Proteomic Analysis and Expression of Selected Genes During the Early Somatic Embryogenesis of Jatropha curcas L.
Abstract
1. Introduction
2. Results and Discussion
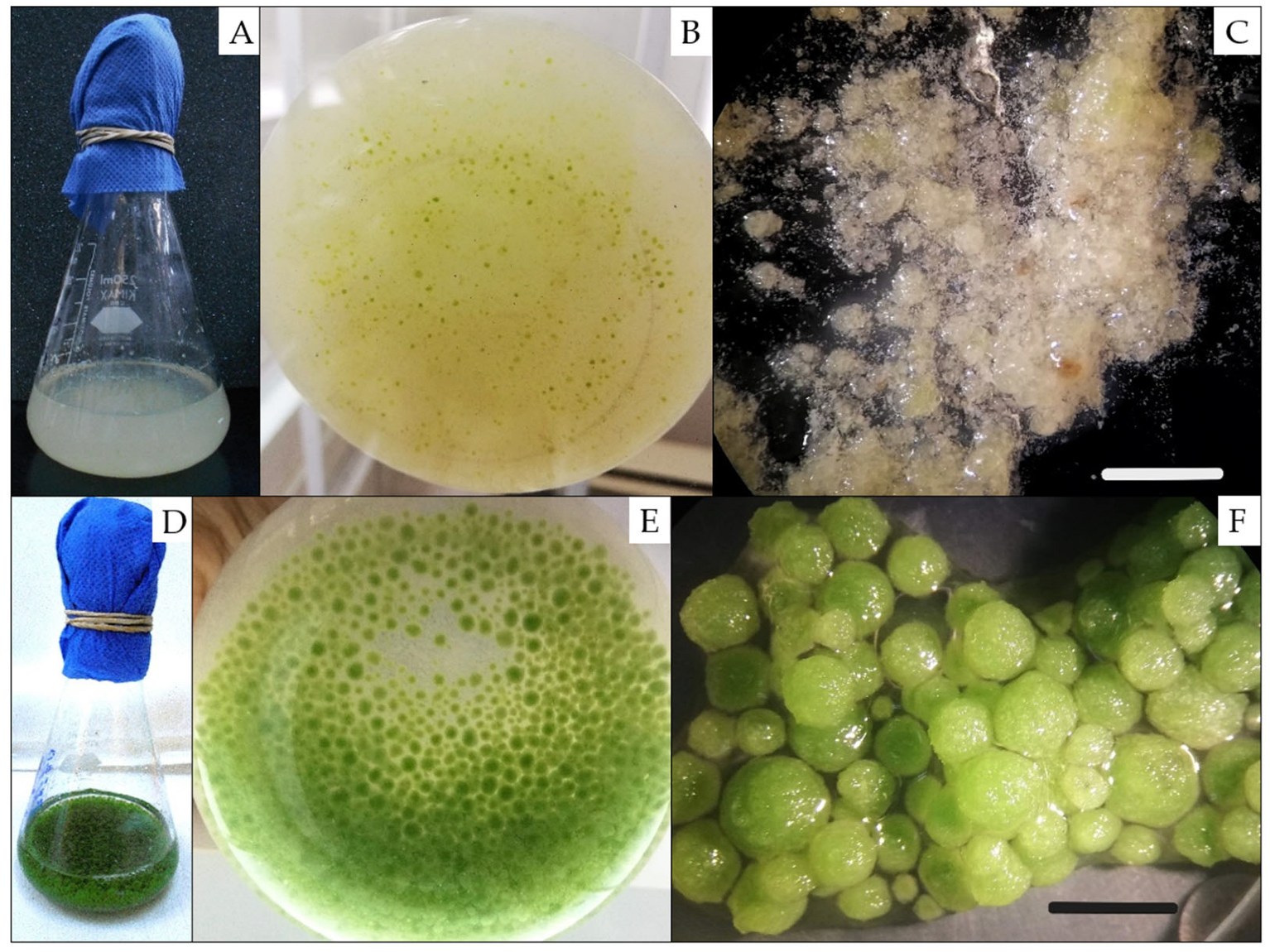
2.1. Obtaining Two Developmental Stages of J. curcas Somatic Embryos in Liquid Medium
2.2. Two-Dimensional Proteome Analysis of Pre-Globular and Globular Somatic Embryos
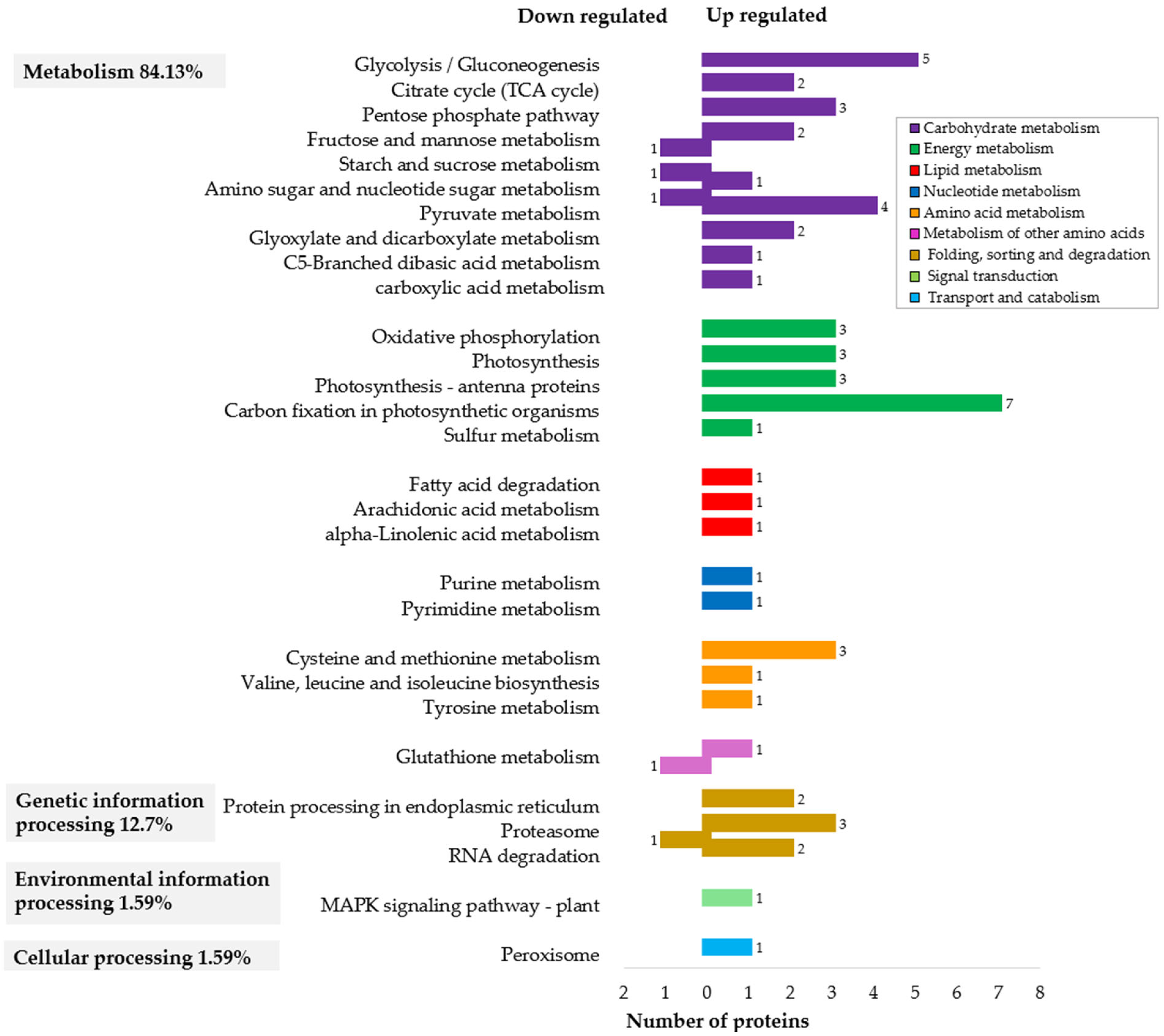
2.3. Functional Classification of Proteins in Pre-Globular and Globular Somatic Embryos
2.3.1. Enhanced Metabolism and Cellular Activity at the Globular Stage
2.3.2. Photosynthetic Priming for Autotrophic Growth
2.3.3. Protein Folding and Degradation Mechanisms
2.3.4. Translational Regulation: Role of eIF3f
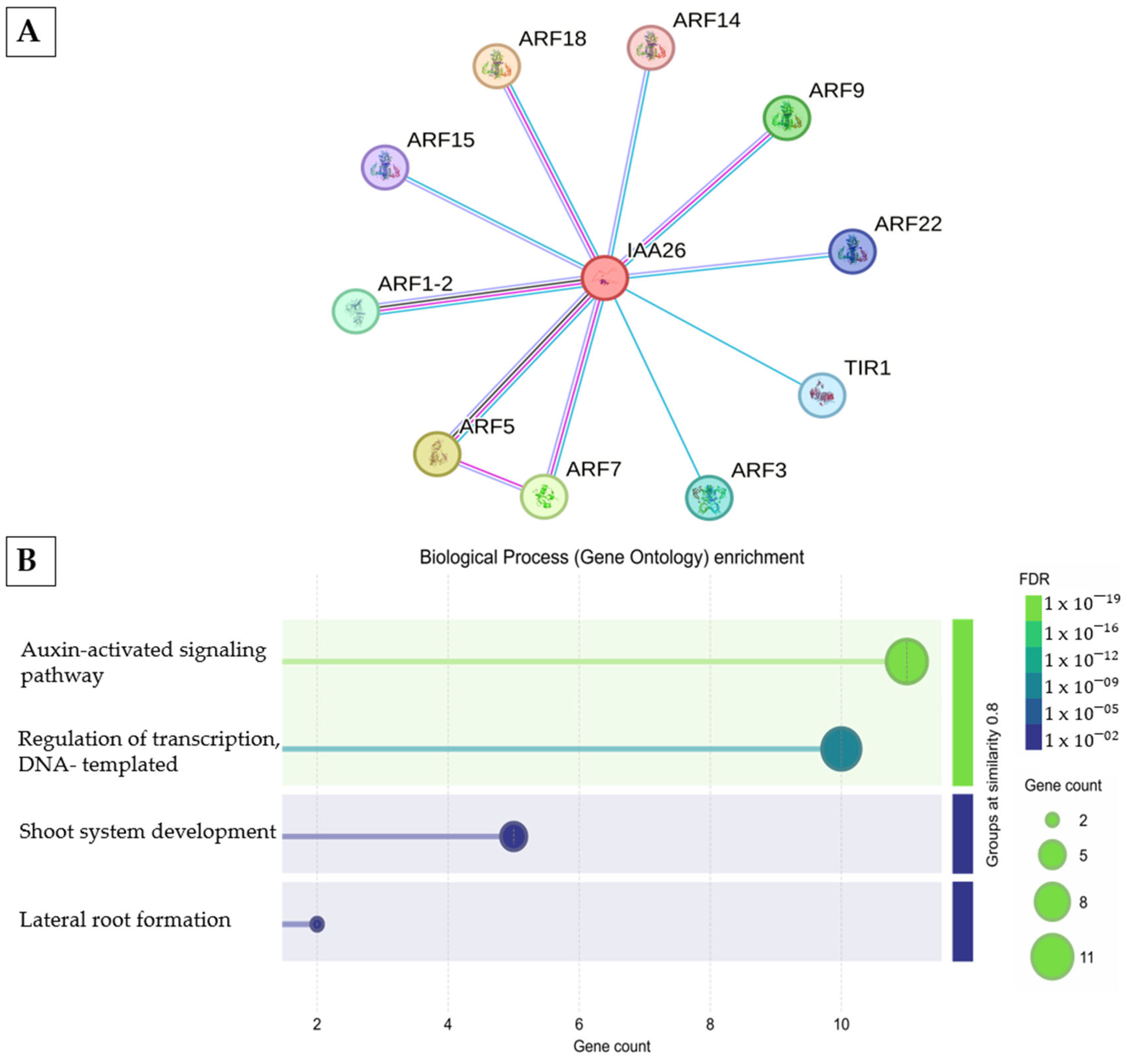
2.3.5. Auxin Signaling and Embryo Developmental Arrest
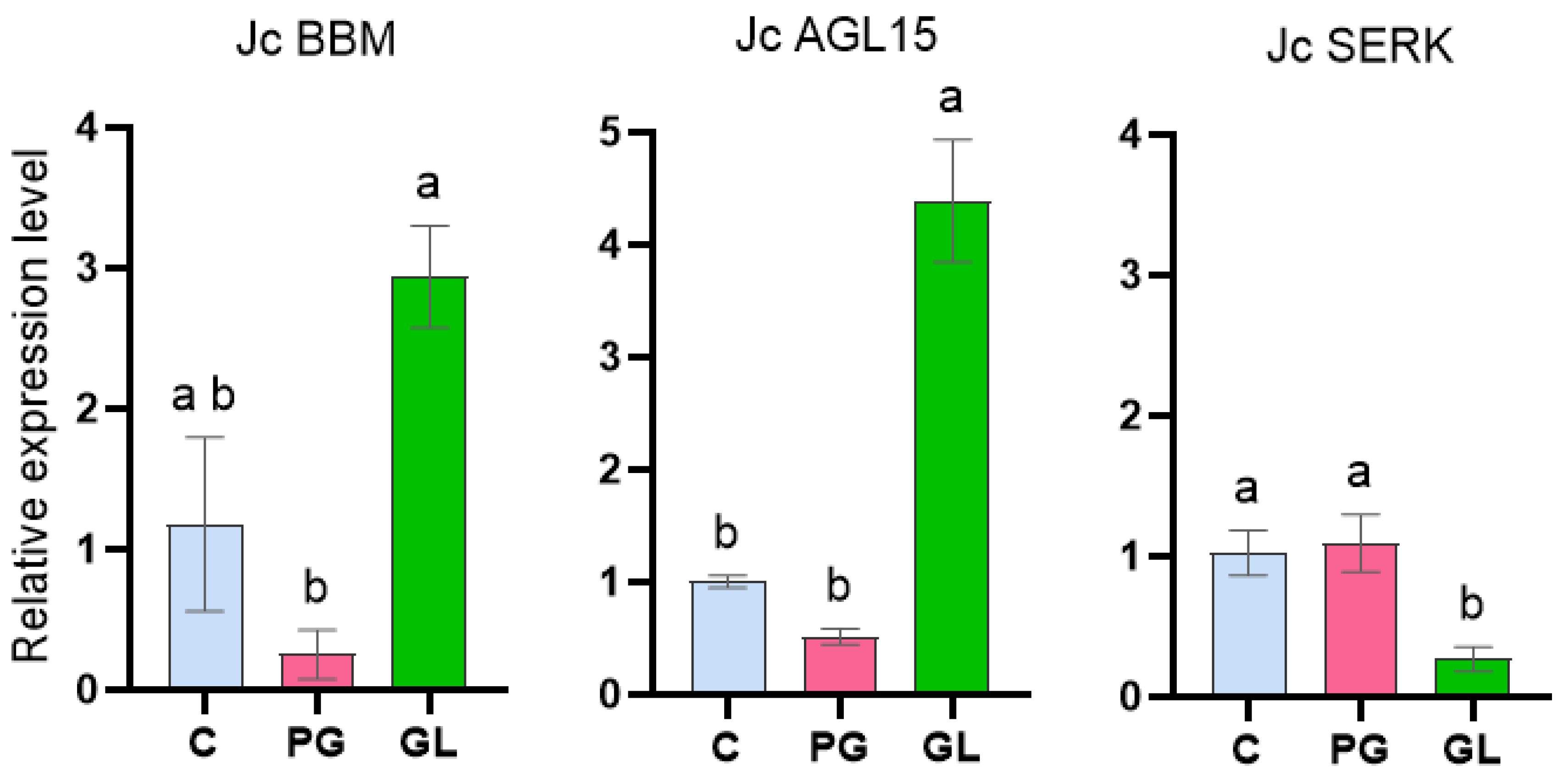
2.4. Gene Expression Analysis in J. curcas
3. Materials and Methods
3.1. Plant Material for Somatic Embryogenesis
3.2. Induction of Callus
3.3. Induction of Somatic Embryos in Liquid Medium
3.4. Protein Extraction
3.5. Two-Dimensional IEF/SDS–PAGE and Protein Staining
3.6. Image Acquisition and Analysis
3.7. In-Gel Digestion and Mass Spectrometry Analysis
3.8. Biological Function
3.9. RNA Extraction and cDNA Synthesis
3.10. Gene Expression Analysis via RT-qPCR
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Khayri, J.M.; Sudheer, W.N.; Preetha, T.R.; Nagella, P.; Rezk, A.A.; Shehata, W.F. Biotechnological research progress in Jatropha, a biodiesel-yielding plant. Plants 2022, 11, 1292. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Srivastava, V.C.; Jha, M.K. Jatropha curcas phytotomy and applications: Development as a potential biofuel plant through biotechnological advancements. Renew. Sustain. Energy Rev. 2016, 59, 818–838. [Google Scholar] [CrossRef]
- Pérez, G.; Islas, J.; Guevara, M.; Suárez, R. The sustainable cultivation of Mexican nontoxic Jatropha curcas to produce biodiesel and food in marginal rural lands. Sustainability 2019, 11, 5823. [Google Scholar] [CrossRef]
- Li, H.; Tsuchimoto, S.; Harada, K.; Yamasaki, M.; Sakai, H.; Wada, N.; Alipour, A.; Sasai, T.; Tsunekawa, A.; Tsujimoto, H.; et al. Genetic tracing of Jatropha curcas L. from its Mesoamerican origin to the world. Front. Plant Sci. 2017, 8, 1539. [Google Scholar] [CrossRef] [PubMed]
- Vandepitte, K.; Valdés-Rodríquez, O.A.; Sánchez-Sánchez, O.; De Kort, H.; Martinez-Herrera, J.; García-Pérez, E.; De Meyer, T.; Pérez-Vázquez, A.; Muys, B.; Honnay, O. High SNP diversity in the non-toxic indigenous Jatropha curcas germplasm widens the potential of this upcoming major biofuel crop species. Ann. Bot. 2019, 124, 645–652. [Google Scholar] [CrossRef]
- Zavala del Angel, I.; García-Pérez, E.; González-Hernández, D.; Pérez-Vázquez, A.; Ávila Reséndiz, C. Genetic diversity of Jatropha curcas L. in Veracruz state, Mexico, and its relationships with the content of phorbol esters. Glob. Adv. Res. J. Agric. Sci. 2016, 5, 149–158. [Google Scholar]
