Abstract
Drought stress is a major abiotic constraint that severely restricts the growth of Medicago falcata L. by inducing the accumulation of reactive oxygen species (ROS) in plants. WRKY transcription factors (TFs) play a key role in regulating plant responses to drought stress. In this study, we investigated the role of the MfWRKY40 gene in drought tolerance. Under mannitol and ABA stress treatments, MfWRKY40-overexpressing lines (OEs) showed significantly longer primary roots, increased lateral roots, and higher fresh weight compared to wild-type (Col) lines, indicating significantly enhanced growth and drought tolerance. Similarly, under soil drought conditions, transgenic Arabidopsis thaliana exhibited enhanced drought tolerance. NBT staining demonstrated decreased ROS accumulation in transgenic lines after stress treatment. Correspondingly, the MfWRKY40-overexpressing lines displayed significantly lower levels of hydrogen peroxide (H2O2), superoxide anion (O2−), and malondialdehyde (MDA) compared to Col, along with elevated activities of superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD), as well as increased proline (Pro) content. Furthermore, MfWRKY40 upregulated the expression of antioxidant enzyme genes (AtPOD3, AtSOD4, and AtCAT1) and modulated the expression of other drought-related genes. In summary, our results demonstrate that MfWRKY40 enhances drought tolerance in A. thaliana by improving ROS scavenging capacity. This study provides a theoretical foundation for further exploration of MfWRKY40’s functional mechanisms in drought stress adaptation.
1. Introduction
During their normal life activities, plants are affected by various abiotic stresses (such as drought, salinity, high temperature and cold) [1]. Among these, drought is one of the most severe abiotic stress factors that limit plant growth and development [2]. Drought stress reduces stomatal conductance, impairs photosynthesis, and induces the accumulation of reactive oxygen species (ROS) [3]. ROS are normal metabolic byproducts in plants and function in oxidative stress response and signal transduction at low levels [4]. However, at elevated concentrations, ROS can damage cellular structures and disrupt normal metabolic processes. Consequently, plants have evolved multiple mechanisms to counteract drought-induced damage. Numerous studies have demonstrated that transcription factors (TFs) play crucial roles in plant responses to various abiotic stresses [5,6,7].
TFs are a class of protein molecules that can bind to specific sequences in DNA, where they either promote or inhibit gene transcription [8], thereby exerting their regulatory functions. In plants, stress-related TFs primarily include five major families: MYB, bZIP, AP2/ERF, WRKY, and NAC [9]. Overexpression of FvMYB114 and FvMYB44 from Fragaria vesca enhances salt and cold tolerance in transgenic Arabidopsis thaliana [10,11]. Similarly, VvERF63 from grape confers cold stress tolerance in transgenic A. thaliana by enhancing photosynthetic capacity and mitigating cellular damage [12].
Among these, WRKY TFs are named after their highly conserved WRKY domain [13]. The N-terminus of the WRKY domain contains a conserved motif, WRKYGQK, while the C-terminus consists of a conserved zinc finger motif [14]. Based on the invariant WRKY motif and varying zinc finger structures, WRKY TFs are categorized into three groups: Group I contains two WRKY domains, while Groups II and III possess only one [15]. Since the first WRKY gene, SPF1, was identified in sweet potato [16], members of the WRKY transcription factor family have been continuously discovered in various species. These studies revealed that many WRKY family members are involved in plant responses to abiotic stress [17,18,19]. For instance, BpWRKY32 in birch enhances salt tolerance by reducing water loss, improving osmotic potential, and decreasing reactive ROS accumulation [20]. Similarly, HpWRKY85 from St. John’s wort and BnWRKY49 from ramie enhance drought tolerance in A. thaliana [21,22]. In apple and tomato, overexpression of MdWRKY56 and SlWRKY17, respectively, results in reduced malondialdehyde (MDA) and ROS accumulation, increased proline (Pro) content, and greater drought tolerance [23,24].
Medicago falcata L. is a perennial leguminous forage grass that harbors important genetic resources for stress resistance, making it a candidate model plant for studying the mechanisms of stress tolerance in legumes [25,26,27]. However, in China, M. falcata is primarily distributed in the northwest and northeast, where drought is one of the major factors affecting alfalfa production. In our previous study, we identified MfWRKY40 as a stress-responsive gene through transcriptomic analysis of drought-stressed M. falcata plants and demonstrated that it exhibits higher expression levels in the leaves and roots of M. falcata and is significantly induced by drought stress [28]. In this study, we utilized heterologous expression in A. thaliana to functionally characterize this gene under both controlled (culture medium) and natural (soil) conditions. Our comprehensive analysis included changes in various components of the ROS scavenging pathway, quantification of physiological parameters (water loss rate, fresh/dry weight), and the expression level of drought-related genes. These findings reveal the molecular mechanism of MfWRKY40-mediated drought tolerance in M. falcata, establishing a crucial foundation for further exploration of this gene.
2. Results
2.1. Generation of Transgenic A. thaliana Overexpressing MfWRKY40
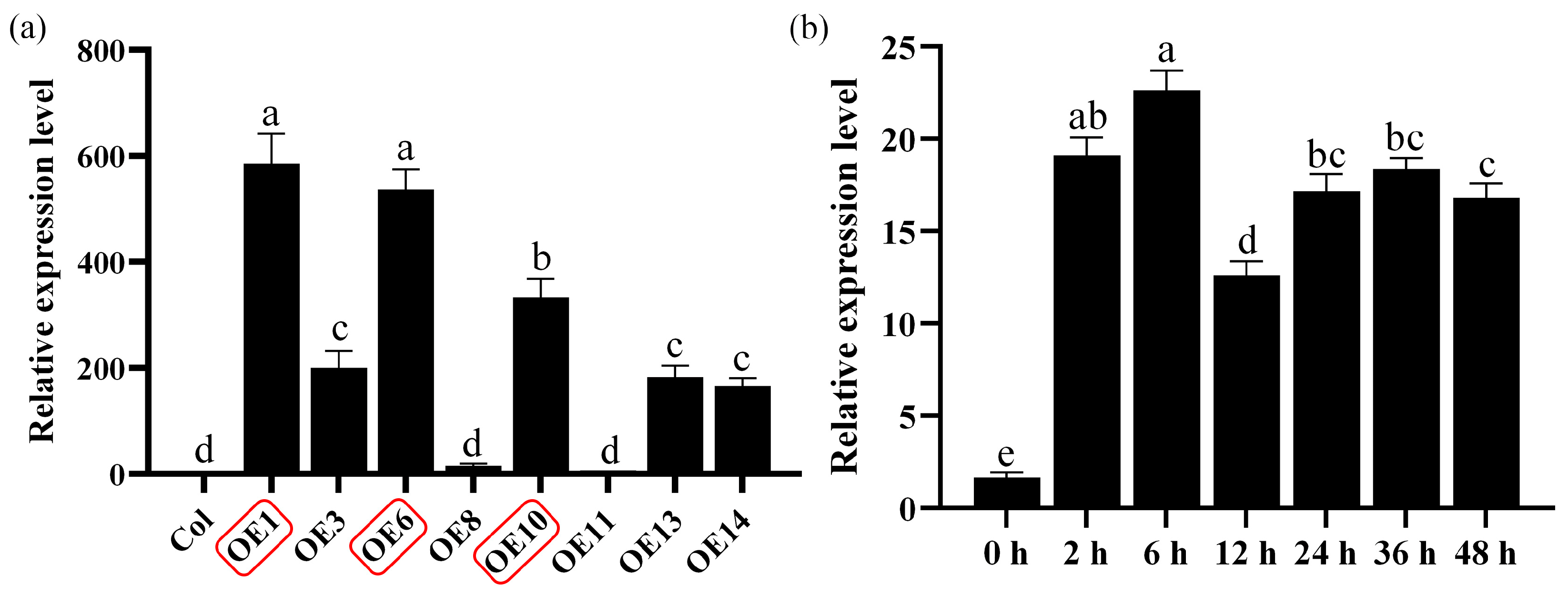
To investigate the functional role of MfWRKY40 in plants, we constructed a pB7WG2RS overexpression vector and introduced it into wild-type A. thaliana plants (Col) using the floral dip method. After 25 d of normal growth, T0 seeds were collected and screened under a fluorescence microscope to select bright red fluorescent seeds. Due to the presence of the RedSeed selection marker, positive transgenic A. thaliana lines (OEs) could be easily screened under a fluorescence microscope. Following three consecutive generations of selection, homozygous A. thaliana lines were identified. Quantitative real-time PCR (qRT-PCR) analysis showed that MfWRKY40 expression levels varied among transgenic lines, with all exhibiting significantly higher expression compared to wild-type lines (Figure 1a). Among the transgenic lines, OE1 exhibited the highest MfWRKY40 expression (584-fold increase relative to wild-type), followed by OE6 (536-fold) and OE10 (333-fold). The remaining lines (OE8, OE11, and OE14) showed basal expression levels comparable to Col (Figure 1a). Based on these results, we selected lines OE1, OE6, and OE10 for further drought tolerance assays.
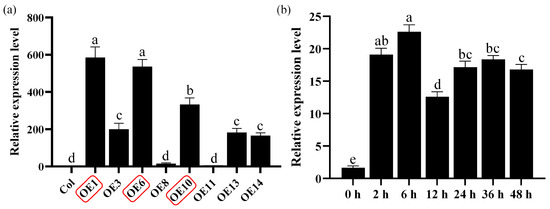
Figure 1.
Relative expression levels of the MfWRKY40 gene in different Arabidopsis thaliana lines and analysis of its expression patterns in Medicago falcata under mannitol stress. (a) Relative expression levels of the MfWRKY40 gene in wild-type (Col) and different transgenic (OEs) A. thaliana lines. (b) Relative expression levels of the MfWRKY40 gene in M. falcata under 300 mM mannitol treatment at 0, 2, 6, 12, 24, 36, and 48 h. The transgenic A. thaliana lines (OE1, OE6, and OE10) with high expression levels (highlighted in red) were selected for further experiments. All values are expressed as the mean ± standard error (n ≥ 3). The different lowercase letters indicate significant differences among various lines (a) and significant differences among different time points after 300 mM mannitol treatment (b) (p < 0.05). Error bars represent the standard error among replicates.
We performed expression pattern analysis of the MfWRKY40 gene in M. falcata, which revealed that its relative expression levels began to increase 2 h after 300 mM mannitol treatment, peaked at 6 h, and subsequently declined (Figure 1b). These findings demonstrate that MfWRKY40 expression is induced by mannitol stress.
