Identification of SUMO Proteins and Their Expression Profile During Induction of Somatic Embryogenesis in Medicago truncatula Gaertn.
Abstract
1. Introduction
2. Results
2.1. Characterization and Identification of SUMO Pathway Proteins in Medicago truncatula
2.2. Expression Profile SUMO Pathway Genes During Induction Phase of Medicago truncatula Somatic Embryogenesis
2.3. SUMOylation Site and SIM Prediction
3. Discussion
4. Materials and Methods
4.1. Tissue Culture Protocol
4.2. Identification of SUMO Pathway Genes in Medicago truncatula Genome
4.3. Gene Structure, Motif Identification, and Chromosomal Distribution
4.4. Gene Expression Analysis
4.5. Prediction of SUMOylation and SIM Sites in Key Proteins Involved in Medicago truncatula Somatic Embryogenesis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DSE | Direct somatic embryogenesis |
| ISE | Indirect somatic embryogenesis |
| M9 | Non-embryogenic line |
| M9-10a | Embryogenic line |
| PEMs | Pro-embryogenic masses |
| PGRs | Plant growth regulators |
| SE | Somatic embryogenesis |
| SIM | SUMO-interacting motif |
| SUMO | Small ubiquitin-like modifier |
References
- Williams, E.G.; Maheswaran, G. Somatic embryogenesis: Fac-tors influencing coordinated behavior of cells as an embryogenic group. Ann. Bot. 1986, 57, 433–462. [Google Scholar] [CrossRef]
- Zimmerman, J.L. Somatic embryogenesis: A model for early development in higher plants. Plant Cell 1993, 5, 1411–1423. [Google Scholar] [CrossRef]
- De-la-Peña, C.; Nic-Can, G.I.; Galaz-Ávalos, R.M.; Avilez-Montalvo, R.; Loyola-Vargas, V.M. The role of chromatin modifications in somatic embryogenesis in plants. Front. Plant Sci. 2015, 6, 635. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowska, B.; Wójcik, A.M.; Gaj, M.D. Epigenetic Regulation of Auxin-Induced Somatic Embryogenesis in Plants. Int. J. Mol. Sci. 2020, 21, 2307. [Google Scholar] [CrossRef]
- Santos, D.; Fevereiro, P. Loss of DNA methylation affects somatic embryogenesis in Medicago truncatula. Plant Cell Tissue Organ Cult. 2002, 70, 155–161. [Google Scholar] [CrossRef]
- Nolan, K.E.; Song, Y.; Liao, S.; Saeed, N.A.; Zhang, X.; Rose, R.J. An unusual abscisic acid and gibberellic acid synergism increases somatic embryogenesis, facilitates its genetic analysis and improves transformation in M. truncatula. PLoS ONE 2014, 9, e99908. [Google Scholar] [CrossRef]
- Orłowska, A.; Igielski, R.; Łagowska, K.; Kępczyńska, E. Identification of LEC1, L1L and Polycomb Repressive Complex 2 genes and their expression during the induction phase of Medicago truncatula Gaertn. somatic embryogenesis. Plant Cell Tissue Organ Cult. 2017, 129, 119–132. [Google Scholar] [CrossRef]