- Ewunie, G.A.; Morken, J.; Lekang, O.I.; Yigezu, Z.D. Factors affecting the potential of Jatropha curcas for sustainable biodiesel production: A critical review. Renew. Sustain. Energy Rev. 2021, 137, 110500. [Google Scholar] [CrossRef]
- Moniruzzaman, M.; Yaakob, Z.; Khatun, R. Biotechnology for Jatropha improvement: A worthy exploration. Renew. Sustain. Energy Rev. 2016, 54, 1262–1277. [Google Scholar] [CrossRef]
- Salaün, C.; Lepiniec, L.; Dubreucq, B. Genetic and molecular control of somatic embryogenesis. Plants 2021, 10, 1467. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Xu, C.; Yan, S.; Xu, B. An efficient somatic embryo liquid culture system for potential use in large-scale and synchronic production of Anthurium andraeanum seedlings. Front. Plant Sci. 2019, 10, 29. [Google Scholar] [CrossRef]
- Wójcikowska, B.; Chwiałkowska, K.; Nowak, K.; Citerne, S.; Morończyk, J.; Wójcik, A.M.; Kiwior-Wesołowska, A.; Francikowski, J.; Kwaśniewski, M.; Gaj, M.D. Transcriptomic profiling reveals histone acetylation-regulated genes involved in somatic embryogenesis in Arabidopsis thaliana. BMC Genom. 2024, 25, 788. [Google Scholar] [CrossRef] [PubMed]
- Kumaravel, M.; Uma, S.; Backiyarani, S.; Saraswathi, M.S. Proteomic analysis of somatic embryo development in Musa Spp. cv. Grand Naine (AAA). Sci. Rep. 2020, 10, 4501. [Google Scholar] [CrossRef] [PubMed]
- Fehér, A. Why somatic plant cells start to form embryos? In Somatic Embryogenesis; Mujib, A., Samaj, J., Eds.; Springer: Berlin, Germany, 2005; pp. 85–101. [Google Scholar] [CrossRef]
- Gulzar, B.; Mujib, A.; Malik, M.Q.; Sayeed, R.; Mamgain, J.; Ejaz, B. Genes, proteins and other networks regulating somatic embryogenesis in plants. J. Genet. Eng. Biotechnol. 2020, 18, 31. [Google Scholar] [CrossRef]
- Karami, O.; Philipsen, C.; Rahimi, A.; Nurillah, A.R.; Boutilier, K.; Offringa, R. Endogenous auxin maintains embryonic cell identity and promotes somatic embryo development in Arabidopsis. Plant J. 2023, 113, 7–22. [Google Scholar] [CrossRef]
- Zaragoza-Martínez, F.; Lucho-Constantino, G.G.; Ponce-Noyola, T.; Esparza-García, F.; Poggi-Varaldo, H.; Cerda-García-Rojas, C.M.; Trejo-Tapia, G.; Ramos-Valdivia, A.C. Jasmonic acid stimulates the oxidative responses and triterpene production in Jatropha curcas cell suspension cultures through mevalonate as biosynthetic precursor. Plant Cell Tiss Organ Cult. 2016, 127, 47–56. [Google Scholar] [CrossRef]
- Marcus, K.; Lelong, C.; Rabilloud, T. What room for two-dimensional gel-based proteomics in a shotgun proteomics world? Proteomes 2020, 8, 17. [Google Scholar] [CrossRef]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified Protein Abundance Index (EmPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, C.; Komatsu, S.; He, M.; Liu, G.; Shen, S. Proteomic analysis of the seed development in Jatropha curcas: From carbon flux to the lipid accumulation. J. Proteom. 2013, 91, 23–40. [Google Scholar] [CrossRef]
