Precision Medicine Study of Post-Exertional Malaise Epigenetic Changes in Myalgic Encephalomyelitis/Chronic Fatigue Patients During Exercise
Abstract
1. Introduction
2. Results
2.1. Two-Day Maximum Repeated Effort CardioPulmonary Exercise Testing (CPET)
Summary of Cardiopulmonary Parameters for the Five ME/CFS Patients
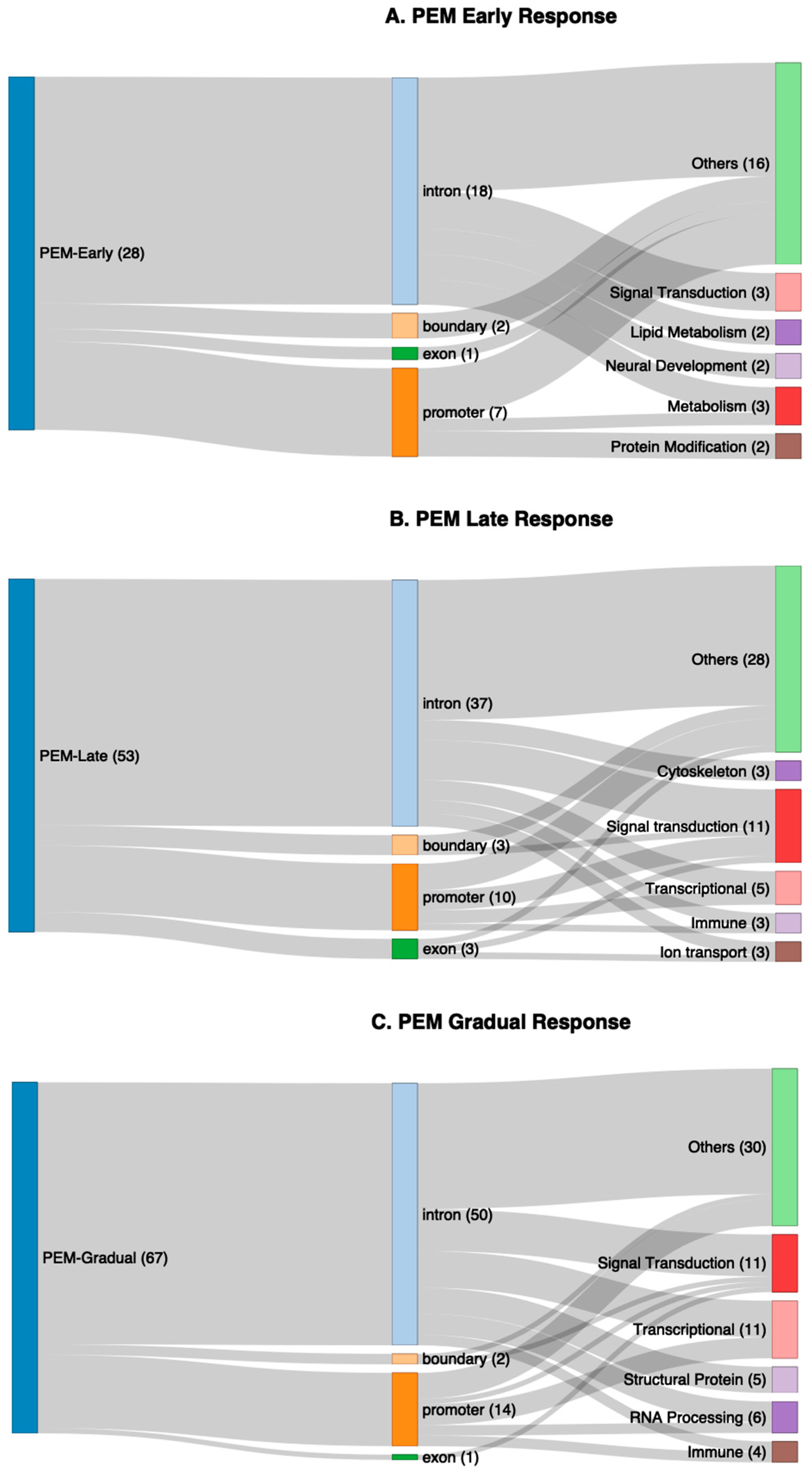
2.2. Overview of Differential Methylation Analysis of Post-Exertional Malaise
2.3. Principal Component Analysis of Differentially Methylated Fragments
2.4. Heatmap Analysis of Differentially Methylated Fragments in Controls and ME/CFS at Each Timepoint of CPET
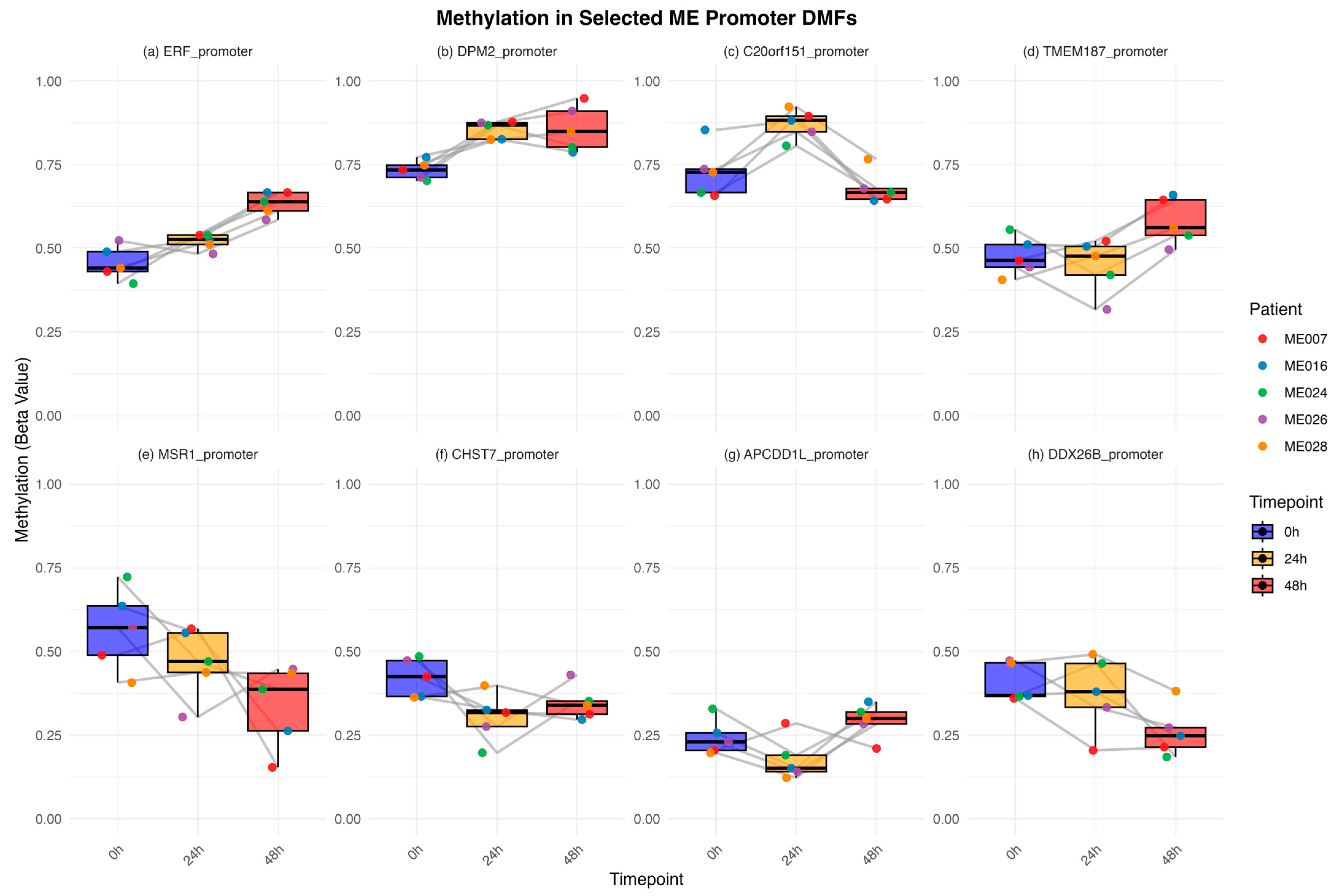
2.5. Methylation Dynamics in ME/CFS-Specific Promoter Fragments at Each Timepoint of the CPET Protocol
- (a)
- Continuous hypermethylation (ERF *, FAM3A, RNPS1, ZNF135, ZN785, OR2V1),
- (b)
- Early hypermethylation (DPM2 *, ACR, APEX2, PRGGR3, ZAP70, ZDHHC9),
- (c)
- Transient hypermethylation (C20orf151 *, C7, NPBWR2, FFAR2, ZG16B),
- (d)
- Late hypermethylation (TMEM187 *, CPEB),
- (e)
- Continuous decrease in hypomethylation (MSR1 *),
- (f)
- Early hypomethylation (CHST7 *),
- (g)
- Transient hypomethylation (APCDD1L *, FAM123B),
