Systematic Analysis of Alternative Splicing in Transcriptomes of Multiple Sclerosis Patient Brain Samples
Abstract
1. Introduction
2. Results
2.1. Differentially Expressed and Alternatively Spliced Genes Showed Low Percentages of Overlap and High Variety in Enriched Pathways
2.2. Diagnosis Comparisons
2.3. Brain Region Comparisons
2.4. Tissue Type Comparisons
2.5. Cell Type Comparisons
2.5.1. Microglia
2.5.2. CD4/CD8 T-Cells
3. Discussion
4. Materials and Methods
4.1. Data Access
4.2. Differential Expression Analysis
4.3. Alternative Splicing Analysis
4.4. Pearson Correlation Analysis
4.5. NEASE (Network Enrichment Method for Alternative Splicing Events) and Domain Interaction Graph Guided ExploreR (DIGGER)
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M.; et al. Mechanism of Alternative Splicing and Its Regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef]
- Chen, S.; Benbarche, S.; Abdel-Wahab, O. Splicing Factor Mutations in Hematologic Malignancies. Blood 2021, 138, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Dam, S.H.; Olsen, L.R.; Vitting-Seerup, K. Expression and Splicing Mediate Distinct Biological Signals. BMC Biol. 2023, 21, 220. [Google Scholar] [CrossRef] [PubMed]
- Innes, P.A.; Goebl, A.M.; Smith, C.C.R.; Rosenberger, K.; Kane, N.C. Gene Expression and Alternative Splicing Contribute to Adaptive Divergence of Ecotypes. Heredity 2024, 132, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Sak, M.; Chariker, J.H.; Park, J.W.; Rouchka, E.C. Gene Expression and Alternative Splicing Analysis in a Large-Scale Multiple Sclerosis Study. Int. J. Mol. Sci. 2024, 25, 11957. [Google Scholar] [CrossRef]
- Bebo, B.; Cintina, I.; LaRocca, N.; Ritter, L.; Talente, B.; Hartung, D.; Ngorsuraches, S.; Wallin, M.; Yang, G. The Economic Burden of Multiple Sclerosis in the United States: Estimate of Direct and Indirect Costs. Neurology 2022, 98, e1810–e1817. [Google Scholar] [CrossRef]
- Walton, C.; King, R.; Rechtman, L.; Kaye, W.; Leray, E.; Marrie, R.A.; Robertson, N.; La Rocca, N.; Uitdehaag, B.; van der Mei, I.; et al. Rising Prevalence of Multiple Sclerosis Worldwide: Insights from the Atlas of Ms, Third Edition. Mult. Scler. J. 2020, 26, 1816–1821. [Google Scholar] [CrossRef]
- International Multiple Sclerosis Genetics Consortium. Multiple Sclerosis Genomic Map Implicates Peripheral Immune Cells and Microglia in Susceptibility. Science 2019, 365, eaav7188. [Google Scholar] [CrossRef]
- Putscher, E.; Hecker, M.; Fitzner, B.; Boxberger, N.; Schwartz, M.; Koczan, D.; Lorenz, P.; Zettl, U.K. Genetic Risk Variants for Multiple Sclerosis Are Linked to Differences in Alternative Pre-Mrna Splicing. Front. Immunol. 2022, 13, 931831. [Google Scholar] [CrossRef]
- Reder, A.T.; Goel, A.; Garcia, T.; Feng, X. Alternative Splicing of Rna Is Excessive in Multiple Sclerosis and Not Linked to Gene Expression Levels: Dysregulation Is Corrected by Ifn-Beta. J. Interferon Cytokine Res. 2024, 44, 355–371. [Google Scholar] [CrossRef]
- Mohseni, S.O.; Au, K.M.; Issa, W.; Ruan, L.; Stuve, O.; Wang, A.Z. Multiple Sclerosis Treatments a Review of Current Biomedical Engineering Approaches. Biomaterials 2025, 313, 122807. [Google Scholar] [CrossRef]
- Ascherio, A.; Munger, K.L. Epidemiology of Multiple Sclerosis: From Risk Factors to Prevention-an Update. Semin. Neurol. 2016, 36, 103–114. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. Ncbi Geo: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- van der Poel, M.; Ulas, T.; Mizee, M.R.; Hsiao, C.C.; Miedema, S.S.M.; Adelia, N.; Schuurman, K.G.; Helder, B.; Tas, S.W.; Schultze, J.L.; et al. Transcriptional Profiling of Human Microglia Reveals Grey-White Matter Heterogeneity and Multiple Sclerosis-Associated Changes. Nat. Commun. 2019, 10, 1139. [Google Scholar] [CrossRef]
- Voskuhl, R.R.; Itoh, N.; Tassoni, A.; Matsukawa, M.A.; Ren, E.; Tse, V.; Jang, E.; Suen, T.T.; Itoh, Y. Gene Expression in Oligodendrocytes during Remyelination Reveals Cholesterol Homeostasis as a Therapeutic Target in Multiple Sclerosis. Proc. Natl. Acad. Sci. USA 2019, 116, 10130–10139. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Lorenzo, S.; Konings, J.; van der Pol, S.; Kamermans, A.; Amor, S.; van Horssen, J.; Witte, M.E.; Kooij, G.; de Vries, H.E. Inflammation of the Choroid Plexus in Progressive Multiple Sclerosis: Accumulation of Granulocytes and T Cells. Acta Neuropathol. Commun. 2020, 8, 9, Erratum in Acta Neuropathol. Commun. 2020, 8, 24. [Google Scholar] [CrossRef]
- Elkjaer, M.L.; Frisch, T.; Reynolds, R.; Kacprowski, T.; Burton, M.; Kruse, T.A.; Thomassen, M.; Baumbach, J.; Illes, Z. Molecular Signature of Different Lesion Types in the Brain White Matter of Patients with Progressive Multiple Sclerosis. Acta Neuropathol. Commun. 2019, 7, 205. [Google Scholar] [CrossRef] [PubMed]
- van Wageningen, T.A.; Gerrits, E.; Brouwer, N.; Breve, J.J.P.; Geurts, J.J.G.; Eggen, B.J.L.; Boddeke, H.; van Dam, A.M. Distinct Gene Expression in Demyelinated White and Grey Matter Areas of Patients with Multiple Sclerosis. Brain Commun. 2022, 4, fcac005. [Google Scholar] [CrossRef] [PubMed]
- Miedema, A.; Gerrits, E.; Brouwer, N.; Jiang, Q.; Kracht, L.; Meijer, M.; Nutma, E.; Peferoen-Baert, R.; Pijnacker, A.T.E.; Wesseling, E.M.; et al. Brain Macrophages Acquire Distinct Transcriptomes in Multiple Sclerosis Lesions and Normal Appearing White Matter. Acta Neuropathol. Commun. 2022, 10, 8. [Google Scholar] [CrossRef]
- Salapa, H.E.; Thibault, P.A.; Libner, C.D.; Ding, Y.; Clarke, J.W.E.; Denomy, C.; Hutchinson, C.; Abidullah, H.M.; Hammond, S.A.; Pastushok, L.; et al. Hnrnp A1 Dysfunction Alters Rna Splicing and Drives Neurodegeneration in Multiple Sclerosis (Ms). Nat. Commun. 2024, 15, 356. [Google Scholar] [CrossRef]
- Jansen, M.; Castorina, A. Transcriptomic Analyses of Normal-Appearing Cns White Matter from Multiple Sclerosis Donors Reveal Subtype-Specific Molecular Signatures of Disease; National Center for Biotechnology Information: Bethesda, MD, USA, 2022.