- Ramzan, A.; Shah, M.; Ullah, N.; Sheheryar; Nascimento, J.R.S.; Campos, F.A.P.; Domont, G.B.; Nogueira, F.C.S.; Abdellattif, M.H. Proteomic analysis of embryo isolated from mature Jatropha curcas L. seeds. Front. Plant Sci. 2022, 13, 843764. [Google Scholar] [CrossRef]
- Wang, F.X.; Shang, G.D.; Wu, L.Y.; Xu, Z.G.; Zhao, X.Y.; Wang, J.W. Chromatin accessibility dynamics and a hierarchical transcriptional regulatory network structure for plant somatic embryogenesis. Dev. Cell 2020, 54, 742–757. [Google Scholar] [CrossRef]
- Wójcik, A.M.; Wójcikowska, B.; Gaj, M.D. Current perspectives on the auxin-mediated genetic network that controls the induction of somatic embryogenesis in plants. Int. J. Mol. Sci. 2020, 21, 1333. [Google Scholar] [CrossRef]
- Gulzar, B.; Mujib, A.; Rajam, M.V.; Frukh, A.; Zafar, N. Identification of somatic embryogenesis (SE) related proteins through label-free shotgun proteomic method and cellular role in Catharanthus Roseus (L.) G. Don. Plant Cell. Tissue Organ Cult. 2019, 137, 225–237. [Google Scholar] [CrossRef]
- Su, Y.H.; Tang, L.P.; Zhao, X.Y.; Zhang, X.S. Plant cell totipotency: Insights into cellular reprogramming. J. Integr. Plant Biol. 2021, 63, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Nolan, K.E.; Irwanto, R.R.; Rose, R.J. Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol. 2003, 133, 218–230. [Google Scholar] [CrossRef]
- El-Sayed, M.; Aly, U.I.; Mohamed, M.S.; Rady, M.R. In vitro regeneration and molecular characterization of Jatropha curcas plant. Bull. Natl. Res. Cent. 2020, 44, 70. [Google Scholar] [CrossRef]
- Mendoza-Peña, C.N.; Hvoslef-Eide, A.K. A novel genotype-independent technique for successful induction of somatic embryogenesis of adult plants of Jatropha curcas L. using petiole transverse Thin Cell Layer (TCL). Afr. J. Biotechnol. 2021, 20, 85–91. [Google Scholar] [CrossRef]
- Su, Y.H.; Liu, Y.B.; Bai, B.; Zhang, X.S. Establishment of embryonic shoot-root axis is involved in auxin and cytokinin response during Arabidopsis somatic embryogenesis. Front. Plant Sci. 2015, 5, 792. [Google Scholar] [CrossRef]
- Amaral-Silva, P.M.; Clarindo, W.R.; Guilhen, J.H.S.; de Jesus Passos, A.B.R.; Sanglard, N.A.; Ferreira, A. Global 5-methylcytosine and physiological changes are triggers of indirect somatic embryogenesis in Coffea canephora. Protoplasma 2021, 258, 45–57. [Google Scholar] [CrossRef]
- Jin, F.; Hu, L.; Yuan, D.; Xu, J.; Gao, W.; He, L.; Yang, X.; Zhang, X. Comparative transcriptome analysis between somatic embryos (SEs) and zygotic embryos in cotton: Evidence for stress response functions in SE development. Plant Biotechnol. J. 2014, 12, 161–173. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, L.; Fan, Y.; Fan, Y.; Tang, Z.; Zeng, F. Dynamic TMT-based quantitative proteomics analysis of critical initiation process of totipotency during cotton somatic embryogenesis transdifferentiation. Int. J. Mol. Sci. 2019, 20, 1691. [Google Scholar] [CrossRef]
