Genetic Diversity of Equid Herpesvirus 5 in Temporal Samples from Mares and Their Foals at Three Polish National Studs
Abstract
1. Introduction
2. Results
2.1. Phylogenetic Analysis
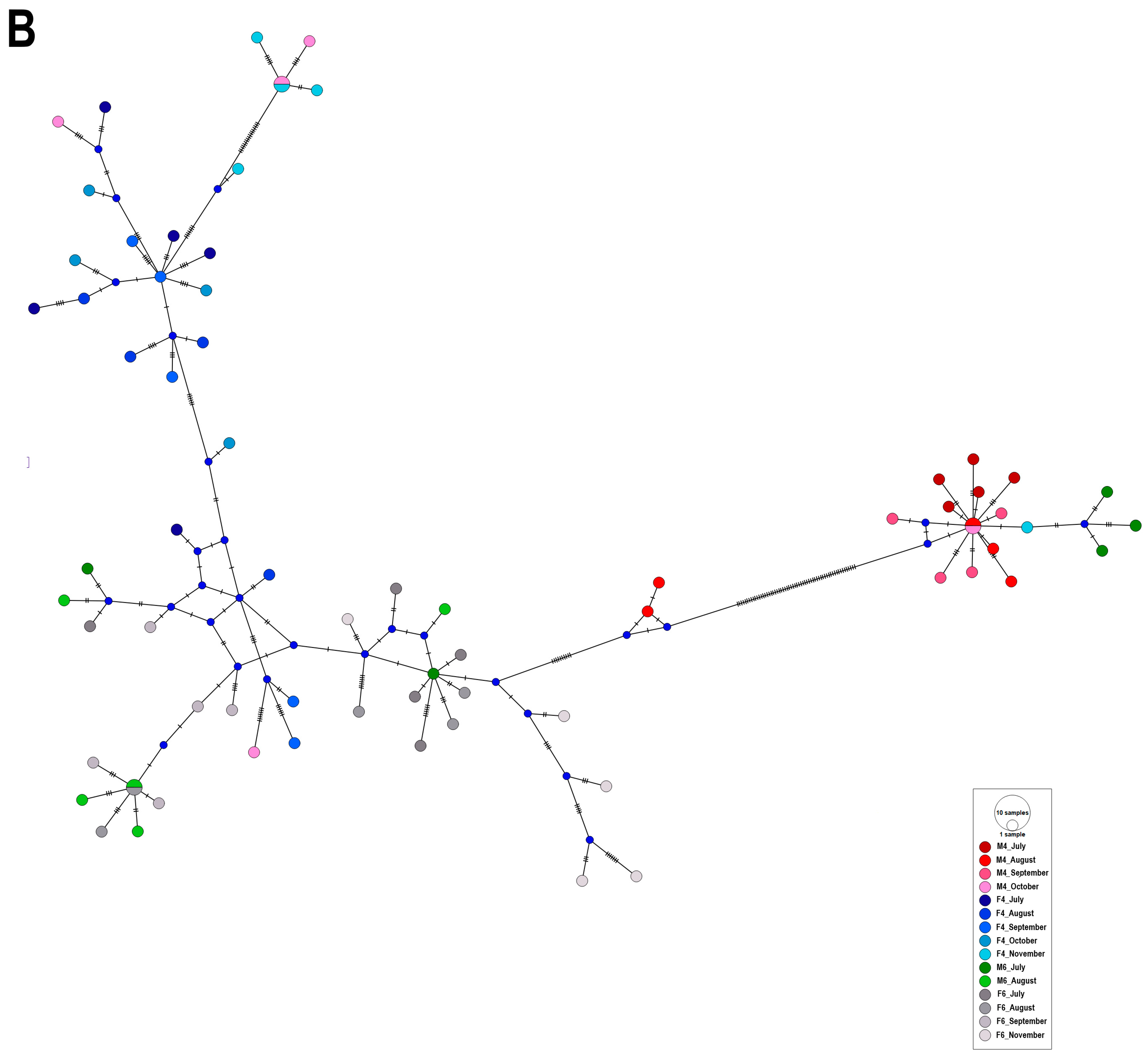
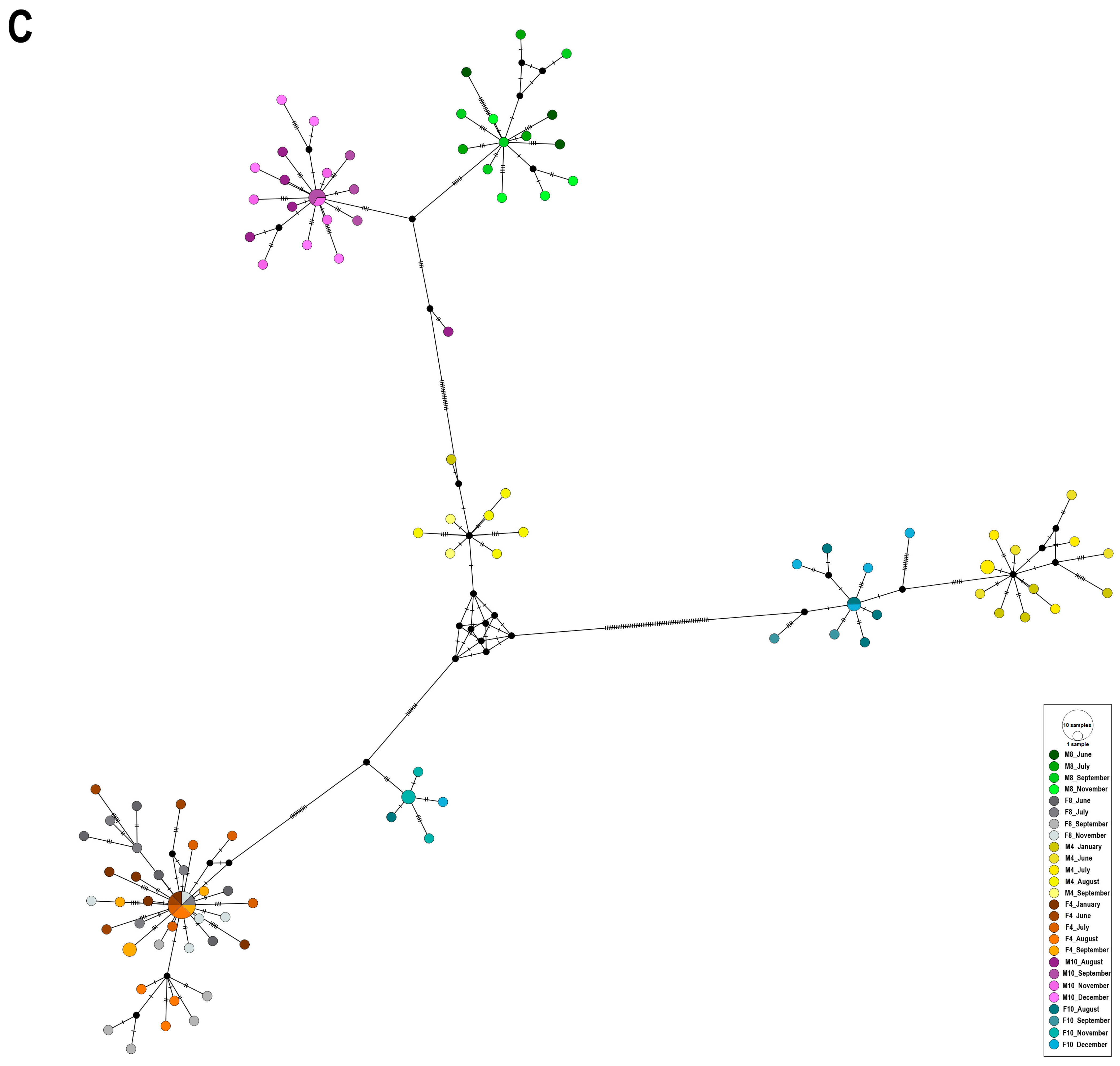
2.2. Haplotype Network Analysis
3. Discussion
4. Materials and Methods
4.1. Source of Samples
4.2. Conventional PCR Assays and Cloning
4.3. Sequence Analyses
4.4. GenBank Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marenzoni, M.L.; Stefanetti, V.; Danzetta, M.L.; Timoney, P.J. Gammaherpesvirus infections in equids: A review. Vet. Med. 2015, 6, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef]
- Telford, E.A.; Studdert, M.J.; Agius, C.T.; Watson, M.S.; Aird, H.C.; Davison, A.J. Equine herpesviruses 2 and 5 are gamma-herpesviruses. Virology 1993, 195, 492–499. [Google Scholar] [CrossRef]
- Wang, L.; Raidal, S.L.; Pizzirani, A.; Wilcox, G.E. Detection of respiratory herpesviruses in foals and adult horses determined by nested multiplex PCR. Vet. Microbiol. 2007, 121, 18–28. [Google Scholar] [CrossRef]
- Dunowska, M.; Wilks, C.R.; Studdert, M.J.; Meers, J. Equine respiratory viruses in foals in New Zealand. N. Z. Vet. J. 2002, 50, 140–147. [Google Scholar] [CrossRef]
- Stasiak, K.; Dunowska, M.; Rola, J. Prevalence and sequence analysis of equid herpesviruses from the respiratory tract of Polish horses. Virol. J. 2018, 15, 106. [Google Scholar] [CrossRef] [PubMed]
- Pusterla, N.; Sandler-Burtness, E.; Barnum, S.; Hill, L.A.; Mendonsa, E.; Khan, R.; Portener, D.; Ridland, H.; Schumacher, S. Frequency of Detection of Respiratory Pathogens in Nasal Secretions From Healthy Sport Horses Attending a Spring Show in California. J. Equine Vet. Sci. 2022, 117, 104089. [Google Scholar] [CrossRef] [PubMed]
- Vander Werf, K.A.; Davis, E.G.; Janardhan, K.; Bawa, B.; Bolin, S.; Almes, K. Identification of Equine Herpesvirus 5 in Horses with Lymphoma. J. Equine Vet. Sci. 2014, 34, 738–741. [Google Scholar] [CrossRef]
- Nordengrahn, A.; Merza, M.; Ros, C.; Lindholmc, A.; Palfl, V.; Hannant, D.; Belak, S. Prevalence of equine herpesvirus types 2 and 5 in horse populations by using type-specific PCR assays. Vet. Res. 2002, 33, 251–259. [Google Scholar] [CrossRef]