- Hsiao, C.C.; Engelenburg, H.J.; Jongejan, A.; Zhu, J.; Zhang, B.; Mingueneau, M.; Moerland, P.D.; Huitinga, I.; Smolders, J.; Hamann, J. Osteopontin Associates with Brain T(Rm)-Cell Transcriptome and Compartmentalization in Donors with and without Multiple Sclerosis. iScience 2023, 26, 105785. [Google Scholar] [CrossRef] [PubMed]
- Tiane, A.; Schepers, M.; Reijnders, R.A.; van Veggel, L.; Chenine, S.; Rombaut, B.; Dempster, E.; Verfaillie, C.; Wasner, K.; Grunewald, A.; et al. From Methylation to Myelination: Epigenomic and Transcriptomic Profiling of Chronic Inactive Demyelinated Multiple Sclerosis Lesions. Acta Neuropathol. 2023, 146, 283–299. [Google Scholar] [CrossRef] [PubMed]
- van den Bosch, A.M.R.; van der Poel, M.; Fransen, N.L.; Vincenten, M.C.J.; Bobeldijk, A.M.; Jongejan, A.; Engelenburg, H.J.; Moerland, P.D.; Smolders, J.; Huitinga, I.; et al. Profiling of Microglia Nodules in Multiple Sclerosis Reveals Propensity for Lesion Formation. Nat. Commun. 2024, 15, 1667. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology Knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Qi, S.; Gao, L.; Huang, G.; Ren, Z.; Li, K.; Peng, Y.; Yi, G.; Guo, J.; et al. Spliceosome-Regulated Rsrp1-Dependent Nf-Kappab Activation Promotes the Glioblastoma Mesenchymal Phenotype. Neuro Oncol. 2021, 23, 1693–1708. [Google Scholar] [CrossRef]
- Lindstrom, M.S.; Nister, M. Silencing of Ribosomal Protein S9 Elicits a Multitude of Cellular Responses Inhibiting the Growth of Cancer Cells Subsequent to P53 Activation. PLoS ONE 2010, 5, e9578. [Google Scholar] [CrossRef]
- Pourhaghighi, R.; Ash, P.E.A.; Phanse, S.; Goebels, F.; Hu, L.Z.M.; Chen, S.; Zhang, Y.; Wierbowski, S.D.; Boudeau, S.; Moutaoufik, M.T.; et al. Brainmap Elucidates the Macromolecular Connectivity Landscape of Mammalian Brain. Cell Syst. 2020, 10, 333–350.E14. [Google Scholar] [CrossRef]
- Zhu, Z.; Ni, S.; Zhang, J.; Yuan, Y.; Bai, Y.; Yin, X.; Zhu, Z. Genome-Wide Analysis of Dysregulated Rna-Binding Proteins and Alternative Splicing Genes in Keloid. Front. Genet. 2023, 14, 1118999. [Google Scholar] [CrossRef]
- Koch, M.W.; Cutter, G.; Stys, P.K.; Yong, V.W.; Metz, L.M. Treatment Trials in Progressive Ms—Current Challenges and Future Directions. Nat. Rev. Neurol. 2013, 9, 496–503. [Google Scholar] [CrossRef]
- Ozturk, A.; Smith, S.A.; Gordon-Lipkin, E.M.; Harrison, D.M.; Shiee, N.; Pham, D.L.; Caffo, B.S.; Calabresi, P.A.; Reich, D.S. Mri of the Corpus Callosum in Multiple Sclerosis: Association with Disability. Mult. Scler. J. 2010, 16, 166–177. [Google Scholar] [CrossRef]
- De Rossi, P.; Buggia-Prevot, V.; Clayton, B.L.; Vasquez, J.B.; van Sanford, C.; Andrew, R.J.; Lesnick, R.; Botte, A.; Deyts, C.; Salem, S.; et al. Predominant Expression of Alzheimer’s Disease-Associated Bin1 in Mature Oligodendrocytes and Localization to White Matter Tracts. Mol. Neurodegener. 2016, 11, 59. [Google Scholar] [CrossRef]
- Lavon, I.; Leykin, I.; Charbit, H.; Binyamin, O.; Brill, L.; Ovadia, H.; Vaknin-Dembinsky, A. Qki-V5 Is Downregulated in Cns Inflammatory Demyelinating Diseases. Mult. Scler. Relat. Disord. 2020, 39, 101881. [Google Scholar] [CrossRef]
- Louadi, Z.; Elkjaer, M.L.; Klug, M.; Lio, C.T.; Fenn, A.; Illes, Z.; Bongiovanni, D.; Baumbach, J.; Kacprowski, T.; List, M.; et al. Functional Enrichment of Alternative Splicing Events with Nease Reveals Insights into Tissue Identity and Diseases. Genome Biol. 2021, 22, 327. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Steinacker, P.; Seubert, S.; Turnescu, T.; Melms, A.; Manzel, A.; Otto, M.; Linker, R.A. Role of Glial 14-3-3 Gamma Protein in Autoimmune Demyelination. J. Neuroinflamm. 2015, 12, 187. [Google Scholar] [CrossRef] [PubMed]
- Agliardi, C.; Guerini, F.R.; Zanzottera, M.; Bolognesi, E.; Picciolini, S.; Caputo, D.; Rovaris, M.; Pasanisi, M.B.; Clerici, M. Myelin Basic Protein in Oligodendrocyte-Derived Extracellular Vesicles as a Diagnostic and Prognostic Biomarker in Multiple Sclerosis: A Pilot Study. Int. J. Mol. Sci. 2023, 24, 894. [Google Scholar] [CrossRef] [PubMed]
- Schlauch, K.A.; Khaiboullina, S.F.; De Meirleir, K.L.; Rawat, S.; Petereit, J.; Rizvanov, A.A.; Blatt, N.; Mijatovic, T.; Kulick, D.; Palotas, A.; et al. Genome-Wide Association Analysis Identifies Genetic Variations in Subjects with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Transl. Psychiatry 2016, 6, e730. [Google Scholar] [CrossRef]