- Quintana-Escobar, A.O.; Bojórquez-Velázquez, E.; Ruiz-May, E.; Loyola-Vargas, V.M. Proteomic approach during the induction of somatic embryogenesis in Coffea canephora. Plants 2023, 12, 4095. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho-Silva, R.; Travassos-Carmo, L.S.; Gomes-Luis, Z.; Silva, L.P.; Scherwinski-Pereira, J.E.; Mehta, A. Proteomic identification of differentially expressed proteins during the acquisition of somatic embryogenesis in oil palm (Elaeis guineensis Jacq.). J. Proteom. 2014, 104, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Noah, A.M.; Niemenak, N.; Sunderhaus, S.; Haase, C.; Omokolo, D.N.; Winkelmann, T.; Braun, H.P. Comparative proteomic analysis of early somatic and zygotic embryogenesis in Theobroma cacao L. J. Proteom. 2013, 78, 123–133. [Google Scholar] [CrossRef]
- Rode, C.; Lindhorst, K.; Braun, H.P.; Winkelmann, T. From callus to embryo: A proteomic view on the development and maturation of somatic embryos in Cyclamen persicum. Planta 2012, 235, 995–1011. [Google Scholar] [CrossRef]
- Xia, C.; Wang, Y.J.; Li, W.Q.; Chen, Y.R.; Deng, Y.; Zhang, X.Q.; Chen, L.Q.; Ye, D. The Arabidopsis eukaryotic translation initiation factor 3, subunit F (AteIF3f), is required for pollen germination and embryogenesis. Plant J. 2010, 63, 189–202. [Google Scholar] [CrossRef]
- Gliwicka, M.; Nowak, K.; Balazadeh, S.; Mueller-Roeber, B.; Gaj, M.D. Extensive modulation of the transcription factor transcriptome during somatic embryogenesis in Arabidopsis thaliana. PLoS ONE 2013, 8, e69261. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowska, B.; Gaj, M.D. Expression profiling of AUXIN RESPONSE FACTOR genes during somatic embryogenesis induction in Arabidopsis. Plant Cell Rep. 2017, 36, 843–858. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.-M.; Van Lammeren, A.A.M.; Miki, B.L.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef]
- Kulinska-Lukaszek, K.; Tobojka, M.; Adamiok, A.; Kurczynska, E.U. Expression of the BBM gene during somatic embryogenesis of Arabidopsis thaliana. Biol. Plant. 2012, 56, 389–394. [Google Scholar] [CrossRef]
- Silva, A.T.; Barduche, D.; do Livramento, K.G.; Paiva, L.V. A putative BABY BOOM-like gene (CaBBM) is expressed in embryogenic calli and embryogenic cell suspension culture of Coffea arabica L. Plant Tissue Cult. 2015, 51, 93–101. [Google Scholar] [CrossRef]
- Awada, R.; Lepelley, M.; Breton, D.; Charpagne, A.; Campa, C.; Berry, V.; Georget, F.; Breitler, J.C.; Léran, S.; Djerrab, D.; et al. Global transcriptome profiling reveals differential regulatory, metabolic and hormonal networks during somatic embryogenesis in Coffea arabica. BMC Genom. 2023, 24, 41. [Google Scholar] [CrossRef] [PubMed]
- Horstman, A.; Li, M.; Heidmann, I.; Weemen, M.; Chen, B.; Muino, J.M.; Angenent, G.C.; Boutilier, K. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 2017, 175, 848–857. [Google Scholar] [CrossRef]
