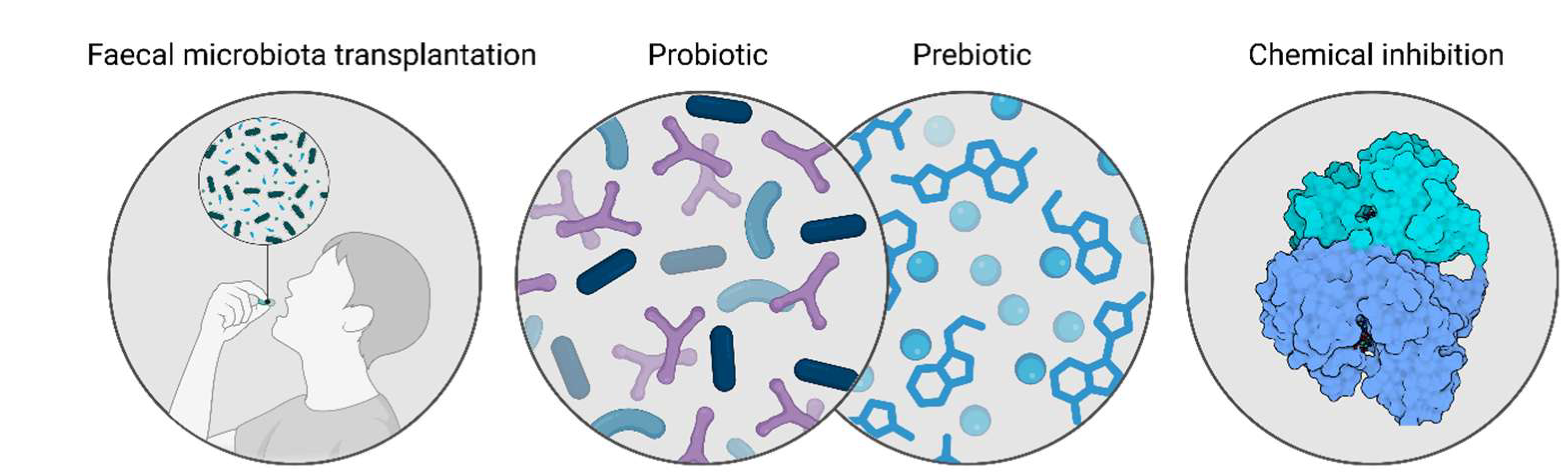
5. Gut Bacteria as a Therapeutic Target for MASLD-MASH
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GM | Gut Microbiome |
| MASLD | Metabolic Dysfunction-Associated Steatotic Liver Disease |
| T2D | Type 2 Diabetes |
| SCFA(s) | Short-Chain Fatty Acids |
| GI | Gastrointestinal |
| MASH | Metabolic Dysfunction-Associated Steatohepatitis |
| ADH | Alcohol Dehydrogenase |
| CYP2E1 | Cytochrome P450 2E1 |
| ROS | Reactive Oxygen Species |
| ALDH | Acetaldehyde Dehydrogenase |
| F/B ratio | Firmicutes To Bacteroidetes Ratio |
| LPS | Lipopolysaccharide |
| EMP | Embden–Meyerhof–Parnas |
| ED | Entner–Doudoroff |
| PP | Pentose Phosphate |
| ABE | Acetone–Butanol–Ethanol |
| SRB | Sulfate-Reducing Bacteria |
| NRB | Nitrate-Reducing Bacteria |
| FMTs | Fecal Microbial Transplantations |
| FA | Fatty Acid |
| HFD | High-Fat Diet |
References
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Ghosh, T.S.; Rampelli, S.; Jeffery, I.B.; Santoro, A.; Neto, M.; Capri, M.; Giampieri, E.; Jennings, A.; Candela, M.; Turroni, S.; et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: The NU-AGE 1-year dietary intervention across five European countries. Gut 2020, 69, 1218–1228. [Google Scholar] [CrossRef]
- Anthony, W.E.; Wang, B.; Sukhum, K.V.; D’Souza, A.W.; Hink, T.; Cass, C.; Seiler, S.; Reske, K.A.; Coon, C.; Dubberke, E.R.; et al. Acute and persistent effects of commonly used antibiotics on the gut microbiome and resistome in healthy adults. Cell Rep. 2022, 39, 110649. [Google Scholar] [CrossRef]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef]
- Kopp, W. How Western Diet and Lifestyle Drive the Pandemic of Obesity and Civilization Diseases. Diabetes Metab. Syndr. Obes. 2019, 12, 2221–2236. [Google Scholar] [CrossRef]
- Martinez, K.B.; Leone, V.; Chang, E.B. Western diets, gut dysbiosis, and metabolic diseases: Are they linked? Gut Microbes 2017, 8, 130–142. (In English) [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Wang, Y.; Zhang, Q.; Zou, R.; Guo, M.; Zheng, H. Characteristics of gut microbiota in people with obesity. PLoS ONE 2021, 16, e0255446. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zheng, Y.; Gao, Y.; Xu, W. Dietary Fiber Intake and Gut Microbiota in Human Health. Microorganisms 2022, 10, 2507. [Google Scholar] [CrossRef]
- Bulsiewicz, W.J. The Importance of Dietary Fiber for Metabolic Health. Am. J. Lifestyle Med. 2023, 17, 639–648. (In English) [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Mann, E.R.; Lam, Y.K.; Uhlig, H.H. Short-chain fatty acids: Linking diet, the microbiome and immunity. Nat. Rev. Immunol. 2024, 24, 577–595. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Cheng, S.; Kou, Y.; Wang, Z.; Jin, R.; Hu, H.; Zhang, X.; Gong, J.F.; Li, J.; Lu, M.; et al. The Gut Microbiome Is Associated with Clinical Response to Anti-PD-1/PD-L1 Immunotherapy in Gastrointestinal Cancer. Cancer Immunol. Res. 2020, 8, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Minnebo, Y.; Delbaere, K.; Goethals, V.; Raes, J.; Van de Wiele, T.; De Paepe, K. Gut microbiota response to in vitro transit time variation is mediated by microbial growth rates, nutrient use efficiency and adaptation to in vivo transit time. Microbiome 2023, 11, 240. [Google Scholar] [CrossRef]
- Konjar, S.; Pavsic, M.; Veldhoen, M. Regulation of Oxygen Homeostasis at the Intestinal Epithelial Barrier Site. Int. J. Mol. Sci. 2021, 22, 9170. [Google Scholar] [CrossRef]
- Procházková, N.; Laursen, M.F.; La Barbera, G.; Tsekitsidi, E.; Jørgensen, M.S.; Rasmussen, M.A.; Raes, J.; Licht, T.R.; Dragsted, L.O.; Roager, H.M. Gut physiology and environment explain variations in human gut microbiome composition and metabolism. Nat. Microbiol. 2024, 9, 3210–3225. [Google Scholar] [CrossRef]