- Nishida, K.; Matsumura, K.; Tamura, M.; Nakamichi, T.; Shimamori, K.; Kuragano, M.; Kabir, A.M.R.; Kakugo, A.; Kotani, S.; Nishishita, N.; et al. Effects of Three Microtubule-Associated Proteins (Map2, Map4 and Tau) on Microtubules’ Physical Properties and Neurite Morphology. Sci. Rep. 2023, 13, 8870. [Google Scholar] [CrossRef]
- Rahmani, B.; Fekrmandi, F.; Ahadi, K.; Ahadi, T.; Alavi, A.; Ahmadiani, A.; Asadi, S. A Novel Nonsense Mutation in Wnk1/Hsn2 Associated with Sensory Neuropathy and Limb Destruction in Four Siblings of a Large Iranian Pedigree. BMC Neurol. 2018, 18, 195. [Google Scholar] [CrossRef]
- Bampton, A.; Gittings, L.M.; Fratta, P.; Lashley, T.; Gatt, A. The Role of Hnrnps in Frontotemporal Dementia and Amyotrophic Lateral Sclerosis. Acta Neuropathol. 2020, 140, 599–623. [Google Scholar] [CrossRef]
- Prat, E.; Tomaru, U.; Sabater, L.; Park, D.M.; Granger, R.; Kruse, N.; Ohayon, J.M.; Bettinotti, M.P.; Martin, R. Hla-Drb5*0101 and -Drb1*1501 Expression in the Multiple Sclerosis-Associated Hla-Dr15 Haplotype. J. Neuroimmunol. 2005, 167, 108–119. [Google Scholar] [CrossRef]
- Lundmark, F.; Duvefelt, K.; Hillert, J. Genetic Association Analysis of the Interleukin 7 Gene (Il7) in Multiple Sclerosis. J. Neuroimmunol. 2007, 192, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Gregory, S.G.; Schmidt, S.; Seth, P.; Oksenberg, J.R.; Hart, J.; Prokop, A.; Caillier, S.J.; Ban, M.; Goris, A.; Barcellos, L.F.; et al. Interleukin 7 Receptor Alpha Chain (Il7r) Shows Allelic and Functional Association with Multiple Sclerosis. Nat. Genet. 2007, 39, 1083–1091. [Google Scholar] [CrossRef]
- Martin, R.; Sospedra, M.; Eiermann, T.; Olsson, T. Multiple Sclerosis: Doubling down on Mhc. Trends Genet. 2021, 37, 784–797. [Google Scholar] [CrossRef]
- Peerlings, D.; Mimpen, M.; Damoiseaux, J. The Il-2–Il-2 Receptor Pathway: Key to Understanding Multiple Sclerosis. J. Transl. Autoimmun. 2021, 4, 100123. [Google Scholar] [CrossRef]
- Pittaluga, A. Ccl5-Glutamate Cross-Talk in Astrocyte-Neuron Communication in Multiple Sclerosis. Front. Immunol. 2017, 8, 1079. [Google Scholar] [CrossRef]
- Salim, A.A.; Alsaimary, I.E.; Alsudany, A.A.K. The Role of Chemokines (Ccl2, Ccl5 and Cxcl10) in the Neuroinflammation among Patients with Multiple Sclerosis. Mult. Scler. Relat. Disord. 2023, 80, 105174. [Google Scholar] [CrossRef]
- Thangapandian, J. Insilico Analysis of Role Played by Cd3d Gene in the Pathogenesis of Multiple Sclerosis. Res. Sq. 2022, preprint. [Google Scholar] [CrossRef]
- Hu, F.; Zhu, Y.; Tian, J.; Xu, H.; Xue, Q. Single-Cell Sequencing Combined with Transcriptome Sequencing Constructs a Predictive Model of Key Genes in Multiple Sclerosis and Explores Molecular Mechanisms Related to Cellular Communication. J. Inflamm. Res. 2024, 17, 191–210. [Google Scholar] [CrossRef]
- Mai, S.; Qu, X.; Li, P.; Ma, Q.; Cao, C.; Liu, X. Global Regulation of Alternative Rna Splicing by the Sr-Rich Protein Rbm39. Biochim. Biophys. Acta 2016, 1859, 1014–1024. [Google Scholar] [CrossRef]
- Amann-Zalcenstein, D.; Avidan, N.; Kanyas, K.; Ebstein, R.P.; Kohn, Y.; Hamdan, A.; Ben-Asher, E.; Karni, O.; Mujaheed, M.; Segman, R.H.; et al. Ahi1, a Pivotal Neurodevelopmental Gene, and C6orf217 Are Associated with Susceptibility to Schizophrenia. Eur. J. Hum. Genet. 2006, 14, 1111–1119, Erratum in Eur. J. Hum. Genet. 2007, 15, 387. [Google Scholar] [CrossRef] [PubMed]
- Paez-Gonzalez, P.; Abdi, K.; Luciano, D.; Liu, Y.; Soriano-Navarro, M.; Rawlins, E.; Bennett, V.; Garcia-Verdugo, J.M.; Kuo, C.T. Ank3-Dependent Svz Niche Assembly Is Required for the Continued Production of New Neurons. Neuron 2011, 71, 61–75. [Google Scholar] [CrossRef]
- Zhou, X.; Qiao, B. Inhibition of Hdac3 and Atxn3 by Mir-25 Prevents Neuronal Loss and Ameliorates Neurological Recovery in Cerebral Stroke Experimental Rats. J. Physiol. Biochem. 2022, 78, 139–149. [Google Scholar] [CrossRef]