- Torres-Ruiz, J.J. Análisis de la Expresión de Genes de Jatropha curcas Involucrados en la Respuesta Regenerativa en Cultivos In Vitro; Cinvestav: Mexico City, México, 2023. [Google Scholar]
- Su, Y.H.; Zhao, X.Y.; Liu, Y.B.; Zhang, C.L.; O’Neill, S.D.; Zhang, X.S. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 2009, 59, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zheng, Y.; Ji, H.; Burnie, W.; Perry, S.E. Gene regulation by the AGL15 transcription factor reveals hormone interactions in somatic embryogenesis. Plant Physiol. 2016, 172, 2374–2387. [Google Scholar] [CrossRef]
- Neves, M.; Correia, S.; Cavaleiro, C.; Canhoto, J. Modulation of organogenesis and somatic embryogenesis by ethylene: An overview. Plants 2021, 10, 1208. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; Boeren, S.; Russinova, E.; Aker, J.; Vervoort, J.; de Vries, S. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1 protein complex includes BRASSINOSTEROID-INTENSITIVE1. Plant Cell 2006, 18, 626–638. [Google Scholar] [CrossRef]
- Pérez-Pascual, D.; Jiménez-Guillen, D.; Villanueva-Alonzo, H.; Souza-Perera, R.; Godoy-Hernández, G.; Zúñiga-Aguilar, J.J. Ectopic expression of the Coffea canephora SERK1 homolog-induced differential transcription of genes involved in auxin metabolism and in the developmental control of embryogenesis. Physiol. Plant. 2018, 163, 530–551. [Google Scholar] [CrossRef]
- Li, H.; Cai, Z.; Wang, X.; Li, M.; Cui, Y.; Cui, N.; Yang, F.; Zhu, M.; Zhao, J.; Du, W.; et al. SERK receptor-like kinases control division patterns of vascular precursors and ground tissue stem cells during embryo development in Arabidopsis. Mol. Plant 2019, 12, 984–1002. [Google Scholar] [CrossRef]
- Hardtke, C.S.; Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998, 17, 1405–1411. [Google Scholar] [CrossRef]
- Raabe, K.; Honys, D.; Michailidis, C. The role of eukaryotic initiation factor 3 in plant translation regulation. Plant Physiol. Biochem. 2019, 145, 75–83. [Google Scholar] [CrossRef]
- Gamborg, O.L.; Miller, R.A.; Ojima, K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Galaz-Ávalos, R.M.; Martínez-Sánchez, H.G.; Loyola-Vargas, V.M. Induction of somatic embryogenesis in Jatropha curcas. In Plant Cell Culture Protocolos, Methods in Molecular Biology; Loyola-Vargas, V.M., Ochoa-Alejo, N., Eds.; Springer: New York, NY, USA, 2018; Volume 1815, pp. 207–214. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Wanqian, L.; Bochu, W.; Chuanren, D.; Biao, L. A Method for isolating functional RNA from callus of Dendrobium candidum contented rich polysaccharides. Colloids Surf. B Biointerfaces 2005, 42, 259–262. [Google Scholar] [CrossRef]
- Rocha, A.J.; Maranhao, P.A.; Silva, R.O.; Pohl, S.; Finteles, C.S.R. Identification of suitable reference genes for gene expression normalization in Jatropha curcas L. during development and under stress conditions using real time quantitative PCR. Brazilian Arch. Biol. Technol. 2016, 59, e16150396. [Google Scholar] [CrossRef]
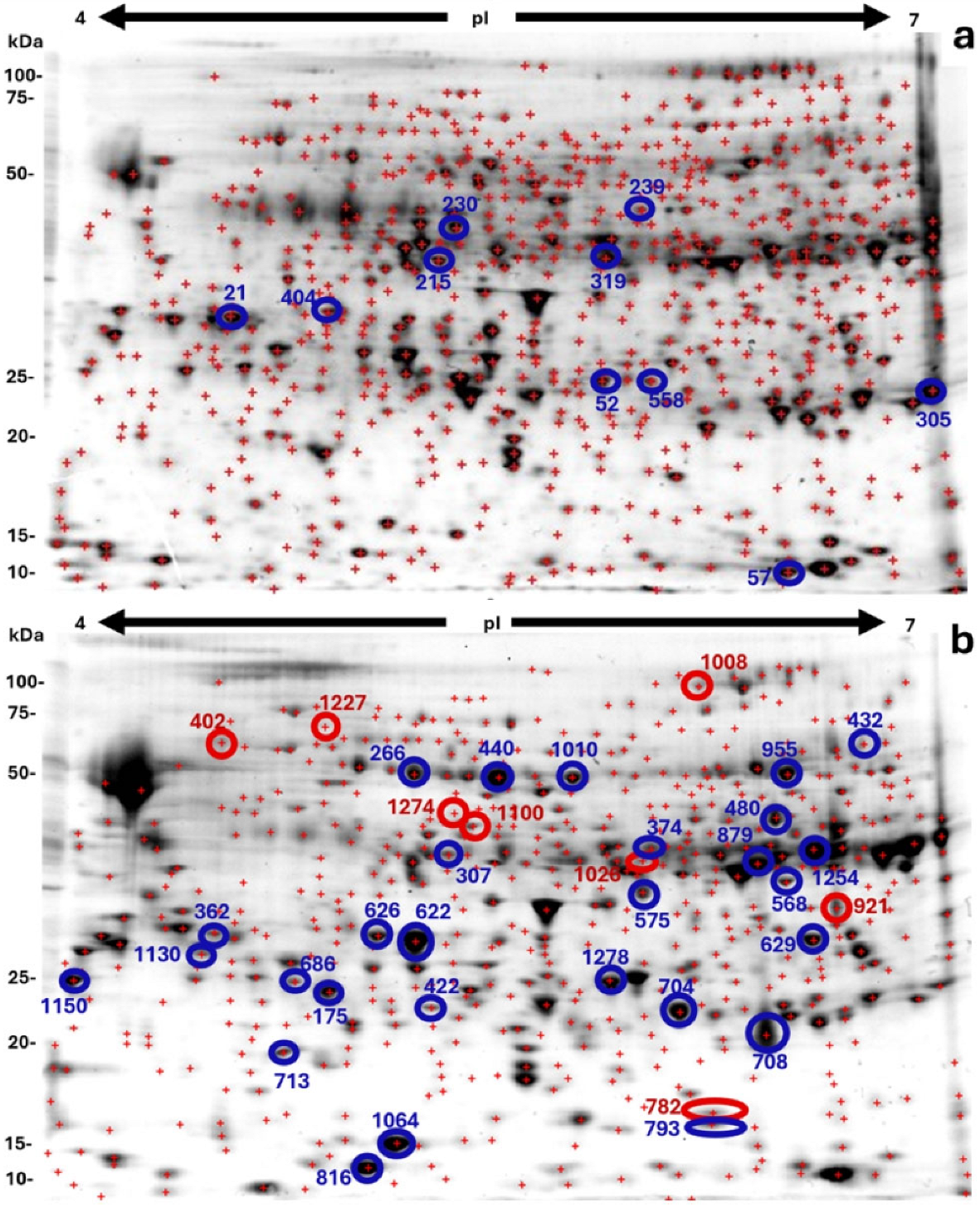

