Molecular Mechanisms of Herbicide Resistance in Rapeseed: Current Status and Future Prospects for Resistant Germplasm Development
Abstract
1. Introduction
2. Modes of Action of Three Major Herbicides and the Molecular Mechanisms of Crop Resistance
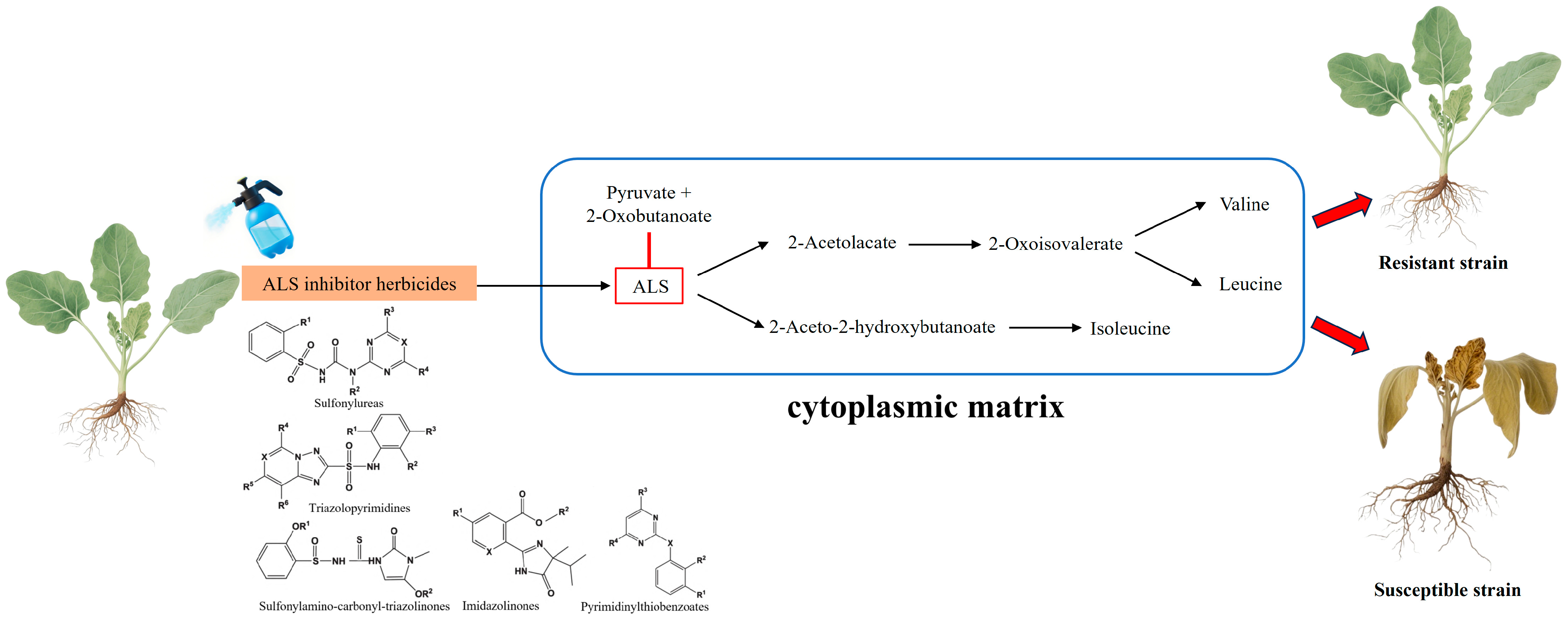
2.1. ALS Inhibitor Herbicides and ALS
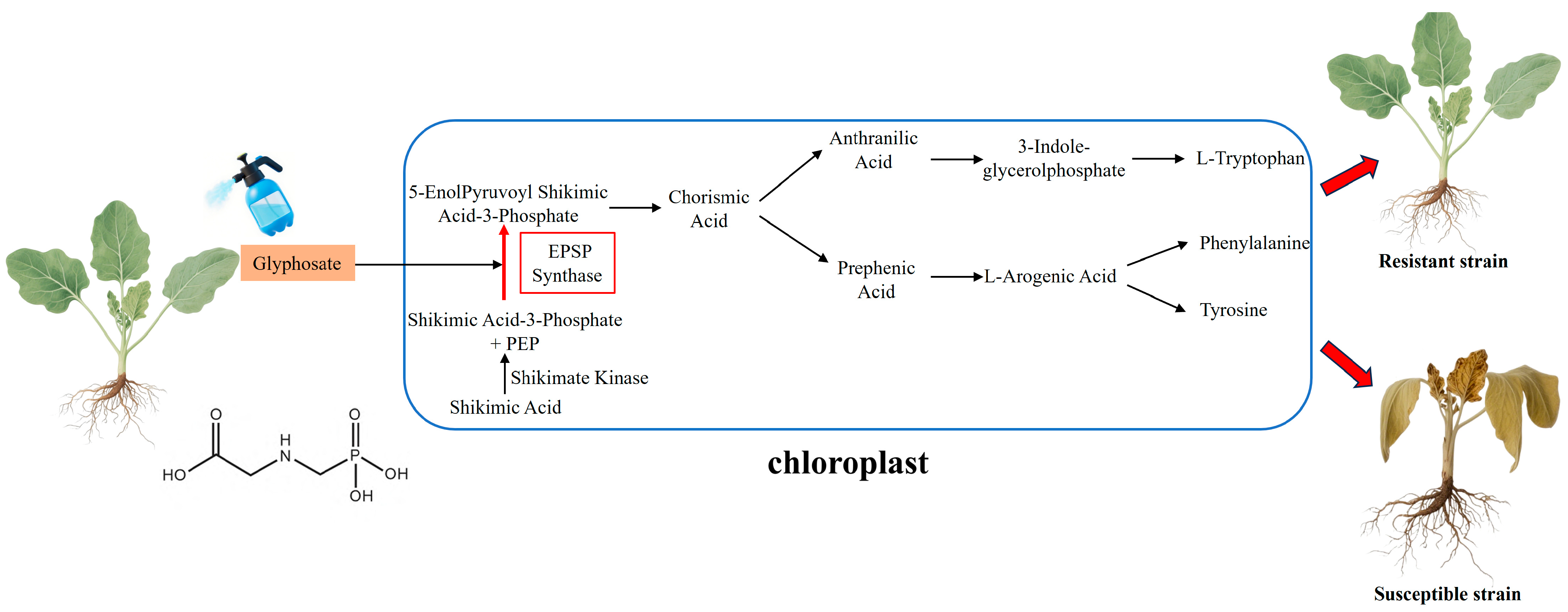
2.2. Glyphosate and EPSPS
2.3. Glufosinate and GS
3. Research Progress in the Development of Herbicide-Resistant Rapeseed Germplasm
3.1. Development of Herbicide-Resistant Rapeseed by Natural Mutation
3.2. Development of Herbicide-Resistant Rapeseed Germplasm Through Artificial Mutagenesis
3.3. Development of Herbicide-Resistant Rapeseed Germplasm by Genetic Engineering
3.3.1. Introduction of an Exogenous Resistance Gene
3.3.2. Endogenous Gene Editing
4. Future Prospects for Herbicide-Resistant Rapeseed Germplasm Development
4.1. Discovery of New Resistance Genes and Development of New Resistant Rapeseed
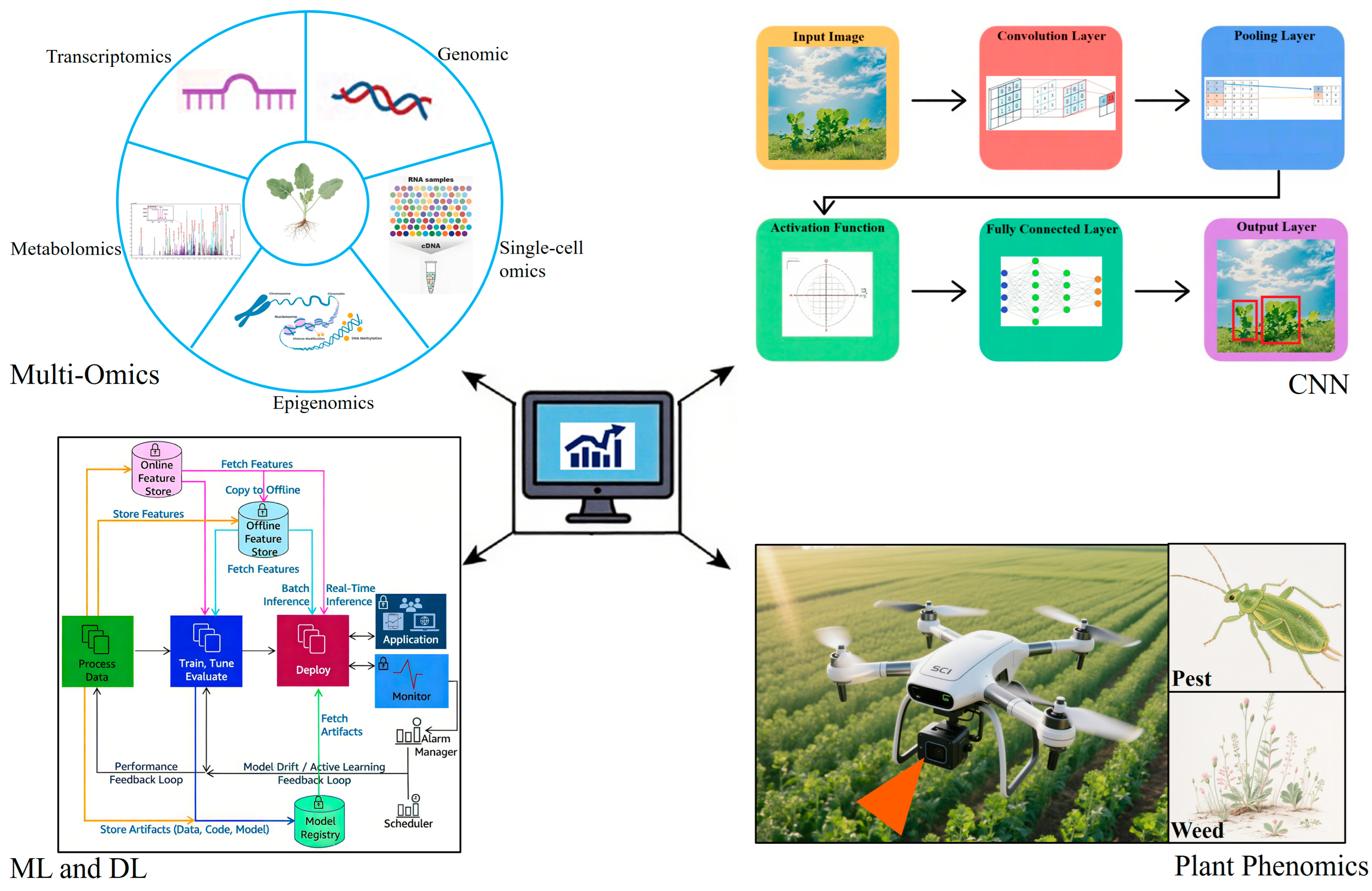
4.2. Integration of Multi-Omics and Artificial Intelligence for Precision Breeding
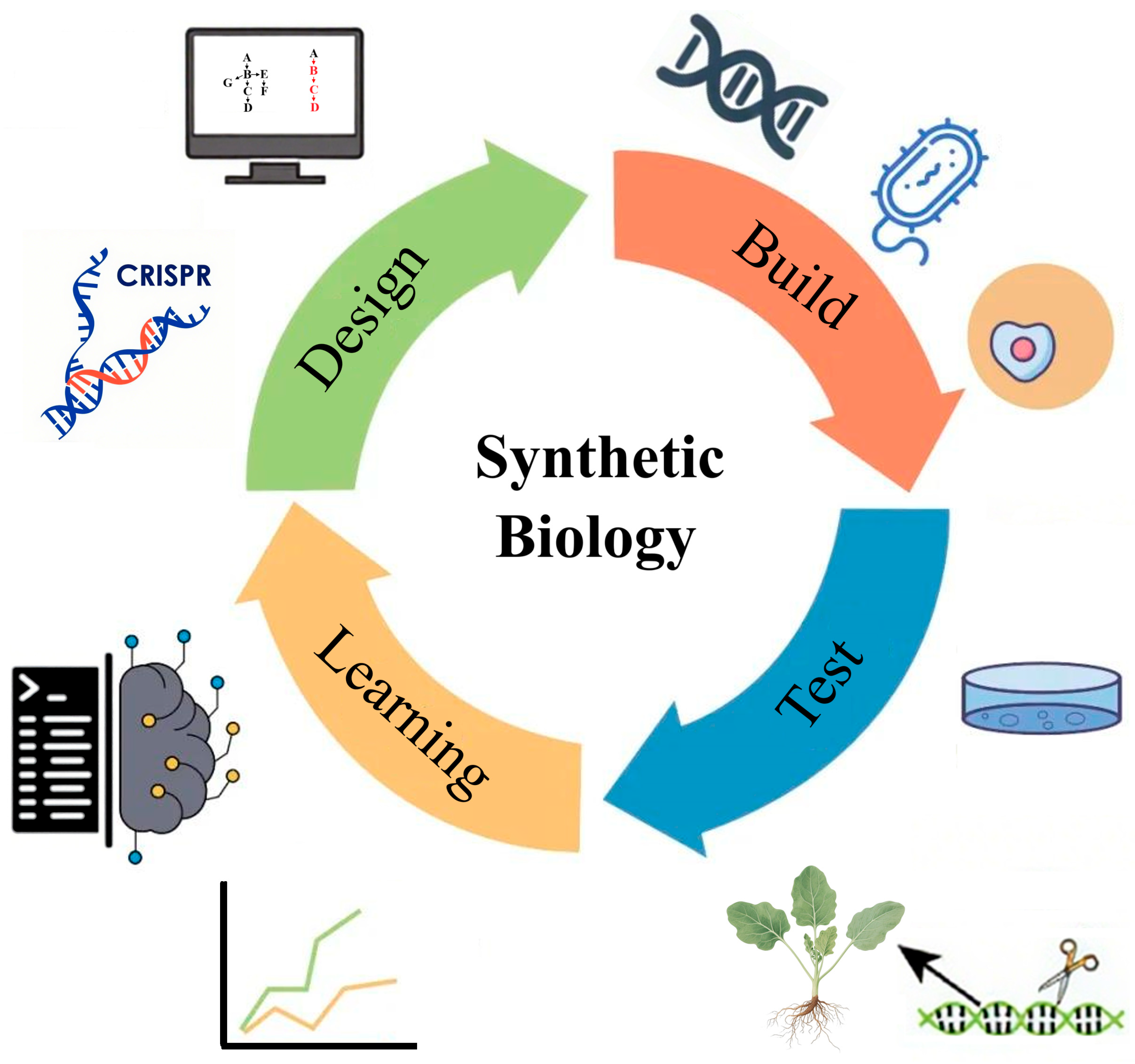
4.3. Exploring New Strategies for Synthetic Biology and Directed Evolution
5. Potential Challenges and Solutions for the Promotion of Herbicide-Resistant Rapeseed
6. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, X.; Huang, T.; Hou, G.; Li, L.; Hou, Y.; Lu, Y. Identification of QTLs for seed quality traits in rapeseed (Brassica napus L.) using recombinant inbred lines (RILs). Euphytica 2016, 210, 1–16. [Google Scholar] [CrossRef]
- Fu, D.; Jiang, L.; Mason, A.S.; Xiao, M.; Zhu, L.; Li, L.; Zhou, Q.; Shen, C.; Huang, C. Research progress and strategies for multifunctional rapeseed: A case study of China. J. Integr. Agric. 2016, 15, 1673–1684. [Google Scholar] [CrossRef]
- Bijanzadeh, E.; Naderi, R.; Behpoori, A. Interrelationships between oilseed rape yield and weeds population under herbicides application. Aust. J. Crop Sci. 2010, 4, 155–162. [Google Scholar]
- Larue, C.T.; Goley, M.; Shi, L.; Evdokimov, A.G.; Sparks, O.C.; Ellis, C.; Wollacott, A.M.; Rydel, T.J.; Halls, C.E.; Van Scoyoc, B.; et al. Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops. Pest Manag. Sci. 2019, 75, 2086–2094. [Google Scholar] [CrossRef]
- Bagherani, N.; Shimi, P. Evaluation of some herbicides for weed control in oilseed rape (Brassica napus L.). J. Agric. Sci. Nat. Resour. 2001, 8, 157–163. [Google Scholar]
- Huang, Q.; Lv, J.; Sun, Y.; Wang, H.; Guo, Y.; Qu, G.; Hu, S. Inheritance and molecular characterization of a novel mutated AHAS gene responsi ble for the resistance of AHAS-inhibiting herbicides in rapeseed (Brassica napus L.). Int. J. Mol. Sci. 2020, 21, 1345. [Google Scholar] [CrossRef]
- Freeman, S.E.; Lutman, P.J.W. The effects of timing of control of weeds on the yield of winter oilseed rape (Brassica napus) in the context of the potential commercialization of herbicide-tolerant winter rape. J. Agric. Sci. 2004, 142, 263–272. [Google Scholar] [CrossRef]
- Bansal, A.; Gopera, S.; Sandal, S.S.; Walia, P.; Singh, A. Herbicide resistance in canola: From cytogenetics to CRISPR-based innovations. J. Adv. Biol. Biotechnol. 2025, 28, 253–267. [Google Scholar] [CrossRef]
- Tan, S.; Evans, R.R.; Dahmer, M.; Singh, B.K.; Shaner, D.L. Imidazolinone-tolerant crops: History, current status and future. Pest Manag. Sci. 2004, 61, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Duggleby, R.G.; McCourt, J.A.; Guddat, L.W. Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol. Biochem. 2008, 46, 309–324. [Google Scholar] [CrossRef]
- Chipman, D.; Barak, Z.; Schloss, J.V. Biosynthesis of 2 aceto-2-hydroxy acids: Acetolactate synthases and ace-tohydroxyacid synthases. Biochim. Biophys. Acta 1998, 1385, 401–419. [Google Scholar] [CrossRef]
- Singh, B.K.; Shaner, D.L. Biosynthesis of branched chain amino acids: From test tube to field. Plant Cell 1995, 7, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Chen, L.; Deng, X.; Tang, X. Development of herbicide resistance genes and their application in rice. Crop J. 2022, 10, 26–35. [Google Scholar] [CrossRef]