2.2. Response of the Transgenic A. thaliana to Mannitol Treatment
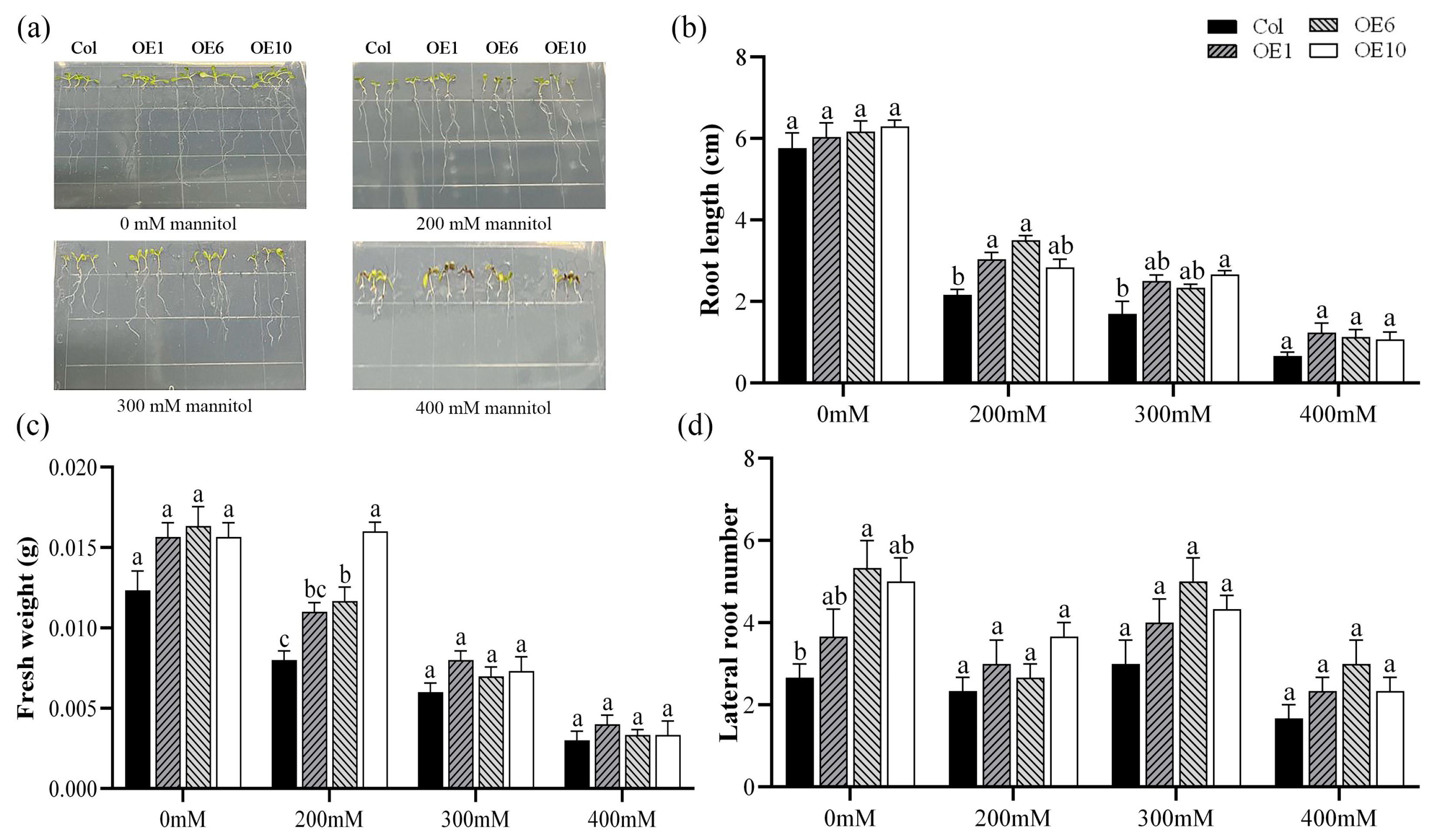
To elucidate the role of MfWRKY40 in drought stress response, Col and transgenic A. thaliana were subjected to stress treatment on media containing 0 mM, 200 mM, 300 mM, and 400 mM mannitol. After 7 d of treatment, root phenotype was photographed and root length, fresh weight, and lateral root number were measured (Figure 2a). The measurements revealed no significant differences in root length among the four lines when grown on normal MS medium (Figure 2b). However, under stress conditions, both Col and transgenic lines displayed progressively reduced primary root length and lateral root number with increasing mannitol concentrations (Figure 2b,d). Fresh weight gradually decreased under increasing stress. Notably, at 200 mM mannitol, transgenic lines exhibited significantly higher fresh weight than Col. Specifically, OE1, OE6, and OE10 exhibited fresh weights of 0.011 g, 0.012 g, and 0.016 g, respectively, compared to 0.008 g in wild-type plants (Figure 2c). These results demonstrate that MfWRKY40 responds to mannitol-induced stress and enhances A. thaliana tolerance to mannitol.
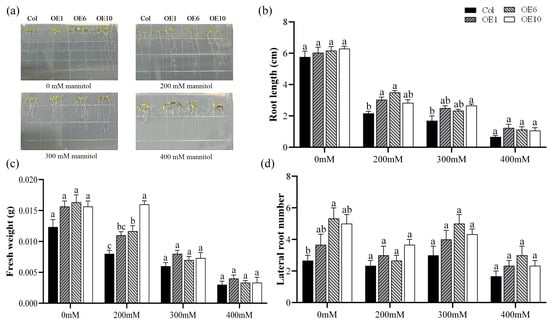
Figure 2.
Phenotype comparison of wild-type (Col) and transgenic (OEs) A. thaliana lines grown on medium supplied with different concentration of mannitol for 7 d. (a) Phenotypic images of A. thaliana seedlings. (b) Root length. (c) Fresh weight. (d) Lateral root number. All values are expressed as the mean ± SE (n ≥ 15). The different lowercase letters at the same mannitol concentration indicate significant differences among the four lines (p < 0.05). Error bars represent the standard error among replicates.
2.3. Response of the MfWRKY40 Gene to ABA
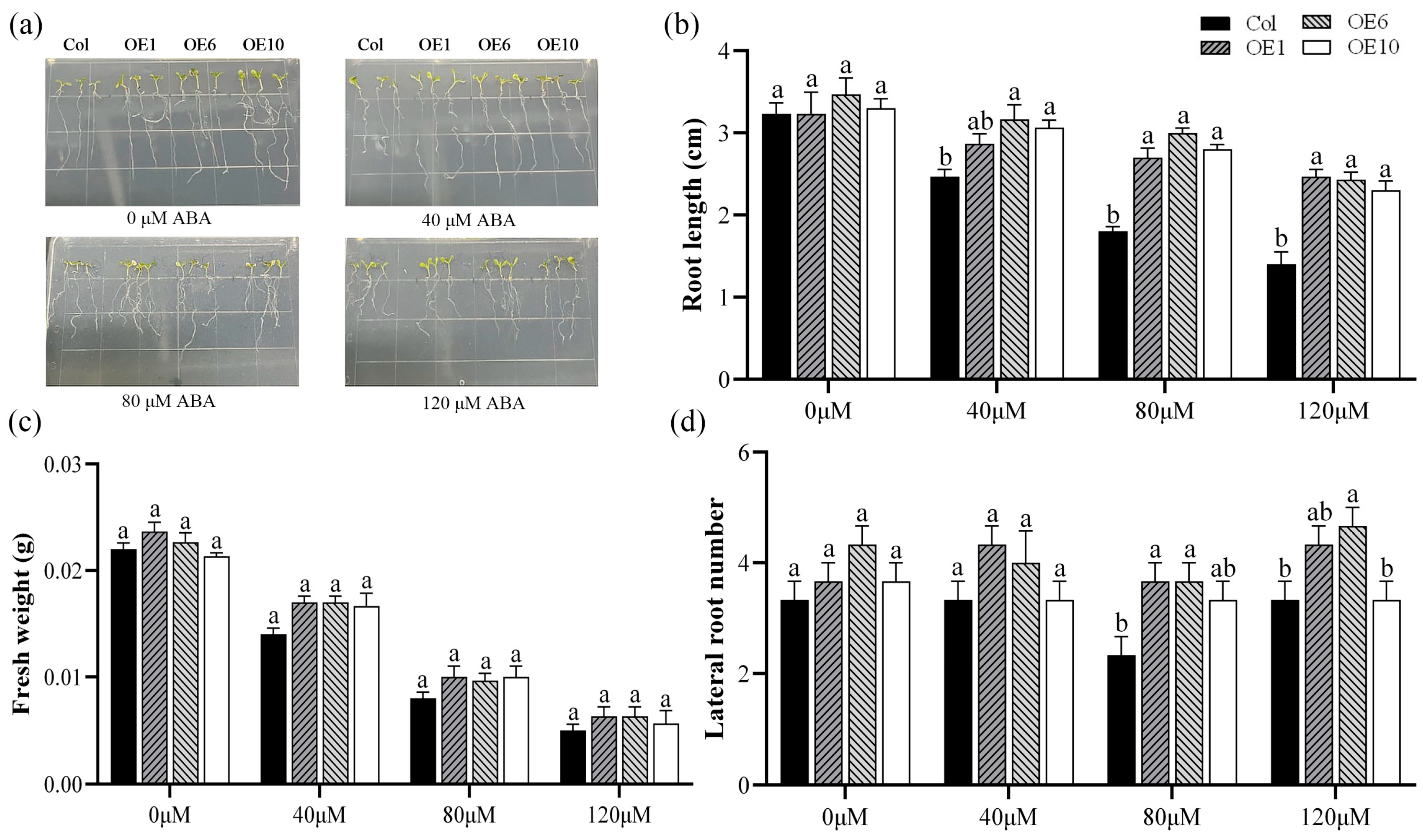
Abscisic acid (ABA) is a crucial phytohormone that plays an essential role in plant stress response and tolerance. To investigate whether the MfWRKY40 gene responds to ABA stress, we treated Col and transgenic A. thaliana with MS medium containing 0 μM, 40 μM, 80 μM, and 120 μM ABA. Our results demonstrated that after 7 d of cultivation on MS medium without ABA (0 μM), no significant differences were observed in root length, lateral root number, or fresh weight between the transgenic lines and Col (Figure 3a). After 7 d of growth on ABA-supplemented media (40 μM, 80 μM, and 120 μM), all lines showed significant reductions in primary root length, with transgenic A. thaliana showing greater root length than Col (Figure 3b). Fresh weight also exhibited a decreasing trend with increasing ABA concentrations, with the fresh weight of Col reaching its lowest value (0.005 g) at 120 μM ABA (Figure 3c,d). These findings strongly indicate that MfWRKY40 plays a responsive role in ABA-mediated stress regulation.