- Orłowska, A.; Kępczyńska, E. Identifcation of Polycomb Repressive Complex1, Trithorax group genes and their simultaneous expression with WUSCHEL, WUSCHEL-related Homeobox5 and SHOOT MERISTEMLESS during the induction phase of somatic embryogenesis in Medicago truncatula Gaertn. Plant Cell Tissue Organ Cult. 2018, 134, 345–356. [Google Scholar] [CrossRef]
- Miura, K.; Rus, A.; Sharkhuu, A.; Yokoi, S.; Karthikeyan, A.S.; Raghothama, K.G.; Baek, D.; Koo, Y.D.; Jin, J.B.; Bressan, R.A.; et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 2005, 102, 7760–7765. [Google Scholar] [CrossRef]
- Zheng, Z.; Liu, D. SIZ1 regulates phosphate deficiency-induced inhibition of primary root growth of Arabidopsis by modulating Fe accumulation and ROS production in its roots. Plant Signal. Behav. 2021, 16, 1946921. [Google Scholar] [CrossRef]
- Han, D.; Chen, C.; Xia, S.; Liu, J.; Shu, J.; Nguyen, V.; Lai, J.; Cui, Y.; Yang, C. Chromatin-associated SUMOylation controls the transcriptional switch between plant development and heat stress responses. Plant Commun. 2020, 2, 100091. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Jin, J.B.; Lee, J.; Yoo, C.Y.; Stirm, V.; Miura, T.; Ashworth, E.N.; Bressan, R.A.; Yun, D.J.; Hasegawa, P.M. SIZ1-Mediated Sumoylation of ICE1 Controls CBF3/DREB1A Expression and Freezing Tolerance in Arabidopsis. Plant Cell 2007, 19, 1403–1414. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Zhang, C.; Yates, G.; Bailey, M.; Brown, A.; Sadanandom, A. SUMO is a critical regulator of salt stress responses in rice. Plant Physiol. 2016, 170, 2378–2391. [Google Scholar] [CrossRef]
- Lee, J.; Nam, J.; Park, H.C.; Na, G.; Miura, K.; Jin, J.B.; Yoo, C.Y.; Baek, D.; Kim, D.H.; Jeong, J.C.; et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007, 49, 79–90. [Google Scholar] [CrossRef]
- Miura, K.; Lee, J.; Miura, T.; Hasegawa, P.M. SIZ1 Controls Cell Growth and Plant Development in Arabidopsis Through Salicylic Acid. Plant Cell Physiol. 2010, 51, 103–113. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, W.Y.; Park, H.C.; Lee, S.Y.; Bohnert, H.J.; Yun, D.J. SUMO and SUMOylation in plants. Mol. Cells 2011, 32, 305–316. [Google Scholar] [CrossRef]
- Ishida, T.; Fujiwara, S.; Miura, K.; Stacey, N.; Yoshimura, M.; Schneider, K.; Adachi, S.; Minamisawa, K.; Umeda, M.; Sugimoto, K. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 2009, 21, 2284–2297. [Google Scholar] [CrossRef] [PubMed]
- Tomanov, K.; Zeschmann, A.; Hermkes, R.; Eifler, K.; Ziba, I.; Grieco, M.; Novatchkova, M.; Hofmann, K.; Hesse, H.; Bachmair, A. Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 2014, 26, 4547–4560. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.J.; Barrett-Wilt, G.A.; Hua, Z.; Vierstra, R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 16512–16517. [Google Scholar] [CrossRef]