- Grohmann, K.; Rossoll, W.; Kobsar, I.; Holtmann, B.; Jablonka, S.; Wessig, C.; Stoltenburg-Didinger, G.; Fischer, U.; Hubner, C.; Martini, R.; et al. Characterization of Ighmbp2 in Motor Neurons and Implications for the Pathomechanism in a Mouse Model of Human Spinal Muscular Atrophy with Respiratory Distress Type 1 (Smard1). Hum. Mol. Genet. 2004, 13, 2031–2042. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Nagaraj, S.; Kucukali, F.; de Fisenne, M.A.; Kosa, A.C.; Doeraene, E.; Gutierrez, L.L.; Brion, J.P.; Leroy, K. Picalm and Alzheimer’s Disease: An Update and Perspectives. Cells 2022, 11, 3994. [Google Scholar] [CrossRef] [PubMed]
- Elleuch, N.; Depienne, C.; Benomar, A.; Hernandez, A.M.; Ferrer, X.; Fontaine, B.; Grid, D.; Tallaksen, C.M.; Zemmouri, R.; Stevanin, G.; et al. Mutation Analysis of the Paraplegin Gene (Spg7) in Patients with Hereditary Spastic Paraplegia. Neurology 2006, 66, 654–659. [Google Scholar] [CrossRef]
- Wirgenes, K.V.; Sonderby, I.E.; Haukvik, U.K.; Mattingsdal, M.; Tesli, M.; Athanasiu, L.; Sundet, K.; Rossberg, J.I.; Dale, A.M.; Brown, A.A.; et al. Tcf4 Sequence Variants and Mrna Levels Are Associated with Neurodevelopmental Characteristics in Psychotic Disorders. Transl. Psychiatry 2012, 2, e112. [Google Scholar] [CrossRef]
- Xuan, X.; Ruan, J.; Wu, C.; Gao, Y.; Li, L.; Lei, X. A Ttc19 Mutation Associated with Progressive Movement Disorders and Peripheral Neuropathy: Case Report and Systematic Review. CNS Neurosci. Ther. 2024, 30, e14425. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Torra, I.; Siddique, S.; Waddington, K.E.; Farrell, R.; Jury, E.C. Disrupted Lipid Metabolism in Multiple Sclerosis: A Role for Liver X Receptors? Front. Endocrinol. 2021, 12, 639757. [Google Scholar] [CrossRef] [PubMed]
- Franklin, R.J.M.; Simons, M. Cns Remyelination and Inflammation: From Basic Mechanisms to Therapeutic Opportunities. Neuron 2022, 110, 3549–3565. [Google Scholar] [CrossRef]
- Camargo, N.; Goudriaan, A.; van Deijk, A.F.; Otte, W.M.; Brouwers, J.F.; Lodder, H.; Gutmann, D.H.; Nave, K.A.; Dijkhuizen, R.M.; Mansvelder, H.D.; et al. Oligodendroglial Myelination Requires Astrocyte-Derived Lipids. PLoS Biol. 2017, 15, e1002605. [Google Scholar] [CrossRef]
- Mironova, Y.A.; Lin, J.P.; Kalinski, A.L.; Huffman, L.D.; Lenk, G.M.; Havton, L.A.; Meisler, M.H.; Giger, R.J. Protective Role of the Lipid Phosphatase Figure 4 in the Adult Nervous System. Hum. Mol. Genet. 2018, 27, 2443–2453. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, M.; Chen, G.; Lin, Y.; Kang, D.; Yu, L. Identifying Msmo1, Elovl6, Aacs, and Cers2 Related to Lipid Metabolism as Biomarkers of Parkinson’s Disease. Sci. Rep. 2024, 14, 17478. [Google Scholar] [CrossRef]
- Degraeve, B.; Sequeira, H.; Mecheri, H.; Lenne, B. Corpus Callosum Damage to Account for Cognitive, Affective, and Social-Cognitive Dysfunctions in Multiple Sclerosis: A Model of Callosal Disconnection Syndrome? Mult. Scler. J. 2023, 29, 160–168. [Google Scholar] [CrossRef]
- Qiu, G.; Cao, L.; Chen, Y.J. Novel Heterozygous Mutation in Alpha-2-Macroglobulin (A2m) Suppressing the Binding of Amyloid-Beta (Abeta). Front. Neurol. 2022, 13, 1090900. [Google Scholar]
- Mohammadi, S.; Sadeghiyan, T.; Rezaei, M.; Azadeh, M. Initial Evaluation of Lncrna A2m-As1 Gene Expression in Multiple Sclerosis Patients. Adv. Biomed. Res. 2024, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Hoch-Kraft, P.; White, R.; Tenzer, S.; Kramer-Albers, E.M.; Trotter, J.; Gonsior, C. Dual Role of the Rna Helicase Ddx5 in Post-Transcriptional Regulation of Myelin Basic Protein in Oligodendrocytes. J. Cell Sci. 2018, 131, jcs204750. [Google Scholar] [CrossRef] [PubMed]
- Mandler, M.D.; Ku, L.; Feng, Y. A Cytoplasmic Quaking I Isoform Regulates the Hnrnp F/H-Dependent Alternative Splicing Pathway in Myelinating Glia. Nucleic Acids Res. 2014, 42, 7319–7329. [Google Scholar] [CrossRef] [PubMed]
- Greene, D.N.; Schmidt, R.L.; Wilson, A.R.; Freedman, M.S.; Grenache, D.G. Cerebrospinal Fluid Myelin Basic Protein Is Frequently Ordered but Has Little Value: A Test Utilization Study. Am. J. Clin. Pathol. 2012, 138, 262–272. [Google Scholar] [CrossRef]
- Herman, S.; Khoonsari, P.E.; Tolf, A.; Steinmetz, J.; Zetterberg, H.; Akerfeldt, T.; Jakobsson, P.J.; Larsson, A.; Spjuth, O.; Burman, J.; et al. Integration of Magnetic Resonance Imaging and Protein and Metabolite Csf Measurements to Enable Early Diagnosis of Secondary Progressive Multiple Sclerosis. Theranostics 2018, 8, 4477–4490. [Google Scholar] [CrossRef]