- Litvak, Y.; Byndloss, M.X.; Baumler, A.J. Colonocyte metabolism shapes the gut microbiota. Science 2018, 362, eaat9076. [Google Scholar] [CrossRef] [PubMed]
- Chikina, A.; Matic Vignjevic, D. At the right time in the right place: How do luminal gradients position the microbiota along the gut? Cells Dev. 2021, 168, 203712. [Google Scholar] [CrossRef]
- Boursier, J.; Diehl, A.M. Nonalcoholic Fatty Liver Disease and the Gut Microbiome. Clin. Liver Dis. 2016, 20, 263–275. [Google Scholar] [CrossRef]
- Meijnikman, A.S.; Davids, M.; Herrema, H.; Aydin, O.; Tremaroli, V.; Rios-Morales, M.; Levels, H.; Bruin, S.; de Brauw, M.; Verheij, J.; et al. Microbiome-derived ethanol in nonalcoholic fatty liver disease. Nat. Med. 2022, 28, 2100–2106. (In English) [Google Scholar] [CrossRef]
- Vallianou, N.; Christodoulatos, G.S.; Karampela, I.; Tsilingiris, D.; Magkos, F.; Stratigou, T.; Kounatidis, D.; Dalamaga, M. Understanding the Role of the Gut Microbiome and Microbial Metabolites in Non-Alcoholic Fatty Liver Disease: Current Evidence and Perspectives. Biomolecules 2021, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; You, H.J.; Bajaj, J.S.; Joo, S.K.; Yu, J.; Park, S.; Kang, H.; Park, J.H.; Kim, J.H.; Lee, D.H.; et al. Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat. Commun. 2020, 11, 4982. [Google Scholar] [CrossRef]
- Lin, Y.C.; Wu, C.C.; Li, Y.E.; Chen, C.L.; Lin, C.R.; Ni, Y.H. Full-length 16S rRNA Sequencing Reveals Gut Microbiome Signatures Predictive of MASLD in children with obesity. BMC Microbiol. 2025, 25, 146. (In English) [Google Scholar] [CrossRef]
- Du, L.; Zhang, K.; Liang, L.; Yang, Y.; Lu, D.; Zhou, Y.; Ren, T.; Fan, J.; Zhang, H.; Wang, Y.; et al. Multi-omics analyses of the gut microbiota and metabolites in children with metabolic dysfunction-associated steatotic liver disease. mSystems 2025, 10, e01148-24. [Google Scholar] [CrossRef]
- Boursier, J.; Mueller, O.; Barret, M.; Machado, M.; Fizanne, L.; Araujo-Perez, F.; Guy, C.D.; Seed, P.C.; Rawls, J.F.; David, L.A.; et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016, 63, 764–775. (In English) [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef]
- Wang, L.J.; Sun, J.G.; Chen, S.C.; Sun, Y.L.; Zheng, Y.; Feng, J.C. The role of intestinal flora in metabolic dysfunction-associated steatotic liver disease and treatment strategies. Front. Med. 2024, 11, 1490929. (In English) [Google Scholar] [CrossRef]
- Rao, G.; Peng, X.; Li, X.; An, K.; He, H.; Fu, X.; Li, S.; An, Z. Unmasking the enigma of lipid metabolism in metabolic dysfunction-associated steatotic liver disease: From mechanism to the clinic. Front. Med. 2023, 10, 1294267. [Google Scholar] [CrossRef]
- Ipsen, D.H.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell Mol. Life Sci. 2018, 75, 3313–3327. (In English) [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, Z.; Li, H.; Zhao, J.; Wei, X.; Lin, W.; Zhao, X.; Jiang, A.; Yuan, J. Endogenous ethanol produced by intestinal bacteria induces mitochondrial dysfunction in non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2020, 35, 2009–2019. [Google Scholar] [CrossRef]
- Hyun, J.; Han, J.; Lee, C.; Yoon, M.; Jung, Y. Pathophysiological Aspects of Alcohol Metabolism in the Liver. Int. J. Mol. Sci. 2021, 22, 5717. (In English) [Google Scholar] [CrossRef]
- Contreras-Zentella, M.L.; Villalobos-García, D.; Hernández-Muñoz, R. Ethanol Metabolism in the Liver, the Induction of Oxidant Stress, and the Antioxidant Defense System. Antioxidants 2022, 11, 1258. (In English) [Google Scholar] [CrossRef]
- Chung, B.S.; Yang, K.; Park, C.; Ryu, T. Prolonged Intestinal Ethanol Absorption and Oxidative Stress: Revisiting the Gut-Liver Axis in Alcohol-Associated Disease. Int. J. Mol. Sci. 2025, 26, 5442. (In English) [Google Scholar] [CrossRef]