- Zhao, L.; Deng, L.; Zhang, Q.; Jing, X.; Ma, M.; Yi, B.; Wen, J.; Ma, C.; Tu, J.; Fu, T.; et al. Autophagy contributes to sulfonylurea herbicide tolerance via GCN2-independent regulation of amino acid homeostasis. Autophagy 2018, 14, 702–714. [Google Scholar] [CrossRef]
- Grandoni, J.A.; Marta, P.T.; Schloss, J.V. Inhibitors of branched-chain amino acid biosynthesis as potential an-tituberculosis agents. J. Antimicrob. Chemother. 1998, 42, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Evans, R.; Singh, B. Herbicidal inhibitors of amino acid biosynthesis and herbicide-tolerant crops. Amino Acids 2006, 30, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Liu, W.; Zhang, Y.; Liu, K. Action mechanisms of acetolactate synthase-inhibiting herbicides. Pestic. Biochem. Phys. 2007, 89, 89–96. [Google Scholar] [CrossRef]
- Yu, Q.; Zhang, X.; Hashem, A.; Walsh, M.J.; Powles, S.B. ALS gene proline (197) mutations confer ALS herbicide resistance in eight separated wild radish (Raphanus raphanistrum) populations. Weed Sci. 2017, 51, 831–838. [Google Scholar] [CrossRef]
- McCourt, J.A.; Pang, S.; King-Scott, J.; Guddat, L.W.; Duggleby, R.G. Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc. Natl. Acad. Sci. USA 2006, 103, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cheng, L.; Long, W.; Gao, J.; Zhang, J.; Chen, S.; Pu, H.; Hu, M. Synergistic mutations of two rapeseed AHAS genes confer high resistance to sulfonylurea herbicides for weed control. Theor. Appl. Genet. 2020, 133, 2811–2824. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, C.; Long, W.; Gao, J.; Zhang, J.; Chen, S.; Pu, H.; Hu, M. Development and molecular analysis of a novel acetohydroxyacid synthase rapeseed mutant with high resistance to sulfonylurea herbicides. Crop J. 2022, 10, 56–66. [Google Scholar] [CrossRef]
- Garcia, M.D.; Nouwens, A.; Lonhienne, T.G.; Guddat, L.W. Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families. Proc. Natl. Acad. Sci. USA 2017, 114, E1091–E1100. [Google Scholar] [CrossRef] [PubMed]
- Lonhienne, T.; Garcia, M.D.; Pierens, G.; Mobli, M.; Nouwens, A.; Guddat, L.W. Structural insights into the mechanism of inhibition of AHAS by herbicides. Proc. Natl. Acad. Sci. USA 2018, 115, E1945–E1954. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Wang, X. Acetohydroxyacid synthases: Evolution, structure, and function. Appl. Microbiol. Biot. 2016, 100, 8633–8649. [Google Scholar] [CrossRef] [PubMed]
- Duke, S.O.; Powles, S.B. Glyphosate: A once-in-a-century herbicide. Pest Manag. Sci. 2008, 64, 319–325. [Google Scholar] [CrossRef]
- Dill, G.M. Glyphosate-resistant crops: History, status and future. Pest Manag. Sci. 2005, 61, 219–224. [Google Scholar] [CrossRef]
- Herrmann, K.M.; Weaver, L.M. The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 473–503. [Google Scholar] [CrossRef]
- Schönbrunn, E.; Eschenburg, S.; Shuttleworth, W.A.; Schloss, J.V.; Amrhein, N.; Evans, J.N.; Kabsch, W. Interaction of the herbicide glyphosate with its target en-zyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. USA 2001, 98, 1376–1380. [Google Scholar] [CrossRef]
- Preston, C.; Wakelin, A.M. Resistance to glyphosate from altered herbicide translocation patterns. Pest Manag. Sci. 2007, 64, 372–376. [Google Scholar] [CrossRef]
- Light, S.H.; Krishna, S.N.; Minasov, G.; Anderson, W.F. An unusual cation-binding site and distinct domain-domain interactions distinguish class II enolpyruvylshikimate-3-phosphate synthases. Biochemistry 2016, 55, 1239–1245. [Google Scholar] [CrossRef]
- Chinnadurai, P.; Stojsin, D.; Liu, K.; Frierdich, G.E.; Glenn, K.C.; Geng, T.; Schapaugh, A.; Huang, K.; Deffenbaugh, A.E.; Liu, Z.; et al. Variability of CP4 EPSPS expression in genetically engineered soybean (Glycine max L. Merrill). Transgenic Res. 2018, 27, 511–524. [Google Scholar] [CrossRef]