| ImageMasterTM 2D Platinum Data | QuickGo Data | ||||
|---|---|---|---|---|---|
| Spot Number a | Protein Number b | Fold Change c | Protein Name d | GO SLIM e | GO TERMS f |
| 21 | 1 | 4.02 | 14-3-3 protein 6 | __ | __ |
| 319 | 1 | 3.63 | Hypothetical protein JCGZ_08406 | __ | __ |
| 558 | 1 | 3.23 | Endo-1,3;1,4-beta-D-glucanase | __ | __ |
| 230 | 3 | 1.94 | Hypothetical protein JCGZ_19572 | __ | __ |
| 215 | 1 | 4.33 | Fructokinase-2 | Carbohydrate metabolic process | Fructose metabolic process; carbohydrate phosphorylation |
| 57 | 2 | 4.43 | Nucleoside diphosphate kinase | Nucleobase-containing compound metabolic process | GTP, UTP, CTP biosynthetic process |
| 52 | 1 | 1.74 | Proteasome subunit alpha type | Protein metabolic process | Ubiquitin-dependent protein catabolic process |
| 305 | 1 | 5.58 | Glutathione S-transferase PARB | Response to chemical stimulus | Glutathione metabolic process; response to toxic substance |
| 57 | 1 | 4.43 | Superoxide dismutase [Cu-Zn] | Response to stress and chemical stimulus | Superoxide metabolic process; removal of superoxide radicals; cellular oxidant detoxification |
| 239 | 1 | 3.64 | LysM domain receptor-like kinase 3 | Response to stress, biotic stimulus; external stimulus | Innate immune response |
| 404 | 1 | 2.92 | Eukaryotic translation initiation factor 3 subunit F | Translation | Formation of cytoplasmic translation initiation complex; cytoplasmic translational initiation |
| ImageMasterTM 2D Platinum Data | QuickGo Data | ||||
|---|---|---|---|---|---|
| Spot Number a | Protein Number b | Fold Change c | Protein Name d | GO SLIM e | GO TERMS f |
| 921 | 1 | unique | PREDICTED: Secoisolariciresinol dehydrogenase-like | __ | __ |
| 1274 | 1 | unique | Actin-7 | __ | _ |
| 1227 | 1 | unique | LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA | __ | _ |
| 1278 | 3 | 3.6 | Secoisolariciresinol dehydrogenase | __ | __ |
| 629 | 1 | 3.47 | PREDICTED: Secoisolariciresinol dehydrogenase-like | __ | __ |
| 362 | 2 | 2.07 | Plastid-lipid-associated protein, chloroplastic | __ | _ |
| 575 | 1 | 3.43 | Cysteine synthase | Biosynthetic process | Cysteine biosynthetic process from serine |
| 1150 | 1 | 2.85 | Acidic endochitinase | Carbohydrate metabolic process | Carbohydrate metabolic process |
| 307 | 1 | 2.55 | Sedoheptulose-1,7-bisphosphatase, chloroplastic | Carbohydrate metabolic process | Carbohydrate metabolic process |
| 422 | 1 | 3.44 | WEB family protein At5g55860 isoform X1 | Cellular component organization | Chloroplast avoidance movement; chloroplast accumulation movement |
| 782 | 1 | Unique | Cytochrome b6-f complex iron-sulfur subunit | Cellular process | Transmembrane transport |
| 793 | 1 | 6.18 | Cytochrome b6-f complex iron-sulfur subunit | Cellular process | Transmembrane transport |
| 266 | 3 | 3.7 | AtpB | Cellular process | ATP metabolic process |
| 1100 | 1 | Unique | PREDICTED: Malate dehydrogenase | Generation of precursor metabolites and energy | Malate metabolic process |