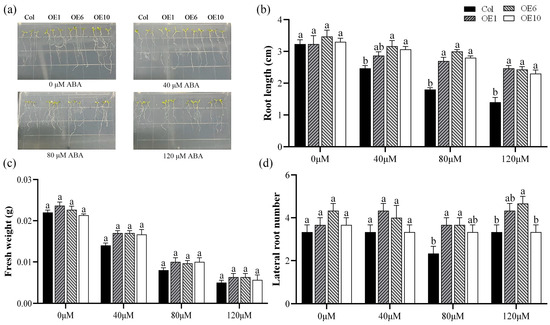
Figure 3.
Phenotype comparison of wild-type (Col) and transgenic (OEs) A. thaliana lines grown on medium supplied with different concentration of ABA for 7 d. (a) Phenotypic images of A. thaliana seedlings. (b) Root length. (c) Fresh weight. (d) Lateral root number. All values are expressed as the mean ± SE (n ≥ 15). The different lowercase letters at the same ABA concentration indicate significant differences among the four lines (p < 0.05). Error bars represent the standard error among replicates.
2.4. Phenotypic Analysis of Transgenic A. thaliana
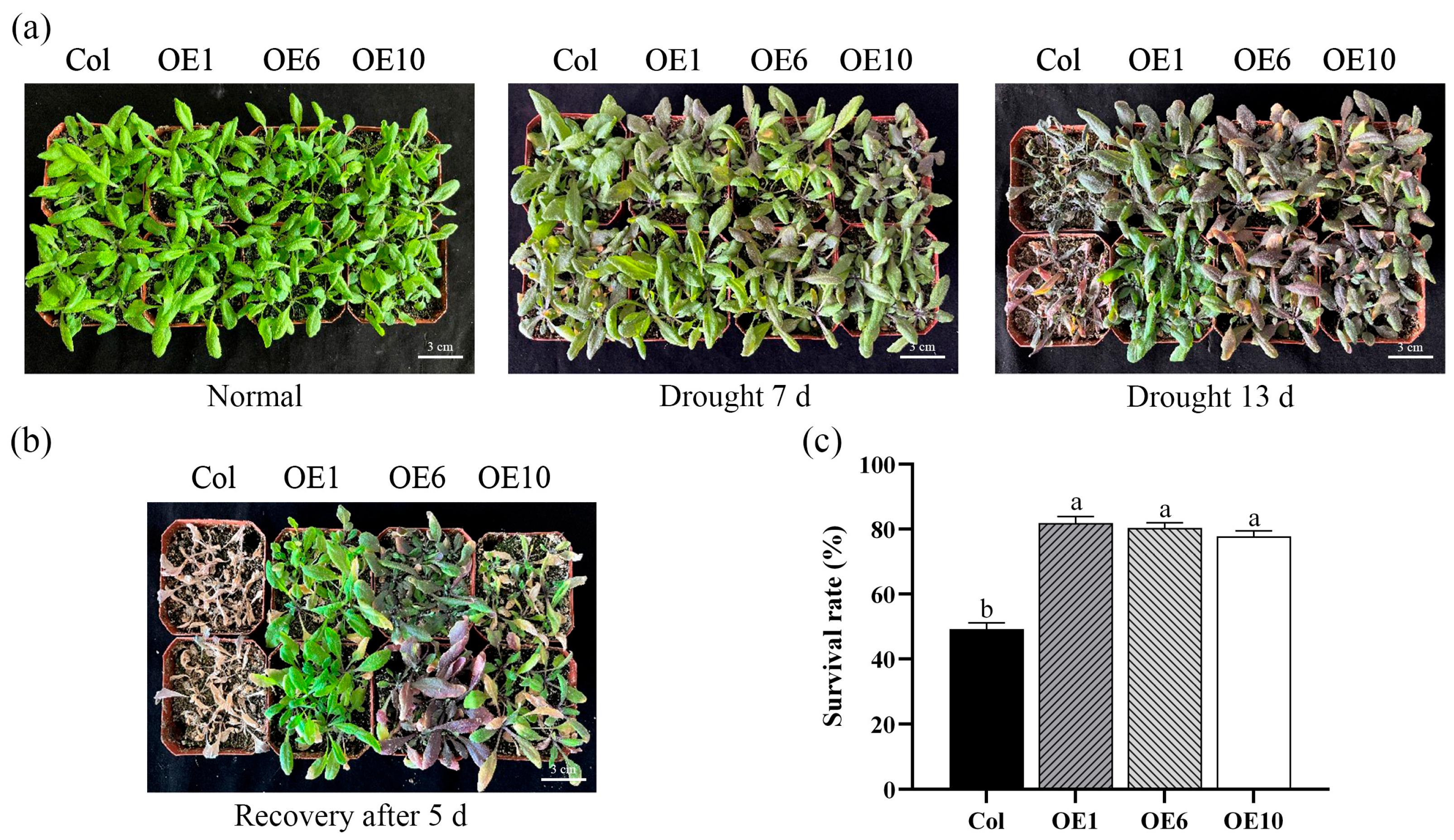
To further investigate the role of MfWRKY40 under drought stress, both the Col and transgenic A. thaliana were subjected to natural drought stress in soil. Under normal watering conditions, no significant growth differences were observed between Col and transgenic lines (Figure 4a). After 7 d of drought treatment, Col were severely affected, with basal leaves turning to purplish-red, while OE1, OE6, and OE10 exhibited only slight purplish-red discoloration and maintained relatively better growth vigor. Following 13 d of drought stress, wild-type A. thaliana stopping growing, while transgenic lines maintained slow growth with partial leaves retaining green color (Figure 4a). Upon rewatering, Col plants exhibited no recovery after 5 d but progressed to severe wilting, whereas transgenic lines developed new green leaves (Figure 4b). The survival rates were 81.8%, 80.3%, and 77.8% for OE1, OE6, and OE10, respectively, compared to only 49.2% for Col (Figure 4c), demonstrating significantly higher survival in transgenic lines. These results indicate that transgenic A. thaliana exhibits generally enhanced drought tolerance and superior recovery capacity after rewatering compared to Col.
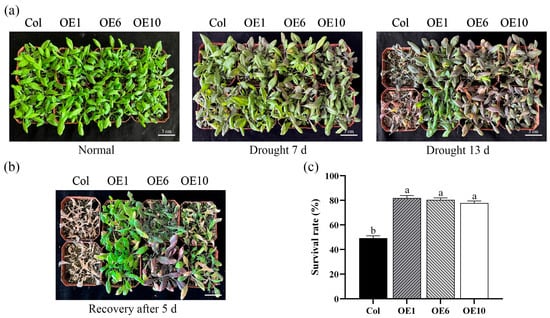
Figure 4.
Phenotypes and survival rate of wild-type (Col) and transgenic (OEs) A. thaliana under drought stress. (a) Phenotypic images of A. thaliana under drought stress. Scale bar = 3 cm. (b) Phenotypic images of A. thaliana after 5 d of rewatering. Scale bar = 3 cm. (c) Survival rate after 5 d of rewatering. All values are expressed as the mean ± SE (n ≥ 3). The different lowercase letters indicate significant differences among the four lines (p < 0.05). Error bars represent the standard error among three replicates.
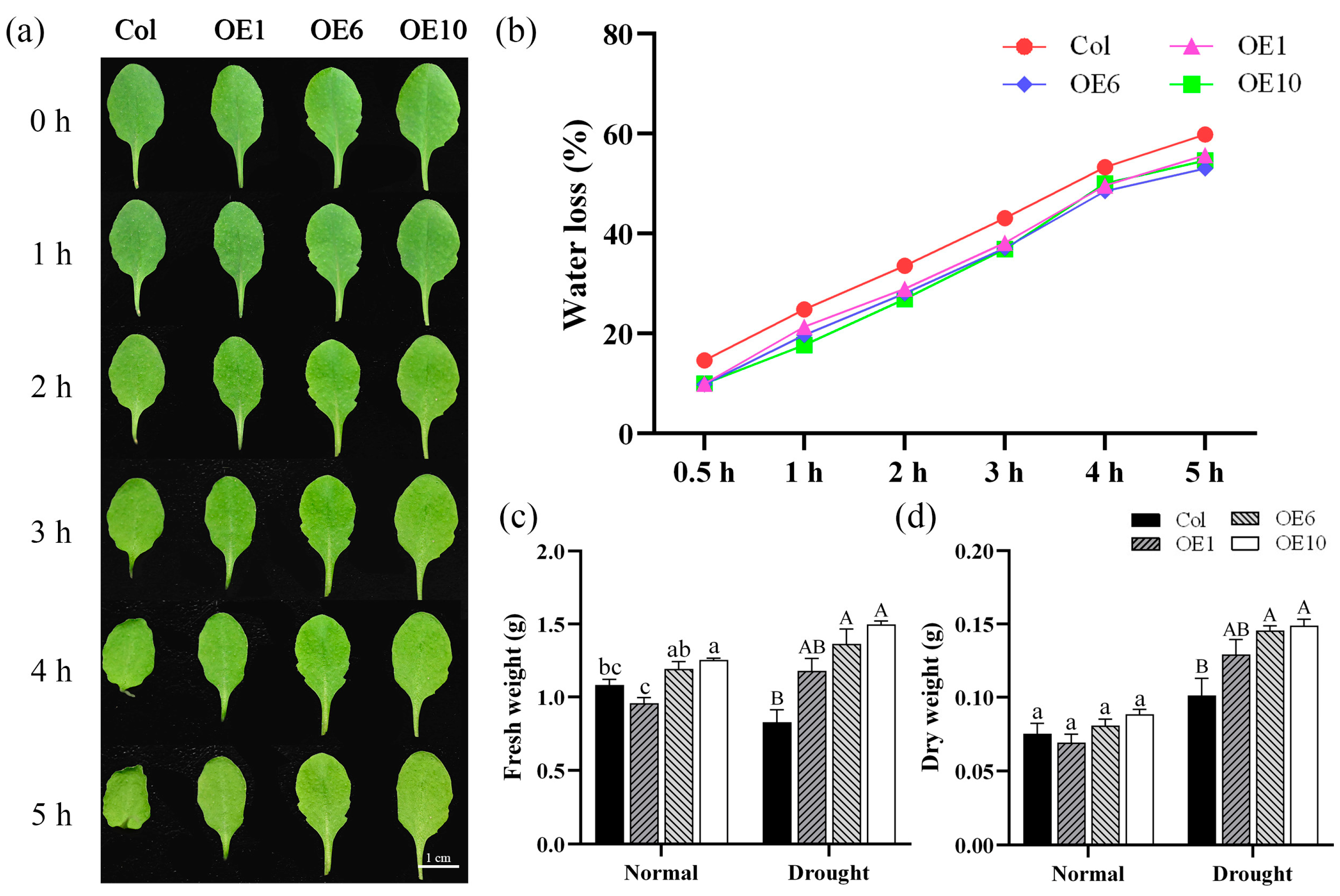
2.5. MfWRKY40 Reduces Water Loss in Transgenic A. thaliana
We also investigated the water loss rates of Col and transgenic A. thaliana under normal conditions. Leaves from Col and OEs under normal growth conditions were collected, and phenotypic photographs were recorded at designated time points. It was observed that starting from 1 h, the petioles of Col leaves began to curl, with the curling becoming more pronounced over time. By 4 h, the entire leaf edges started to curl inward, showing obvious signs of dehydration (Figure 5a). Meanwhile, water loss rate measurements revealed that Col exhibited the most rapid water dissipation, reaching 14.6% at 0.5 h, while the transgenic lines showed significantly lower water loss at merely 9.85% (Figure 5b). Col and transgenic lines were subjected to drought stress, and their fresh and dry weights were measured before and after the treatment. The results showed that after stress, the transgenic lines maintained significantly higher fresh and dry weights than Col (Figure 5c,d), indicating that transgenic A. thaliana sustained less damage and retained more biomass under drought conditions. Therefore, we concluded that transgenic A. thaliana exhibited slower water loss than Col, and that MfWRKY40 overexpression confers reduced water loss capacity.