- (h)
- Late hypomethylation (DDX26B).
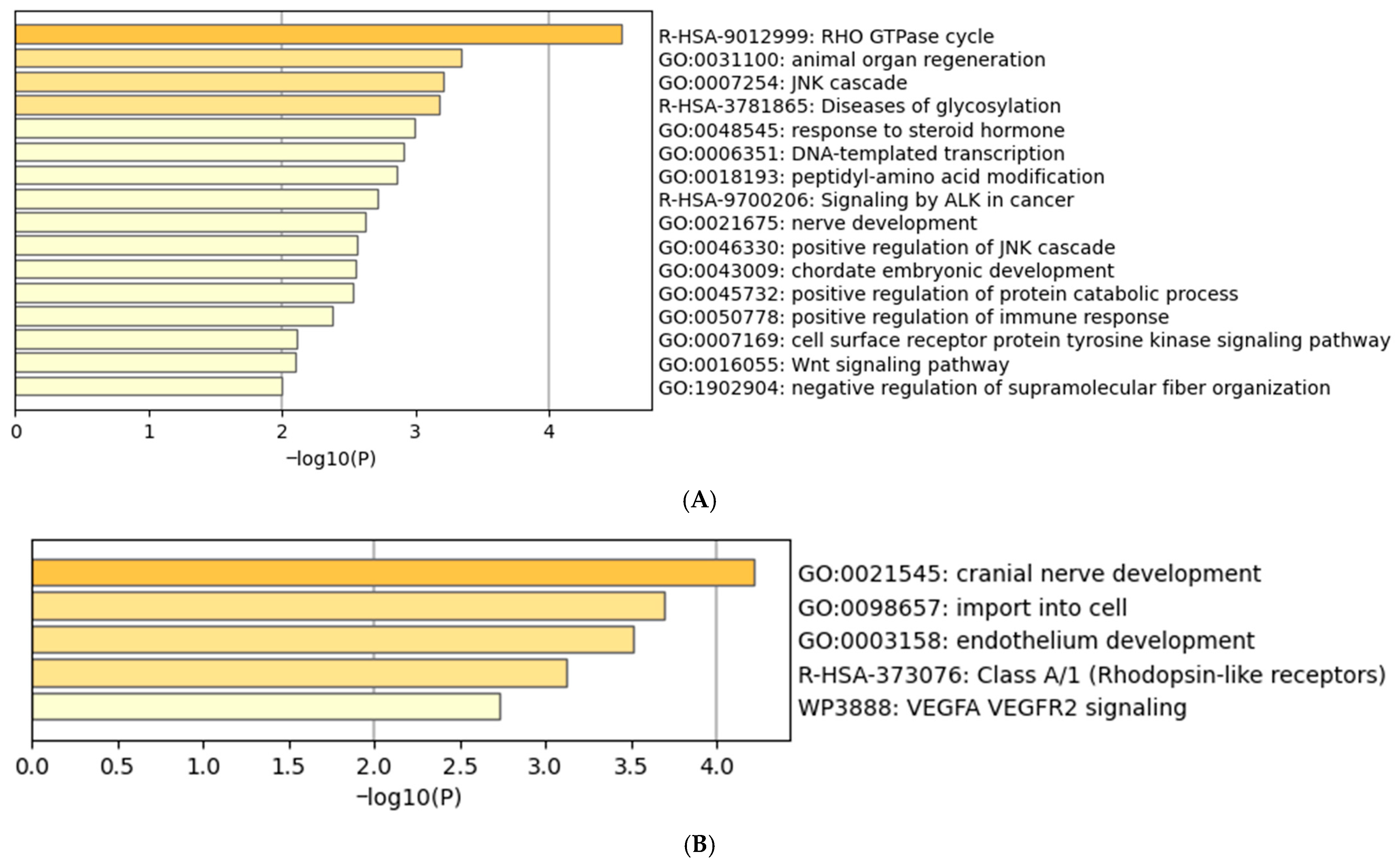
2.6. Functional Enrichment of Genes Associated with Differentially Methylated Fragments
3. Discussion
4. Materials and Methods
4.1. The Cardiopulmonary Exercise Training (CPET) Protocol
4.2. Peripheral Blood Mononuclear Cell (PBMC) Isolation
4.3. DNA Extraction
4.4. Reduced Representation Bisulphite Sequencing
4.4.1. DNA Sequencing
4.4.2. Statistical Analyses
5. Conclusions
6. Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shepherd, C. ME/CFS/PVFS: The MEA Clinical & Research Guide: An Exploration of the Key Clinical Issues; ME Association: Buckingham, UK, 2022. [Google Scholar]
- Das, S.; Taylor, K.; Kozubek, J.; Sardell, J.; Gardner, S. Genetic risk factors for ME/CFS identified using combinatorial analysis. J. Transl. Med. 2022, 20, 598. [Google Scholar] [CrossRef]
- Jiang, Z.; Rajamanickam, S.; Justice, N.J. CRF signaling between neurons in the paraventricular nucleus of the hypothalamus (PVN) coordinates stress responses. Neurobiol. Stress 2019, 11, 100192. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C. Jarred Younger Finds a VERY Inflamed Brain in ME/CFS. 2025. Available online: https://www.healthrising.org/blog/2025/08/07/younger-inflamed-brain-me-cfs/ (accessed on 9 August 2025).