- Ortega, J.L.; Rajapakse, W.; Bagga, S.; Apodaca, K.; Lucero, Y.; Sengupta-Gopalan, C. An intragenic approach to confer glyphosate resistance in chile (Capsicum annuum) by in-troducing an in vitro mutagenized chile EPSPS gene encoding for a glyphosate resistant EPSPS protein. PLoS ONE 2018, 13, e0194666. [Google Scholar] [CrossRef] [PubMed]
- Timmers, L.F.S.M.; Neto, A.M.S.; Montalvao, R.W.; Basso, L.A.; Santos, D.S.; de Souza, O.N. EPSP synthase flexibility is determinant to its function: Computational molecular dynamics and metadynamics studies. J. Mol. Model. 2017, 23, 197. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Borphukan, B.; Fartyal, D.; Ram, B.; Singh, J.; Manna, M.; Sheri, V.; Panditi, V.; Yadav, R.; Achary, V.M.M.; et al. Concurrent overexpression of OsGS1;1 and OsGS2 genes in transgenic rice (Oryza sativa L.): Impact on tolerance to abiotic stresses. Front. Plant Sci. 2018, 9, 786. [Google Scholar] [CrossRef] [PubMed]
- Risch, N.; Merikangas, K. The future of genetic studies of complex human diseases. Science 1996, 273, 1516–1517. [Google Scholar] [CrossRef]
- Avila-Garcia, W.V.; Sanchez-Olguin, E.; Hulting, A.G.; Mallory-Smith, C. Target-site mutation associated with glufosinate resistance in Italian ryegrass (Lolium perenne L. ssp multiflorum). Pest Manag. Sci. 2012, 68, 1248–1254. [Google Scholar] [CrossRef]
- Goodman, H.; DasSarma, S.; Tischer, E.; Peterman, T.K. Expression of Wild Type and Mutant Glutamine Synthetase in Foreign Hosts. U.S. Patent US5098838A, 24 July 1990. [Google Scholar]
- Deng, L.; Zhang, Z.; Lu, Y.; Fu, Y.; Tang, Y.; Xiang, R.; Feng, X.; Xu, N. Application and Cultivation of Glutamine Synthase Mutants with Glyphosate Resistance. China Patent, CN110229794A, 1 July 2019. [Google Scholar]
- Takano, H.K.; Fernandes, V.N.A.; Adegas, F.S.; Oliveira, R.S.; Westra, P.; Gaines, T.A.; Dayana, F.E. A novel TIPT double mutation in EPSPS conferring glyphosate resistance in tetraploid. Pest Manag. Sci. 2020, 76, 95–102. [Google Scholar] [CrossRef]
- Yang, Q.; Deng, W.; Wang, S.; Liu, H.; Li, X.; Zheng, M. Effects of resistance mutations of Pro197, Asp376 and Trp574 on the characteristics of acetohydroxyacid synthase (AHAS) isozymes. Pest Manag. Sci. 2018, 74, 1870–1879. [Google Scholar] [CrossRef]
- Perotti, V.E.; Larran, A.S.; Palmieri, V.E.; Martinatto, A.K.; Alvarez, C.E.; Tuesca, D.; Permingeat, H.R. A novel triple amino acid substitution in the EPSPS found in a high-level glyphosate-resistant Amaranthus hybridus population from Argentina. Pest Manag. Sci. 2019, 75, 1242–1251. [Google Scholar] [CrossRef]
- Rutledge, R.G.; Quellet, T.; Hattori, J.; Miki, B.L. Molecular characterization and genetic origin of the Brassica napus acetohydroxyacid synthase multigene family. Mol. Genet. Genom. 1991, 229, 31–40. [Google Scholar] [CrossRef]
- Wiersma, P.A.; Schmiemann, M.G.; Condie, J.A.; Crosby, W.L.; Moloney, M.M. Isolation, expression and phylogenetic inheritance of an acetolactate synthase gene from Brassica napus. Mol. Genet. Genom. 1989, 219, 413–420. [Google Scholar] [CrossRef]
- Ouellet, T.; Rutledge, R.G.; Miki, B.L. Members of the acetohydroxyacid synthase multigene family of Brassica napus have divergent patterns of expression. Plant J. 1992, 2, 321–330. [Google Scholar] [CrossRef]
- Huang, B.; Bird, S.; Kemble, R.; Simmonds, D.; Keller, W.; Miki, B. Effects of culture density, conditioned medium and feeder cultures on mi-crospore embryogenesis in Brassica napus L. cv. Topas. Plant Cell Rep. 1990, 8, 594–597. [Google Scholar] [CrossRef]
- Hu, M.; Pu, H.; Kong, L.; Gao, J.; Long, W.; Chen, S.; Zhang, J.; Qi, C. Molecular characterization and detection of a spontaneous mutation conferring imidazolinone resistance in rapeseed and its application in hybrid rapeseed production. Mol. Breeding 2015, 35, 46. [Google Scholar] [CrossRef]
- Xin, X.; Qu, G.; Zhang, R.; Pang, H.; Wu, Q.; Wang, F.; Hu, S. Identification of the tribenuron-methyl tolerance in different rapeseed genotypes. Acta Agric. Boreal.-Occident. Sin. 2014, 23, 68–74. [Google Scholar]