- Cichoż-Lach, H.; Michalak, A. Oxidative stress as a crucial factor in liver diseases. World J. Gastroenterol. 2014, 20, 8082–8091. (In English) [Google Scholar] [CrossRef]
- Burger, K.; Jung, F.; Staufer, K.; Ladurner, R.; Trauner, M.; Baumann, A.; Brandt, A.; Bergheim, I. MASLD is related to impaired alcohol dehydrogenase (ADH) activity and elevated blood ethanol levels: Role of TNFα and JNK. Redox Biol. 2024, 71, 103121. (In English) [Google Scholar] [CrossRef] [PubMed]
- Abdollahiyan, S.; Nabavi-Rad, A.; Keshavarz Azizi Raftar, S.; Monnoye, M.; Salarieh, N.; Farahanie, A.; Asadzadeh Aghdaei, H.; Zali, M.R.; Hatami, B.; Gérard, P.; et al. Characterization of gut microbiome composition in Iranian patients with nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Sci. Rep. 2023, 13, 20584. [Google Scholar] [CrossRef] [PubMed]
- Dissayabutra, T.; Chuaypen, N.; Somnark, P.; Boonkaew, B.; Udomkarnjananun, S.; Kittiskulnam, P.; Charoenchittang, P.; Prombutara, P.; Tangkijvanich, P. Characterization of gut dysbiosis and intestinal barrier dysfunction in patients with metabolic dysfunction-associated steatotic liver disease and chronic kidney disease: A comparative study. Sci. Rep. 2025, 15, 15481. [Google Scholar] [CrossRef] [PubMed]
- Zazueta, A.; Valenzuela-Pérez, L.; Ortiz-López, N.; Pinto-León, A.; Torres, V.; Guiñez, D.; Aliaga, N.; Merino, P.; Sandoval, A.; Covarrubias, N.; et al. Alteration of Gut Microbiota Composition in the Progression of Liver Damage in Patients with Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD). Int. J. Mol. Sci. 2024, 25, 4387. [Google Scholar] [CrossRef]
- Jiang, W.; Wu, N.; Wang, X.; Chi, Y.; Zhang, Y.; Qiu, X.; Hu, Y.; Li, J.; Liu, Y. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci. Rep. 2015, 5, 8096. [Google Scholar] [CrossRef]
- Ren, M.; Pan, H.; Zhou, X.; Yu, M.; Ji, F. Alterations of the duodenal mucosal microbiome in patients with metabolic dysfunction-associated steatotic liver disease. Sci. Rep. 2024, 14, 9124. [Google Scholar] [CrossRef]
- Leitman, M.; Zhang, D.; Pawar, S.; Shera, S.; Hernandez, L.; Jacobs, J.P.; Dong, T.S. The Association Between Prevotella copri and Advanced Fibrosis in the Progression of Metabolic Dysfunction-Associated Steatotic Liver Disease. bioRxiv 2024. [Google Scholar] [CrossRef]
- Yuan, H.; Zhou, J.; Wu, X.; Wang, S.; Park, S. Enterotype-stratified gut microbial signatures in MASLD and cirrhosis based on integrated microbiome data. Front. Microbiol. 2025, 16, 1568672. (In English) [Google Scholar] [CrossRef]
- Börnigen, D.; Morgan, X.C.; Franzosa, E.A.; Ren, B.; Xavier, R.J.; Garrett, W.S.; Huttenhower, C. Functional profiling of the gut microbiome in disease-associated inflammation. Genome Med. 2013, 5, 65. (In English) [Google Scholar] [CrossRef]
- Dantas, G.; Sommer, M.O.; Degnan, P.H.; Goodman, A.L. Experimental approaches for defining functional roles of microbes in the human gut. Annu. Rev. Microbiol. 2013, 67, 459–475. (In English) [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, P.; Wu, T.; Ruan, H. Bioethanol Production Based on Saccharomyces cerevisiae: Opportunities and Challenges. Fermentation 2023, 9, 709. [Google Scholar] [CrossRef]
- Ruchala, J.; Kurylenko, O.O.; Dmytruk, K.V.; Sibirny, A.A. Construction of advanced producers of first- and second-generation ethanol in Saccharomyces cerevisiae and selected species of non-conventional yeasts (Scheffersomyces stipitis, Ogataea polymorpha). J. Ind. Microbiol. Biotechnol. 2020, 47, 109–132. [Google Scholar] [CrossRef]
- de Oliveira Lino, F.S.; Garg, S.; Li, S.S.; Misiakou, M.A.; Kang, K.; Vale da Costa, B.L.; Beyer-Pedersen, T.S.; Giacon, T.G.; Basso, T.O.; Panagiotou, G.; et al. Strain dynamics of contaminating bacteria modulate the yield of ethanol biorefineries. Nat. Commun. 2024, 15, 5323. (In English) [Google Scholar] [CrossRef] [PubMed]