- Nakatomi, Y.; Mizuno, K.; Ishii, A.; Wada, Y.; Tanaka, M.; Tazawa, S.; Onoe, K.; Fukuda, S.; Kawabe, J.; Takahashi, K.; et al. Neuroinflammation in Patients with Chronic Fatigue Syndrome/Myalgic Encephalomyelitis: An 11C-(R)-PK11195 PET Study. J. Nucl. Med. 2014, 55, 945–950. [Google Scholar] [CrossRef] [PubMed]
- Carruthers, B.M.; Jain, A.K.; De Meirleir, K.L.; Peterson, D.L.; Klimas, N.G.; Lerner, A.M.; Bested, A.C.; Flor-Henry, P.; Joshi, P.; Powles, A.C.P.; et al. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. J. Chronic Fatigue Syndr. 2003, 11, 7–115. [Google Scholar] [CrossRef]
- Carruthers, B.M.; van de Sande, M.I.; De Meirleir, K.L.; Klimas, N.G.; Broderick, G.; Mitchell, T.; Staines, D.; Powles, A.C.; Speight, N.; Vallings, R.; et al. Myalgic encephalomyelitis: International Consensus Criteria. J. Intern. Med. 2011, 270, 327–338, Erratum in J. Intern. Med. 2017, 282, 353. [Google Scholar] [CrossRef]
- CDC. IOM 2015 Diagnostic Criteria. Available online: https://www.cdc.gov/me-cfs/hcp/diagnosis/iom-2015-diagnostic-criteria-1.html (accessed on 1 July 2025).
