Genetic Diversity Evaluation and Population Structure Analysis of the Genus Paphiopedilum in Guangxi: Promoting the Selection and Breeding of New Species
Abstract
1. Introduction
2. Results
2.1. Cross-Species Amplification and Microsatellite Polymorphism
2.2. Genetic Diversity
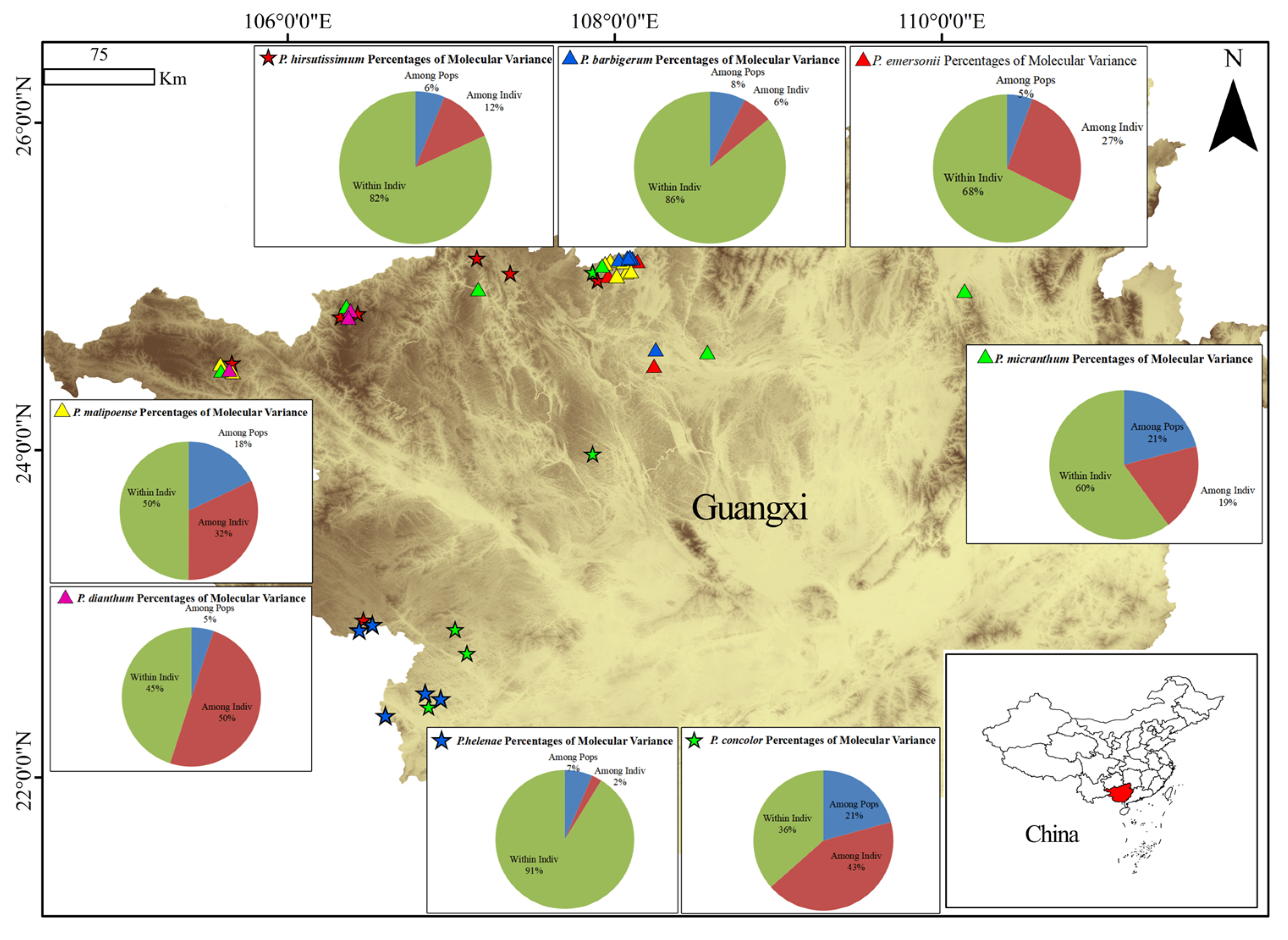
2.3. Genetic Differentiation and Species Relationships
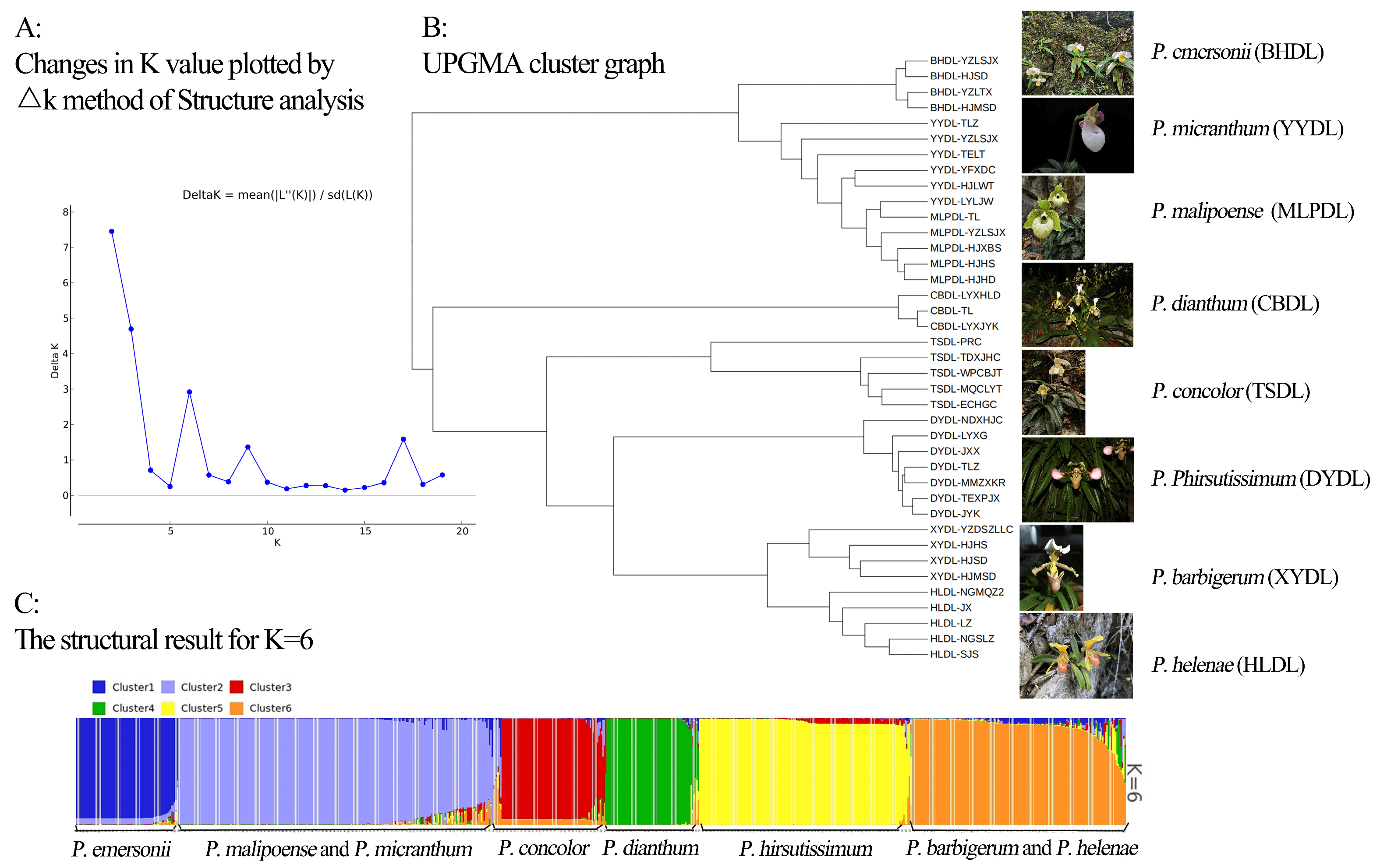
2.4. Genetic Structure and Cluster Analysis
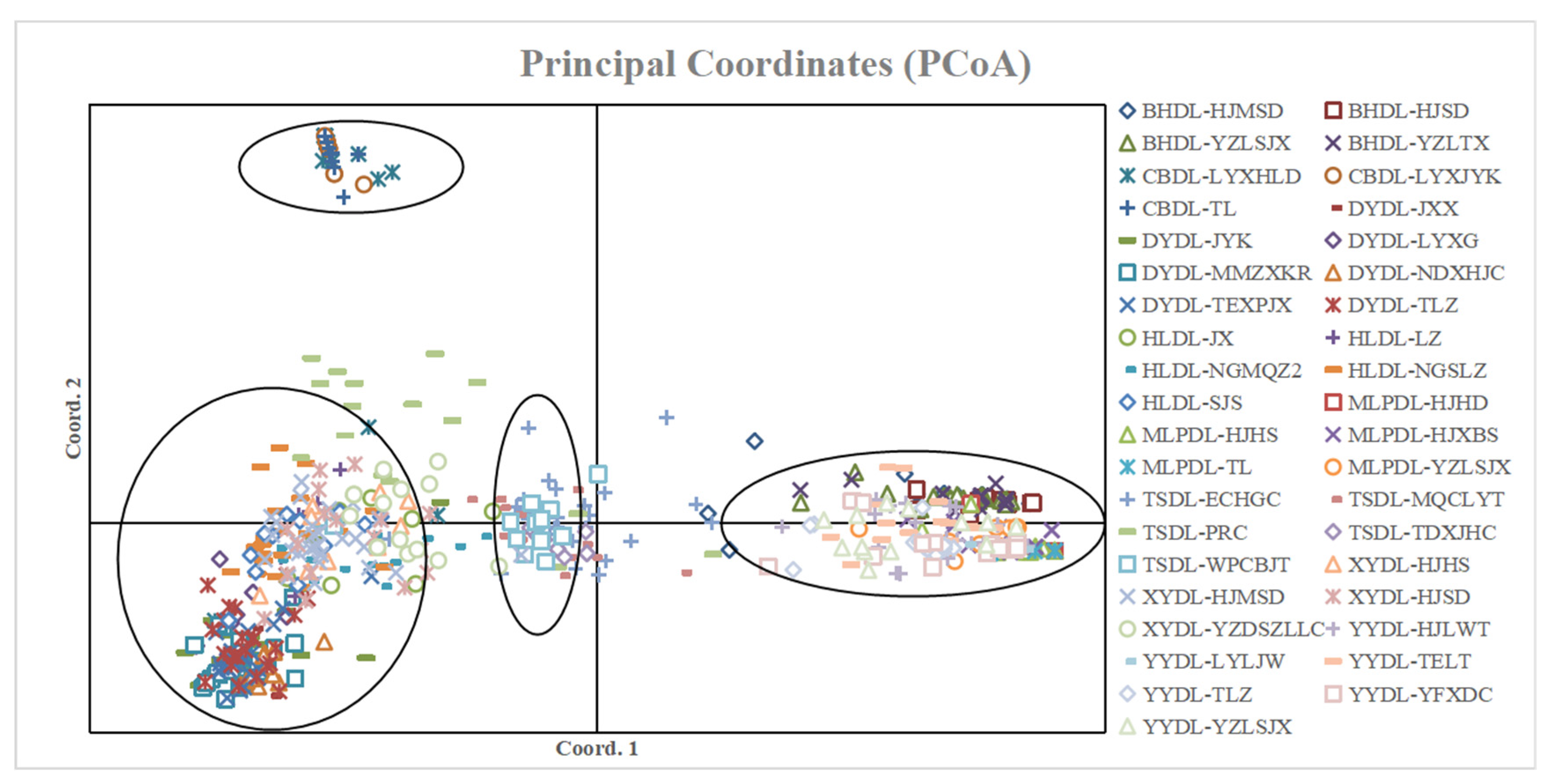
2.5. PCoA Analysis
2.6. Genetic Distance and Geographic Distances Analysis
3. Discussion
4. Materials and Methods
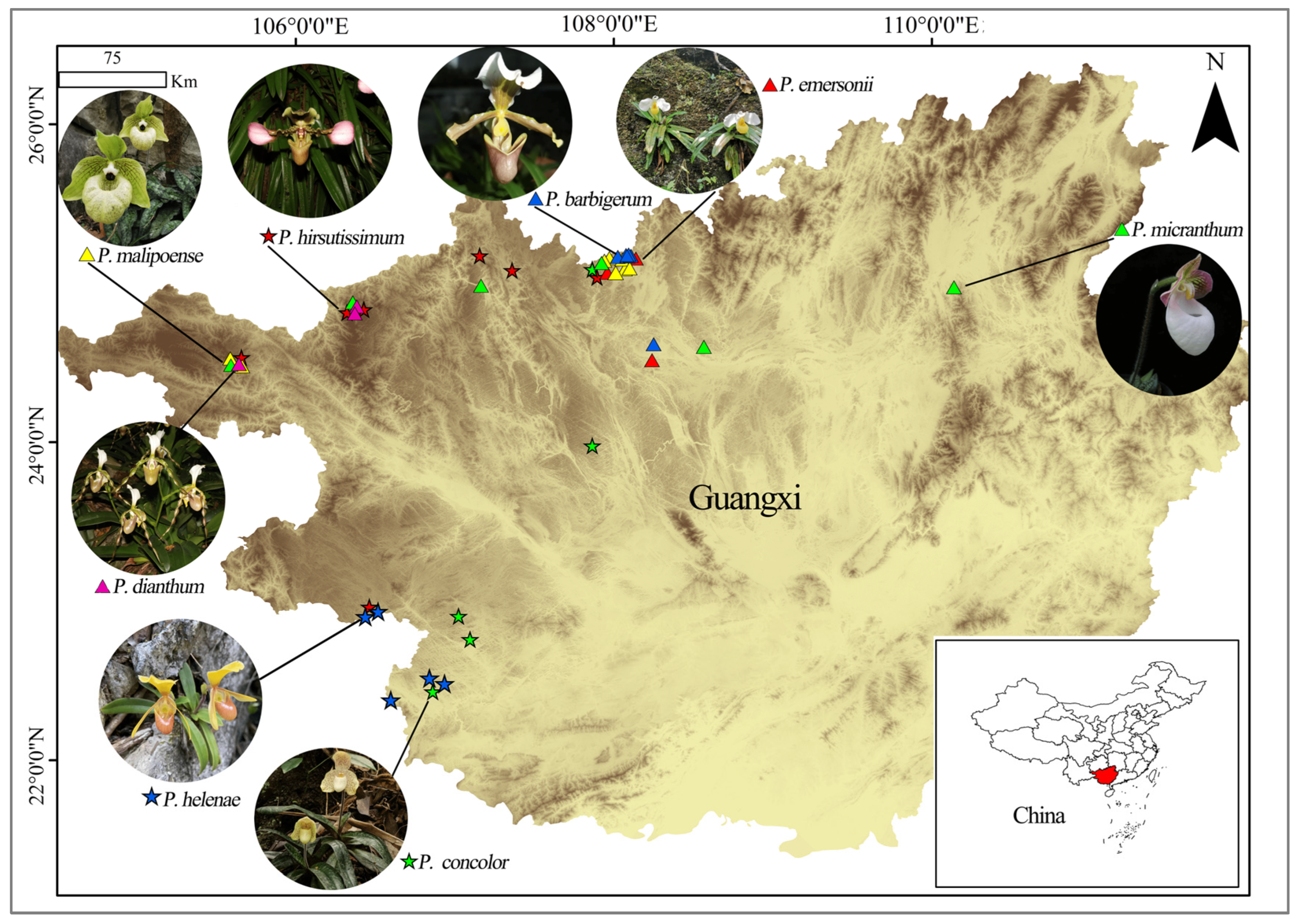
4.1. Plant Material and Sampling Strategy
4.2. Primer Development
4.3. Microsatellite Analyses
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Y.J.; Huang, J.L.; Hu, H.; Zhang, S.B. Progress in the conservation and utilization of germplasm resources of Chinese tulips. West. For. Sci. 2021, 50, 108–112+119. [Google Scholar] [CrossRef]
- Chen, Q.W.; Guo, Y.Q. Paphiopedilum plants in China: Scopes and review. Guangxi Agric. Sci. 2010, 41, 818–821. [Google Scholar]
- Cribb, P.J. The Genus Paphiopedilum, 2nd ed.; Borneo Natural History Publications: Kota Kinabalu, Malaysia, 1998. [Google Scholar]
- Yuan, H.; Gu, W.B.; Liu, L.A.; Sun, G.F. Research on the resources and domestication of the genus Tulipa in China. In Proceedings of the Sixteenth Symposium of the Botanical Garden Branch of the Botanical Society of China, Shenzhen, China, 6–7 December 2023; pp. 157–164. [Google Scholar]
- Lu, S.C. The Phalaenopsis orchids of China. J. Bot. 1988, 6, 16–18. [Google Scholar]
- Long, B.; Long, C.L. Amazing Paphiopedilum and Its Research Status. Nat. J. 2006, 6, 341–344. [Google Scholar]
- Wang, Z.; Cong, L.; Liu, Y. A Review of Paphiopedilum Research. For. Sci. 2006, 42, 113–119. [Google Scholar]
- Wang, D.G.; Deng, K.Y.; Wei, C.J. Current status and outlook of the genus Dulcis in Guizhou. Anhui Agric. Sci. 2009, 37, 2469–2470. [Google Scholar] [CrossRef]
- Yang, Z.J.; Zhu, G.F.; Lu, F.B.; Zhang, X.; Wang, B.Q. Studies on the Karyotypes of Eight Species of Paphiopedilum subgenes brachypetalum. J. Hortic. 2006, 33, 1015–1020. [Google Scholar] [CrossRef]
- Zeng, S.J.; Chen, Z.L.; Li, L.N.; Wu, K.L.; Duan, J. Enchanting Chinese Paphiopedilum Plants. Guangdong Gard. 2010, 32, 71–76. [Google Scholar]
- Zeng, S.J.; Tian, R.S.; Chen, Z.L.; Wu, K.L.; Duan, J. Research Progress on Cross Breeding of Paphiopedilum. J. Trop. Subtrop. Bot. 2010, 18, 459–468. [Google Scholar]