- Wang, Q.; Cui, C.; Ye, S.; Cui, M.; Zhao, Y.; Lin, N.; Tang, Z.; Li, J.; Zhou, Q. Screening and comprehensive evaluation of germplasm resources with tribe nuron-methyl tolerance at germination stage in rapeseed (Brassica napus L.). Acta Agron. Sin. 2018, 44, 1169–1184. [Google Scholar] [CrossRef]
- Li, L.; Fu, Y.; Xiao, G.; Guan, C. Selection and identification of herbicide resistan mutants in rapeseed. Chin. J. Oil Crop Sci. 2008, 30, 361–365. [Google Scholar]
- Chen, Z.; Wang, C.; Liu, W.; Tang, X.; Deng, X. Rice Herbicide-Resistant Protein and Its Application in Plant Breeding. China Patent, CN102586215B, 20 February 2012. [Google Scholar]
- Chen, L.; Gu, G.; Wang, C.; Chen, Z.; Yan, W.; Jin, M.; Xie, G.; Zhou, J.; Deng, X.; Tang, X. Trp548Met mutation of acetolactate synthase in rice confers resistance to a broad spectrum of ALS-inhibiting herbicides. Crop J. 2021, 9, 750–758. [Google Scholar] [CrossRef]
- Swanson, E.B.; Herrgesell, M.J.; Arnoldo, M.; Sippell, D.W.; Wong, R.S.C. Micro-spore mutagenesis and selection: Canola plants with field tolerance to the imidazolinones. Theor. Appl. Genet. 1989, 78, 525–530. [Google Scholar] [CrossRef]
- Tonnemaker, K.A.; Auld, D.L.; Thill, D.C.; Mallory-Smith, C.A.; Erickson, D.A. Develop-ment of sulfonylurea-resistant rapeseed using chemical mutagenesis. Crop Sci. 1992, 32, 1387–1391. [Google Scholar] [CrossRef]
- Hu, M.; Pu, H.; Gao, J.; Long, W.; Feng, C.; Zhou, X.; Wei, Z.; Qi, P.; Song, C.; Zhang, J. Inheritance and molecular characterization of resistance to AHAS-inhibiting herbicides in rapeseed. J. Integr. Agric. 2017, 16, 2421–2433. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Zhao, B.; Wang, J.; Yi, L.; Liu, C.; Wu, J.; King, G.J.; Liu, K. Generation and characterization of tribenuron-methyl herbicide resistant rapeseed (Brasscia napus) for hybrid seed production using chemically induced male sterility. Theor. Appl. Genet. 2015, 128, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Huang, Q.; Sun, Y.; Qu, G.; Guo, Y.; Zhang, X.; Zhao, H.; Hu, S. Male sterility of an AHAS-mutant induced by tribenuron-methyl solution correlated with the decrease of AHAS activity in Brassica napus L. Front. Plant. Sci. 2018, 9, 1014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, Q.; Wang, S.; Gao, L.; Qu, G.; Guo, Y.; Hu, Z.; Hu, S. Genetic characterization of the AHAS mutant line K4 with resistance to AHAS-inhibitor herbicides in rapeseed (Brassica napus L.). Correction in Stress Biology 2025, 5, 16. [Google Scholar]
- Wan, L.; Wang, Z.; Xin, Q.; Dong, F.; Hong, D.; Yang, G. Study on glyphosate-resistant GOX-CP4EPSP transformation of Brassica napus L. Hubei Agric. Sci. 2016, 55, 2661–2666. [Google Scholar]
- He, M.; Zeng, H.; Xu, P.; Ye, C. Cloning and optimisation of the aroA gene. Acta Sci. Nat. Univ. Sunyatseni 2002, 41, 76–79. [Google Scholar]
- Wang, J.; Zhao, F.; Xu, P.; Tian, Y. Development of transgenic oilseed plants resistant to glyphosate and insects. Acta Genet. Sin. 2005, 32, 1293–1300. [Google Scholar]
- Kahrizi, D.; Salmanian, A.H.; Afshari, A.; Moieni, A.; Mousavi, A. Simultaneous substitution of Gly96 to Ala and Ala183 to Thr in 5-enolpyruvylshikimate-3-phosphate synthase gene of E. coli (k12) and transformation of rapeseed (Brassica napus L.) in order to make tolerance to glyphosate. Plant Cell Rep. 2006, 26, 95–104. [Google Scholar] [CrossRef]
- Wu, X.; Yi, L.; Hou, F.; Wu, J.; Yao, X.; Liu, K. Generation of tribenuron-methyl herbicide resistant rapeseed (Brasscia napus) plants expressing mutated gene DsALS-108 of flixweed (Descurainia sophia). J. Agric. Biotechnol. 2016, 24, 469–477. [Google Scholar]
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011, 45, 247–271. [Google Scholar] [CrossRef]
- Wu, J.; Chen, C.; Xian, G.; Liu, D.; Lin, L.; Yin, S.; Sun, Q.; Fang, Y.; Zhang, H.; Wang, Y. Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnol. J. 2020, 18, 1857–1859. [Google Scholar] [CrossRef]