- Komaroff, A.L.; Lipkin, W.I. Insights from myalgic encephalomyelitis/chronic fatigue syndrome may help unravel the pathogenesis of postacute COVID-19 syndrome. Trends Mol. Med. 2021, 27, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Marshall-Gradisnik, S.; Eaton-Fitch, N. Understanding myalgic encephalomyelitis. Science 2022, 377, 1150–1151. [Google Scholar] [CrossRef]
- Karen Leslie, N.C.-B.; Hilliard, N.; Bull, M. A Physiotherapist's Guide to Understanding and Managing ME/CFS; Jessica Kingsley Publishers: Philadelphia, PA, USA, 2023. [Google Scholar]
- Chu, L.; Valencia, I.J.; Garvert, D.W.; Montoya, J.G. Deconstructing post-exertional malaise in myalgic encephalomyelitis/ chronic fatigue syndrome: A patient-centered, cross-sectional survey. PLoS ONE 2018, 13, e0197811. [Google Scholar] [CrossRef]
- Davenport, T.E.; Stevens, S.R.; Stevens, J.; Snell, C.R.; Van Ness, J.M. Properties of measurements obtained during cardiopulmonary exercise testing in individuals with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Work 2020, 66, 247–256. [Google Scholar] [CrossRef]
- VanNess, J.M.; Stevens, S.R.; Bateman, L.; Stiles, T.L.; Snell, C.R. Postexertional malaise in women with chronic fatigue syndrome. J. Women’s Health 2010, 19, 239–244. [Google Scholar] [CrossRef]
- Vermeulen, R.C.; Kurk, R.M.; Visser, F.C.; Sluiter, W.; Scholte, H.R. Patients with chronic fatigue syndrome performed worse than controls in a controlled repeated exercise study despite a normal oxidative phosphorylation capacity. J. Transl. Med. 2010, 8, 93. [Google Scholar] [CrossRef]
- Barbara, S.; Brice, C.; Gina, N.; Angelique, G.; Snigdha, C.; Avindra, N.; Brian, W. Mixed methods system for the assessment of post-exertional malaise in myalgic encephalomyelitis/chronic fatigue syndrome: An exploratory study. BMJ Neurol. Open 2024, 6, e000529. [Google Scholar] [CrossRef]
- Helliwell, A.M.; Stockwell, P.A.; Edgar, C.D.; Chatterjee, A.; Tate, W.P. Dynamic Epigenetic Changes during a Relapse and Recovery Cycle in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Int. J. Mol. Sci. 2022, 23, 11852. [Google Scholar] [CrossRef]
- Helliwell, A.M.; Sweetman, E.C.; Stockwell, P.A.; Edgar, C.D.; Chatterjee, A.; Tate, W.P. Changes in DNA methylation profiles of myalgic encephalomyelitis/chronic fatigue syndrome patients reflect systemic dysfunctions. Clin. Epigenet. 2020, 12, 167. [Google Scholar] [CrossRef] [PubMed]
- Peppercorn, K.; Sharma, S.; Edgar, C.D.; Stockwell, P.A.; Rodger, E.J.; Chatterjee, A.; Tate, W.P. Comparing DNA Methylation Landscapes in Peripheral Blood from Myalgic Encephalomyelitis/Chronic Fatigue Syndrome and Long COVID Patients. Int. J. Mol. Sci. 2025, 26, 6631. [Google Scholar] [CrossRef]
- Vrije Universiteit Brussel. Epigenetics of Post-Exertional Malaise in Patients with ME/CFS (EPIME). 2023. Available online: https://clinicaltrials.gov/study/NCT04378634 (accessed on 14 July 2025).