- Parveen, I.; Singh, H.K.; Raghuvanshi, S.; Pradhan, U.C.; Babbar, S.B. DNA barcoding of endangered Indian Paphiopedilum species. Mol. Ecol. Resour. 2012, 12, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Wu, Y.L.; Peng, K. Micro-morphological Characters of Leaf Epidermis of Ten Species in Genus Paphiopedilum. Plant Res. 2014, 34, 723–729. [Google Scholar]
- Wu, R.H.; Zhang, X. Research Advance on Reproductive Biology of Paphiopedilum. Henan Agric. Sci. 2013, 42, 6–10. [Google Scholar] [CrossRef]
- Charlesworth, D.; Wright, S.I. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 2001, 11, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Cole, C. Genetic variation in rare and common plants. Annu. Rev. Ecol. Syst. 2003, 34, 213–237. [Google Scholar] [CrossRef]
- Gitzendanner, M.A.; Soltis, P.S. Patterns of genetic variation in rare and widespread plant congeners. Am. J. Bot. 2000, 87, 783–792. [Google Scholar] [CrossRef]
- Luo, L.L.; Wang, Q.; Li, X.L.; Xu, D.L.; Hu, H. Population Genetic Diversity and Species Distribution Evaluation of Bletilla striata (Orchidaceae) in Southwest China Using SSR Markers. Ecol. Evol. 2025, 15, e72043. [Google Scholar] [CrossRef]
- Zhu, G.F.; Guo, E. Progress in molecular biology of important ornamental orchids. Bot. Bull. 2004, 21, 471–477. [Google Scholar]
- Gao, L.X.; Qin, G.L.; Yi, G.P. SRAP analysis of genetic diversity in leaf-bearing tulips. South. J. Agric. 2014, 45, 1734–1738. [Google Scholar]
- Obara-Okeyo, P.; Kako, S. Genetic diversity and identification of Cymbidium cultivars as measured by amplified polymorphic DNA (RAPD) markers. Euphytica 1998, 99, 95–101. [Google Scholar] [CrossRef]
- Gao, L.X. Screening of ISSR primers and optimization of reaction system in Paphiopedilum malipoense. Anhui Agric. Sci. 2014, 42, 6553–6555. [Google Scholar] [CrossRef]
- Zhu, Y.Y.; Wang, G.; Hou, N.; Wang, L.H. SRAP genetic diversity analysis of wild tulips in southern Guizhou. J. Southwest For. Univ. 2017, 37, 10–14. [Google Scholar]
- Qin, H.Z.; Pan, B.; Zhao, J.; Zou, R.; Wei, X.; Tang, F.L. ISSR genetic diversity analysis of very small populations of the wild plant Dulcis alba. Guangxi Sci. 2022, 29, 1134–1140. [Google Scholar] [CrossRef]
- Li, Z.Y.; Guan, M.Y.; Li, J.; Li, M.Y. Genetic Diversity of Paphiopedilum micranthum Detected by ISSR Data. Northwest J. Bot. 2016, 36, 1351–1356. [Google Scholar]
- Xu, Y.; Chen, Z.G.; Xu, Y.F.; Ge, H.; Yang, S.H.; Zhao, X.; Kou, Y.P.; Yu, X.N.; Jia, R.D. Genetic Diversity of Wild Paphiopedilum hirsutissimum Populations in Southwest China with SSR Markers J. Trop. Crops 2023, 44, 2208–2218. [Google Scholar]