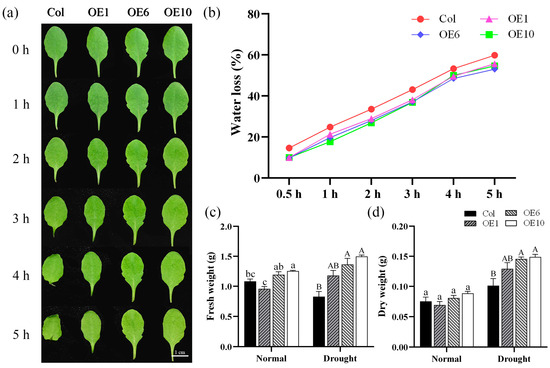
Figure 5.
Water loss rate and fresh/dry weight of wild-type (Col) and transgenic (OEs) A. thaliana. (a) Water loss in detached leaves of A. thaliana at 0, 1, 2, 3, 4, and 5 h. Scale bar = 1 cm. (b) Water loss rate. (c) Fresh weight of A. thaliana before drought treatment and after 10 d of treatment. (d) Dry weight. All values are expressed as the mean ± SE (n ≥ 10). The different lowercase letters indicate significant differences among the four lines before stress treatment (p < 0.05), while different uppercase letters indicate significant differences among the four lines after stress treatment (p < 0.05). Error bars represent the standard error among replicates.
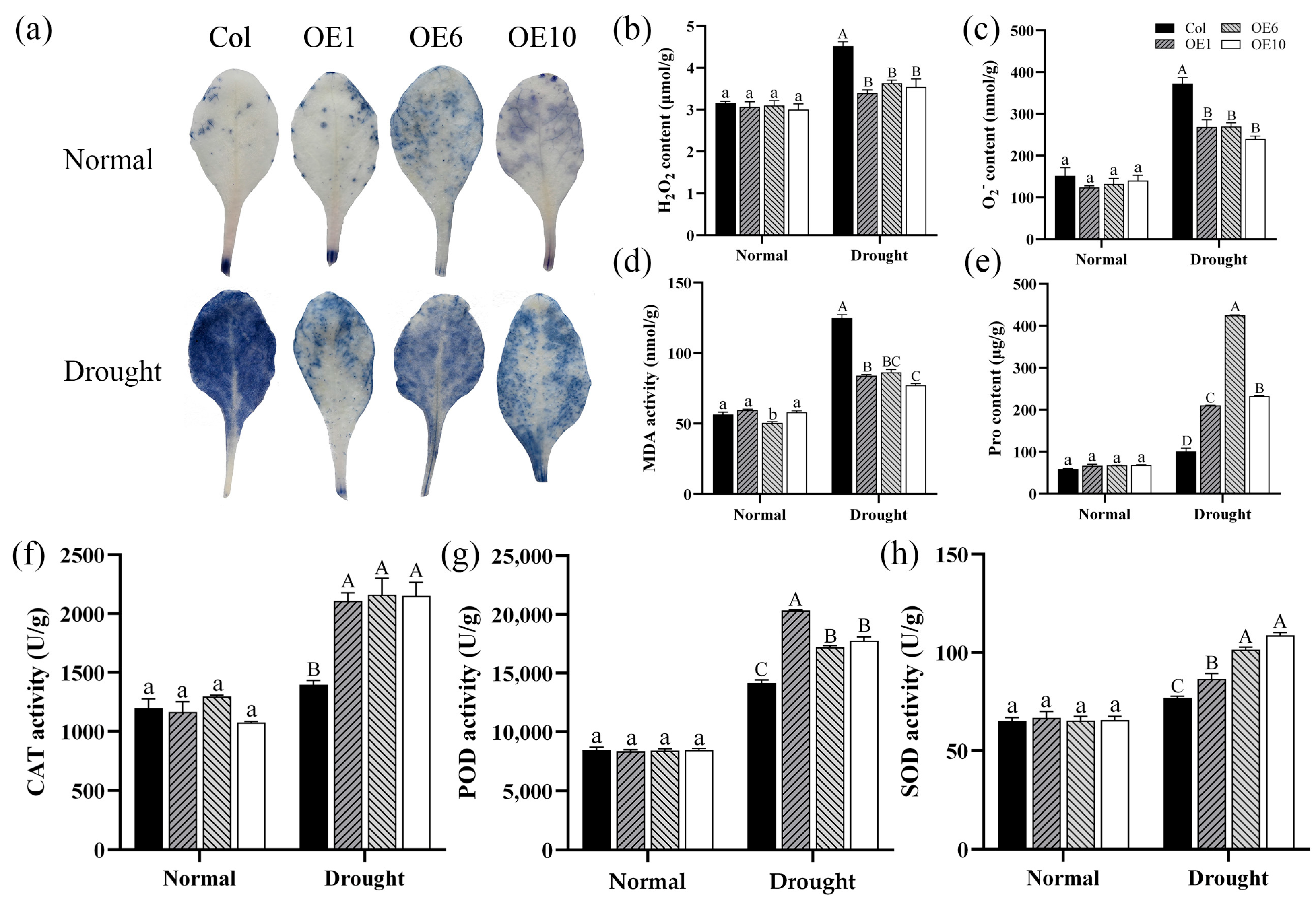
2.6. Effects of MfWRKY40 on Physiological Indicators
To investigate the impact of MfWRKY40 on physiological indicators in A. thaliana, we conducted NBT staining and measured the content of hydrogen peroxide (H2O2) and superoxide anion (O2−), and MDA, as well as the activity of key antioxidant enzymes. The results showed that before drought stress, no significant differences were observed in the content of these substances between Col and transgenic lines (Figure 6). However, under drought stress, Col accumulated significantly higher levels of H2O2, O2−, and MDA compared to transgenic lines. Specifically, O2− content reached 372.48 nmol/g and MDA levels increased to 124.95 nmol/g, both markedly higher than in the transgenic line (Figure 6b–d). Additionally, NBT staining revealed more extensive formazan deposition (indicated by larger and darker blue areas) in Col compared to transgenic lines, demonstrating greater oxidative damage in the Col (Figure 6a). Comparative analysis demonstrated that although both the activities of the antioxidant enzymes superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) and Pro content increased in Col under stress conditions, the magnitude of the increase was significantly smaller than that in transgenic lines (Figure 6e–h). Based on these findings, it can be concluded that the transgenic lines enhance drought tolerance in A. thaliana by reducing the accumulation of ROS such as H2O2 and O2− and by increasing the activity of antioxidant enzymes.
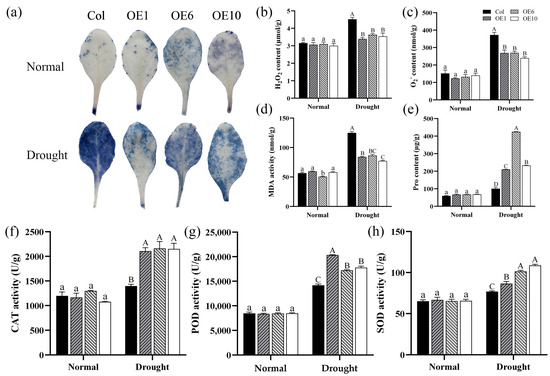
Figure 6.
Physiological indices of wild-type (Col) and transgenic (OEs) A. thaliana under drought stress. (a) NBT staining of A. thaliana before drought treatment and after 5 d of drought stress. (b) Hydrogen peroxide (H2O2) content. (c) Superoxide anion (O2−) content. (d) Malondialdehyde (MDA) content. (e) Proline (Pro) content. (f) Catalase (CAT) activity. (g) Peroxidase (POD) activity. (h) Superoxide dismutase (SOD) activity. All values are expressed as the mean ± SE (n ≥ 3). The different lowercase letters indicate significant differences among the four lines before stress treatment (p < 0.05), while different uppercase letters indicate significant differences among the four lines after stress treatment (p < 0.05). Error bars represent the standard error among replicates.