- Marenzoni, M.L.; Coppola, G.; Maranesi, M.; Passamonti, F.; Cappelli, K.; Capomaccio, S.; Verini Supplizi, A.; Thiry, E.; Coletti, M. Age-dependent prevalence of equid herpesvirus 5 infection. Vet. Res. Commun. 2010, 34, 703–708. [Google Scholar] [CrossRef]
- Hue, E.S.; Fortier, G.D.; Fortier, C.I.; Leon, A.M.; Richard, E.A.; Legrand, L.J.; Pronost, S.L. Detection and quantitation of equid gammaherpesviruses (EHV-2, EHV-5) in nasal swabs using an accredited standardised quantitative PCR method. J. Virol. Methods 2014, 198, 18–25. [Google Scholar] [CrossRef]
- Stasiak, K.; Dunowska, M.; Rola, J. Kinetics of the Equid Herpesvirus 2 and 5 Infections among Mares and Foals from Three Polish National Studs. Viruses 2022, 14, 713. [Google Scholar] [CrossRef]
- Stasiak, K.; Dunowska, M.; Trewick, S.; Rola, J. Genetic Variation in the Glycoprotein B Sequence of Equid Herpesvirus 5 among Horses of Various Breeds at Polish National Studs. Pathogens 2021, 10, 322. [Google Scholar] [CrossRef] [PubMed]
- Mekuria, Z.H.; El-Hage, C.; Ficorilli, N.P.; Washington, E.A.; Gilkerson, J.R.; Hartley, C.A. Mapping B lymphocytes as major reservoirs of naturally occurring latent equine herpesvirus 5 infection. J. Gen. Virol. 2017, 98, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Hartley, C.A.; Dynon, K.J.; Mekuria, Z.H.; El-Hage, C.M.; Holloway, S.A.; Gilkerson, J.R. Equine gammaherpesviruses: Perfect parasites? Vet. Microbiol. 2013, 167, 86–92. [Google Scholar] [CrossRef]
- Ackermann, M. Pathogenesis of gammaherpesvirus infections. Vet. Microbiol. 2006, 113, 211–222. [Google Scholar] [CrossRef]
- Williams, K.J. Gammaherpesviruses and pulmonary fibrosis: Evidence from humans, horses, and rodents. Vet. Pathol. 2014, 51, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.J.; Robinson, N.E.; Lim, A.; Brandenberger, C.; Maes, R.; Behan, A.; Bolin, S.R. Experimental induction of pulmonary fibrosis in horses with the gammaherpesvirus equine herpesvirus 5. PLoS ONE 2013, 8, e77754. [Google Scholar] [CrossRef]