- Xu, Z.Y.; Chang, R.Z. Comparison of the information content of different molecular marker techniques. Plant Genet. Resour. Sci. 2000, 1, 41–46. [Google Scholar]
- Ping, J.; Feng, P.; Li, J.; Zhang, R.; Su, Y.; Wang, T. Molecular evolution and SSRs analysis based on the chloroplast genome of Callitropsis funebris. Ecol. Evol. 2021, 11, 4786–4802. [Google Scholar] [CrossRef] [PubMed]
- Tambarussi, E.; de Souza, A.P. Population genetic structure, introgression, and hybridization in the genus Rhizophora along the Brazilian coast. Ecol. Evol. 2018, 8, 3491–3504. [Google Scholar] [CrossRef]
- Ellis, J.R.; Pashley, C.H.; Burke, J.M.; McCauley, D.E. High genetic diversity in a rare and endangered sunflower as compared to a common congener. Mol. Ecol. 2006, 15, 2345–2355. [Google Scholar] [CrossRef]
- Furches, M.S.; Wallace, L.E.; Helenurm, K. High genetic divergence characterizes populations of the endemic plant Lithophragma maximum (Saxifragaceae) on San Clemente Island. Conserv. Genet. 2009, 10, 115–126. [Google Scholar] [CrossRef]
- Riley, L.; McGlaughlin, M.E.; Helenurm, K. Genetic diversity following demographic recovery in the insular endemic plant Galium catalinense subspecies acrispum. Conserv. Genet. 2010, 11, 2015–2025. [Google Scholar] [CrossRef]
- Dunbar-Co, S.; Wieczorek, A.M. Genetic structure among populations in the endemic Hawaiian Plantago lineage: Insights from microsatellite variation. Plant Species Biol. 2011, 26, 134–144. [Google Scholar] [CrossRef]
- Kim, C.; Jung, J.; Choi, H.-K. Molecular identification of Schoenoplectiella species (Cyperaceae) by use of microsatellite markers. Plant Syst. Evol. 2012, 298, 811–817. [Google Scholar] [CrossRef]
- Ou, T.Y.; Wu, Z.N.; Tian, C.Y.; Yang, Y.; Gong, W.; Niu, J.; Li, Z. Development of Genome-Wide SSR Markers in Leymus chinensis with Genetic Diversity Analysis and DNA Fingerprints. Int. J. Mol. Sci. 2025, 26, 918. [Google Scholar] [CrossRef]
- Yan, J.Y.; Zheng, B.; Wang, S.B.; Xu, W.; Qian, M.; Ma, X.; Wu, H. Genetic Diversity and Fingerprinting of 231 Mango Germplasm Using Genome SSR Markers. Int. J. Mol. Sci. 2024, 25, 13625. [Google Scholar] [CrossRef]
- Fan, J.Z.; Li, X.L.; He, J.Z.; Zeng, Y.H.; Wang, F.S.; Bu, C.Y. Studies on the Phenotypic Diversity and the Genetic Relationships of 29 Species of Paphiopedilum. J. Plant Genet. Resour. 2023, 24, 680–691. [Google Scholar] [CrossRef]