- Elshaghabee, F.M.; Bockelmann, W.; Meske, D.; de Vrese, M.; Walte, H.G.; Schrezenmeir, J.; Heller, K.J. Ethanol Production by Selected Intestinal Microorganisms and Lactic Acid Bacteria Growing under Different Nutritional Conditions. Front. Microbiol. 2016, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Hackmann, T.J. The vast landscape of carbohydrate fermentation in prokaryotes. FEMS Microbiol. Rev. 2024, 48, fuae016. [Google Scholar] [CrossRef]
- Mbaye, B.; Borentain, P.; Wasfy, R.; Tidjani Alou, M.; Armstrong, N.; Mottola, G.; Meddeb, L.; Ranque, S.; Gérolami, R.; Million, M.; et al. Endogenous Ethanol and Triglyceride Production by Gut Pichia kudriavzevii, Candida albicans and Candida glabrata Yeasts in Non-Alcoholic Steatohepatitis. Cells 2022, 11, 3390. [Google Scholar] [CrossRef]
- Bayoumy, A.B.; Mulder, C.J.J.; Mol, J.J.; Tushuizen, M.E. Gut fermentation syndrome: A systematic review of case reports. United Eur. Gastroenterol. J. 2021, 9, 332–342. (In English) [Google Scholar] [CrossRef] [PubMed]
- Song, C.W.; Park, J.M.; Chung, S.C.; Lee, S.Y.; Song, H. Microbial production of 2,3-butanediol for industrial applications. J. Ind. Microbiol. Biotechnol. 2019, 46, 1583–1601. [Google Scholar] [CrossRef] [PubMed]
- Ward, B. Chapter 11—Bacterial Energy Metabolism. In Molecular Medical Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar] [CrossRef]
- Li, N.N.; Li, W.; Feng, J.X.; Zhang, W.W.; Zhang, R.; Du, S.H.; Liu, S.Y.; Xue, G.H.; Yan, C.; Cui, J.H.; et al. High alcohol-producing Klebsiella pneumoniae causes fatty liver disease through 2,3-butanediol fermentation pathway in vivo. Gut Microbes 2021, 13, 1979883. [Google Scholar] [CrossRef]
- Falony, G.; Vlachou, A.; Verbrugghe, K.; De Vuyst, L. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 2006, 72, 7835–7841. (In English) [Google Scholar] [CrossRef]
- de Vries, W.; Stouthamer, A.H. Fermentation of glucose, lactose, galactose, mannitol, and xylose by bifidobacteria. J. Bacteriol. 1968, 96, 472–478. [Google Scholar] [CrossRef]
- Mbaye, B.; Magdy Wasfy, R.; Borentain, P.; Tidjani Alou, M.; Mottola, G.; Bossi, V.; Caputo, A.; Gerolami, R.; Million, M. Increased fecal ethanol and enriched ethanol-producing gut bacteria Limosilactobacillus fermentum, Enterocloster bolteae, Mediterraneibacter gnavus and Streptococcus mutans in nonalcoholic steatohepatitis. Front. Cell Infect. Microbiol. 2023, 13, 1279354. (In English) [Google Scholar] [CrossRef]
- Magdy Wasfy, R.; Mbaye, B.; Borentain, P.; Tidjani Alou, M.; Murillo Ruiz, M.L.; Caputo, A.; Andrieu, C.; Armstrong, N.; Million, M.; Gerolami, R. Ethanol-Producing Enterocloster bolteae Is Enriched in Chronic Hepatitis B-Associated Gut Dysbiosis: A Case-Control Culturomics Study. Microorganisms 2023, 11, 2437. (In English) [Google Scholar] [CrossRef]
- Yuan, J.; Chen, C.; Cui, J.; Lu, J.; Yan, C.; Wei, X.; Zhao, X.; Li, N.; Li, S.; Xue, G.; et al. Fatty Liver Disease Caused by High-AlcoholProducing Klebsiella pneumoniae. Cell Metab. 2019, 30, 675–688.e7, Correction in Cell Metab. 2019, 30, 1172. [Google Scholar] [CrossRef]
- Engevik, M.; Ruan, W.; Visuthranukul, C.; Shi, Z.; Engevik, K.A.; Engevik, A.C.; Fultz, R.; Schady, D.A.; Spinler, J.K.; Versalovic, J. Limosilactobacillus reuteri ATCC 6475 metabolites upregulate the serotonin transporter in the intestinal epithelium. Benef. Microbes 2021, 12, 583–599. [Google Scholar] [CrossRef] [PubMed]
- Fusco, V.; Quero, G.M.; Cho, G.S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M. The genus Weissella: Taxonomy, ecology and biotechnological potential. Front. Microbiol. 2015, 6, 155. (In English) [Google Scholar] [CrossRef]
- Law, R.C.; Nurwono, G.; Park, J.O. A parallel glycolysis provides a selective advantage through rapid growth acceleration. Nat. Chem. Biol. 2024, 20, 314–322. [Google Scholar] [CrossRef]