- Polli, A.; Ghosh, M.; Bakusic, J.; Ickmans, K.; Monteyne, D.; Velkeniers, B.; Bekaert, B.; Godderis, L.; Nijs, J. DNA Methylation and Brain-Derived Neurotrophic Factor Expression Account for Symptoms and Widespread Hyperalgesia in Patients with Chronic Fatigue Syndrome and Comorbid Fibromyalgia. Arthritis Rheumatol. 2020, 72, 1936–1944. [Google Scholar] [CrossRef]
- Polli, A.; Hendrix, J.; Ickmans, K.; Bakusic, J.; Ghosh, M.; Monteyne, D.; Velkeniers, B.; Bekaert, B.; Nijs, J.; Godderis, L. Genetic and epigenetic regulation of Catechol-O-methyltransferase in relation to inflammation in chronic fatigue syndrome and Fibromyalgia. J. Transl. Med. 2022, 20, 487. [Google Scholar] [CrossRef]
- Brenu, E.W.; Staines, D.; Marshall-Gradisnik, S. Methylation Profile of CD4+ T Cells in Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. J. Clin. Cell. Immunol. 2014, 5, 3. [Google Scholar] [CrossRef]
- de Vega, W.C.; Vernon, S.D.; McGowan, P.O. DNA methylation modifications associated with chronic fatigue syndrome. PLoS ONE 2014, 9, e104757. [Google Scholar] [CrossRef]
- de Vega, W.C.; Herrera, S.; Vernon, S.D.; McGowan, P.O. Epigenetic modifications and glucocorticoid sensitivity in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). BMC Med. Genom. 2017, 10, 11. [Google Scholar] [CrossRef]
- de Vega, W.C.; Erdman, L.; Vernon, S.D.; Goldenberg, A.; McGowan, P.O. Integration of DNA methylation & health scores identifies subtypes in myalgic encephalomyelitis/chronic fatigue syndrome. Epigenomics 2018, 10, 539–557. [Google Scholar] [CrossRef]
- Herrera, S.; de Vega, W.C.; Ashbrook, D.; Vernon, S.D.; McGowan, P.O. Genome-epigenome interactions associated with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Epigenetics 2018, 13, 1174–1190. [Google Scholar] [CrossRef]
- Trivedi, M.S.; Oltra, E.; Sarria, L.; Rose, N.; Beljanski, V.; Fletcher, M.A.; Klimas, N.G.; Nathanson, L. Identification of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome-associated DNA methylation patterns. PLoS ONE 2018, 13, e0201066. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, J.N.; Coe, S.; Izadi, H. A retrospective outcome study of 42 patients with Chronic Fatigue Syndrome, 30 of whom had Irritable Bowel Syndrome. Half were treated with oral approaches, and half were treated with Faecal Microbiome Transplantation. Hum. Microbiome J. 2019, 13, 100061. [Google Scholar] [CrossRef]
- Stockwell, P.A.; Rodger, E.J.; Gimenez, G.; Morison, I.M.; Chatterjee, A. DMAP2: A Pipeline for Analysis of Whole-Genome-Scale DNA Methylation Sequencing Data. Curr. Protoc. 2024, 4, e70003. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Kobayashi, H.; Yamada, K.; Morita, S.; Hiura, H.; Fukuda, A.; Kagami, M.; Ogata, T.; Hata, K.; Sotomaru, Y.; Kono, T. Identification of the mouse paternally expressed imprinted gene Zdbf2 on chromosome 1 and its imprinted human homolog ZDBF2 on chromosome 2. Genomics 2009, 93, 461–472. [Google Scholar] [CrossRef]