- Gu, R.; Xu, R.; Wei, S.; Ingvarsson, P.K.; Fan, S.; Liu, G. Reduced representation sequencing reveals genetic diversity and adaptive genetic divergence in Calamus rhabdocladus. Sci. Rep. 2025, 15, 21848. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.Y.; Li, S.Y.; Ma, B.Y.; Liu, C.L.; Qi, Y.K.; Pang, C.H. Genetic Diversity Analysis Core Collection Construction of Ancient Sophora japonica L. Using SSR Markers. Int. J. Mol. Sci. 2024, 25, 12776. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.C.; Chen, C.; Liu, N.; Liu, F.F.; Su, X.H.; Liu, C.G.; Huang, Q.J. Genetic Diversity and Association Analysis of Traits Related to Water-Use Efficiency and Nitrogen-Use Efficiency of Populus deltoides Based on SSR Markers. Int. J. Mol. Sci. 2024, 25, 11515. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, K.P.; Appiah, R.K.; Cicalese, J.J.; Vorsa, N.; Reddy, U.K.; Elavarthi, S.; Melmaiee, K. Generation and characterization of novel transcriptome-derived SSR markers for genetic applications in blueberry. Sci. Rep. 2025, 15, 25050. [Google Scholar] [CrossRef]
- Li, J.; Zhang, X.F.; Chen, J.Y.; Wu, L.Q.; Xu, L.L. Genetic Diversity of Cypripedium tibeticum Populations Revealed by ISSR Analysis. Acta Bot. Boreali-Occident. Sin. 2020, 40, 0669–0977. [Google Scholar]
- Nybom, H. Comparison of Different Nuclear DNA Markers for Estimating Genetic Diversity in Plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Q.; Tang, X.; Huang, C.Y.; Deng, J.L.; Li, X.; Lu, J.S.; Li, X.M.; Zhang, Z.B. Genetic diversity analysis and DNA fingerprint construction for 22 Dendrobium species. Mol. Plant Breed. 2021, 19, 3005–3014. [Google Scholar]
- Xu, Y.; Jia, R.; Zhou, Y.; Cheng, H.; Zhao, X.; Ge, H. Development and characterization of polymorphic EST-SSR markers for Paphiopedilum henryanum (Orchidaceae). Appl. Plant Sci. 2018, 6, e01152. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Evolution in Mendelian populations. Genetics 1931, 16, 97–159. [Google Scholar] [CrossRef] [PubMed]

| Locus | Na | Ne | I | Ho | He | F | PIC | Signif |
|---|---|---|---|---|---|---|---|---|
| DL014 | 5 | 2.68 | 1.133 | 0.172 | 0.627 | 0.726 | 0.555 | *** |
| DL020 | 17 | 5.545 | 2.049 | 0.469 | 0.82 | 0.428 | 0.802 | *** |
| DL021 | 6 | 2.144 | 0.942 | 0.23 | 0.534 | 0.568 | 0.474 | *** |
| DL023 | 17 | 4.011 | 1.728 | 0.182 | 0.751 | 0.757 | 0.72 | *** |
| DL030 | 6 | 2.536 | 1.074 | 0.127 | 0.606 | 0.79 | 0.531 | *** |
| DL032 | 17 | 9.676 | 2.455 | 0.42 | 0.897 | 0.532 | 0.888 | *** |
| DL034 | 14 | 3.242 | 1.462 | 0.166 | 0.692 | 0.76 | 0.644 | *** |
| DL036 | 6 | 3.385 | 1.341 | 0.153 | 0.705 | 0.783 | 0.656 | *** |