- Jugulam, M.; Shyam, C. Non-target-site resistance to herbicides: Recent developments. Plants 2019, 8, 417. [Google Scholar] [CrossRef]
- Rangani, G.; Rouse, C.E.; Saski, C.; Noorai, R.E.; Shankar, V.; Lawton-Rauh, A.L.; Werle, I.S.; Roma-Burgos, N. High resistance to quinclorac in multiple-resistant echinochloa colona associated with elevated stress tolerance gene expression and enriched xenobiotic detoxification pathway. Genes 2022, 13, 515. [Google Scholar] [CrossRef]
- Hu, M.; Pu, H.; Gao, J.Q.; Long, W.; Chen, F.; Zhang, W.; Zhou, X.; Peng, Q.; Chen, S.; Zhang, J. Comparative analysis of miRNAs of two rapeseed genotypes in response to acetohydroxyacid synthase-inhibiting herbicides by high-throughput sequencing. PLoS ONE 2017, 12, e0184917. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, T.; Lu, X.; Li, W.; Lv, X.; Peng, Q.; Zhang, J.; Gao, J.; Hu, M. Comparative genome-wide analysis of circular RNAs in Brassica napus L.: Target-site versus non-target-site resistance to herbicide stress. Theor. Appl. Genet. 2024, 137, 176. [Google Scholar] [CrossRef] [PubMed]
- Vega, T.; Gil, M.; Martin, G.; Moschen, S.; Picardi, L.; Nestares, G. Stress response and detoxification mechanisms involved in non-target-site herbicide resistance in sunflower. Crop Sci. 2020, 60, 1809–1822. [Google Scholar] [CrossRef]
- Délye, C. Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: A major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 2012, 69, 176–187. [Google Scholar] [CrossRef]
- Powles, S.B.; Yu, Q. Evolution in action: Plants resistant to herbicides. Annu. Rev. Plant. Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef]
- Maeda, H.; Murata, K.; Sakuma, N.; Takei, S.; Yamazaki, A.; Karim, M.D.; Kawata, M.; Hirose, S.; Kobayashi, M.K.; Tanigushi, Y.; et al. A rice gene that confers broad-spectrum resistance to b-triketone herbicides. Science 2019, 365, 393–396. [Google Scholar] [CrossRef]
- Su, D. Transformtiom of the glyphosate oxidoreductase gene to rape (Brasscia napus L.). Master’s Thesis, Inner Mongolia Agricultural University, Huhehaote, China, 2008. [Google Scholar]
- Block, M.D.; Brouwer, D.D.; Tenning, P. Transformation of Brassica napus and Brassica oleracea using Agrobacte-rium tumefaciens and the expression of the bar and neo genes in the transgenic plants. Plant Physiol. 1989, 91, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R. Molecular marker assisted breeding of transgenic herbicide-resistant lines in rapeseed (Brassica napus L.). Master’s Thesis, Northwest A&F University, Yangling, China, 2015. [Google Scholar]
- Yuan, J.; Tranel, P.J.; Stewart, C.N. Non-target-site herbicide resistance: A family business. Trends Plant Sci. 2007, 12, 6–13. [Google Scholar] [CrossRef]
- Pan, G.; Zhang, X.; Liu, K.; Zhang, J.; Wu, X.; Zhu, J.; Tu, J. Map-based cloning of a novel rice cytochrome P450 gene CYP81A6 that confers resistance to two different classes of herbicides. Plant Mol. Biol. 2006, 61, 933–943. [Google Scholar] [CrossRef]
- Saika, H.; Horita, J.; Taguchi-Shiobara, F.; Nonaka, S.; Nishizawa-Yokoi, A.; Iwakami, S.; Hori, K.; Matsumoto, T.; Tanaka, T.; Itoh, T.; et al. A novel rice cytochrome P450 Gene, CYP72A31, confers tolerance to acetolactate synthase-inhibiting herbicides in rice and Arabidopsis. Plant Physiol. 2014, 166, 1232–1240. [Google Scholar] [CrossRef]
- Siminszky, B.; Corbin, F.T.; Ward, E.R.; Fleischmann, T.J.; Dewey, R.E. Expression of a soybean cytochrome P450 monooxygenase cDNA in yeast and tobacco enhances the metabolism of phenylurea herbicides. Proc. Natl. Acad. Sci. USA 1999, 96, 1750–1755. [Google Scholar] [CrossRef]