- Colucci, M.; Roccatagliata, L.; Capello, E.; Narciso, E.; Latronico, N.; Tabaton, M.; Mancardi, G.L. The 14-3-3 Protein in Multiple Sclerosis: A Marker of Disease Severity. Mult. Scler. J. 2004, 10, 477–481. [Google Scholar] [CrossRef]
- Morales, D.; Hechavarria, R.; Wojna, V.; Acevedo, S.F. Ywhae/14-3-3epsilon: A Potential Novel Genetic Risk Factor and Csf Biomarker for Hiv Neurocognitive Impairment. J. Neurovirol. 2013, 19, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Denomme-Pichon, A.S.; Collins, S.C.; Bruel, A.L.; Mikhaleva, A.; Wagner, C.; Vancollie, V.E.; Thomas, Q.; Chevarin, M.; Weber, M.; Prada, C.E.; et al. Ywhae Loss of Function Causes a Rare Neurodevelopmental Disease with Brain Abnormalities in Human and Mouse. Genet. Med. 2023, 25, 100835. [Google Scholar] [CrossRef] [PubMed]
- Friese, M.A.; Fugger, L. Pathogenic Cd8(+) T Cells in Multiple Sclerosis. Ann. Neurol. 2009, 66, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Tuovinen, E.A.; Gronholm, J.; Ohman, T.; Poysti, S.; Toivonen, R.; Kreutzman, A.; Heiskanen, K.; Trotta, L.; Toiviainen-Salo, S.; Routes, J.M.; et al. Novel Hemizygous Il2rg P.(Pro58ser) Mutation Impairs Il-2 Receptor Complex Expression on Lymphocytes Causing X-Linked Combined Immunodeficiency. J. Clin. Immunol. 2020, 40, 503–514. [Google Scholar] [CrossRef]
- Scholz, E.M.; Marcilla, M.; Daura, X.; Arribas-Layton, D.; James, E.A.; Alvarez, I. Human Leukocyte Antigen (Hla)-Drb1*15:01 and Hla-Drb5*01:01 Present Complementary Peptide Repertoires. Front. Immunol. 2017, 8, 984. [Google Scholar] [CrossRef]
- Hecker, M.; Ruge, A.; Putscher, E.; Boxberger, N.; Rommer, P.S.; Fitzner, B.; Zettl, U.K. Aberrant Expression of Alternative Splicing Variants in Multiple Sclerosis—A Systematic Review. Autoimmun. Rev. 2019, 18, 721–732. [Google Scholar] [CrossRef]
- Evsyukova, I.; Somarelli, J.A.; Gregory, S.G.; Garcia-Blanco, M.A. Alternative Splicing in Multiple Sclerosis and Other Autoimmune Diseases. RNA Biol. 2010, 7, 462–473. [Google Scholar] [CrossRef]
- Parnell, G.P.; Gatt, P.N.; McKay, F.C.; Schibeci, S.; Krupa, M.; Powell, J.E.; Visscher, P.M.; Montgomery, G.W.; Lechner-Scott, J.; Broadley, S.; et al. Ribosomal Protein S6 Mrna Is a Biomarker Upregulated in Multiple Sclerosis, Downregulated by Interferon Treatment, and Affected by Season. Mult. Scler. J. 2014, 20, 675–685. [Google Scholar] [CrossRef]
- Miller, R.M.; Jordan, B.T.; Mehlferber, M.M.; Jeffery, E.D.; Chatzipantsiou, C.; Kaur, S.; Millikin, R.J.; Dai, Y.; Tiberi, S.; Castaldi, P.J.; et al. Enhanced Protein Isoform Characterization through Long-Read Proteogenomics. Genome Biol. 2022, 23, 69. [Google Scholar] [CrossRef]
- Sinitcyn, P.; Richards, A.L.; Weatheritt, R.J.; Brademan, D.R.; Marx, H.; Shishkova, E.; Meyer, J.G.; Hebert, A.S.; Westphall, M.S.; Blencowe, B.J.; et al. Global Detection of Human Variants and Isoforms by Deep Proteome Sequencing. Nat. Biotechnol. 2023, 41, 1776–1786. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast Universal Rna-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. Htseq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for Rna-Seq Data with Deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. Rmats: Robust and Flexible Detection of Differential Alternative Splicing from Replicate Rna-Seq Data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, P. Pearson’s Correlation Coefficient. BMJ Br. Med. J. 2012, 345, e4483. [Google Scholar] [CrossRef]
- Louadi, Z.; Yuan, K.; Gress, A.; Tsoy, O.; Kalinina, O.V.; Baumbach, J.; Kacprowski, T.; List, M. Digger: Exploring the Functional Role of Alternative Splicing in Protein Interactions. Nucleic Acids Res. 2021, 49, D309–D318. [Google Scholar] [CrossRef]
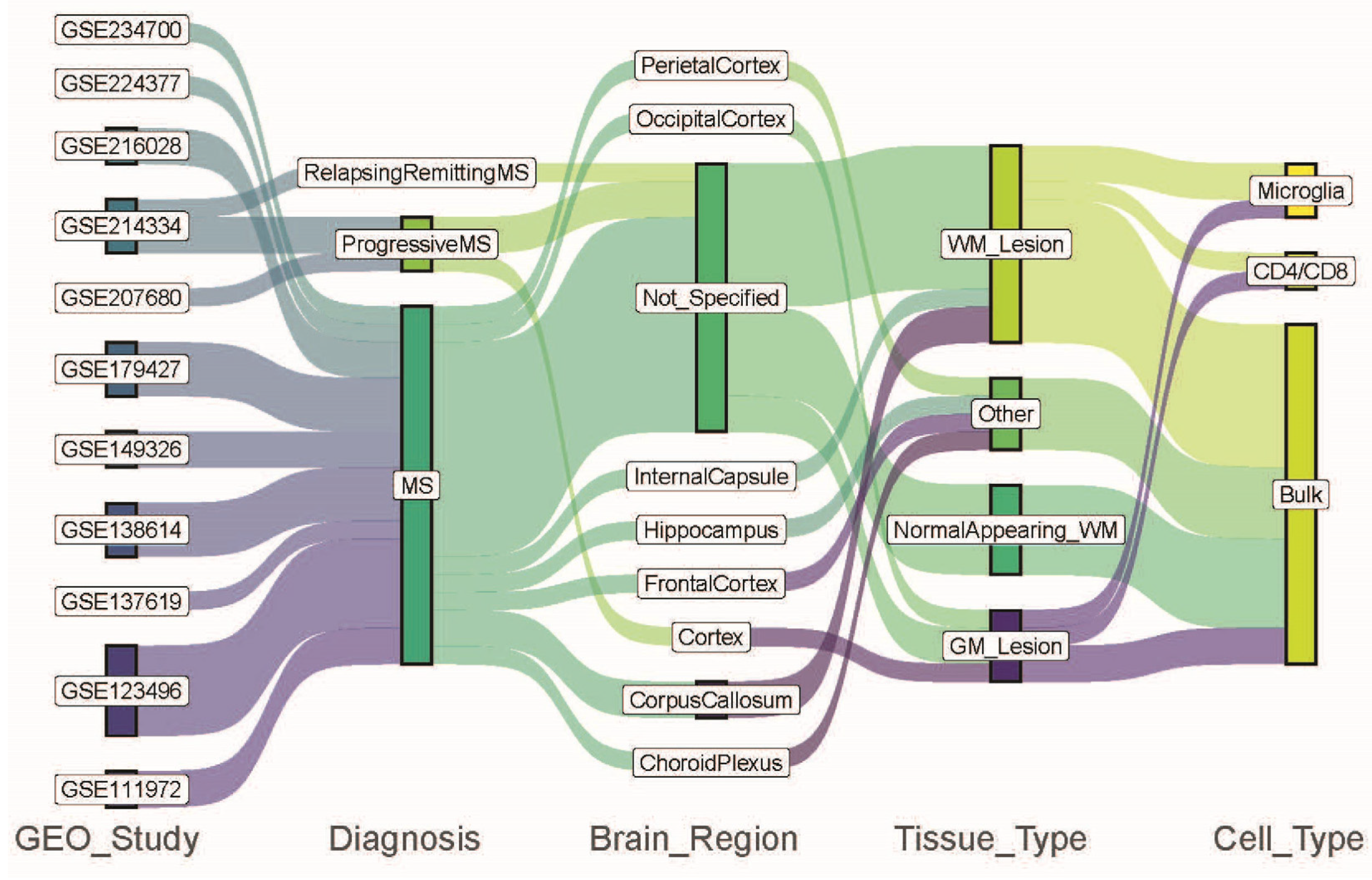

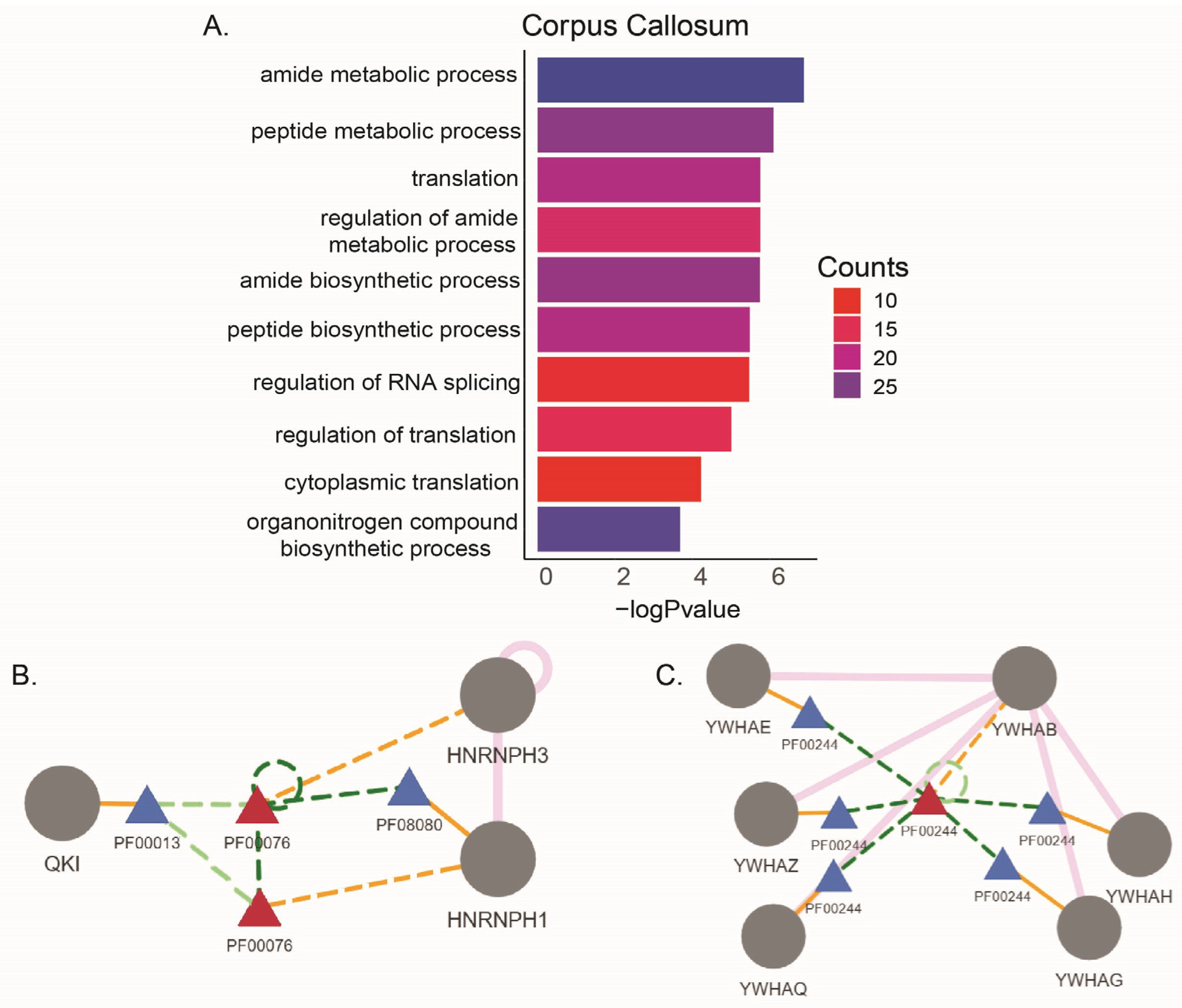
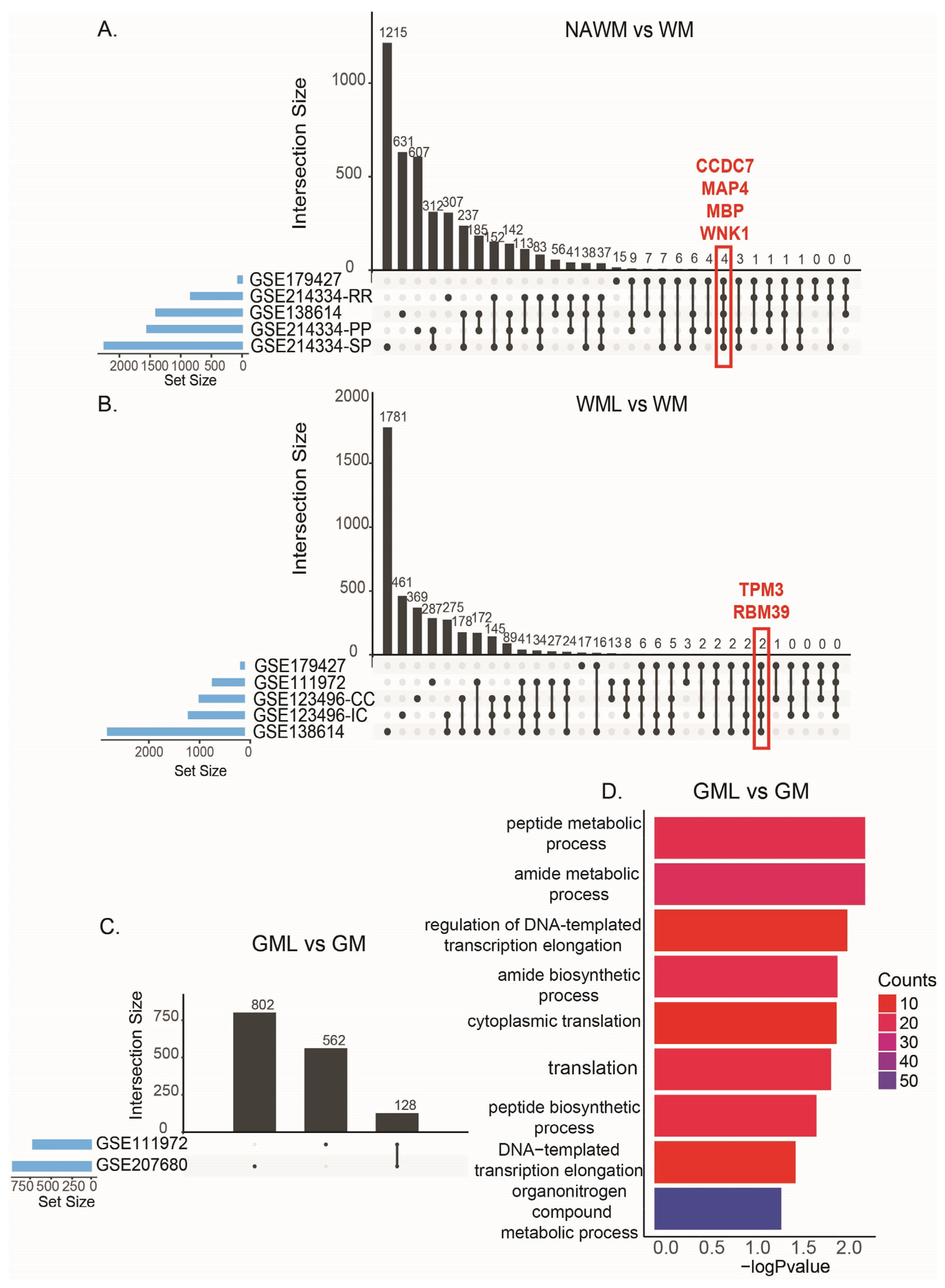
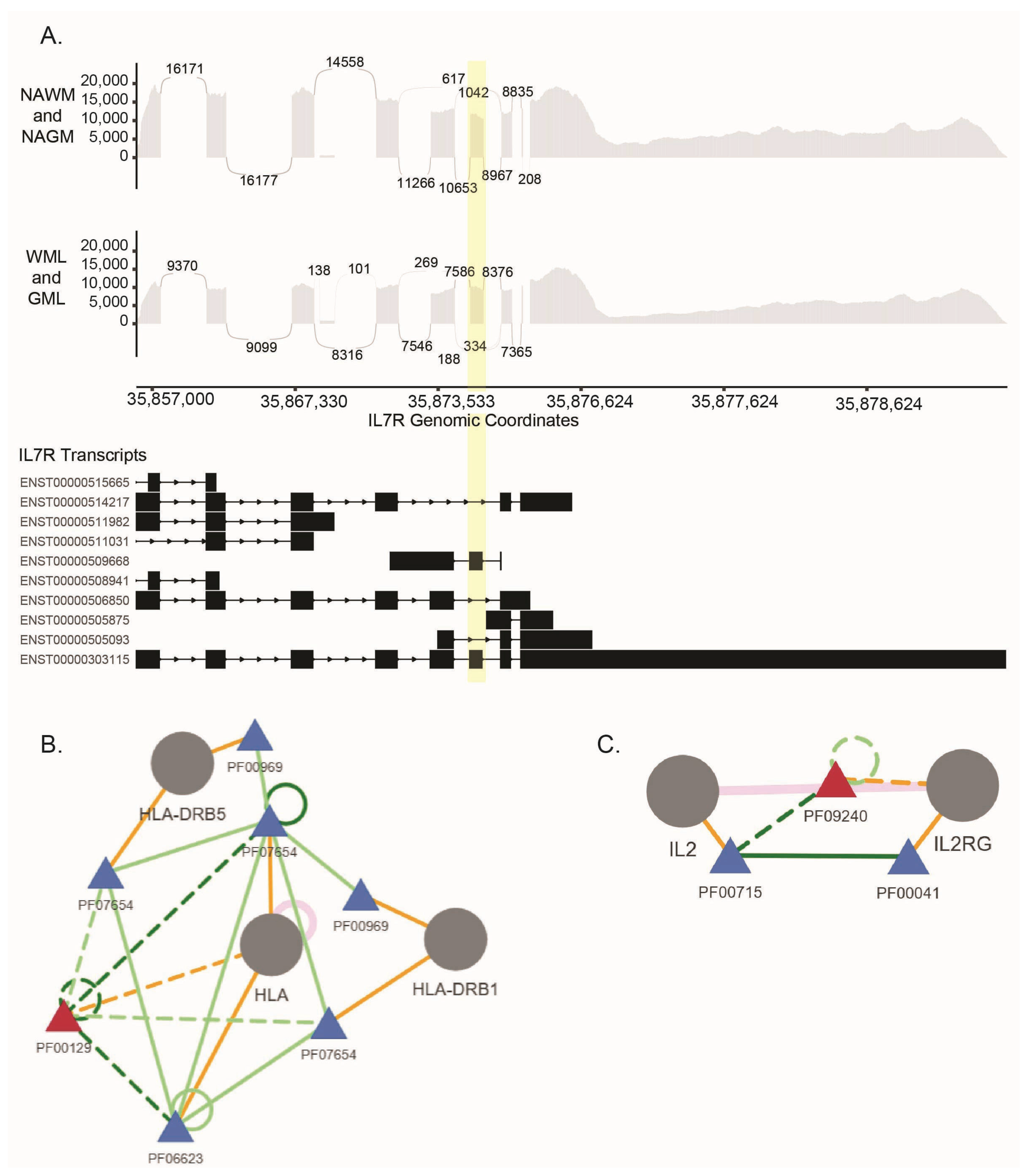
| Study ID | Summary | Study Site | Reference |
|---|---|---|---|
| GSE111972 | Microglia from occipital cortex from non-MS controls (GM, n = 5) and MS patients GML (n = 5). Microglia from corpus callosum from non-MS controls (WM, n = 11) and MS patients (WML, n = 11) | Germany | [14] |