| DL039 | 15 | 4.563 | 1.96 | 0.357 | 0.781 | 0.543 | 0.763 | *** |
| DL040 | 6 | 3.38 | 1.372 | 0.267 | 0.704 | 0.62 | 0.657 | *** |
| Mean | 10.9 | 4.116 | 1.552 | 0.254 | 0.712 | 0.651 | 0.669 | |
| St Dev | 5.466 | 2.197 | 0.485 | 0.121 | 0.107 | 0.129 |
| Species | Pop | Na | Ne | I | Ho | He | F |
|---|---|---|---|---|---|---|---|
| P. emersonii | BHDL-HJMSD | 2.600 | 1.281 | 0.360 | 0.148 | 0.186 | 0.202 |
| BHDL-HJSD | 1.700 | 1.242 | 0.240 | 0.129 | 0.140 | 0.023 | |
| BHDL-YZLSJX | 2.500 | 1.441 | 0.366 | 0.181 | 0.196 | 0.054 | |
| BHDL-YZLTX | 1.900 | 1.270 | 0.275 | 0.171 | 0.166 | −0.043 | |
| Mean | 2.175 | 1.308 | 0.310 | 0.157 | 0.172 | 0.059 | |
| P. dianthum | CBDL-LYXHLD | 2.600 | 1.197 | 0.313 | 0.060 | 0.154 | 0.696 |
| CBDL-LYXJYK | 1.200 | 1.112 | 0.092 | 0.063 | 0.061 | 0.367 | |
| CBDL-TL | 1.400 | 1.124 | 0.132 | 0.083 | 0.079 | −0.064 | |
| Mean | 1.733 | 1.144 | 0.179 | 0.069 | 0.098 | 0.333 | |
| P. hirsutissimum | DYDL-JXX | 2.500 | 1.730 | 0.600 | 0.330 | 0.349 | 0.087 |
| DYDL-JYK | 3.100 | 1.665 | 0.640 | 0.289 | 0.349 | 0.134 | |
| DYDL-LYXG | 2.900 | 1.736 | 0.634 | 0.353 | 0.359 | 0.029 | |
| DYDL-MMZXKR | 2.700 | 1.745 | 0.597 | 0.351 | 0.348 | −0.048 | |
| DYDL-NDXHJC | 2.600 | 1.895 | 0.604 | 0.331 | 0.339 | −0.021 | |
| DYDL-TEXPJX | 3.300 | 1.793 | 0.661 | 0.326 | 0.362 | 0.030 | |
| DYDL-TLZ | 2.800 | 1.981 | 0.722 | 0.402 | 0.411 | 0.017 | |
| Mean | 2.843 | 1.792 | 0.637 | 0.340 | 0.360 | 0.032 | |
| P. helenae | HLDL-JX | 3.400 | 2.513 | 0.961 | 0.550 | 0.537 | −0.044 |
| HLDL-LZ | 3.100 | 2.267 | 0.876 | 0.600 | 0.507 | −0.143 | |
| HLDL-NGMQZ2 | 3.300 | 2.210 | 0.789 | 0.490 | 0.442 | −0.114 | |
| HLDL-NGSLZ | 3.800 | 2.057 | 0.785 | 0.409 | 0.425 | 0.007 | |
| HLDL-SJS | 3.500 | 2.274 | 0.775 | 0.425 | 0.409 | 0.030 | |
| Mean | 3.420 | 2.264 | 0.837 | 0.495 | 0.464 | −0.053 | |
| P. malipoense | MLPDL-HJHD | 1.100 | 1.018 | 0.029 | 0.000 | 0.015 | 1.000 |
| MLPDL-HJHS | 1.700 | 1.158 | 0.174 | 0.043 | 0.099 | 0.504 | |
| MLPDL-HJXBS | 2.100 | 1.241 | 0.274 | 0.086 | 0.152 | 0.318 | |
| MLPDL-TL | 1.200 | 1.147 | 0.123 | 0.078 | 0.085 | 0.112 | |
| MLPDL-YZLSJX | 2.600 | 1.515 | 0.421 | 0.195 | 0.216 | 0.073 | |
| Mean | 1.740 | 1.216 | 0.204 | 0.080 | 0.113 | 0.401 | |
| P. concolor | TSDL-ECHGC | 3.900 | 1.783 | 0.671 | 0.220 | 0.340 | 0.402 |
| TSDL-MQCLYT | 3.100 | 1.727 | 0.513 | 0.189 | 0.252 | 0.291 | |
| TSDL-PRC | 3.400 | 2.132 | 0.842 | 0.252 | 0.472 | 0.483 | |
| TSDL-TDXJHC | 1.700 | 1.453 | 0.327 | 0.230 | 0.204 | −0.016 | |