- Brazier-Hicks, M.; Franco-Ortega, S.; Watson, P.; Rougemont, B.; Cohn, J.; Dale, R.; Hawkes, T.R.; Goldberg-Cavalleri, A.; Onkokesung, N.; Edwards, R. Characterization of cytochrome P450s with key roles in determining herbicide selectivity in maize. ACS Omega 2022, 7, 17416–17431. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Yu, Q.; Wang, J.; Han, H.; Mao, L.; Nyporko, A.; Maguza, A.; Fan, L.; Bai, L.; Powles, S. An ABCC-type transporter endowing glyphosate resistance in plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2100136118. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Powles, S. Metabolism-based herbicide resistance and cross-resistance in crop weeds: A threat to herbicide sustainability and global crop production. Plant Physiol. 2014, 166, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhang, H.; Kong, L.; Ma, J.; Wang, T.; Lu, X.; Guo, Y.; Zhang, J.; Guan, R.; Chu, P. Comparative proteomic and physiological analyses reveal tribenuron-methyl phytotoxicity and nontarget-site resistance mechanisms in Brassica napus. Plant Cell Environ. 2023, 46, 2255–2272. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, L.; Li, H.; He, J.; Chao, H.; Yan, S.; Yin, Y.; Zhao, W.; Li, M. 3D genome structural variations play important roles in regulating seed oil content of Brassica napus. Plant Commun. 2024, 5, 100666. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Z.; He, Y.; Liu, D.; Liu, Y.; Liang, C.; Xie, M.; Jia, Y.; Ke, Q.; Zhou, Y.; et al. Structural variation reshapes population gene expression and trait variation in 2,105 Brassica napus accessions. Nat. Genet. 2024, 56, 2538–2550. [Google Scholar] [CrossRef]
- Hu, J.; Chen, B.; Zhao, J.; Zhang, F.; Xie, T.; Xu, K.; Gao, G.; Yan, G.; Li, H.; Li, L.; et al. Genomic selection and genetic architecture of agronomic traits during modern rapeseed breeding. Nat. Genet. 2022, 54, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.A.; Gao, S.; Hassan, M.A.; Huang, Z.; Rasheed, A.; Hearne, S.; Prasanna, B.; Li, X.; Li, H. Artificial intelligence in plant breeding. Trends Genet. 2024, 40, 891–908. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Sun, G.; Xiao, J.; He, X.; Jiang, H.; Zhang, Z.; Zhang, Q.; Li, K.; Zhang, S.; Shi, X.; et al. AlphaFold-guided redesign of a plant pectin methylesterase inhibitor for broad-spectrum disease resistance. Mol. Plant 2024, 17, 1344–1368. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Yan, Z.; Bernardo, M.D.; Sgrizzi, S.R.; Villiger, L.; Kayabolen, A.; Kim, B.J.; Carscadden, J.K.; Hiraizumi, M.; Nishimasu, H.; et al. Rapid in silico directed evolution by a protein language model with EVOLVEpro. Science 2025, 387, eadr6006. [Google Scholar] [CrossRef]
- Ubbens, J.R.; Stavness, I. Deep plant phenomics: A deep learning platform for complex plant phenotyping tasks. Correction in Front. Plant Sci. 2017, 8, 276225. [Google Scholar] [CrossRef]
- Geetharamani, G.; Pandian, A. Identification of plantleaf diseases using a nine-layer deep convolutional neural network. Comput. Electr. Eng. 2019, 76, 323–338. [Google Scholar]
- Zhang, G.; Zhou, J.; Peng, Y.; Tan, Z.; Zhang, Y.; Zhao, H.; Liu, D.; Liu, X.; Li, L.; Yu, L.; et al. High-throughput phenotyping-based quantitative trait loci mapping reveals the genetic architecture of the salt stress tolerance of Brassica napus. Plant Cell Environ. 2023, 46, 549–566. [Google Scholar] [CrossRef]
- Juliana, P.; Poland, J.; Huerta-Espino, J.; Shrestha, S.; Crossa, J.; Crespo-Herrera, L.; Toledo, F.H.; Govindan, V.; Mondal, S.; Kumar, U.; et al. Improving grain yield, stress resilienceand quality of bread wheat using large-scale genomics. Nat. Genet. 2019, 51, 1530–1539. [Google Scholar] [CrossRef]
- Romero Navarro, J.A.; Phillips-Mora, W.; Arciniegas-Leal, A.; Mata-Quirós, A.; Haiminen, N.; Mustiga, G.; Livingstone Iii, D.; van Bakel, H.; Kuhn, D.N.; Parida, L.; et al. Application of genome wide association and genomic prediction for improvement of cacao productivity and resistance to black and frosty pod diseases. Front. Plant Sci. 2017, 8, 1905. [Google Scholar] [CrossRef]
- Liu, W.; Stewart, C.N. Plant synthetic biology. Trends Plant Sci. 2015, 20, 309–317. [Google Scholar] [CrossRef]
- Zhang, D.; Xu, F.; Wang, F.; Le, L.; Pu, L. Synthetic biology and artificial intelligence in crop improvement. Plant Commun. 2025, 6, 101220. [Google Scholar] [CrossRef]