- Bowdish, D.M.E.; Gordon, S. Conserved domains of the class A scavenger receptors: Evolution and function. Immunol. Rev. 2009, 227, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Kools, P.; Van Imschoot, G.; van Roy, F. Characterization of three novel human cadherin genes (CDH7, CDH19, and CDH20) clustered on chromosome 18q22-q23 and with high homology to chicken cadherin-7. Genomics 2000, 68, 283–295. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Ong, E.S.; Dyck, J.A.; Evans, R.M. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature 1990, 345, 224–229. [Google Scholar] [CrossRef]
- Steinberg, S.J.; Morgenthaler, J.; Heinzer, A.K.; Smith, K.D.; Watkins, P.A. Very Long-chain Acyl-CoA Synthetases: Human “bubblegum” represents a new family of proteins capable of activating very long-chain fatty acids. J. Biol. Chem. 2000, 275, 35162–35169. [Google Scholar] [CrossRef] [PubMed]
- Catterall, W.A.; Goldin, A.L.; Waxman, S.G. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol. Rev. 2005, 57, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Byers, R.J.; Currie, T.; Tholouli, E.; Rodig, S.J.; Kutok, J.L. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood 2011, 118, 2857–2867. [Google Scholar] [CrossRef]
- Dhanoa, B.S.; Cogliati, T.; Satish, A.G.; Bruford, E.A.; Friedman, J.S. Update on the Kelch-like (KLHL) gene family. Human. Genom. 2013, 7, 13. [Google Scholar] [CrossRef]
- Tillander, V.; Arvidsson Nordström, E.; Reilly, J.; Strozyk, M.; Van Veldhoven, P.P.; Hunt, M.C.; Alexson, S.E. Acyl-CoA thioesterase 9 (ACOT9) in mouse may provide a novel link between fatty acid and amino acid metabolism in mitochondria. Cell Mol. Life Sci. 2014, 71, 933–948. [Google Scholar] [CrossRef]
- Choi, J.; Clement, K.; Huebner, A.J.; Webster, J.; Rose, C.M.; Brumbaugh, J.; Walsh, R.M.; Lee, S.; Savol, A.; Etchegaray, J.P.; et al. DUSP9 Modulates DNA Hypomethylation in Female Mouse Pluripotent Stem Cells. Cell Stem Cell 2017, 20, 706–719.e707. [Google Scholar] [CrossRef]
- Li, Y.; Chen, R.; Zhou, Q.; Xu, Z.; Li, C.; Wang, S.; Mao, A.; Zhang, X.; He, W.; Shu, H.-B. LSm14A is a processing body-associated sensor of viral nucleic acids that initiates cellular antiviral response in the early phase of viral infection. Proc. Natl. Acad. Sci. USA 2012, 109, 11770–11775. [Google Scholar] [CrossRef]
- Zhang, H.; Meng, L.H.; Pommier, Y. Mitochondrial topoisomerases and alternative splicing of the human TOP1mt gene. Biochimie 2007, 89, 474–481. [Google Scholar] [CrossRef]
- Collet, P.; Domenjoud, L.; Devignes, M.D.; Murad, H.; Schohn, H.; Dauça, M. The human semaphorin 6B gene is down regulated by PPARs. Genomics 2004, 83, 1141–1150. [Google Scholar] [CrossRef]
- American College of Sports Medicine. ACSM's Resource Manual for Guidelines for Exercise Testing and Prescription; Lippincott Williams and WIlkins: Philadelphia, PA, USA, 2014. [Google Scholar]
- Borg, G.A. Psychophysical bases of perceived exertion. Med. Sci. Sports Exerc. 1982, 14, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Wasserman, K.; Beaver, W.L.; Whipp, B.J. Gas exchange theory and the lactic acidosis (anaerobic) threshold. Circulation 1990, 81 (Suppl. 1), Ii14–Ii30. [Google Scholar]