| 1008 | 1 | Unique | PREDICTED: NADH dehydrogenase | Generation of precursor metabolites and energy | ATP synthesis coupled electron transport |
| 432 | 1 | 8.51 | ATP synthase subunit beta | Generation of precursor metabolites and energy | ATP biosynthetic process; ATP metabolic process |
| 955 | 1 | 5.06 | PREDICTED: Enolase-like | Generation of precursor metabolites and energy | Glycolytic process |
| 1010 | 1 | 4.59 | PREDICTED: Enolase | Generation of precursor metabolites and energy | Glycolytic process |
| 1254 | 2 | 4.13 | Malate dehydrogenase | Generation of precursor metabolites and energy | Tricarboxylic acid cycle; malate metabolic process |
| 1254 | 1 | 4.13 | Fructose-bisphosphate aldolase | Generation of precursor metabolites and energy | Glycolytic process |
| 374 | 1 | 2.85 | Fructose-bisphosphate aldolase | Generation of precursor metabolites and energy | Glycolytic process |
| 175 | 1 | 2.18 | Probable ribose-5-phosphate isomerase 3, chloroplastic | Generation of precursor metabolites and energy | Pentose-phosphate shunt, non-oxidative branch |
| 440 | 1 | 1.73 | ATP synthase subunit beta | Generation of precursor metabolites and energy | ATP biosynthetic process; ATP metabolic process |
| 568 | 1 | 1.52 | PREDICTED: Malate dehydrogenase, chloroplastic | Generation of precursor metabolites and energy | Tricarboxylic acid cycle; malate metabolic process |
| 422 | 4 | 3.44 | Type I inositol polyphosphate 5-phosphatase 13 isoform X1 | Lipid metabolic process | Phosphatidylinositol dephosphorylation |
| 1026 | 1 | Unique | L-lactate dehydrogenase | Metabolic process | Lactate metabolic process; pyruvate metabolic process |
| 879 | 1 | 4.57 | L-lactate dehydrogenase | Metabolic process | Lactate metabolic process; pyruvate metabolic process |
| 713 | 2 | 2.22 | 3-isopropylmalate dehydratase small subunit 3 | Metabolic process | Oxoacid metabolic process |
| 1064 | 3 | 3.68 | 4-hydroxy-4-methyl-2-oxoglutarate aldolase | Nucleic acids metabolic process | Regulation of RNA metabolic process |
| 686 | 1 | 13.68 | PREDICTED: Chlorophyll a-b binding protein 8, Chloroplastic | Photosynthesis | Photosynthesis, light harvesting |
| 708 | 1 | 9.59 | PREDICTED: Oxygen-evolving enhancer protein 2, chloroplastic | Photosynthesis | Photosynthesis |
| 704 | 1 | 5.42 | PREDICTED: Chlorophyll a-b binding protein of LHCII (light harvesting complex II) type 1 | Photosynthesis | Photosynthesis, light harvesting |
| 626 | 1 | 4.7 | PREDICTED: Oxygen-evolving enhancer protein 1, chloroplastic | Photosynthesis | Photosystem II assembly |
| 422 | 5 | 3.44 | Chlorophyll a-b binding protein 8, chloroplastic | Photosynthesis | Photosynthesis, light harvesting |
| 622 | 1 | 2.74 | PREDICTED: Oxygen-evolving enhancer protein 1, chloroplastic | Photosynthesis | Photosystem II assembly |
| 402 | 2 | unique | PREDICTED: Ubiquitin domain-containing protein DSK2a-like | Protein metabolic process | Ubiquitin-dependent protein catabolic process |