- Xu, J.; Lei, Y.; Zhang, X.; Li, J.; Lin, Q.; Wu, X.; Jiang, Y.; Zhang, W.; Qian, R.; Xiong, S.; et al. Design of CoQ10 crops based on evolutionary history. Cell 2025, 188, 1941–1954.e15. [Google Scholar] [CrossRef]
- Prywes, N.; Phillips, N.R.; Oltrogge, L.; Lindner, S.; Taylor-Kearney, L.; Tsai, Y.C.; de Pins, B.; Cowan, A.E.; Chang, H.; Wang, R.; et al. A map of the rubisco biochemical landscape. Correction in Nature 2025, 638, 823–828. [Google Scholar] [CrossRef]
- Wang, X.; Pan, W.; Sun, C.; Yang, H.; Cheng, Z.; Yan, F.; Ma, G.; Shang, Y.; Zhang, R.; Gao, C.; et al. Creating large-scale genetic diversity in Arabidopsis via base editing-mediated deep artificial evolution. Genome Biol. 2024, 25, 215. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Qu, Y.; Li, M.; Song, X.; Huang, Y. Genetic diversity, genetic structure and migration routes of wild Brassica juncea in China assessed by SSR markers. Genet. Resour. Crop Ev. 2018, 65, 1581–1590. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, W.; Ma, K.P.; Darmency, H. Backcrosses to Brassica napus of hybrids between B. juncea and B. napus as a source of herbicide-resistant volunteer-like feral populations. Plant Sci. 2010, 179, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, Z.; Zuo, J.; Huangfu, C.; Qiang, S. Potential gene flow of two herbicide-tolerant transgenes from oilseed rape to wild, B. juncea var. gracilis. Theor. Appl. Genet. 2010, 120, 1501–1510. [Google Scholar] [CrossRef]
- Yoshimura, Y.; Beckie, H.; Matsuo, K. Transgenic oilseed rape along transportation routes and port of Vancouver in western Canada. Environ. Biosaf. Res. 2006, 5, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Knispel, A.L.; McLachlan, S.M. Landscape-scale distribution and persistence of genetically modified oilseed rape (Brassica napus) in Manitoba, Canada. Environ. Sci. Pollut. R. 2010, 17, 13–25. [Google Scholar] [CrossRef]
- Beckie, H.J.; Harker, K.N.; Hall, L.M.; Holm, F.A.; Gulden, R.H. Risk assessment of glyphosate resistance in western Canada. Weed Technol. 2011, 25, 159–164. [Google Scholar] [CrossRef]
- Munier, D.J.; Brittan, K.L.; Lanini, W.T. Seed bank persistence of genetically modified canola in California. Environ. Sci. Pollut. R. 2012, 19, 2281–2284. [Google Scholar] [CrossRef] [PubMed]
- Ureta, M.S.; Carbonell, F.T.; Pandolfo, C.; Presotto, A.D.; Cantamutto, M.A.; Poverene, M. IMI resistance associated with crop-weed hybridization in a natural Brassica rapa population: Characterization and fate. Environ. Monit. Assess. 2017, 189, 101. [Google Scholar] [CrossRef] [PubMed]
- Pandolfo, C.E.; Presotto, A.; Carbonell, F.T.; Ureta, S.; Poverene, M.; Cantamutto, M. Transgenic glyphosate-resistant oilseed rape (Brassica napus) as an invasive weed in Argentina: Detection, characterization, and control alternatives. Environ. Sci. Pollut. R. 2016, 23, 24081–24091. [Google Scholar] [CrossRef]
- Busi, R.; Powles, S.B. Transgenic glyphosate-resistant canola (Brassica napus) can persist outside agricultural fields in Australia. Agr. Ecosyst. Environ. 2016, 220, 28–34. [Google Scholar] [CrossRef]
- Aono, M.; Wakiyama, S.; Nagatsu, M.; Nakajima, N.; Tamaoki, M.; Kubo, A.; Saji, H. Detection of feral transgenic oilseed rape with multiple-herbicide resistance in Japan. Environ. Biosaf. Res. 2006, 5, 77–87. [Google Scholar] [CrossRef]
- Devos, Y.; Hails, R.S.; Messéan, A.; Perry, J.N.; Squire, G.R. Feral genetically modified herbicide tolerant oilseed rape from seed import spills: Are concerns scientifically justified? Transgenic Res. 2012, 21, 1–21. [Google Scholar] [CrossRef]
- Schulze, J.; Frauenknecht, T.; Brodmann, P.; Bagutti, C. Unexpected diversity of feral genetically modified oilseed rape (Brassica napus L.) despite a cultivation and import ban in Switzerland. PLoS ONE 2014, 9, e114477. [Google Scholar] [CrossRef]
- Heap, I. The International Survey of Herbicide Resistant Weeds. Available online: www.weedscience.org (accessed on 1 March 2024).