| GSE123496 | Corpus callosum (WM n = 5, WML n = 5), frontal cortex (n = 5, n = 5), parietal cortex (n = 5, n = 5), hippocampus (n = 5, n = 5), and internal capsule (WM n = 5, WML n = 5) from non-MS control and MS patients | USA | [15] |
| I confiGSE137619 | Choroid plexus from non-MS control (n = 6) and MS patients (n = 6) | Netherlands | [16] |
| GSE138614 | WM (n = 25) from non-MS controls, NAWM (n = 21), and WML (n = 52) from MS patients | Denmark | [17] |
| GSE149326 | NAGM (n = 11), GML (n = 11), NAWM (n = 11), WML (n = 10) from MS patients | Netherlands | [18] |
| GSE179427 | NAWM (n = 31) and WML (n = 11) from MS patients, WM (n = 26) from non-MS controls | Netherlands | [19] |
| GSE207680 | GM Cortex from non-MS controls (n = 3) and from PMS patients (n = 3) | Canada | [20] |
| GSE214334 | NAWM from non-MS control (n = 7), RRMS (n = 3), SPMS (n = 4), and PPMS (n = 4) patients | Australia | [21] |
| GSE216028 | NAGM (n = 11), GML (n = 5), NAWM (n = 14), WML (n = 10) CD4/CD8 T-cells from MS patients | Netherlands | [22] |
| GSE224377 | NAWM and MS from MS patients (n = 9) | Belgium | [23] |
| GSE234700 | Microglia from NAWM and WML from MS patients (n = 7) | Netherlands | [24] |
| Study ID | Experimental Group Diagnosis | Control Group Diagnosis | Brain Region | Tissue Type | Cell Type | Comparison Number |
|---|---|---|---|---|---|---|
| GSE111972 | MS | Non-MS | Corpus Callosum | White Matter | Microglia | C1 |
| GSE111972 | MS | Non-MS | Occipital Cortex | Gray Matter | Microglia | C2 |
| GSE123496 | MS | Non-MS | Corpus Callosum | White Matter | Bulk | C3 |
| GSE123496 | MS | Non-MS | Internal Capsule | White Matter | Bulk | C4 |
| GSE123496 | MS | Non-MS | Frontal Cortex | Other | Bulk | C5 |
| GSE123496 | MS | Non-MS | Parietal Cortex | Other | Bulk | C6 |
| GSE123496 | MS | Non-MS | Hippocampus | Other | Bulk | C7 |
| GSE137619 | MS | Non-MS | Choroid Plexus | Other | Bulk | C8 |
| GSE138614 | MS | Non-MS | Not Specified | White Matter (AL) | Bulk | C9 |
| GSE138614 | MS | Non-MS | Not Specified | White Matter (RL) | Bulk | C10 |
| GSE138614 | MS | Non-MS | Not Specified | White Matter (IL) | Bulk | C11 |
| GSE138614 | MS | Non-MS | Not Specified | White Matter (CA) | Bulk | C12 |
| GSE138614 | MS | Non-MS | Not Specified | White Matter (NAWM) | Bulk | C13 |
| GSE149326 | MS | MS * | Not specified | White Matter | Bulk | C14 |
| GSE149326 | MS | MS * | Not specified | Gray Matter | Bulk | C15 |
| GSE179427 | MS | Non-MS | Not specified | White Matter (WML) | Bulk | C16 |
| GSE179427 | MS | Non-MS | Not specified | White Matter (NAWM) | Bulk | C17 |
| GSE207680 | PMS | Non-MS | Cortex | Gray Matter | Bulk | C18 |
| GSE214334 | PPMS | Non-MS | Not Specified | White Matter (NAWM) | Bulk | C19 |
| GSE214334 | SPMS | Non-MS | Not Specified | White Matter (NAWM) | Bulk | C20 |
| GSE214334 | RRMS | Non-MS | Not Specified | White Matter (NAWM) | Bulk | C21 |
| GSE216028 | MS | MS * | Not Specified | White Matter | T-cells (CD4+ and CD8+) | C22 |
| GSE216028 | MS | MS * | Not Specified | Gray Matter | T-cells (CD4+ and CD8+) | C23 |
| GSE224377 | MS | MS * | Not specified | White Matter | Bulk | C24 |
| GSE234700 | MS | MS * | Not Specified | White Matter | Microglia | C25 |
| Study ID | Comparison | DEG Only (Gene Count) | DEG and ASE (Gene Count—Percentage) | ASE Only (Gene Count) |
|---|---|---|---|---|
| GSE111972 | C1 | 808 | 70—(4.46%) | 691 |
| GSE111972 | C2 | 264 | 17—(1.59%) | 691 |
| GSE123496 | C3 | 445 | 53—(3.32%) | 1097 |
| GSE123496 | C4 | 444 | 26—(1.92%) | 883 |
| GSE123496 | C5 | 10 | 0—(0%) | 790 |
| GSE123496 | C6 | 4 | 0—(0%) | 878 |
| GSE123496 | C7 | 47 | 0—(0%) | 571 |
| GSE137619 | C8 | 11 | 0—(0%) | 798 |
| GSE138614 | C9 | 6125 | 304—(4.27%) | 687 |
| GSE138614 | C10 | 2907 | 263—(5.97%) | 1233 |
| GSE138614 | C11 | 4937 | 401—(6.2%) | 1126 |
| GSE138614 | C12 | 5731 | 234—(3.48%) | 764 |
| GSE138614 | C13 | 719 | 32—(1.49%) | 1403 |
| GSE149326 | C14 | 32 | 0—(0%) | 4 |
| GSE149326 | C15 | 61 | 0—(0%) | 4 |
| GSE179427 | C16 | 104 | 0—(0%) | 64 |
| GSE179427 | C17 | 351 | 2—(0.48%) | 65 |
| GSE207680 | C18 | 80 | 8—(0.72%) | 1022 |
| GSE214334 | C19 | 1163 | 129—(4.19%) | 1786 |
| GSE214334 | C20 | 4593 | 1355—(16.03%) | 2504 |
| GSE214334 | C21 | 6 | 0—(0%) | 912 |
| GSE216028 | C22 | 2 | 1—(0.32%) | 458 |
| GSE216028 | C23 | 142 | 1—(0.29%) | 200 |