- Kim, J.; Kim, G.L.; Norambuena, J.; Boyd, J.M.; Parker, D. Impact of the pentose phosphate pathway on metabolism and pathogenesis of Staphylococcus aureus. PLoS Pathog. 2023, 19, e1011531. (In English) [Google Scholar] [CrossRef]
- Thuy, C.X.; Pham, V.T.; Nguyen, T.T.N.H.; Nguyen, T.T.N.; Ton, N.T.A.; Tuu, T.T.; Vu, N.D. Effect of Fermentation Conditions (Dilution Ratio, Medium pH, Total Soluble Solids, and Saccharomyces cerevisiae Yeast Ratio) on the Ability to Ferment Cider from Tamarillo (Solanum betaceum) Fruit. J. Food Process. Preserv. 2024, 2024, 8841207. [Google Scholar] [CrossRef]
- Liu, X.; Jia, B.; Sun, X.; Ai, J.; Wang, L.; Wang, C.; Zhao, F.; Zhan, J.; Huang, W. Effect of Initial PH on Growth Characteristics and Fermentation Properties of Saccharomyces cerevisiae. J. Food Sci. 2015, 80, M800–M808. [Google Scholar] [CrossRef] [PubMed]
- Forster, A.H.; Gescher, J. Metabolic Engineering of Escherichia coli for Production of Mixed-Acid Fermentation End Products. Front. Bioeng. Biotechnol. 2014, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Voidarou, C.; Antoniadou, Μ.; Rozos, G.; Tzora, A.; Skoufos, I.; Varzakas, T.; Lagiou, A.; Bezirtzoglou, E. Fermentative Foods: Microbiology, Biochemistry, Potential Human Health Benefits and Public Health Issues. Foods 2020, 10, 69. (In English) [Google Scholar] [CrossRef]
- Nguyen, B.D.; Sintsova, A.; Schubert, C.; Sichert, A.; Scheidegger, C.; Naf, J.; Huttman, J.; Lentsch, V.; Keys, T.; Rutschmann, C.; et al. Salmonella Typhimurium screen identifies shifts in mixed-acid fermentation during gut colonization. Cell Host Microbe 2024, 32, 1758–1773.e4. [Google Scholar] [CrossRef]
- Palaiogeorgou, A.M.; Papanikolaou, S.; de Castro, A.M.; Freire, D.M.G.; Kookos, I.K.; Koutinas, A.A. A newly isolated Enterobacter sp. strain produces 2,3-butanediol during its cultivation on low-cost carbohydrate-based substrates. FEMS Microbiol. Lett. 2019, 366, fny280. [Google Scholar] [CrossRef]
- Xue, G.; Feng, J.; Zhang, R.; Du, B.; Sun, Y.; Liu, S.; Yan, C.; Liu, X.; Du, S.; Feng, Y.; et al. Three Klebsiella species as potential pathobionts generating endogenous ethanol in a clinical cohort of patients with auto-brewery syndrome: A case control study. eBioMedicine 2023, 91, 104560. [Google Scholar] [CrossRef]
- Alberto Garcia Mogollon, C.; Carlos Quintero Diaz, J.; Omar Gil Posada, J. Production of acetone, butanol, and ethanol by electro-fermentation with Clostridium saccharoperbutylacetonicum N1-4. Bioelectrochemistry 2023, 152, 108414. [Google Scholar] [CrossRef] [PubMed]
- Buehler, E.A.; Mesbah, A. Kinetic Study of Acetone-Butanol-Ethanol Fermentation in Continuous Culture. PLoS ONE 2016, 11, e0158243. [Google Scholar] [CrossRef]
- Riaz, S.; Mazhar, S.; Abidi, S.H.; Syed, Q.; Abbas, N.; Saleem, Y.; Nadeem, A.A.; Maryam, M.; Essa, R.; Ashfaq, S. Biobutanol production from sustainable biomass process of anaerobic ABE fermentation for industrial applications. Arch. Microbiol. 2022, 204, 672. [Google Scholar] [CrossRef]
- Li, S.; Huang, L.; Ke, C.; Pang, Z.; Liu, L. Pathway dissection, regulation, engineering and application: Lessons learned from biobutanol production by solventogenic clostridia. Biotechnol. Biofuels 2020, 13, 39. [Google Scholar] [CrossRef]
- Pony, P.; Rapisarda, C.; Terradot, L.; Marza, E.; Fronzes, R. Filamentation of the bacterial bi-functional alcohol/aldehyde dehydrogenase AdhE is essential for substrate channeling and enzymatic regulation. Nat. Commun. 2020, 11, 1426. [Google Scholar] [CrossRef] [PubMed]
- Pech-Canul, A.; Hammer, S.K.; Ziegler, S.J.; Richardson, I.D.; Sharma, B.D.; Maloney, M.I.; Bomble, Y.J.; Lynd, L.R.; Olson, D.G. The role of AdhE on ethanol tolerance and production in Clostridium thermocellum. J. Biol. Chem. 2024, 300, 107559. [Google Scholar] [CrossRef]