- Peppercorn, K.; Edgar, C.D.; Al Momani, S.; Rodger, E.J.; Tate, W.P.; Chatterjee, A. Application of DNA Methylome Analysis to Patients with ME/CFS. In Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS): Methods and Protocols; Tate, W., Peppercorn, K., Eds.; Springer: New York, NY, USA, 2025; pp. 141–160. [Google Scholar] [CrossRef]
- Ludgate, J.L.; Wright, J.; Stockwell, P.A.; Morison, I.M.; Eccles, M.R.; Chatterjee, A. A streamlined method for analysing genome-wide DNA methylation patterns from low amounts of FFPE DNA. BMC Med. Genom. 2017, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Rodger, E.J.; Stockwell, P.A.; Almomani, S.; Eccles, M.R.; Chatterjee, A. Protocol for generating high-quality genome-scale DNA methylation sequencing data from human cancer biospecimens. STAR Protoc. 2023, 4, 102714. [Google Scholar] [CrossRef]
- Stockwell, P.A.; Chatterjee, A.; Rodger, E.J.; Morison, I.M. DMAP: Differential methylation analysis package for RRBS and WGBS data. Bioinformatics 2014, 30, 1814–1822. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Heyward, V. Advanced Fitness Assessment and Exercise Prescription, 6th ed.; Human Kinetics: Champaign, IL, USA, 2010; Volume xiii. [Google Scholar]
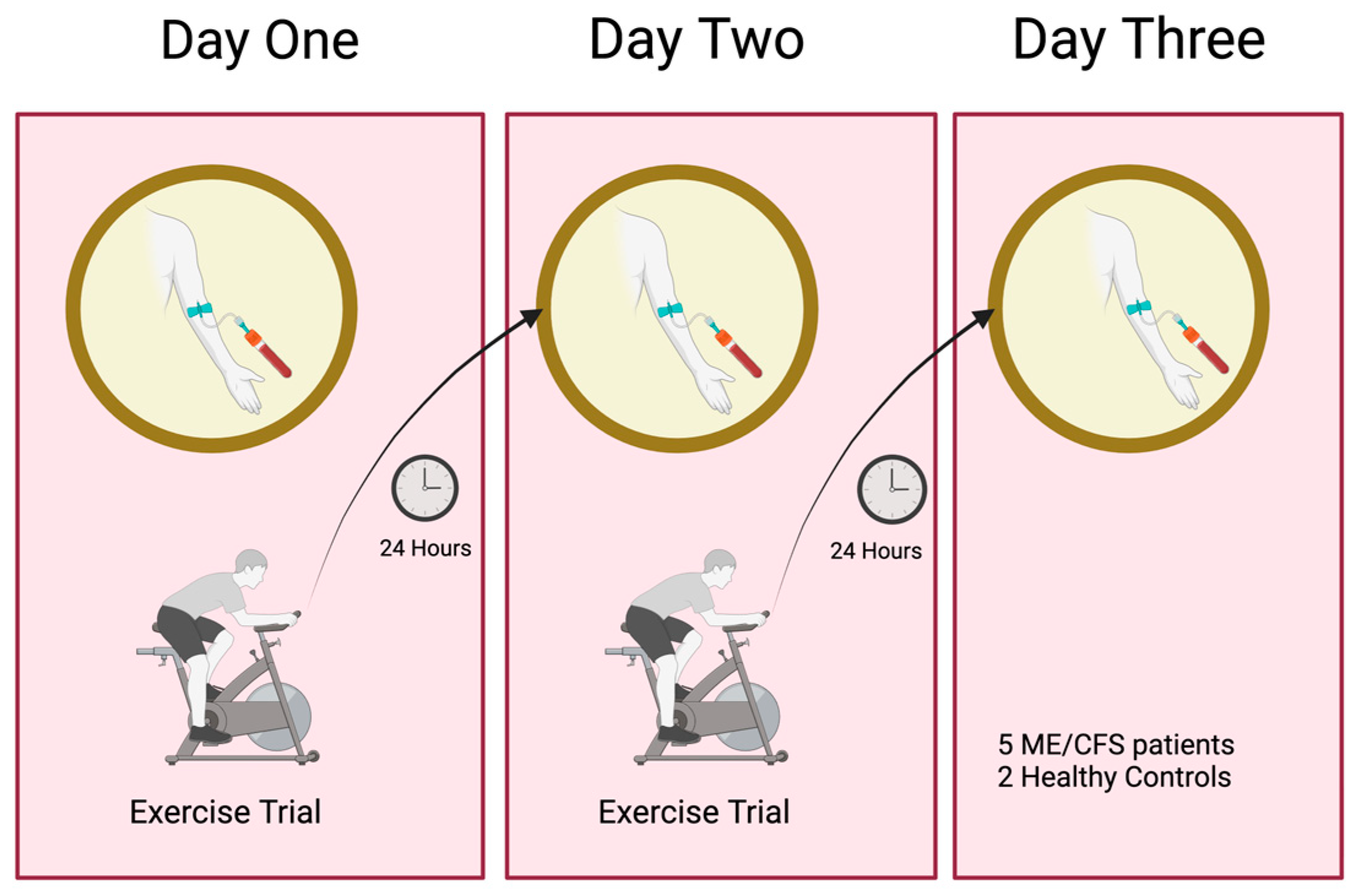
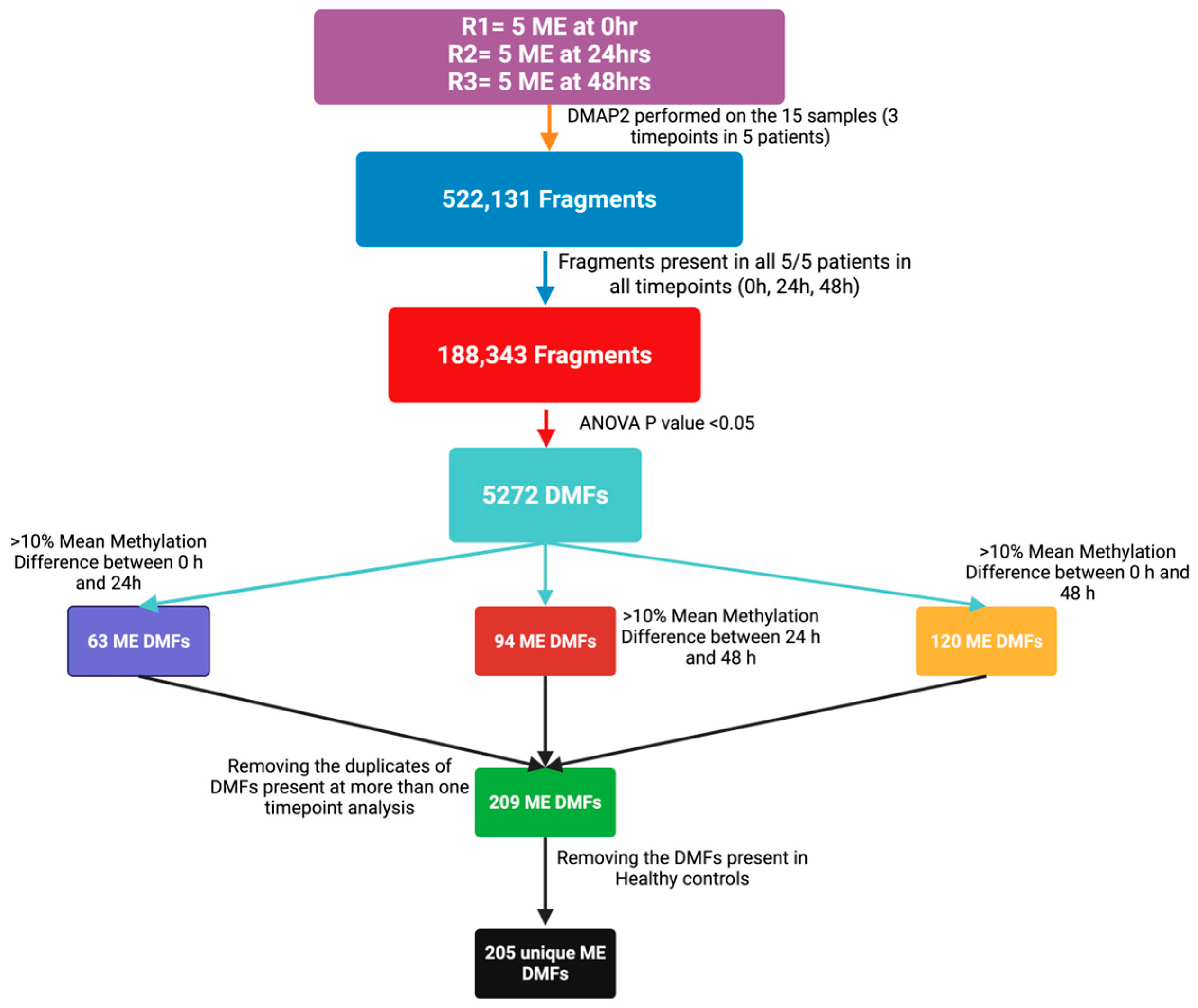
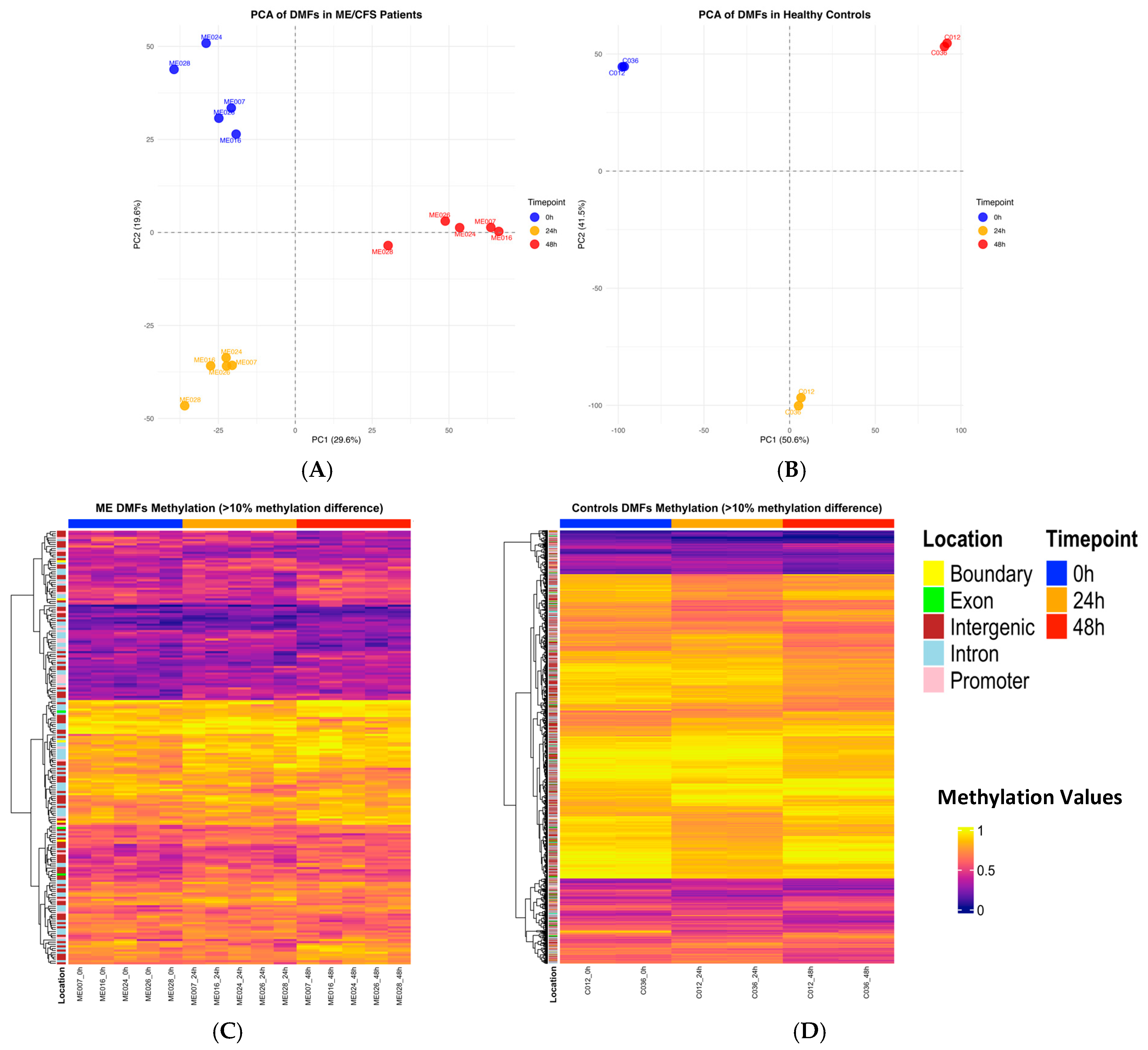
| CPET Parameter | ME007 | ME016 | ME024 | ME026 | ME028 | CO12 | C036 |
|---|---|---|---|---|---|---|---|
| Day One Maximum HR (bpm) | 178 | 184 | 187 | 197 | 157 | 188 | 183 |
| Day Two Maximum HR (bpm) | 172 * | 188 | 171 * | 189 * | 138 * | 191 | 181 |
| Day One Max.Workload (Watts) | 135 | 195 | 135 | 210 | 180 | 285 | 165 |
| Day Two Max.Workload (Watts) | 120 # | 195 | 135 | 180 # | 165 # | 285 | 165 |
| Day One VO2 peak (mL/kg/min) | 25.4 | 34.4 | 29.1 | 30.1 | 35.1 | 49.4 | 21.8 |
| Day Two VO2 peak (mL/kg/min) | 26.1 | 33.2 ++ | 31.6 | 29.0 ++ | 31.1 ++ | 50.7 | 22.0 |
| Day One RER | 1.24 | 1.09 | 1.12 | 1.13 | 1.25 | 1.20 | 1.04 |
| Day Two RER | 1.13 | 1.11 | 1.19 | 1.04 | 1.30 | 1.17 | 1.17 |
| Day One AT Workload (Watts) | 60 | 105 | 75 | 135 | 90 | 195 | 75 |
| Day Two AT Workload (Watts) | 45 ** | 60 ** | 75 | 150 ## | 120 ## | 210 ## | 75 |
| Chr | Start | End | CpG | p-Value | Mean % Methylation (0 h) | Mean % Methylation (24 h) | Mean % Methylation (48 h) | GeneID | Functional Category |
|---|---|---|---|---|---|---|---|---|---|
| 16 | 2880299 | 2880358 | 3 | 0.042 | 31 | 37 | 27 | ZG16B | Cell Migration |
| X | 55026095 | 55026179 | 3 | 0.014 | 26 | 35 | 38 | APEX2 | DNA Repair |
| 5 | 40909532 | 40909656 | 1 | 0.009 | 28 | 38 | 20 | C7 | Immune |
| 2 | 98329337 | 98329462 | 3 | 0.042 | 25 | 34 | 37 | ZAP70 | Immune |
| 8 | 16425403 | 16425502 | 1 | 0.033 | 57 | 47 | 38 | MSR1 | Immune |
| 9 | 130700685 | 130700757 | 6 | 0.001 | 74 | 85 | 85 | DPM2 | Metabolism |
| 19 | 35939798 | 35939864 | 3 | 0.015 | 29 | 36 | 23 | FFAR2 | Metabolism |
| X | 153743994 | 153744125 | 7 | 0.04 | 26 | 30 | 36 | FAM3A | Metabolism |
| X | 46433376 | 46433503 | 18 | 0.029 | 42 | 30 | 36 | CHST7 | Metabolism |