| GSE224377 | C24 | 34 | 0—(0%) | 3 |
| GSE234700 | C25 | 1053 | 0—(0%) | 3 |
| MS Lesions vs. Non-MS | Progressive MS vs. Non-MS | RRMS vs. Non-MS | |||
|---|---|---|---|---|---|
| Study ID | Comparison Number | Study ID | Comparison Number | Study ID | Comparison Number |
| GSE111972 | C1 | GSE207680 | C18 | GSE214334 | C21 |
| GSE111972 | C2 | GSE214334 | C19 | ||
| GSE123496 | C3 | GSE214334 | C20 | ||
| GSE123496 | C4 | ||||
| GSE123496 | C5 | ||||
| GSE123496 | C6 | ||||
| GSE123496 | C7 | ||||
| GSE138614 | C9 | ||||
| GSE138614 | C10 | ||||
| GSE138614 | C11 | ||||
| GSE138614 | C12 | ||||
| GSE149326 | C14 | ||||
| GSE149326 | C15 | ||||
| GSE179427 | C16 | ||||
| GSE207680 | C18 | ||||
| Corpus Callosum | IC, FC, PC, Hipp | Occipital Cortex | Choroid Plexus | ||||
|---|---|---|---|---|---|---|---|
| Study ID | Comparison Number | Study ID | Comparison Number (Respectively IC, FC, PC, Hipp) | Study ID | Comparison Number | Study ID | Comparison Number |
| GSE111972 | C1 | GSE123496 | C4 | GSE111972 | C2 | GSE137619 | C8 |
| GSE123496 | C3 | GSE123496 | C5 | ||||
| GSE123496 | C6 | ||||||
| GSE123496 | C7 | ||||||
| Corpus Callosum | ||
|---|---|---|
| Gene Symbol | Gene Description | ASE |
| A2M | alpha-2-macroglobulin | RI |
| ACSL1 | acyl-CoA synthetase long chain family member 1 | MXE |
| ADAM28 | ADAM metallopeptidase domain 28 | SE |
| AKAP8L | A-kinase anchoring protein 8 like | RI |
| AMPD3 | adenosine monophosphate deaminase 3 | SE |
| BIN1 | bridging integrator 1 | SE |
| CLK1 | CDC like kinase 1 | SE, RI |
| COX4I1 | cytochrome c oxidase subunit 4I1 | RI |
| DDX5 | DEAD-box helicase 5 | SE |
| DENND5A | DENN domain containing 5A | SE |
| DNAJB2 | DnaJ heat shock protein family (Hsp40) member B2 | RI |
| EEF1D | eukaryotic translation elongation factor 1 delta | RI |
| EPB41L2 | erythrocyte membrane protein band 4.1 like 2 | SE |
| FTH1 | ferritin heavy chain 1 | RI |
| HEXA | hexosaminidase subunit alpha | SE |
| HNRNPH1 | heterogeneous nuclear ribonucleoprotein H1 | SE, RI |
| HNRNPH3 | heterogeneous nuclear ribonucleoprotein H3 | SE |
| INTS6 | integrator complex subunit 6 | SE |
| LZTS2 | leucine zipper tumor suppressor 2 | SE |
| NDUFV3 | NADH:ubiquinone oxidoreductase subunit V3 | SE |
| PHB2 | prohibitin 2 | SE |
| QKI | KH domain containing RNA binding | A3SS |
| RGS2 | regulator of G protein signaling 2 | SE |
| RPL10A | ribosomal protein L10a | RI |
| RPL28 | ribosomal protein L28 | RI |
| RPS9 | ribosomal protein S9 | A3SS |
| SNHG1 | small nucleolar RNA host gene 1 | RI |
| SPP1 | secreted phosphoprotein 1 | SE |
| TMEM59 | transmembrane protein 59 | SE |
| TPM3 | tropomyosin 3 | SE |
| TPP1 | tripeptidyl peptidase 1 | RI |
| YBX3 | Y-box binding protein 3 | RI |
| YWHAB | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | RI |
| NAWM vs. WM | WML vs. WM | GML vs. GM | |||
|---|---|---|---|---|---|
| Study ID | Comparison Number | Study ID | Comparison Number | Study ID | Comparison Number |
| GSE138614 | C13 | GSE111972 | C1 | GSE111972 | C2 |
| GSE179427 | C17 | GSE123496-CC | C3 | GSE207680 | C18 |
| GSE214334-PP | C19 | GSE123496-IC | C4 | ||
| GSE214334-SP | C20 | GSE138614 | C9–C12 | ||
| GSE214334-RR | C21 | ||||
| Microglia | T-Cells (CD4+/CD8+) | ||
|---|---|---|---|
| Study ID | Comparison Number | Study ID | Comparison Number |
| GSE111972 | C1 | GSE216028 | C22 |
| GSE111972 | C2 | GSE216028 | C23 |
| GSE234700 | C25 | ||
| CD8/CD4 T-Cells | ||
|---|---|---|
| Gene Symbol | Gene Description | ASE |
| AL590764.2 | NA | SE |
| ARGLU1 | arginine and glutamate rich 1 | RI |
| ARHGEF1 | Rho guanine nucleotide exchange factor 1 | A3SS |
| ARL6IP4 | ADP ribosylation factor like GTPase 6 interacting protein 4 | RI |
| ARPC2 | actin related protein 2/3 complex subunit 2 | SE |
| BIN2 | bridging integrator 2 | SE |
| C9orf78 | chromosome 9 open reading frame 78 | SE |
| CD37 | CD37 molecule | SE, A3SS |
| CD96 | CD96 molecule | SE |
| CD99 | CD99 molecule (Xg blood group) | SE, A3SS |
| CDK5RAP3 | CDK5 regulatory subunit associated protein 3 | RI |
| CENPT | centromere protein T | A3SS, RI |
| CHURC1 | Churchill domain containing 1 | SE |
| CIRBP | cold-inducible RNA binding protein | SE |
| COX5B | cytochrome c oxidase subunit 5B | SE |