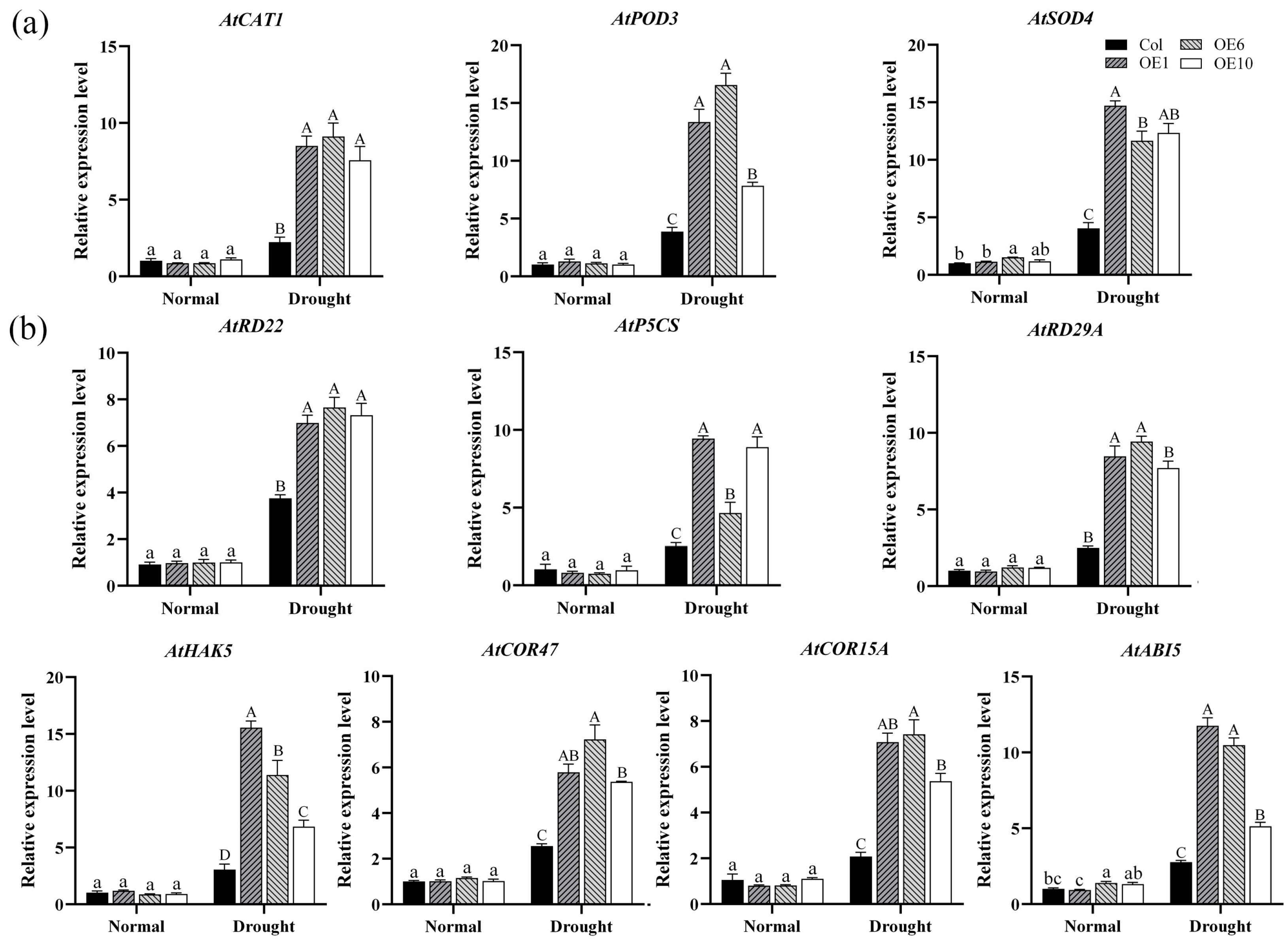
2.7. Regulation of Enzymes and Drought-Related Genes by MfWRKY40
Transcription factors typically exert their roles by regulating the expression of downstream target genes in a coordinated manner. In our study, we analyzed the effect of the MfWRKY40 gene on the expression levels of antioxidant enzyme genes and drought-related genes. The results demonstrated that under normal conditions, both Col and transgenic lines exhibited low expression levels of AtPOD3, AtSOD4, and AtCAT1. However, after drought stress, their relative expression levels showed differential upregulation patterns. Notably, the expression of AtPOD3 and AtSOD4 in transgenic A. thaliana exhibited more pronounced changes, while the increase in AtCAT1 was relatively modest but still significantly higher than Col (Figure 7a). These results suggests that MfWRKY40 enhances ROS-scavenging capacity through transcriptional upregulation of these three genes, consequently modulating antioxidant activity in transgenic lines.
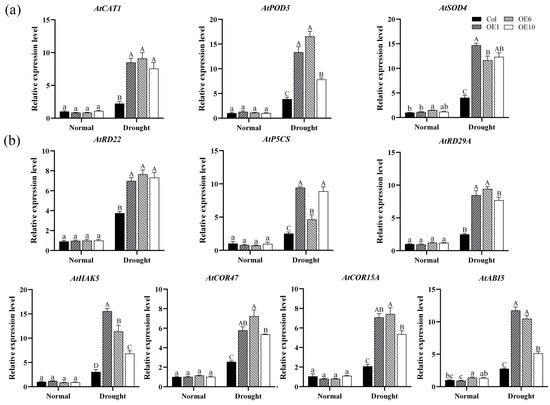
Figure 7.
Relative expression levels of enzyme and drought-related genes in wild-type (Col) and transgenic (OEs) A. thaliana under drought stress. (a) Relative expression levels of enzyme-related genes in A. thaliana before drought treatment and after 5 d of drought stress. (b) Relative expression levels of drought-related genes. All values are expressed as the mean ± SE (n ≥ 3). The different lowercase letters indicate significant differences among the four lines before stress treatment (p < 0.05), while different uppercase letters indicate significant differences among the four lines after stress treatment (p < 0.05). Error bars represent the standard error among three replicates.
A similar trend was observed for drought-related genes (AtHAK5, AtCOR47, AtCOR15A, AtABI5, AtRD22, AtP5CS, and AtRD29A): under drought stress conditions, overexpression of MfWRKY40 induced the expression of all seven tested genes, with their expression levels being significantly higher in transgenic lines compared to Col (Figure 7b). These findings demonstrate that MfWRKY40 modulates the drought stress response in A. thaliana by regulating the expression of these key genes.
3. Discussion
WRKY TFs are one of the largest families of transcription factors in plants and play a crucial role in plant responses to abiotic stress [29]. Numerous studies have demonstrated that WRKY TFs can enhance stress tolerance in various plants, such as TaWRKY76, TaWRKY2, and TaWRKY24 in wheat [30,31,32], OsWRKY97 in rice [33], and ZmWRKY25 and ZmWRKY30 in maize [34,35], which confer stress tolerance through multiple regulatory pathways. In alfalfa, MsWRKY11 is induced by PEG and ABA, and enhances drought tolerance in alfalfa by reducing leaf stomatal density and improving water use efficiency [36]. Similarly, MsWRKY42 exhibits the highest expression levels in roots and leaves, with its expression being induced by PEG stress [37]. However, studies on the drought tolerance of the WRKY family have primarily focused on alfalfa, while research on their counterparts in M. falcata remains limited. Therefore, in our study, the MfWRKY40 gene from M. falcata was investigated to explore its role in drought tolerance. Using the floral dip method, we introduced MfWRKY40 into A. thaliana, and subsequently treated the transgenic lines with mannitol and ABA. The results showed that under these treatments, MfWRKY40-overexpressing A. thaliana exhibited significantly better root length performance compared to Col. Notably, under 200 mM treatment, the transgenic lines showed significantly higher fresh weight than wild-type (Figure 2 and Figure 3). Similarly, overexpression of the Iris germanica genes IgWRKY50 and IgWRKY32, along with wheat TaWRKY46, in A. thaliana resulted in transgenic lines exhibiting higher germination rates and longer root lengths on 1/2 MS medium containing mannitol [38,39]. Roots, as the primary sensors of abiotic stress [40,41], develop enhanced growth under drought conditions to facilitate water uptake. The study by Reicosky and Deaton (1979) demonstrated that cultivars with well-developed root systems exhibit enhanced drought tolerance [42]. These results preliminarily suggest that the MfWRKY40 gene plays a role in root growth and drought tolerance in A. thaliana.
Drought stress induces excessive accumulation of reactive ROS in plants, including typical forms such as H2O2 and O2− [43,44]. While high concentrations of ROS can cause oxidative damage, ROS within a certain range also serve as crucial signaling molecules involved in various physiological and biochemical processes [4]. MDA, a primary byproduct of membrane lipid peroxidation, serves as a reliable indicator of oxidative stress severity. Elevated MDA levels directly correlate with the extent of cellular oxidative damage [45]. Antioxidant enzymes such as SOD, CAT, and POD play essential roles in scavenging excess ROS [46], and their activities are indicative of plant stress tolerance under abiotic stress [47,48,49]. Pro, an important osmotic regulator, helps to maintain cellular osmotic balance and protects membrane integrity [13]. Previous studies demonstrated that ItfWRKY70 from Ipomoea trifida increases Pro content, enhances SOD and POD activities, and reduces MDA and H2O2 levels [50]. In parallel, SlWRKY8 from Solanum lycopersicum was shown to regulate ROS scavenging pathways, conferring drought tolerance through enhanced ROS elimination [51]. In our study, under drought stress, MfWRKY40-overexpressing A. thaliana lines exhibited significantly higher antioxidant enzyme activities and Pro content compared to wild-type, along with lower levels of MDA, H2O2, and O2− (Figure 6). Additionally, we quantified the expression levels of key antioxidant enzyme-related genes, including AtPOD3, AtSOD4, and AtCAT1. The results consistently showed that the gene expression levels in transgenic A. thaliana were significantly higher than those in wild-type under drought stress (Figure 7a). These findings demonstrate that MfWRKY40 enhances drought tolerance by participating in ROS scavenging pathways.
The plant’s response to abiotic stress is a complex physiological process orchestrated by multiple genes functioning synergistically. WRKY TFs recognize and bind to W-box cis-elements in drought-responsive gene promoters [51], thereby regulating their expression and enhancing plant drought tolerance. P5CS1 is a key gene in Pro biosynthesis, which modulates plant adaptive responses to abiotic stress through Pro accumulation [52]. RD29B and RD29A are dehydration-inducible genes in A. thaliana that encode hydrophilic proteins [53]. The RD29A gene product mitigates damage caused by various abiotic stresses including drought, salinity, and cold [54]. Other drought-responsive genes exhibit similar functions, where upregulated expression enhances stress tolerance. Research demonstrates that WRKY TFs regulate drought-responsive gene networks through coordinated modulation of multiple signaling pathways, thereby affecting plant drought adaptation. PheWRKY86 from Phyllostachys edulis is induced by both drought and ABA treatments and enhances drought tolerance by upregulating NCED1 expression [55]. The MbWRKY53 gene from apple improves cold and drought adaptation in transgenic A. thaliana by modulating the expression of SOS1, COR47, DREB2A, P5CS1, COR6.6, and RD29b [13]. Similarly, overexpression of MbWRKY40 in apple alters the expression levels of AtKIN1, AtDREB2A, AtRD29A, AtERD10, AtCOR47A, and AtRD29B in A. thaliana, thereby enhancing stress tolerance [56]. In our study, the drought-related genes AtABI5, AtCOR15A, AtCOR47, AtHAK5, AtP5CS, AtRD22, and AtRD29A were similarly regulated by MfWRKY40, exhibiting higher expression levels in transgenic A. thaliana (Figure 7b). This suggests that MfWRKY40 may specifically bind to the W-box elements of these genes to enhance drought tolerance in A. thaliana.
4. Materials and Methods
4.1. Plant Materials
The seeds of M. falcata were provided by the College of Grassland Science, Xinjiang Agricultural University (Urumqi, China), while the A. thaliana ecotype Columbia-0 (Col-0) seeds were supplied by the Forage Germplasm Resources Conservation and Utilization Innovation Team at the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (Beijing, China).