| TSDL-WPCBJT | 2.500 | 1.624 | 0.519 | 0.251 | 0.287 | 0.241 | |
| Mean | 2.920 | 1.744 | 0.574 | 0.228 | 0.311 | 0.280 | |
| P. barbigerum | XYDL-HJHS | 3.400 | 2.215 | 0.796 | 0.503 | 0.421 | −0.089 |
| XYDL-HJMSD | 4.600 | 2.799 | 1.024 | 0.493 | 0.514 | 0.015 | |
| XYDL-HJSD | 3.600 | 2.568 | 0.913 | 0.453 | 0.484 | 0.058 | |
| XYDL-YZDSZLLC | 3.900 | 2.583 | 0.960 | 0.498 | 0.501 | 0.011 | |
| Mean | 3.875 | 2.541 | 0.923 | 0.487 | 0.480 | −0.001 | |
| P. micranthum | YYDL-HJLWT | 3.700 | 1.773 | 0.586 | 0.252 | 0.281 | 0.115 |
| YYDL-LYLJW | 1.900 | 1.362 | 0.282 | 0.171 | 0.160 | 0.057 | |
| YYDL-TELT | 3.100 | 1.888 | 0.630 | 0.308 | 0.333 | 0.063 | |
| YYDL-TLZ | 2.600 | 1.506 | 0.404 | 0.064 | 0.204 | 0.640 | |
| YYDL-YFXDC | 2.700 | 1.790 | 0.541 | 0.262 | 0.285 | 0.124 | |
| YYDL-YZLSJX | 2.600 | 1.838 | 0.570 | 0.286 | 0.311 | 0.074 | |
| Mean | 2.767 | 1.693 | 0.502 | 0.224 | 0.262 | 0.179 |
| Locus | Fis | Fit | Fst | Nm |
|---|---|---|---|---|
| DL014 | −0.004 | 0.711 | 0.712 | 0.101 |
| DL020 | −0.024 | 0.395 | 0.409 | 0.361 |
| DL021 | −0.070 | 0.530 | 0.560 | 0.196 |
| DL023 | 0.028 | 0.729 | 0.721 | 0.097 |
| DL030 | 0.188 | 0.782 | 0.732 | 0.091 |
| DL032 | 0.196 | 0.526 | 0.410 | 0.359 |
| DL034 | 0.319 | 0.766 | 0.657 | 0.130 |
| DL036 | 0.169 | 0.759 | 0.710 | 0.102 |
| DL039 | 0.051 | 0.525 | 0.500 | 0.250 |
| DL040 | −0.028 | 0.588 | 0.599 | 0.167 |
| Mean | 0.083 | 0.631 | 0.601 | 0.186 |
| SE | 0.040 | 0.043 | 0.040 | 0.033 |
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among Pops | 38 | 3169.945 | 83.420 | 2.097 | 57% |
| Among Indiv | 721 | 1394.600 | 1.934 | 0.354 | 10% |
| Within Indiv | 760 | 932.500 | 1.227 | 1.227 | 33% |
| Total | 1519 | 5497.044 | 5497.045 | 3.678 | 100% |
| Species | P. Code | Location | No. | E | N | H |
|---|---|---|---|---|---|---|
| P. emersonii | BHDL-YZLSJX | Yizhou District, Hechi City, Guangxi | 20 | 108°34′ | 24°36′ | 437 |
| BHDL-HJMSD | Huanjiang County, Hechi City, Guangxi | 20 | 108°03′ | 25°09′ | 577 | |
| BHDL-YZLTX | Yizhou District, Hechi City, Guangxi | 20 | 108°14′ | 24°30′ | 218 | |
| BHDL-HJSD | Huanjiang County, Hechi City, Guangxi | 14 | 108°02′ | 25°09′ | 592 | |
| P. micranthum | YYDL-LYLJW | Leye County, Baise City, Guangxi | 28 | 106°22′ | 24°50′ | 640 |
| YYDL-YZLSJX | Yizhou District, Hechi City, Guangxi | 14 | 108°34′ | 24°36′ | 285 | |
| YYDL-TELT | Tian′e County, Hechi City, Guangxi | 26 | 107°09′ | 24°59′ | 870 | |
| YYDL-HJMLT | Huanjiang County, Hechi City, Guangxi | 29 | 108°02′ | 25°08′ | 977 | |