- Kang, S.; Long, J.; Park, M.S.; Ji, G.E.; Ju, Y.; Ku, S. Investigating human-derived lactic acid bacteria for alcohol resistance. Microb. Cell Fact. 2024, 23, 118. (In English) [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Sharma, A.; Wu, W.; Gomi, R.; Sung, B.; Hospodsky, D.; Angenent, L.T.; Worgall, S. The fermentation product 2,3-butanediol alters P. aeruginosa clearance, cytokine response and the lung microbiome. ISME J. 2016, 10, 2978–2983. [Google Scholar] [CrossRef] [PubMed]
- Zetterström, C.E.; Uusitalo, P.; Qian, W.; Hinch, S.; Caraballo, R.; Grundström, C.; Elofsson, M. Screening for Inhibitors of Acetaldehyde Dehydrogenase (AdhE) from Enterohemorrhagic Escherichia coli (EHEC). SLAS Discov. 2018, 23, 815–822. (In English) [Google Scholar] [CrossRef]
- Lin, G.H.; Hsieh, M.C.; Shu, H.Y. Role of Iron-Containing Alcohol Dehydrogenases in Acinetobacter baumannii ATCC 19606 Stress Resistance and Virulence. Int. J. Mol. Sci. 2021, 22, 9921. (In English) [Google Scholar] [CrossRef]
- Kavanaugh, D.W.; Porrini, C.; Dervyn, R.; Ramarao, N. The pathogenic biomarker alcohol dehydrogenase protein is involved in Bacillus cereus virulence and survival against host innate defence. PLoS ONE 2022, 17, e0259386. (In English) [Google Scholar] [CrossRef]
- Hallin, S.; Throbäck, I.N.; Dicksved, J.; Pell, M. Metabolic profiles and genetic diversity of denitrifying communities in activated sludge after addition of methanol or ethanol. Appl. Environ. Microbiol. 2006, 72, 5445–5452. (In English) [Google Scholar] [CrossRef] [PubMed]
- Tsuruya, A.; Kuwahara, A.; Saito, Y.; Yamaguchi, H.; Tenma, N.; Inai, M.; Takahashi, S.; Tsutsumi, E.; Suwa, Y.; Totsuka, Y.; et al. Major Anaerobic Bacteria Responsible for the Production of Carcinogenic Acetaldehyde from Ethanol in the Colon and Rectum. Alcohol. Alcohol. 2016, 51, 395–401. [Google Scholar] [CrossRef]
- Tagaino, R.; Washio, J.; Abiko, Y.; Tanda, N.; Sasaki, K.; Takahashi, N. Metabolic property of acetaldehyde production from ethanol and glucose by oral Streptococcus and Neisseria. Sci. Rep. 2019, 9, 10446. [Google Scholar] [CrossRef]
- Martino, C.; Zaramela, L.S.; Gao, B.; Embree, M.; Tarasova, J.; Parker, S.J.; Wang, Y.; Chu, H.; Chen, P.; Lee, K.-C.; et al. Acetate reprograms gut microbiota during alcohol consumption. Nat. Commun. 2022, 13, 4630. [Google Scholar] [CrossRef]
- Crocker, A.; Harty, C.; Hammond, J.; Willger, S.; Salazar, P.; Botelho, N.; Jacobs, N.; Hogan, D. Pseudomonas aeruginosa Ethanol Oxidation by AdhA in Low-Oxygen Environments. J. Bacteriol. 2019, 201, e00393-19. [Google Scholar] [CrossRef]
- Willis, C.L.; Cummings, J.H.; Neale, G.; Gibson, G.R. Nutritional Aspects of Dissimilatory Sulfate Reduction in the Human Large Intestine. Curr. Microbiol. 1997, 35, 294–298. [Google Scholar] [CrossRef]
- Wrighton, K.C.; Castelle, C.J.; Wilkins, M.J.; Hug, L.A.; Sharon, I.; Thomas, B.C.; Handley, K.M.; Mullin, S.W.; Nicora, C.D.; Singh, A.; et al. Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J. 2014, 8, 1452–1463. (In English) [Google Scholar] [CrossRef]
- Little, A.S.; Younker, I.T.; Schechter, M.S.; Bernardino, P.N.; Méheust, R.; Stemczynski, J.; Scorza, K.; Mullowney, M.W.; Sharan, D.; Waligurski, E.; et al. Dietary—And host-derived metabolites are used by diverse gut bacteria for anaerobic respiration. Nat. Microbiol. 2024, 9, 55–69, Correction in Nat. Microbiol. 2025, 10, 1550. [Google Scholar] [CrossRef]
- Bowland, A.C.; Melin, A.D.; Hosken, D.J.; Hockings, K.J.; Carrigan, M.A. The evolutionary ecology of ethanol. Trends Ecol. Evol. 2025, 40, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Gao, A.X.; Liu, X.; Bai, Z.; Wang, P.; Ledesma-Amaro, R. Microbial conversion of ethanol to high-value products: Progress and challenges. Biotechnol. Biofuels Bioprod. 2024, 17, 115. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Mu, H.; Guo, J.; Liu, H.; Zhang, R.; Liu, W.; Xian, M.; Liu, H. Metabolic engineering of Escherichia coli for the utilization of ethanol. J. Biol. Res. 2020, 27, 1. (In English) [Google Scholar] [CrossRef] [PubMed]