Seven-day-old M. falcata seedlings were transferred to hydroponic boxes containing Hoagland’s nutrient solution [36], with the solution replaced every two days. After 10 d of cultivation, seedlings were subjected to 300 mM mannitol treatment. Leaf samples were collected at 0, 2, 6, 12, 24, 36, and 48 h after treatment, immediately frozen in liquid nitrogen, and stored at −80 °C for subsequent analysis of MfWRKY40 relative expression levels.
4.2. Cloning of the MfWRKY40 Gene and Construction of the Overexpression Vector
Fresh leaves of M. falcata were collected, and total RNA was extracted using the Eastep® Super Total RNA Extraction Kit (LS1040, Promega, Shanghai, China). The extracted RNA was then reverse-transcribed into cDNA using the 5× FastKing-RT Super Mix kit (KR118, Tiangen, Beijing, China), with reaction conditions and master mix compositions detailed in Tables S2 and S3. Gene-specific primers were designed using GeneRunner software (version 5.1.06, Gennady A. Peskin, Moscow, Russia), and the sequences are listed in Table S1. PCR amplification was performed using high-fidelity KOD polymerase (Toyobo, Osaka, Japan) with cDNA as the template, following the reaction system described in Table S4 and the thermal cycling program outlined in Table S5. The amplified product was gel-purified, and the recovered DNA fragment was ligated into the pENTR entry vector (Table S6). The reaction mixture was transformed into DH5α competent E. coli cells (Biomed Gene, Beijing, China). Positive clones were selected on Kanamycin (Kan)-containing medium, and the plasmid DNA was extracted. The MfWRKY40 was subcloned from the pENTR vector into the pB7WG2RS overexpression vector through the LR reaction. The LR reaction was performed under the conditions specified in Table S7, with incubation at 25 °C for 1 h, followed by treatment with Proteinase K at 37 °C for 10 min. The pB7WG2RS overexpression vector was a modified version constructed by cloning the RedSeed selection marker (pNAP::DsRed) from pKGW-RedSeed into the pB7WG2 vector [57], using XbaI and KpnI restriction sites. This vector was provided by the Forage Germplasm Resources Conservation and Utilization Innovation Team at the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (Beijing, China). The reaction products were directly transformed into DH5α competent E. coli cells (Biomed Gene, Beijing, China) and cultured on a medium containing spectinomycin (Spec) tolerance. Single colonies were screened and verified by PCR analysis. Colonies showing clear bands of the expected size were then expanded in large-scale culture. Plasmids were extracted from these cultures and further verified by conventional PCR using gene-specific primers to confirm construct integrity. If the plasmid PCR shows a bright band of the expected size, it indicates successful construction of the overexpression vector.
The successfully constructed pB7WG2RS overexpression vector was introduced into Agrobacterium tumefaciens GV3101 (Biomed Gene, Beijing, China) culture. The transformed bacteria were grown in LB medium supplemented with kanamycin and rifampicin, and the OD value was adjusted to 0.6–0.8 by centrifugation and resuspension in buffer. The resulting Agrobacterium suspension was then ready for subsequent A. thaliana transformation experiments.
4.3. Generation of Transgenic A. thaliana
When A. thaliana plants initiated bolting and floral bud formation, newly emerged inflorescences were pruned promptly to promote subsequent flowering. After flowering, siliques were excised, and the plants were watered adequately in preparation for inoculation and infection the following day. T0-generation A. thaliana seeds were obtained using the floral dip method [57]. Transgenic seeds displaying red fluorescence were selected under a fluorescence microscope (Leica M205 FA, Wetzlar, Germany) [58], then cultivated to maturity. Progeny seeds were repeatedly screened until homozygous T3 transgenic A. thaliana lines were obtained.
4.4. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
The samples were flash-frozen in liquid nitrogen for 30 s, then ground into a fine powder. This freeze–grinding cycle was repeated at least three times. Total RNA was subsequently extracted following the protocol in Section 4.2, followed by cDNA synthesis. Using cDNA as the template and the A. thaliana AtActin-qPCR-F/R primers as the reference gene [59], the relative expression levels of enzyme-related genes (AtPOD3, AtSOD4, and AtCAT1) and drought-related genes (AtHAK5, AtCOR47, AtCOR15A, AtABI5, AtRD22, AtP5CS, and AtRD29A) were quantified by qRT-PCR with the SuperReal PreMix Plus kit (FP205, Tiangen, Beijing, China). Primer sequences are listed in Table S1, while reaction components and cycling conditions are detailed in Supplementary Tables S8 and S9. Three biological replicates were performed, with each biological replicate including three technical replicates. Finally, the experimental data were analyzed, and relative gene expression levels were calculated using the 2−ΔΔCt method [60].
4.5. Stress Treatment of Transgenic A. thaliana
By measuring the expression levels of the MfWRKY40 gene in different transgenic lines (Figure 1a), three A. thaliana lines with high expression were selected for further study. For the mannitol and ABA treatment experiment, seeds of Col and transgenic A. thaliana were surface-sterilized and evenly plated on 1/2 MS medium, followed by cold stratification at 4 °C in complete darkness for 3 d before being transferred to a plant growth chamber. After 4 d of cultivation on 1/2 MS medium at 22 °C under a 16-hour light/8-hour dark photoperiod with a light intensity of 400 μmol·m−2·s−1, the seedlings were transferred to MS medium containing different concentrations of mannitol (0, 200, 300, and 400 mM) or ABA (0, 40, 80, and 120 μM) for stress treatment. On the 7th day, the Col and transgenic lines were photographed, and parameters including root length, lateral root number, and fresh weight were recorded.
For the drought experiment, Col and transgenic A. thaliana seeds were vernalized in a 4 °C refrigerator in the dark for 3 d, then cultured in a light incubator for 7 d until two small leaves emerged. The A. thaliana seedlings were transplanted into small pots (6 cm diameter) containing a mixture of nutrient soil, vermiculite, and soil conditioner (3:1:1 ratio), with 9 plants per pot, and grown in a light-controlled chamber. After 30 d of cultivation, the plants were subjected to stress treatment by withholding water to simulate natural drought. The phenotypes of each line were observed under drought conditions. When most leaves turned purplish-red with yellow edges, rehydration was performed, and the phenotype were further observed. Photographs were taken before drought stress, as well as at 7 d, 13 d of stress treatment, and 5 d after rehydration. Survival rates were recorded before stress treatment and after rehydration.
4.6. Determination of Water Loss Rate and Fresh/Dry Weight
For wild-type and transgenic A. thaliana plants grown under normal conditions, detached leaves were sampled and photographed at 0, 1, 2, 3, 4, and 5 h to observe phenotypic changes between wild-type and transgenic lines. Concurrently, 0.5 g leaf samples from four different lines were weighed at 0.5, 1, 2, 3, 4, and 5 h to calculate the water loss rate, thereby comparing water dissipation between wild-type and transgenic A. thaliana [61]. Prior to drought stress and after 10 d of stress treatment, all A. thaliana lines (9 plants per pot) were harvested to measure fresh weight. The samples were then placed in paper envelopes, dried at 105 °C for 30 min, and further oven-dried at 80 °C until constant weight was achieved for dry weight.
4.7. NBT Staining and Determination of Physiological Indices
Leaf samples were collected from identical positions on both wild-type and transgenic A. thaliana lines before drought stress and after 5 d of stress treatment. One portion of the leaves was stained using the NBT Staining Kit (G4816, Solarbio, Beijing, China), while the other portion was homogenized in either the specified extraction buffer or acetone, followed by centrifugation at 4 °C for 10 min. The resulting supernatant was collected for subsequent physiological assays. The activities of the three antioxidant enzymes were determined using the SOD Assay Kit (YH1202, Angle Gene, Nanjing, China), POD Assay Kit (YH1210, Angle Gene, Nanjing, China), and CAT Assay Kit (YH1208, Angle Gene, Nanjing, China), respectively. Additionally, the contents of MDA, Pro, O2−, and H2O2 were determined using the MDA Assay Kit (YH1217, Angle Gene, Nanjing, China), Pro Assay Kit (YH1231, Angle Gene, Nanjing, China), O2− Assay Kit (G0116F, Grace Biotechnology, Suzhou, China), and H2O2 Assay Kit (G0112F, Grace Biotechnology, Suzhou, China).
4.8. Data Analysis
Data organization was performed using Microsoft Excel (Version 2016, Redmond, WA, USA). Statistical significance was analyzed using IBM SPSS software (Version 25, Armonk, NY, USA). One-way ANOVA followed by Tukey’s Honestly Significant Difference (HSD) test was applied, with the significance level set at p < 0.05. All values are presented as the mean ± standard error (SE). The different lowercase letters indicate significant differences among the four lines before stress treatment (p < 0.05), while different uppercase letters indicate significant differences among the four lines after stress treatment (p < 0.05). Figures were generated using GraphPad Prism (version 9, San Diego, CA, USA). All experiments were conducted with three biological replicates and three technical replicates.