- Wilkie, G.S.; Kerr, K.; Stewart, J.P.; Studdert, M.J.; Davison, A.J. Genome sequences of equid herpesviruses 2 and 5. Genome Announc. 2015, 3, e00119-15. [Google Scholar] [CrossRef]
- Sharma, V.; Mobeen, F.; Prakash, T. Comparative Genomics of Herpesviridae Family to Look for Potential Signatures of Human Infecting Strains. Int. J. Genom. 2016, 2016, 9543274. [Google Scholar] [CrossRef]
- Pereira, L. Function of glycoprotein B homologues of the family herpesviridae. Infect. Agents Dis. 1994, 3, 9–28. [Google Scholar]
- Sathiyamoorthy, K.; Chen, J.; Longnecker, R.; Jardetzky, T.S. The COMPLEXity in herpesvirus entry. Curr. Opin. Virol. 2017, 24, 97–104. [Google Scholar] [CrossRef]
- Holloway, S.A.; Lindquester, G.J.; Studdert, M.J.; Drummer, H.E. Identification, sequence analysis and characterisation of equine herpesvirus 5 glycoprotein B. Arch. Virol. 1999, 144, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Holloway, S.A.; Studdert, M.J.; Drummer, H.E. Characterization of glycoprotein B of the gammaherpesvirus equine herpesvirus-2. J. Gen. Virol. 1998, 79 Pt 7, 1619–1629. [Google Scholar] [CrossRef]
- Fortier, G.; van Erck, E.; Pronost, S.; Lekeux, P.; Thiry, E. Equine gammaherpesviruses: Pathogenesis, epidemiology and diagnosis. Vet. J. 2010, 186, 148–156. [Google Scholar] [CrossRef]
- Bell, S.A.; Balasuriya, U.B.; Gardner, I.A.; Barry, P.A.; Wilson, W.D.; Ferraro, G.L.; MacLachlan, N.J. Temporal detection of equine herpesvirus infections of a cohort of mares and their foals. Vet. Microbiol. 2006, 116, 249–257. [Google Scholar] [CrossRef]
- Lee, S.K.; Lee, I. The Molecular Detection of Equine Herpesviruses 2 and 5 in Genital Swabs From Clinically Normal Thoroughbred Mares in South Korea. J. Equine Vet. Sci. 2019, 79, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Back, H.; Ullman, K.; Leijon, M.; Soderlund, R.; Penell, J.; Stahl, K.; Pringle, J.; Valarcher, J.F. Genetic variation and dynamics of infections of equid herpesvirus 5 in individual horses. J. Gen. Virol. 2016, 97, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Dunowska, M.; Holloway, S.A.; Wilks, C.R.; Meers, J. Genomic variability of equine herpesvirus-5. Arch. Virol. 2000, 145, 1359–1371. [Google Scholar] [CrossRef]