- Extance, J.; Danson, M.J.; Crennell, S.J. Structure of an acetylating aldehyde dehydrogenase from the thermophilic ethanologen Geobacillus thermoglucosidasius. Protein Sci. 2016, 25, 2045–2053. (In English) [Google Scholar] [CrossRef]
- Benito-Vaquerizo, S.; Parera Olm, I.; de Vroet, T.; Schaap, P.J.; Sousa, D.Z.; Martins Dos Santos, V.A.P.; Suarez-Diez, M. Genome-scale metabolic modelling enables deciphering ethanol metabolism via the acrylate pathway in the propionate-producer Anaerotignum neopropionicum. Microb. Cell Fact. 2022, 21, 116. (In English) [Google Scholar] [CrossRef]
- Singh, S.B.; Carroll-Portillo, A.; Lin, H.C. Desulfovibrio in the Gut: The Enemy within? Microorganisms 2023, 11, 1772. (In English) [Google Scholar] [CrossRef]
- Sim, M.S.; Skennerton, C.T.; Orphan, V.J. Physiological, genomic, and sulfur isotopic characterization of methanol metabolism by Desulfovibrio carbinolicus. PLoS ONE 2021, 16, e0245069. [Google Scholar] [CrossRef]
- Yoshii, T.; Asanuma, N.; Hino, T. Effect of ethanol on nitrate and nitrite reduction and methanogenesis in the ruminal microbiota. Anim. Sci. J. 2005, 76, 37–42. [Google Scholar] [CrossRef]
- Lee, J.Y.; Tsolis, R.M.; Bäumler, A.J. The microbiome and gut homeostasis. Science 2022, 377, eabp9960. (In English) [Google Scholar] [CrossRef]
- Bertsch, J.; Siemund, A.L.; Kremp, F.; Müller, V. A novel route for ethanol oxidation in the acetogenic bacterium Acetobacterium woodii: The acetaldehyde/ethanol dehydrogenase pathway. Environ. Microbiol. 2016, 18, 2913–2922. [Google Scholar] [CrossRef] [PubMed]
- Salaspuro, V.; Nyfors, S.; Heine, R.; Siitonen, A.; Salaspuro, M.; Jousimies-Somer, H. Ethanol oxidation and acetaldehyde production in vitro by human intestinal strains of Escherichia coli under aerobic, microaerobic, and anaerobic conditions. Scand. J. Gastroenterol. 1999, 34, 967–973. (In English) [Google Scholar] [CrossRef] [PubMed]
- Frimmer, U.; Widdel, F. Oxidation of ethanol by methanogenic bacteria. Arch. Microbiol. 1989, 152, 479–483. [Google Scholar] [CrossRef]
- Haveman, S.A.; DiDonato, R.J., Jr.; Villanueva, L.; Shelobolina, E.S.; Postier, B.L.; Xu, B.; Liu, A.; Lovley, D.R. Genome-wide gene expression patterns and growth requirements suggest that Pelobacter carbinolicus reduces Fe(III) indirectly via sulfide production. Appl. Environ. Microbiol. 2008, 74, 4277–4284. (In English) [Google Scholar] [CrossRef]
- Elshaer, A.; Chascsa, D.M.H.; Lizaola-Mayo, B.C. Exploring Varied Treatment Strategies for Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD). Life 2024, 14, 844. [Google Scholar] [CrossRef]
- Zhang, P.-p.; Li, L.-l.; Han, X.; Li, Q.-w.; Zhang, X.-h.; Liu, J.J.; Wang, Y. Fecal microbiota transplantation improves metabolism and gut microbiome composition in db/db mice. Acta Pharmacol. Sin. 2020, 41, 678–685. [Google Scholar] [CrossRef]
- Hou, P.F.; Yao, Y.; Wu, Y.; Yu, H.T.; Qin, Y.; Yi, L.; Mi, M.T. Fecal microbiota transplantation improves hepatic steatosis induced by HFD in a mouse model associated with liver ILC1 regulation and indole-3-carbinol level. Front. Nutr. 2025, 12, 1500293. (In English) [Google Scholar] [CrossRef]
- Kwan, S.Y.; Gonzales, K.A.; Jamal, M.A.; Stevenson, H.L.; Tan, L.; Lorenzi, P.L.; Futreal, P.A.; Hawk, E.T.; McCormick, J.B.; Fisher-Hoch, S.P.; et al. Protection against fibrosis by a bacterial consortium in metabolic dysfunction-associated steatohepatitis and the role of amino acid metabolism. Gut Microbes 2024, 16, 2399260. (In English) [Google Scholar] [CrossRef]