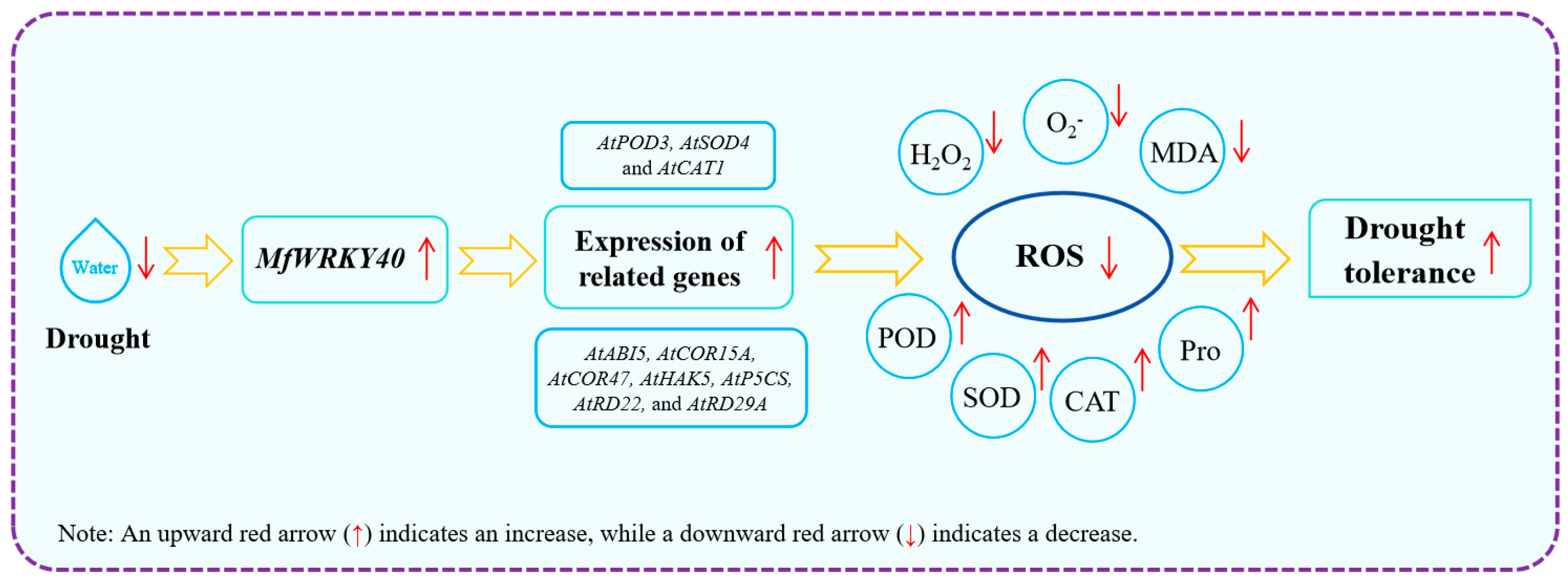
5. Conclusions
In this study, Col and transgenic A. thaliana were first treated with stress media (mannitol and ABA), revealing that transgenic lines exhibited superior root length and fresh weight compared to Col under identical conditions. Subsequent natural drought treatment further demonstrated enhanced recovery capacity in overexpression lines. Analysis revealed significantly higher water loss rates in wild-type compared to transgenic lines, accompanied by reduced biomass accumulation (both fresh and dry weight) under stress conditions. Meanwhile, MfWRKY40-overexpressing lines displayed higher antioxidant enzyme activities and Pro content, along with reduced levels of H2O2, O2−, and MDA. Additionally, the expression of antioxidant enzyme genes (AtPOD3, AtSOD4, and AtCAT1) and drought-responsive genes (AtABI5, AtCOR15A, AtCOR47, AtHAK5, AtP5CS, AtRD22, and AtRD29A) was induced by MfWRKY40 in the transgenic lines. Our findings demonstrate that MfWRKY40 acts as a positive regulator of drought tolerance in A. thaliana by enhancing the activities of antioxidant enzymes (POD, SOD, and CAT) while reducing H2O2, O2−, and MDA, thereby promoting ROS scavenging (Figure 8). In subsequent studies, we plan to generate transgenic Medicago sativa (since the M. falcata transformation system is not yet fully established) to further comprehensively evaluate the functional role of MfWRKY40 in drought stress responses, thereby providing a foundation for molecular breeding in alfalfa.
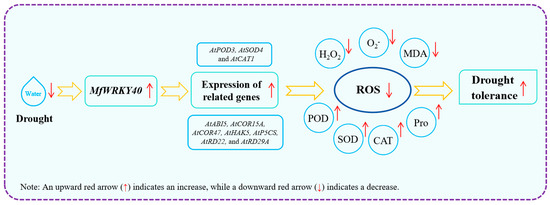
Figure 8.
Regulatory model of MfWRKY40 gene in A. thaliana under drought stress.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms26178495/s1.
Author Contributions
Conceptualization, Q.L. and X.Z.; methodology, Q.L. and X.Z.; software, W.D.; validation, X.Z., W.D., and Z.J.; formal analysis, X.Z.; investigation, W.D.; resources, W.D. and Z.J.; data curation, X.Z. and Z.J.; writing—original draft preparation, X.Z.; writing—review and editing, Q.L. and Y.W.; funding acquisition, Q.L. and Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “National Natural Science Foundation of China” (32460349), Natural Science Foundation of the Autonomous Region-2023D01B35, the Key Research and Development Program of the Xinjiang Uygur Autonomous Region (2023B02031), and the Postdoctoral Mobile Station for Crop Science and Autonomous Region Talent Development Fund “Tianchi Talent” Introduction Plan Project.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article or the Supplementary Materials.
Acknowledgments
In this study, we gratefully acknowledge the Forage Germplasm Resources Conservation and Utilization Innovation Team at the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (Beijing, China), for providing the A. thaliana seeds and pB7WG2RS overexpression vector. We also extend our appreciation to the College of Grassland Science at Xinjiang Agricultural University for their support of this research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Salt Stress Signaling and Mechanisms of Plant Salt Tolerance. Genet. Eng. 2006, 27, 141–177. [Google Scholar]
- Liu, W.; Wei, J.W.; Shan, Q.; Liu, M.; Xu, J.; Gong, B. Genetic Engineering of Drought-and Salt-Tolerant Tomato via Δ1-Pyrroline-5-Carboxylate Reductase S-Nitrosylation. Plant Physiol. 2024, 195, 1038–1052. [Google Scholar] [CrossRef]
- Yang, R.; Sun, Y.; Zhao, Y.; Bai, C.; Liu, Y.; Sun, J.; Wang, Z.; Yuan, F.; Wang, X.; Liu, W.; et al. Overexpression of MtNAC33 Enhances Biomass Yield and Drought Tolerance in Alfalfa. Plant Biotechnol. J. 2025, 23, 1452–1454. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, Z.; Huang, Y.; He, D.; Lu, L.; Wei, M.; Lin, S.; Luo, W.; Liao, X.; Jin, S.; et al. WRKY45 Positively Regulates Salinity and Osmotic Stress Responses in Arabidopsis. Plant Physiol. Biochem. 2025, 219, 109408. [Google Scholar] [CrossRef]
- Ahmad, M.; Alabd, A.; Gao, Y.; Yu, W.; Jamil, W.; Wang, X.; Wei, J.; Ni, J.; Teng, Y.; Bai, S. Three Stress-Responsive NAC Transcription Factors, Pp-SNACs, Differentially and Synergistically Regulate Abiotic Stress in Pear. Sci. Hortic. 2022, 305, 111393. [Google Scholar] [CrossRef]
- Aslam, M.; Fakher, B.; Ashraf, M.A.; Cheng, Y.; Wang, B.; Qin, Y. Plant Low-Temperature Stress: Signaling and Response. Agronomy 2022, 12, 702. [Google Scholar] [CrossRef]
- Wang, Y.; He, W.; Wang, L.; Lan, Y.; Wu, M. TCP Transcription Factor Identification in Pecan (Carya illinoensis) and Salt Tolerance Function Analysis of CiTCP8. Sci. Hortic. 2024, 330, 113051. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef]
- Li, W.; Li, P.; Chen, H.; Zhong, J.; Liang, X.; Wei, Y.; Zhang, L.; Wang, H.; Han, D. Overexpression of a Fragaria vesca 1R-MYB Transcription Factor Gene (FvMYB114) Increases Salt and Cold Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 5261. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wei, Y.; Zhang, L.; Wang, Y.; Song, P.; Li, X.; Han, D. FvMYB44, a Strawberry R2R3-MYB Transcription Factor, Improved Salt and Cold Stress Tolerance in Transgenic Arabidopsis. Agronomy 2023, 13, 1051. [Google Scholar] [CrossRef]
- Li, B.; Wang, X.; Wang, X.; Xi, Z. An AP2/ERF Transcription Factor VvERF63 Positively Regulates Cold Tolerance in Arabidopsis and Grape Leaves. Environ. Exp. Bot. 2023, 205, 105124. [Google Scholar] [CrossRef]
- Liu, W.; Wang, T.; Liang, X.; Ye, Q.; Wang, Y.; Han, J.; Han, D. MbWRKY53, a M. baccata WRKY Transcription Factor, Contributes to Cold and Drought Stress Tolerance in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 7626. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cheng, X.; Yin, D.; Chen, D.; Luo, C.; Liu, H.; Huang, C. Advances in the Research on Plant WRKY Transcription Factors Responsive to External Stresses. Curr. Issues Mol. Biol. 2023, 45, 2861–2880. [Google Scholar] [CrossRef]
- Ma, Z.; Hu, L. WRKY Transcription Factor Responses and Tolerance to Abiotic Stresses in Plants. Int. J. Mol. Sci. 2024, 25, 6845. [Google Scholar] [CrossRef]
- Glöckner, G.; Eichinger, L.; Szafranski, K.; Pachebat, J.A.; Bankier, A.T.; Dear, P.H.; Lehmann, R.; Baumgart, C.; Parra, G.; Abril, J.F.; et al. Sequence and Analysis of Chromosome 2 of Dictyostelium discoideum. Nature 2002, 418, 79–85. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Li, M.; Jia, H.; Wei, F.; Xia, Z.; Zhang, X.; Chang, J.; Wang, Z. Overexpression of The WRKY Transcription Factor Gene NtWRKY65 Enhances Salt Tolerance in Tobacco (Nicotiana tabacum). BMC Plant Biol. 2024, 24, 326. [Google Scholar] [CrossRef]
- Zheng, Y.; Ge, J.; Bao, C.; Chang, W.; Liu, J.; Shao, J.; Liu, X.; Su, L.; Pan, L.; Zhou, D.X. Histone Deacetylase HDA9 and WRKY53 Transcription Factor Are Mutual Antagonists in Regulation of Plant Stress Response. Mol. Plant 2020, 13, 598–611. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Wu, Q.; Wang, A.; Li, Q.; Dong, Q.; Yang, J.; Zhao, H.; Wang, X.; Chen, H.; Li, C.A. WRKY Transcription Factor, FtWRKY46, From Tartary buckwheat Improves Salt Tolerance in Transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 147, 43–53. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, P.; Wang, Z.; Wang, C.; Wang, Y. Birch WRKY Transcription Factor, BpWRKY32, Confers Salt Tolerance by Mediating Stomatal Closing, Proline Accumulation, and Reactive Oxygen Species Scavenging. Plant Physiol. Biochem. 2024, 210, 108599. [Google Scholar] [CrossRef]
- Zhou, W.; Yang, S.; Yang, L.; Xiao, R.; Chen, S.; Wang, D.; Wang, S.; Wang, Z. Genome-Wide Identification of the Hypericum perforatum WRKY Gene Family Implicates HpWRKY85 in Drought Resistance. Int. J. Mol. Sci. 2022, 24, 352. [Google Scholar] [CrossRef]
- Bao, Y.; Zou, Y.; An, X.; Liao, Y.; Dai, L.; Liu, L.; Peng, D.; Huang, X.; Wang, B. Overexpression of a Ramie (Boehmaeria nivea L. Gaud) Group I WRKY Gene, BnWRKY49, Increases Drought Resistance in Arabidopsis thaliana. Plants 2024, 13, 379. [Google Scholar] [CrossRef]
- Duan, D.; Yi, R.; Ma, Y.; Dong, Q.; Mao, K.; Ma, F. Apple WRKY Transcription Factor MdWRKY56 Positively Modulates Drought Stress Tolerance. Environ. Exp. Bot. 2023, 212, 105400. [Google Scholar] [CrossRef]
- Li, W.; Li, D.; Li, H.; Wang, M.; Wang, Z.; Liu, J. The Tomato WRKY Transcription Factor SlWRKY17 Positively Regulates Drought Stress Tolerance in Transgenic Tobacco Plants. Russ. J. Plant Physiol. 2023, 69, 154. [Google Scholar] [CrossRef]