| CPNE1 | copine 1 | RI |
| DDX5 | DEAD-box helicase 5 | RI |
| DENND2D | DENN domain containing 2D | SE |
| EIF1 | eukaryotic translation initiation factor 1 | SE |
| ELOB | elongin B | RI |
| EMP3 | epithelial membrane protein 3 | SE |
| EXOSC8 | exosome component 8 | A3SS |
| GAS5 | growth arrest specific 5 | RI |
| GLIPR1 | GLI pathogenesis related 1 | SE |
| GMFG | glia maturation factor gamma | SE |
| GSTK1 | glutathione S-transferase kappa 1 | SE |
| GTF3A | general transcription factor IIIA | SE |
| GZMA | granzyme A | SE |
| H3-3B | H3.3 histone B | RI |
| HLA-A | major histocompatibility complex, class I, A | RI |
| HLA-B | major histocompatibility complex, class I, B | RI |
| HNRNPA1 | heterogeneous nuclear ribonucleoprotein A1 | SE |
| HNRNPC | heterogeneous nuclear ribonucleoprotein C | A3SS |
| HNRNPU | heterogeneous nuclear ribonucleoprotein U | RI |
| HSPB1 | heat shock protein family B (small) member 1 | A5SS |
| HSPE1 | heat shock protein family E (Hsp10) member 1 | SE |
| IL2RG | interleukin 2 receptor subunit gamma | RI |
| IL32 | interleukin 32 | SE, A3SS |
| IL7R | interleukin 7 receptor | SE |
| ILF3 | interleukin enhancer binding factor 3 | SE |
| ISCU | iron–sulfur cluster assembly enzyme | SE |
| LCK | LCK proto-oncogene, Src family tyrosine kinase | A3SS |
| LIMD2 | LIM domain containing 2 | A3SS, RI |
| MYL6 | myosin light chain 6 | SE, A5SS, A3SS, RI |
| NACA | nascent polypeptide-associated complex subunit alpha | RI |
| NDUFA11 | NADH:ubiquinone oxidoreductase subunit A11 | SE, RI |
| NDUFA3 | NADH:ubiquinone oxidoreductase subunit A3 | SE |
| OAZ1 | ornithine decarboxylase antizyme 1 | RI |
| PABPC1 | poly(A) binding protein cytoplasmic 1 | A5SS |
| PCED1B-AS1 | PCED1B antisense RNA 1 | SE, A5SS |
| PFDN5 | prefoldin subunit 5 | RI |
| PPIA | peptidylprolyl isomerase A | SE |
| PTPN6 | protein tyrosine phosphatase non-receptor type 6 | A3SS |
| RACK1 | receptor for activated C kinase 1 | RI |
| RBM39 | RNA binding motif protein 39 | RI |
| RPL10 | ribosomal protein L10 | RI |
| RPL10A | ribosomal protein L10a | RI |
| RPL13A | ribosomal protein L13a | RI |
| RPL28 | ribosomal protein L28 | RI |
| RPL3 | ribosomal protein L3 | RI |
| RPL31 | ribosomal protein L31 | RI |
| RPL4 | ribosomal protein L4 | RI |
| RPL41 | ribosomal protein L41 | A5SS |
| RPLP1 | ribosomal protein lateral stalk subunit P1 | SE |
| RPS11 | ribosomal protein S11 | RI |
| RPS12 | ribosomal protein S12 | A5SS |
| RPS15 | ribosomal protein S15 | A3SS |
| RPS2 | ribosomal protein S2 | RI |
| RPS20 | ribosomal protein S20 | RI |
| RPS28 | ribosomal protein S28 | RI |
| RPS3 | ribosomal protein S3 | SE, A5SS |
| RPS9 | ribosomal protein S9 | SE |
| SKAP1 | src kinase associated phosphoprotein 1 | SE |
| SNRPN | small nuclear ribonucleoprotein polypeptide N | SE, A3SS |
| SPSB3 | splA/ryanodine receptor domain and SOCS box containing 3 | RI |
| SRRM1 | serine and arginine repetitive matrix 1 | RI |
| SYF2 | SYF2 pre-mRNA splicing factor | SE |
| TPM3 | tropomyosin 3 | SE |
| TPT1 | tumor protein, translationally-controlled 1 | SE |
| UQCRB | ubiquinol-cytochrome c reductase binding protein | SE |
| VPS29 | VPS29 retromer complex component | SE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sak, M.; Chariker, J.H.; Rouchka, E.C. Systematic Analysis of Alternative Splicing in Transcriptomes of Multiple Sclerosis Patient Brain Samples. Int. J. Mol. Sci. 2025, 26, 8195. https://doi.org/10.3390/ijms26178195
Sak M, Chariker JH, Rouchka EC. Systematic Analysis of Alternative Splicing in Transcriptomes of Multiple Sclerosis Patient Brain Samples. International Journal of Molecular Sciences. 2025; 26(17):8195. https://doi.org/10.3390/ijms26178195
Chicago/Turabian StyleSak, Müge, Julia H. Chariker, and Eric C. Rouchka. 2025. "Systematic Analysis of Alternative Splicing in Transcriptomes of Multiple Sclerosis Patient Brain Samples" International Journal of Molecular Sciences 26, no. 17: 8195. https://doi.org/10.3390/ijms26178195
APA StyleSak, M., Chariker, J. H., & Rouchka, E. C. (2025). Systematic Analysis of Alternative Splicing in Transcriptomes of Multiple Sclerosis Patient Brain Samples. International Journal of Molecular Sciences, 26(17), 8195. https://doi.org/10.3390/ijms26178195