| YYDL-YFXDX | Yongfu County, Guilin City, Guangxi | 16 | 110°08′ | 24°58′ | 363 | |
| YYDL-TLZ | Tianlin County, Baise City, Guangxi | 15 | 105°38′ | 24°30′ | 1122 | |
| P. barbigerum | XYDL-YZDSZLLC | Yizhou District, Hechi City, Guangxi | 16 | 108°15′ | 24°37′ | 268 |
| XYDL-HJHS | Huanjiang County, Hechi City, Guangxi | 30 | 108°03′ | 25°09′ | 533 | |
| XYDL-HJSD | Huanjiang County, Hechi City, Guangxi | 17 | 108°02′ | 25°09′ | 541 | |
| XYDL-HJMSD | Huanjiang County, Hechi City, Guangxi | 10 | 108°03′ | 25°09′ | 537 | |
| P. malipoense | MLPDL-YZLSJX | Yizhou District, Hechi City, Guangxi | 15 | 108°34′ | 24°36′ | 285 |
| MLPDL-HJHS | Huanjiang County, Hechi City, Guangxi | 30 | 108°03′ | 25°09′ | 398 | |
| MLPDL-HJHD | Huanjiang County, Hechi City, Guangxi | 24 | 108°03′ | 25°08′ | 377 | |
| MLPDL-HJXBS | Huanjiang County, Hechi City, Guangxi | 20 | 107°56′ | 25°08′ | 709 | |
| MLPDL-TL | Tianlin County, Baise City, Guangxi | 9 | 105°38′ | 24°30′ | 1122 | |
| P. concolor | TSDL-PRC | Zuozhou District, Chongzuo City, Guangxi | 25 | 107°57′ | 25°06′ | 732 |
| TSDL-WPCBJT | Dahua County, Hechi City, Guangxi | 20 | 107°51′ | 23°58′ | 567 | |
| TSDL-TDXJHC | Tiandeng County, Chongzuo City, Guangxi | 9 | 107°01′ | 22°54′ | 384 | |
| TSDL-ECHGC | Daxin County, Chongzuo City, Guangxi | 33 | 107°05′ | 22°45′ | 356 | |
| TSDL-MQCLYT | Longzhou County, Chongzuo City, Guangxi | 22 | 106°53′ | 22°26′ | 404 | |
| P. dianthum | CBDL-LYXJYK | Leye County, Baise City, Guangxi | 19 | 106°24′ | 24°50′ | 400 |
| CBDL-LYXHJD | Leye County, Baise City, Guangxi | 27 | 106°22′ | 24°48′ | 700 | |
| CBDL-TL | Tianlin County, Baise City, Guangxi | 20 | 105°38′ | 24°30′ | 1110 | |
| P. hirsutissimum | DYDL-PEXPJX | Tian′e County, Hechi City, Guangxi | 29 | 107°08′ | 25°11′ | 375 |
| DYDL-JYK | Leye County, Baise City, Guangxi | 23 | 106°24′ | 24°50′ | 400 | |
| DYDL-LYXG | Leye County, Baise City, Guangxi | 15 | 106°19′ | 24°48′ | 820 | |
| DYDL-NDXHJC | Nandan County, Hechi City, Guangxi | 20 | 107°21′ | 25°05′ | 386 | |
| DYDL-MMZXKR | Huanjiang County, Hechi City, Guangxi | 30 | 107°57′ | 25°06′ | 732 | |
| DYDL-JXX | Jinxi City, Guangxi | 10 | 106°29′ | 22°55′ | 750 | |
| DYDL-TLZ | Tianlin County, Baise City, Guangxi | 24 | 105°38′ | 24°30′ | 1122 | |
| P. helenae | HLDL-NGSLZ | Longzhou County, Chongzuo City, Guangxi | 22 | 106°50′ | 22°31′ | 547 |
| HLDL-LZ | Longzhou County, Chongzuo City, Guangxi | 11 | 106°35′ | 22°22′ | 429 | |
| HLDL-NGMQZ2 | Longzhou County, Chongzuo City, Guangxi | 19 | 106°54′ | 22°27′ | 430 | |