- Vallianou, N.G.; Kounatidis, D.; Psallida, S.; Vythoulkas-Biotis, N.; Adamou, A.; Zachariadou, T.; Kargioti, S.; Karampela, I.; Dalamaga, M. NAFLD/MASLD and the Gut-Liver Axis: From Pathogenesis to Treatment Options. Metabolites 2024, 14, 366. (In English) [Google Scholar] [CrossRef]
- Dong, Y.; Wu, Z.; Gai, Z.; Han, M. Bifidobacterium longum subsp. longum BL21 ameliorates alcoholic liver disease in mice through enhancement of the hepatic antioxidant capacity and modulation of the gut microbiota. J. Appl. Microbiol. 2023, 134, lxad251. (In English) [Google Scholar] [CrossRef]
- Hao, J.; Zhang, Y.; Wu, T.; Liu, R.; Sui, W.; Zhu, J.; Fang, S.; Geng, J.; Zhang, M. The antidiabetic effects of Bifidobacterium longum subsp. longum BL21 through regulating gut microbiota structure in type 2 diabetic mice. Food Funct. 2022, 13, 9947–9958. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Lang, G.; Wang, Q.; Zhao, W.; Shi, D.; Zhou, Z.; Shen, Y.; Xia, H.; Han, S.; Li, L. Lactobacillus helveticus attenuates alcoholic liver injury via regulation of gut microecology in mice. Microb. Biotechnol. 2024, 17, e70016. (In English) [Google Scholar] [CrossRef] [PubMed]
- Mitrović, M.; Dobrosavljević, A.; Odanović, O.; Knežević-Ivanovski, T.; Kralj, Đ.; Erceg, S.; Perućica, A.; Svorcan, P.; Stanković-Popović, V. The effects of synbiotics on the liver steatosis, inflammation, and gut microbiome of metabolic dysfunction-associated liver disease patients-randomized trial. Rom. J. Intern. Med. 2024, 62, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Setshedi, M.; Wands, J.R.; Monte, S.M. Acetaldehyde adducts in alcoholic liver disease. Oxid. Med. Cell. Longev. 2010, 3, 178–185. [Google Scholar] [CrossRef] [PubMed]
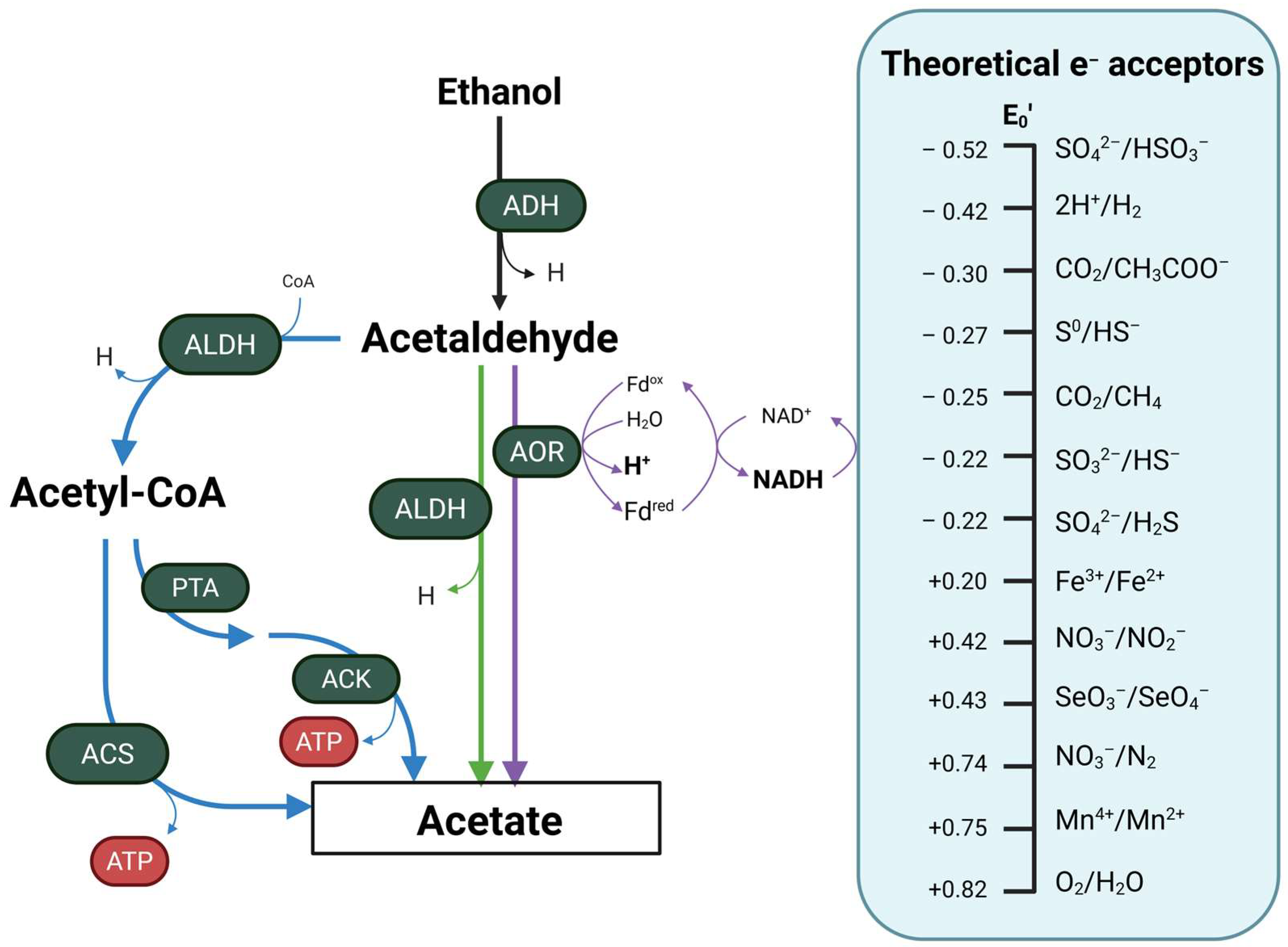
| Bacterial Strain | Origin of Isolation | Reaction | e− Acceptor | Oxygen Tolerance | References |
|---|---|---|---|---|---|
| Acetobacterium woodii | Mud | EtOH → Acetyl-CoA | CO2 | Anaerobe | [99] |
| Collinsella spp. | Fecal | EtOH → AcH | O2 | Aerotolerant | [83] |
| Escherichia coli | Fecal | EtOH → AcH | O2 | Facultative anaerobe | [100] |
| Methanogenium organophilum | Sewage | EtOH → Ac | Sulfide | Anaerobe | [101] |
| Neisseria mucosa | Oral | EtOH → AcH | O2 | Aerobe | [84] |
| Neisseria sicca | Oral | EtOH → AcH | O2 | Microaerophile | [84] |
| Pseudomonas aeruginosa PA14 | Wound | EtOH → Ac | O2 and nitrate | Facultative anaerobe | [86] |
| Ruminococcus spp. | Fecal | EtOH → AcH | n.s. | Obligate anaerobe | [83] |
| Streptococcus mitis | Oral | EtOH → AcH | O2 | Microaerophile | [84] |
| Streptococcus salivarius | Oral | EtOH → AcH | O2 | Microaerophile | [84] |
| Syntrophotalea carbinolica | Sludge | EtOH → Ac | Fe(III) | Facultative anaerobe | [102] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farràs Solé, N.; Wydh, S.; Alizadeh Bahmani, A.H.; Bui, T.P.N.; Nieuwdorp, M. Endogenous Ethanol Metabolism and Development of MASLD-MASH. Int. J. Mol. Sci. 2025, 26, 8609. https://doi.org/10.3390/ijms26178609
Farràs Solé N, Wydh S, Alizadeh Bahmani AH, Bui TPN, Nieuwdorp M. Endogenous Ethanol Metabolism and Development of MASLD-MASH. International Journal of Molecular Sciences. 2025; 26(17):8609. https://doi.org/10.3390/ijms26178609
Chicago/Turabian StyleFarràs Solé, Núria, Sander Wydh, Amir Hossein Alizadeh Bahmani, Thi Phuong Nam Bui, and Max Nieuwdorp. 2025. "Endogenous Ethanol Metabolism and Development of MASLD-MASH" International Journal of Molecular Sciences 26, no. 17: 8609. https://doi.org/10.3390/ijms26178609
APA StyleFarràs Solé, N., Wydh, S., Alizadeh Bahmani, A. H., Bui, T. P. N., & Nieuwdorp, M. (2025). Endogenous Ethanol Metabolism and Development of MASLD-MASH. International Journal of Molecular Sciences, 26(17), 8609. https://doi.org/10.3390/ijms26178609