| Materials | Methods | Genes | Herbicides Resistance | Amino Acid Resistant Mutations | Resistance Level Data | Reference |
|---|---|---|---|---|---|---|
| PM1 | ENU | ALS1 | IMI | Ser653Asp | 50 g ha−1 | [52] |
| PM2 | ENU | ALS3 | IMI, SU, TP | Trp574Leu | 300 g ha−1 | [52] |
| M9 | Spontaneous mutation | ALS1R | IMI | Ser653Asn | 90 g ha−1 | [46] |
| PN19 | EMS | ALS1 | SU, SCT | Trp574Leu | 50 g ha−1 | [20] |
| M342 | EMS | ALS3R | SU | Trp574Leu | 90 g ha−1 | [54] |
| 5N | M342 × PN19 | ALS1 | SU | Trp574Leu | 360 g ha−1 | [20] |
| ALS3 | ||||||
| K5 | EMS | ALS1 | SU | Pro197Ser | 15 g ha−1 | [57] |
| DS3 | EMS | ALS1 | SU | Pro197Ser | 270–360 g ha−1 | [21] |
| ALS3 | Trp574Leu | |||||
| 12WH318 | Introduction | ALS1 | IMI | Ser653Asp | 72–108 g ha−1 | |
| M45, K1, K4 | EMS | ALS3 | SU | Pro197Ser/Leu | 3, 0.6, 0.6 g ha−1 | [56] |
| ALS1 mutation | In vitro mutation | ALS1 | IMI | Ser653 deletion | - | |
| RCS-5 | Spontaneous mutation | - | IMI, SU | - | 10 g ha−1 | |
| M-37, M-42 | ENU | - | SU | - | 30 g ha−1 |
| Materials | Genes | Transgenic Source | Herbicides Resistance | Reference |
|---|---|---|---|---|
| CT73/GT200 | CP4-EPSPS+goxv247 | Agrobacterium sp. CP4+Pseudomonas sp. | Glyphosate | [58] |
| MON88302 | CP4-EPSPS | Agrobacterium sp. CP4 | Glyphosate | [58] |
| XT | aroAM12 | Salmonella typhimurium | Glyphosate | [59] |
| LB-FLFK | AtALS | Arabidopsis thaliana | IMI | [61] |
| DsALS-108-26 | DsALS-108 | Descurainia sophia | TBM | [62] |
| P197S mutants | BnALS1 | Brassica napus | TBM | [64] |
| BnALS1/BnALS3 | Brassica napus | TBM | [64] |
| Development Stage | Breeding Strategies | Benefits | Challenges |
|---|---|---|---|
| First | Natural mutation | Germplasm can be directly used | Un-predictability |
| Second | Artificial mutation | Targeted improvement of a single trait | Lengthy genetic improvement cycles |
| Third | Genetic modification | Simplicity and efficiency | Introduction of foreign genes |
| Gene editing | High efficiency and short cycle | Off-target risk | |
| Fourth | Artificial intelligence | Efficient and intelligent | The technology is still immature |
| Materials | Genes | Transgenic Source | Herbicides Resistance | Reference |
|---|---|---|---|---|
| 61061/73496 | gat4621 | Bacillus licheniformis | Glyphosate | |
| Transgenic plant | gox | Pseudomonas sp. | Glyphosate | [73] |
| MS8 | bar | Streptomyces hygroscopicus | Glufosinate | |
| RF1 | bar | Streptomyces hygroscopicus | Glufosinate | |
| MS1 | bar | Streptomyces hygroscopicus | Glufosinate | |
| PGS1 | bar | Streptomyces hygroscopicus | Glufosinate | |
| HCN10 | bar | Streptomyces hygroscopicus | Glufosinate | |
| PHY14 | bar | Streptomyces hygroscopicus | Glufosinate | |
| 15A | bar | Streptomyces hygroscopicus | Glufosinate | |
| 742R | bar | Streptomyces hygroscopicus | Glufosinate | [63] |
| T45 | pat | Streptomyces viridochromo | Glufosinate | [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Yu, S.; Ji, B.; Peng, Q.; Gao, J.; Zhang, J.; Guo, Y.; Hu, M. Molecular Mechanisms of Herbicide Resistance in Rapeseed: Current Status and Future Prospects for Resistant Germplasm Development. Int. J. Mol. Sci. 2025, 26, 8292. https://doi.org/10.3390/ijms26178292
Liu D, Yu S, Ji B, Peng Q, Gao J, Zhang J, Guo Y, Hu M. Molecular Mechanisms of Herbicide Resistance in Rapeseed: Current Status and Future Prospects for Resistant Germplasm Development. International Journal of Molecular Sciences. 2025; 26(17):8292. https://doi.org/10.3390/ijms26178292
Chicago/Turabian StyleLiu, Decai, Shicheng Yu, Biaojun Ji, Qi Peng, Jianqin Gao, Jiefu Zhang, Yue Guo, and Maolong Hu. 2025. "Molecular Mechanisms of Herbicide Resistance in Rapeseed: Current Status and Future Prospects for Resistant Germplasm Development" International Journal of Molecular Sciences 26, no. 17: 8292. https://doi.org/10.3390/ijms26178292
APA StyleLiu, D., Yu, S., Ji, B., Peng, Q., Gao, J., Zhang, J., Guo, Y., & Hu, M. (2025). Molecular Mechanisms of Herbicide Resistance in Rapeseed: Current Status and Future Prospects for Resistant Germplasm Development. International Journal of Molecular Sciences, 26(17), 8292. https://doi.org/10.3390/ijms26178292