| HLDL-JX | Jinxi City, Guangxi | 7 | 106°29′ | 22°55′ | 790 | |
| HLDL-SJS | Jinxi City, Guangxi | 12 | 106°28′ | 22°54′ | 700 |
| Locus | Repeat Motif | Primer Sequence (F) | Primer Sequence (R) | Ta (°C) | GenBank No. |
|---|---|---|---|---|---|
| DL014 | (CTC)6 | TTCCTTCCCTACCCTTTCCA | CAGCGGTGTCGTTGATGTT | 60 | MG333725 |
| DL020 | (GCC)6 | GGCCAAGTACATGCACCCAT | TTCCCACCTCGGTTATGCAC | 60 | MG333700 |
| DL021 | (CAG)6 | GCAAATCCATTCAGCCCTGC | CGACATGGTCTGAGAGGAGC | 60 | MG333701 |
| DL023 | (AGA)6 | CTTGGGACTCTTTCCTCGGC | CAGCACCTCTTCGCGTAAGA | 60 | MG333703 |
| DL030 | (CCG)6 | CAGGTTGACAGCAATGTCGC | GCCGCAGCTTTTCGGATAAG | 60 | MG333710 |
| DL032 | (AAAC)5 | AGCGTGTTTGGACTAGAGCA | TCGGGGATGCACATGGAAAA | 60 | MG333712 |
| DL034 | (CGG)6 | GGGTGGGGAGAGTAGGAGTT | GCCACAACTTGTTTTCCCGG | 60 | MG333714 |
| DL036 | (CGT)6 | CCACGTGTGACAGAATCCCA | GGCTCCCGACGAGGAATTAC | 60 | MG333716 |
| DL039 | (ATC)6 | CCACCAGCTTTCATATCCTCCA | GCCCATGCTGTGCAAAAAGA | 60 | MG333719 |
| DL040 | (TCT)6 | AAGAAGTGGCTTCCATGGCA | GCAAAACCAAGGTGTCGTCC | 60 | MG333720 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.; Xian, K.; Su, J.; Lu, L.; Cai, X.; Yang, Y.; Pan, B.; Ding, T.; Zhu, X.; Chai, S.; et al. Genetic Diversity Evaluation and Population Structure Analysis of the Genus Paphiopedilum in Guangxi: Promoting the Selection and Breeding of New Species. Int. J. Mol. Sci. 2025, 26, 8543. https://doi.org/10.3390/ijms26178543
Tang J, Xian K, Su J, Lu L, Cai X, Yang Y, Pan B, Ding T, Zhu X, Chai S, et al. Genetic Diversity Evaluation and Population Structure Analysis of the Genus Paphiopedilum in Guangxi: Promoting the Selection and Breeding of New Species. International Journal of Molecular Sciences. 2025; 26(17):8543. https://doi.org/10.3390/ijms26178543
Chicago/Turabian StyleTang, Jianmin, Kanghua Xian, Jiang Su, Li Lu, Xinru Cai, Yishan Yang, Bo Pan, Tao Ding, Xianliang Zhu, Shengfeng Chai, and et al. 2025. "Genetic Diversity Evaluation and Population Structure Analysis of the Genus Paphiopedilum in Guangxi: Promoting the Selection and Breeding of New Species" International Journal of Molecular Sciences 26, no. 17: 8543. https://doi.org/10.3390/ijms26178543
APA StyleTang, J., Xian, K., Su, J., Lu, L., Cai, X., Yang, Y., Pan, B., Ding, T., Zhu, X., Chai, S., Zou, R., & Wei, X. (2025). Genetic Diversity Evaluation and Population Structure Analysis of the Genus Paphiopedilum in Guangxi: Promoting the Selection and Breeding of New Species. International Journal of Molecular Sciences, 26(17), 8543. https://doi.org/10.3390/ijms26178543