| 1278 | 2 | 3.6 | Proteasome subunit alpha type | Protein metabolic process | Ubiquitin-dependent protein catabolic process |
| 1278 | 1 | 3.6 | Proteasome subunit alpha type | Protein metabolic process | Ubiquitin-dependent protein catabolic process |
| 1130 | 1 | 3.11 | PREDICTED: Low-temperature-induced cysteine proteinase-like | Protein metabolic process | Proteolysis |
| 1130 | 2 | 3.11 | Proteasome subunit alpha type | Protein metabolic process | Proteasome-mediated ubiquitin-dependent protein catabolic process |
| 402 | 1 | unique | Protein disulfide-isomerase | Protein metabolic process; response to stress | Protein folding; response to endoplasmic reticulum stress |
| 480 | 1 | 4.4 | Alcohol dehydrogenase | Response to chemical stimulus | Formaldehyde catabolic process |
| 713 | 1 | 2.22 | Glutathione peroxidase | Response to stress and chemical stimulus | Response to oxidative stress |
| 1064 | 1 | 3.68 | Major allergen Pru ar 1 | Response to stress, chemical stimulus, endogenous stimulus; signal transduction | Defense response; abscisic acid-activated signaling pathway |
| 816 | 1 | 5.44 | Major allergen Pru ar 1-like | Response to stress and chemical stimulus, endogenous stimulus; signal transduction | Defense response; abscisic acid-activated signaling pathway; negative regulation of phosphoprotein phosphatase activity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medina-Hernández, A.E.; Vera-Reyes, I.; Ríos-Castro, E.; Torres-Ruiz, J.J.; Ponce-Noyola, T.; Trejo-Tapia, G.; Garay-Arroyo, A.; Barrera-Cortés, J.; Ramos-Valdivia, A.C. Proteomic Analysis and Expression of Selected Genes During the Early Somatic Embryogenesis of Jatropha curcas L. Int. J. Mol. Sci. 2025, 26, 6384. https://doi.org/10.3390/ijms26136384
Medina-Hernández AE, Vera-Reyes I, Ríos-Castro E, Torres-Ruiz JJ, Ponce-Noyola T, Trejo-Tapia G, Garay-Arroyo A, Barrera-Cortés J, Ramos-Valdivia AC. Proteomic Analysis and Expression of Selected Genes During the Early Somatic Embryogenesis of Jatropha curcas L. International Journal of Molecular Sciences. 2025; 26(13):6384. https://doi.org/10.3390/ijms26136384
Chicago/Turabian StyleMedina-Hernández, Anamarel Edzná, Ileana Vera-Reyes, Emmanuel Ríos-Castro, Juan José Torres-Ruiz, Teresa Ponce-Noyola, Gabriela Trejo-Tapia, Adriana Garay-Arroyo, Josefina Barrera-Cortés, and Ana C. Ramos-Valdivia. 2025. "Proteomic Analysis and Expression of Selected Genes During the Early Somatic Embryogenesis of Jatropha curcas L." International Journal of Molecular Sciences 26, no. 13: 6384. https://doi.org/10.3390/ijms26136384
APA StyleMedina-Hernández, A. E., Vera-Reyes, I., Ríos-Castro, E., Torres-Ruiz, J. J., Ponce-Noyola, T., Trejo-Tapia, G., Garay-Arroyo, A., Barrera-Cortés, J., & Ramos-Valdivia, A. C. (2025). Proteomic Analysis and Expression of Selected Genes During the Early Somatic Embryogenesis of Jatropha curcas L. International Journal of Molecular Sciences, 26(13), 6384. https://doi.org/10.3390/ijms26136384