| X | 128977778 | 128977938 | 12 | 0.003 | 26 | 36 | 40 | ZDHHC9 | Protein Modification |
| X | 150863035 | 150863196 | 7 | 0.046 | 56 | 68 | 66 | PRRG3 | Protein Modification |
| 22 | 51172813 | 51172946 | 4 | 0.014 | 61 | 73 | 70 | ACR | Reproduction |
| X | 134655182 | 134655341 | 14 | 0.046 | 40 | 36 | 28 | DDX26B | RNA processing |
| 16 | 2322947 | 2323056 | 4 | 0.046 | 57 | 64 | 69 | RNPS1 | RNA processing |
| 5 | 180554560 | 180554676 | 1 | 0.003 | 84 | 88 | 96 | OR2V1 | Sensory Perception |
| 20 | 62738880 | 62738959 | 4 | 0.004 | 16 | 23 | 12 | NPBWR2 | Signal Transduction |
| 20 | 57090702 | 57090793 | 2 | 0.025 | 23 | 17 | 30 | APCDD1L | Signal Transduction |
| X | 63425502 | 63425652 | 11 | 0.041 | 31 | 29 | 42 | FAM123B | Signal Transduction |
| 19 | 42760491 | 42760639 | 3 | 0.00003 | 46 | 52 | 62 | ERF | Transcription |
| 20 | 48805866 | 48805958 | 1 | 0.014 | 79 | 75 | 92 | CEBPB | Transcription |
| 16 | 30597821 | 30597994 | 7 | 0.025 | 39 | 43 | 53 | ZNF785 | Transcription |
| 19 | 58571275 | 58571401 | 5 | 0.027 | 30 | 34 | 41 | ZNF135 | Transcription |
| 20 | 61005667 | 61005710 | 1 | 0.0008 | 74 | 86 | 69 | C20orf151 | Unknown |
| X | 153233106 | 153233193 | 3 | 0.030 | 48 | 45 | 56 | TMEM187 | Unknown |
| Characteristics | ME/CFS Mean +/− SD | Controls Mean +/− SD |
|---|---|---|
| Sex | 5 females | 2 females |
| Age (years) | 22 ± 3.63 | 25 ± 0 |
| Height (m) | 1.70 ± 0.03 | 1.62 ± 0.57 |
| Weight (kg) | 62.97 ± 9.93 | 75.1 ± 14 |
| BMI (kg·m2) | 21.84 ± 3.87 | 28.86 ± 7.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, S.; Hodges, L.D.; Peppercorn, K.; Davis, J.; Edgar, C.D.; Rodger, E.J.; Chatterjee, A.; Tate, W.P. Precision Medicine Study of Post-Exertional Malaise Epigenetic Changes in Myalgic Encephalomyelitis/Chronic Fatigue Patients During Exercise. Int. J. Mol. Sci. 2025, 26, 8563. https://doi.org/10.3390/ijms26178563
Sharma S, Hodges LD, Peppercorn K, Davis J, Edgar CD, Rodger EJ, Chatterjee A, Tate WP. Precision Medicine Study of Post-Exertional Malaise Epigenetic Changes in Myalgic Encephalomyelitis/Chronic Fatigue Patients During Exercise. International Journal of Molecular Sciences. 2025; 26(17):8563. https://doi.org/10.3390/ijms26178563
Chicago/Turabian StyleSharma, Sayan, Lynette D. Hodges, Katie Peppercorn, Jemma Davis, Christina D. Edgar, Euan J. Rodger, Aniruddha Chatterjee, and Warren P. Tate. 2025. "Precision Medicine Study of Post-Exertional Malaise Epigenetic Changes in Myalgic Encephalomyelitis/Chronic Fatigue Patients During Exercise" International Journal of Molecular Sciences 26, no. 17: 8563. https://doi.org/10.3390/ijms26178563
APA StyleSharma, S., Hodges, L. D., Peppercorn, K., Davis, J., Edgar, C. D., Rodger, E. J., Chatterjee, A., & Tate, W. P. (2025). Precision Medicine Study of Post-Exertional Malaise Epigenetic Changes in Myalgic Encephalomyelitis/Chronic Fatigue Patients During Exercise. International Journal of Molecular Sciences, 26(17), 8563. https://doi.org/10.3390/ijms26178563