- Lopez-Munoz, A.D.; Rastrojo, A.; Martin, R.; Alcami, A. Herpes simplex virus 2 (HSV-2) evolves faster in cell culture than HSV-1 by generating greater genetic diversity. PLoS Pathog. 2021, 17, e1009541. [Google Scholar] [CrossRef]
- Ortigas-Vasquez, A.; Szpara, M. Embracing Complexity: What Novel Sequencing Methods Are Teaching Us About Herpesvirus Genomic Diversity. Annu. Rev. Virol. 2024, 11, 67–87. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, L.N.; Bowen, C.D.; Renner, D.W.; Pandey, U.; Della Fera, A.N.; Kimberlin, D.W.; Prichard, M.N.; Whitley, R.J.; Weitzman, M.D.; Szpara, M.L. Genotypic and Phenotypic Diversity of Herpes Simplex Virus 2 within the Infected Neonatal Population. mSphere 2019, 4, e00590-18. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Roumagnac, P.; Varsani, A.; Martin, D.P. Sequence Demarcation Tool (SDT), a Free User-Friendly Computer Program Using Pairwise Gentic Identity Calculations to Classify Nucleotide or Amino Acid Sequences. Methods Mol. Biol. 2025, 2912, 71–79. [Google Scholar]

| Test/Breed | Variation Within Populations | Variation Between Populations | PhiPT | p 1 |
|---|---|---|---|---|
| Individual sample | ||||
| Stud I | 32.6% | 67.4% | 0.67 | <0.001 |
| Stud II | 37.3% | 62.7% | 0.63 | <0.001 |
| Stud III | 13.1% | 86.9% | 0.87 | <0.001 |
| Individual animal | ||||
| Stud I | 55.6% | 44.4% | 0.44 | <0.001 |
| Stud II | 58.3% | 41.7% | 0.42 | <0.001 |
| Stud III | 36.1% | 63.9% | 0.64 | <0.001 |
| Mare–foal pair | ||||
| Stud I 2 | 84.3% | 15.7% | 0.16 | <0.001 |
| Stud II 3 | 93.6% | 6.4% | 0.06 | 0.03 |
| Stud III 4 | 86.3% | 13.7% | 0.14 | <0.001 |
| Source of the virus (mare/foal) | ||||
| Stud I | 55.6% | 44.4% | 0.44 | <0.001 |
| Stud II | 58.3% | 41.7% | 0.42 | <0.001 |
| Stud III | 36.1% | 63.9% | 0.64 | <0.001 |
| Sampling (month) | ||||
| Stud I | 93.2 | 6.8 | 0.07 | 0.003 |
| Stud II | 96.8 | 3.2 | 0.03 | 0.154 |
| Stud III | 92.6 | 7.4 | 0.07 | 0.012 |
| Animal | Sampling Month | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| May | Jun | Jul | Aug | Sep | Oct | Nov | Dec | Jan | Feb | |