- Ahkami, A.H.; Wang, W.; Wietsma, T.W.; Winkler, T.; Lange, I.; Jansson, C.; Lange, M.; McDowell, N.G. Metabolic Shifts Associated with Drought-Induced Senescence in Brachypodium. Plant Sci. 2019, 289, 110278. [Google Scholar] [CrossRef]
- Duan, M.; Zhang, R.; Zhu, F.; Zhang, Z.; Gou, L.; Wen, J.; Dong, J.; Wang, T. A Lipid-Anchored NAC Transcription Factor Is Translocated into the Nucleus and Activates Glyoxalase I Expression during Drought Stress. Plant cell 2017, 29, 1748. [Google Scholar] [CrossRef]
- Vyšniauskienė, R.; Naugžemys, D.; Patamsytė, J.; Rančelienė, V.; Čėsnienė, T.; Žvingila, D. ISSR and Chloroplast DNA Analyses Indicate Frequent Hybridization of Alien Medicago sativa subsp. sativa and Native M. sativa subsp. falcata. Plant Syst. Evol. 2015, 301, 2341–2350. [Google Scholar] [CrossRef]
- Jiang, Z. Cloning and Functional Analysis of the MfWRKY40 Gene in Medicago falcata L. Master’s Thesis, Xinjiang Agricultural University, Urumqi, China, 2024. [Google Scholar]
- Javed, T.; Gao, S.J. WRKY Transcription Factors in Plant Defense. Trends Genet. 2023, 39, 787–801. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Ma, C.; Wang, Z.; Shi, X.; Duan, W.; Fu, X.; Liu, J.; Guo, C.; Xiao, K. Transcription Factor Gene TaWRKY76 Confers Plants Improved Drought and Salt Tolerance Through Modulating Stress Defensive-Associated Processes in Triticum aestivum L. Plant Physiol. Biochem. 2024, 216, 109147. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Y.; Xu, P.; Zhang, Z. Overexpression of a WRKY Transcription Factor TaWRKY2 Enhances Drought Stress Tolerance in Transgenic wheat. Front. Plant Sci. 2018, 9, 997. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; He, L.; Wu, Y. Wheat WRKY Transcription Factor TaWRKY24 Confers Drought and Salt Tolerance in Transgenic Plants. Plant Physiol. Biochem. 2023, 205, 108137. [Google Scholar] [CrossRef]
- Lv, M.; Hou, D.; Wan, J.; Ye, T.; Zhang, L.; Fan, J.; Li, C.; Dong, Y.; Chen, W.; Rong, S.; et al. OsWRKY97, an Abiotic Stress-Induced Gene of Rice, Plays a Key Role in Drought Tolerance. Plants 2023, 12, 3338. [Google Scholar] [CrossRef]
- Fei, J.; Liu, Z.; Wang, P.; Qu, J.; Liu, S.; Guan, S.; Ma, Y. The Maize WRKY Transcription Factor ZmWRKY25 Respond Drought Stress in Transgenic Tobacco. Phyton-Int. J. Exp. Bot. 2024, 93, 3617–3635. [Google Scholar]
- Gu, L.; Chen, X.; Hou, Y.; Cao, Y.; Wang, H.; Zhu, B.; Du, X.; Wang, H. ZmWRKY30 Modulates Drought Tolerance in Maize by Influencing myo-Inositol and Rreactive Oxygen Species Homeostasis. Physiol. Plant. 2024, 176, e14423. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Wang, R.; Su, L.; Lv, A.; Zhou, P.; An, Y. MsWRKY11, Activated by MsWRKY22, Functions in Drought Tolerance and Modulates Lignin Biosynthesis in Alfalfa (Medicago sativa L.). Environ. Exp. Bot. 2021, 184, 104373. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X.; Ma, L.; Cui, M.; Cai, X.; Zhao, W. Isolation and Characterization of MsWRKY42 From Alfalfa (Medicago sativa) and Its Response to Abiotic Stresses. Sci. Agric. Sin. 2020, 53, 3455–3466. [Google Scholar]
- Zhang, J.; Huang, D.; Zhao, X.; Zhang, M.; Wang, Q.; Hou, X.; Di, D.; Su, B.; Wang, S.; Sun, P. Drought-Responsive WRKY transcription factor genes IgWRKY50 and IgWRKY32 from Iris germanica Enhance Drought Resistance in Transgenic Arabidopsis. Front. Plant Sci. 2022, 13, 983600. [Google Scholar] [CrossRef]
- Li, X.; Tang, Y.; Zhou, C.; Zhang, L.; Lv, J. A Wheat WRKY Transcription Factor TaWRKY46 Enhances Tolerance to Osmotic Stress in Transgenic Arabidopsis Plants. Int. J. Mol. Sci. 2020, 21, 1321. [Google Scholar] [CrossRef] [PubMed]
- Lorens, G.F.; Bennett, J.M.; Loggale, L.B. Differences in Drought Resistance between Two Corn Hybrids. I. Water Relations and Root Length Density 1. Agron. J. 1987, 79, 802–807. [Google Scholar] [CrossRef]
- Ekanayake, I.J.; O’Toole, J.C.; Garrity, D.P.; Masajo, T.M. Inheritance of Root Characters and their Relations to Drought Resistance in Rice 1. Crop Sci. 1985, 25, 927–933. [Google Scholar] [CrossRef]
- Reicosky, D.C.; Deaton, D.E. Soybean Water Extraction, Leaf Water Potential, and Evapotranspiration during Drought 1. Agron. J. 1979, 71, 45–50. [Google Scholar] [CrossRef]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and Concepts in Quantifying Resistance to Drought, Salt and Freezing, Abiotic Stresses That Affect Plant Water Status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Tansley Review No. 112. Oxygen Processing in Photosynthesis: Regulation and Signalling. New Phytol. 2000, 146, 359–388. [Google Scholar] [CrossRef]
- Kumar, A.; Prasad, A.; Sedlářová, M.; Pospíšil, P. Malondialdehyde Enhances PsbP Protein Release during Heat Stress in Arabidopsis. Plant Physiol. Biochem. 2023, 202, 107984. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhao, Y.; Yang, Y.; Guan, Q. Roles of Superoxide Dismutase in Plant Response to Drought, Salinity and Cold Stress. J. Plant Res. 2024, 44, 481–491. [Google Scholar]
- Yadav, S.; Gill, S.S.; Passricha, N.; Gill, R.; Badhwar, P.; Anjum, N.A.; Francisco, J.B.J.; Tuteja, N. Genome-Wide Analysis and Transcriptional Expression Pattern-Assessment of Superoxide Dismutase (SOD) in Rice and Arabidopsis under Abiotic Stresses. Plant Gene 2019, 17, 100165. [Google Scholar] [CrossRef]
- Ampofo, J.O.; Ngadi, M. Stimulation of the Phenylpropanoid Pathway and Antioxidant Capacities by Biotic and Abiotic Elicitation Strategies in Common Bean (Phaseolus vulgaris) Sprouts. Process Biochem. 2021, 100, 98–106. [Google Scholar] [CrossRef]
- Zhong, M.; Song, R.; Wang, Y.; Shu, S.; Sun, J.; Guo, S. TGase Regulates Salt Stress Tolerance through Enhancing Bound Polyamines Mediated Antioxidant Enzymes Activity in Tomato. Environ. Exp. Bot. 2020, 179, 104191. [Google Scholar] [CrossRef]
- Sun, S.; Li, X.; Gao, S.; Nie, N.; Zhang, H.; Yang, Y.; He, S.; Liu, Q.; Zhai, H. A Novel WRKY Transcription Factor from Ipomoea trifida, ItfWRKY70, Confers Drought Tolerance in Sweet Potato. Int. J. Mol. Sci. 2022, 23, 686. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, J.; Yang, F.; Zhang, G.; Wang, D.; Zhang, L.; Ou, Y.; Yao, Y. The WRKY Transcription Factor WRKY8 Promotes Resistance to Pathogen Infection and Mediates Drought and Salt Stress Tolerance in Solanum lycopersicum. Physiol. Plant. 2020, 168, 98–117. [Google Scholar] [CrossRef] [PubMed]
- Akram, W.; Yasin, N.A.; Shah, A.A.; Khan, W.U.; Li, G.; Ahmad, A.; Ahmed, S.; Hussaan, M.; Rizwan, M.; Ali, S. Exogenous Application of Liquiritin Alleviated Salt Stress and Improved Growth of Chinese Kale Plants. Sci. Hortic. 2022, 294, 110762. [Google Scholar] [CrossRef]
- Han, J.; Li, X.; Li, W.; Yao, A.; Niu, C.; Hou, R.; Liu, W.; Wang, Y.; Zhang, L.; Han, D. Overexpression of Malus baccata WRKY40 (MbWRKY40) Enhances Stress Tolerance in Arabidopsis Subjected to Cold and Drought. Plant Stress 2023, 10, 100209. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between Two cis-Acting Elements, ABRE and DRE, in ABA-dependent Expression of Arabidopsis rd29A Gene in Response to Dehydration and High-Salinity Stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Wu, M.; Zhang, K.; Xu, Y.; Wang, L.; Liu, H.; Qin, Z.; Xiang, Y. The Moso Bamboo WRKY Transcription Factor, PheWRKY86, Regulates Drought Tolerance in Transgenic Plants. Plant Physiol. Biochem. 2022, 170, 180–191. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAYTM Vectors for Agrobacterium-Mediated Plant Transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method for Agrobacterium-Mediated Transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Shimada, T.L.; Shimada, T.; Hara-Nishimura, I. A Rapid and Non-Destructive Screenable Marker, FAST, for Identifying Transformed Seeds of Arabidopsis thaliana. Plant J. 2010, 61, 519–528. [Google Scholar] [CrossRef]
- Wang, X.; Gao, M.; Kong, Y.; Yu, Q.; Yao, L.; Li, X.; Li, W.; Liu, W.; Hou, R.; Zhang, L. Overexpression of a Malus baccata (L.) Borkh WRKY Factor Gene MbWRKY33 Increased High Salinity Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2025, 26, 5833. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mei, F.; Chen, B.; Li, F.; Zhang, Y.; Kang, Z.; Wang, X.; Mao, H. Overexpression of the Wheat NAC Transcription Factor TaSNAC4-3A Gene Confers Drought Tolerance in Transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 160, 37–50. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).