| Stud I | ||||||||||
| M4 | nd | nd | + | + | − | − | + | + | + | + |
| F4 | nd | nd | − | − | − | + | + | + | + | + |
| M11 | nd | nd | + | − | − | − | + | + | + | + |
| F11 | nd | nd | + | + | + | + | + | + | + | + |
| M14 | nd | nd | − | − | − | − | + | + | − | − |
| F14 | nd | nd | − | + | + | + | + | + | + | + |
| Stud II | ||||||||||
| M4 | nd | + | + | + | + | + | + | + | nd | nd |
| F4 | nd | + | + | + | + | + | + | + | nd | nd |
| M6 | nd | − | + | + | + | − | − | − | nd | nd |
| F6 | nd | + | + | + | + | + | + | + | nd | nd |
| Stud III | ||||||||||
| M1 | + | + | − | + | nd | + | + | nd | + | nd |
| F1 | − | − | − | + | nd | + | + | nd | + | nd |
| M4 | + | + | + | + | nd | + | + | nd | + | nd |
| F4 | − | + | + | + | nd | + | + | nd | + | nd |
| M8 | − | + | + | + | nd | + | + | nd | − | nd |
| F8 | − | + | + | + | nd | + | + | nd | + | nd |
| M10 | + | + | + | + | nd | + | + | nd | + | nd |
| F10 | − | − | − | + | nd | + | + | nd | + | nd |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stasiak, K.; Dunowska, M.; Rola, J. Genetic Diversity of Equid Herpesvirus 5 in Temporal Samples from Mares and Their Foals at Three Polish National Studs. Int. J. Mol. Sci. 2025, 26, 8298. https://doi.org/10.3390/ijms26178298
Stasiak K, Dunowska M, Rola J. Genetic Diversity of Equid Herpesvirus 5 in Temporal Samples from Mares and Their Foals at Three Polish National Studs. International Journal of Molecular Sciences. 2025; 26(17):8298. https://doi.org/10.3390/ijms26178298
Chicago/Turabian StyleStasiak, Karol, Magdalena Dunowska, and Jerzy Rola. 2025. "Genetic Diversity of Equid Herpesvirus 5 in Temporal Samples from Mares and Their Foals at Three Polish National Studs" International Journal of Molecular Sciences 26, no. 17: 8298. https://doi.org/10.3390/ijms26178298
APA StyleStasiak, K., Dunowska, M., & Rola, J. (2025). Genetic Diversity of Equid Herpesvirus 5 in Temporal Samples from Mares and Their Foals at Three Polish National Studs. International Journal of Molecular Sciences, 26(17), 8298. https://doi.org/10.3390/ijms26178298