- Rytz, T.C.; Miller, M.J.; McLoughlin, F.; Augustine, R.C.; Marshall, R.S.; Juan, Y.T.; Charng, Y.Y.; Scalf, M.; Smith, L.M.; Vierstra, R.D. SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. Plant Cell 2018, 30, 1077–1099. [Google Scholar] [CrossRef]
- Jentsch, S.; Psakhye, I. Control of Nuclear Activities by Substrate-Selective and Protein-Group SUMOylation. Annu. Rev. Genet. 2013, 47, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Saracco, S.A.; Miller, M.J.; Kurepa, J.; Vierstra, R.D. Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007, 145, 119–134. [Google Scholar] [CrossRef]
- van den Burg, H.A.; Kini, R.K.; Schuurink, R.C.; Takken, F.L.W. Arabidopsis Small Ubiquitin-Like Modifier Paralogs Have Distinct Functions in Development and Defense. Plant Cell 2010, 22, 1998–2016. [Google Scholar] [CrossRef]
- Li, Y.; Wang, G.; Xu, Z.; Li, J.; Sun, M.; Guo, J.; Ji, W. Organization and Regulation of Soybean SUMOylation System under Abiotic Stress Conditions. Front. Plant Sci. 2017, 8, 1458. [Google Scholar] [CrossRef] [PubMed]
- Colby, T.; Matthai, A.; Boeckelmann, A.; Stuible, H.P. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006, 142, 318–332. [Google Scholar] [CrossRef]
- Budhiraja, R.; Hermkes, R.; Muller, S.; Schmidt, J.; Colby, T.; Panigrahi, K.; Coupland, G.; Bachmair, A. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol. 2009, 149, 1529–1540. [Google Scholar] [CrossRef]
- Barreto, H.G.; Sagio, S.A.; Chalfun-Junior, A.; Fevereiro, P.; Benedito, V.A. Transcriptional profiling of the AFL subfamily of B3-type transcription factors during the in vitro induction of somatic embryogenesis in the model legume Medicago truncatula. PCTOC 2019, 139, 327–337. [Google Scholar] [CrossRef]
- Neves, L.O.; Duque, S.R.L.; Almeida, J.S.; Fevereiro, P.S. Repetitive somatic embryogenesis in Medicago truncatula ssp. Narbonensis and M. truncatula Gaertn cv. Jemolong. Plant Cell Rep. 1999, 18, 398–405. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 3, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef] [PubMed]
- Gou, Y.; Liu, D.; Chen, M.; Wei, Y.; Huang, X.; Han, C.; Feng, Z.; Zhang, C.; Lu, T.; Peng, D.; et al. GPS-SUMO 2.0: An updated online service for the prediction of SUMOylation sites and SUMO-interacting motifs. Nucleic Acids Res. 2024, 52, W238–W247. [Google Scholar] [CrossRef] [PubMed]
- Beauclair, G.; Bridier-Nahmias, A.; Zagury, J.F.; Saïb, A.; Zamborlini, A. JASSA: A comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics 2015, 31, 3483–3491. [Google Scholar] [CrossRef] [PubMed]
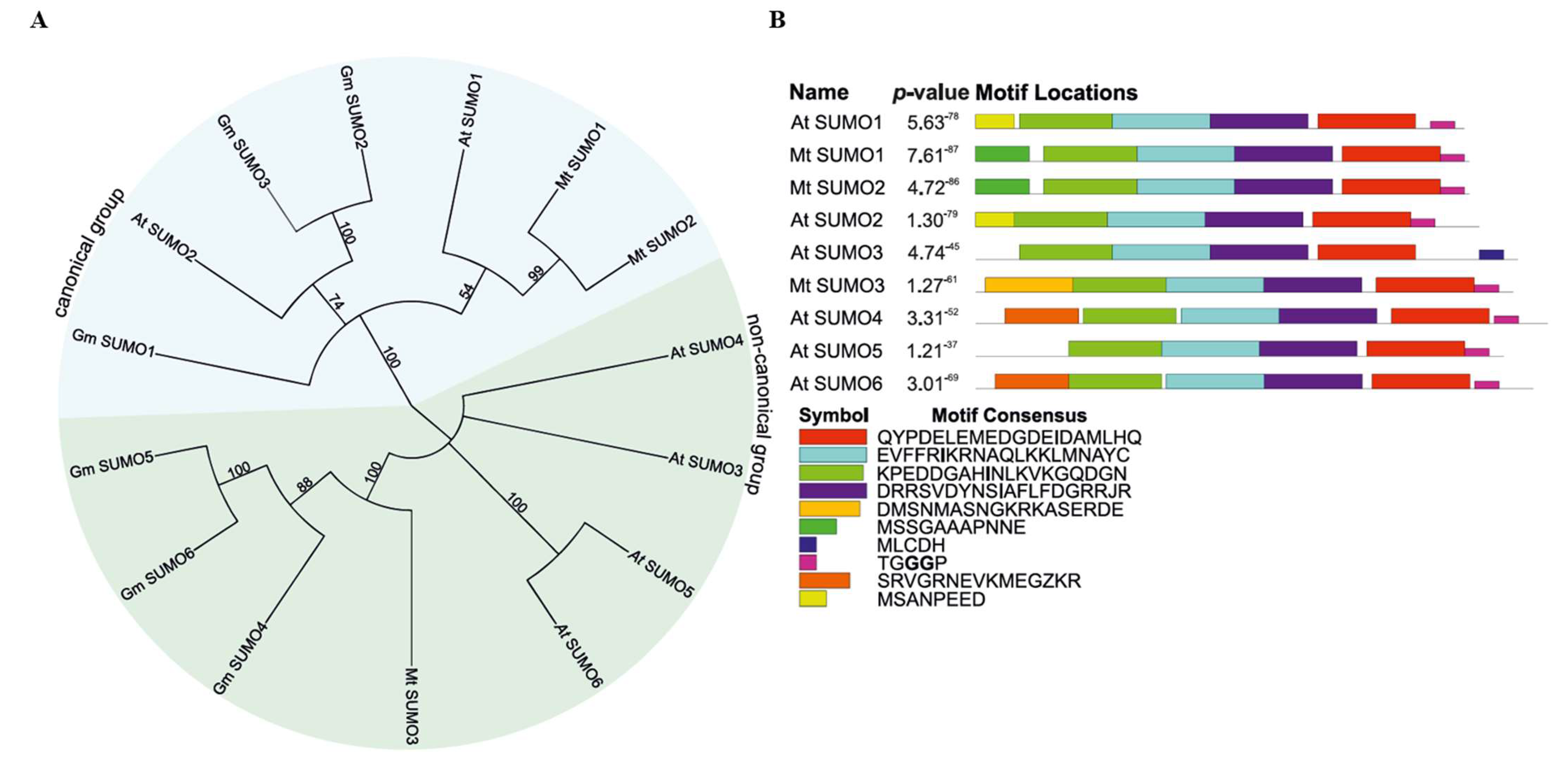

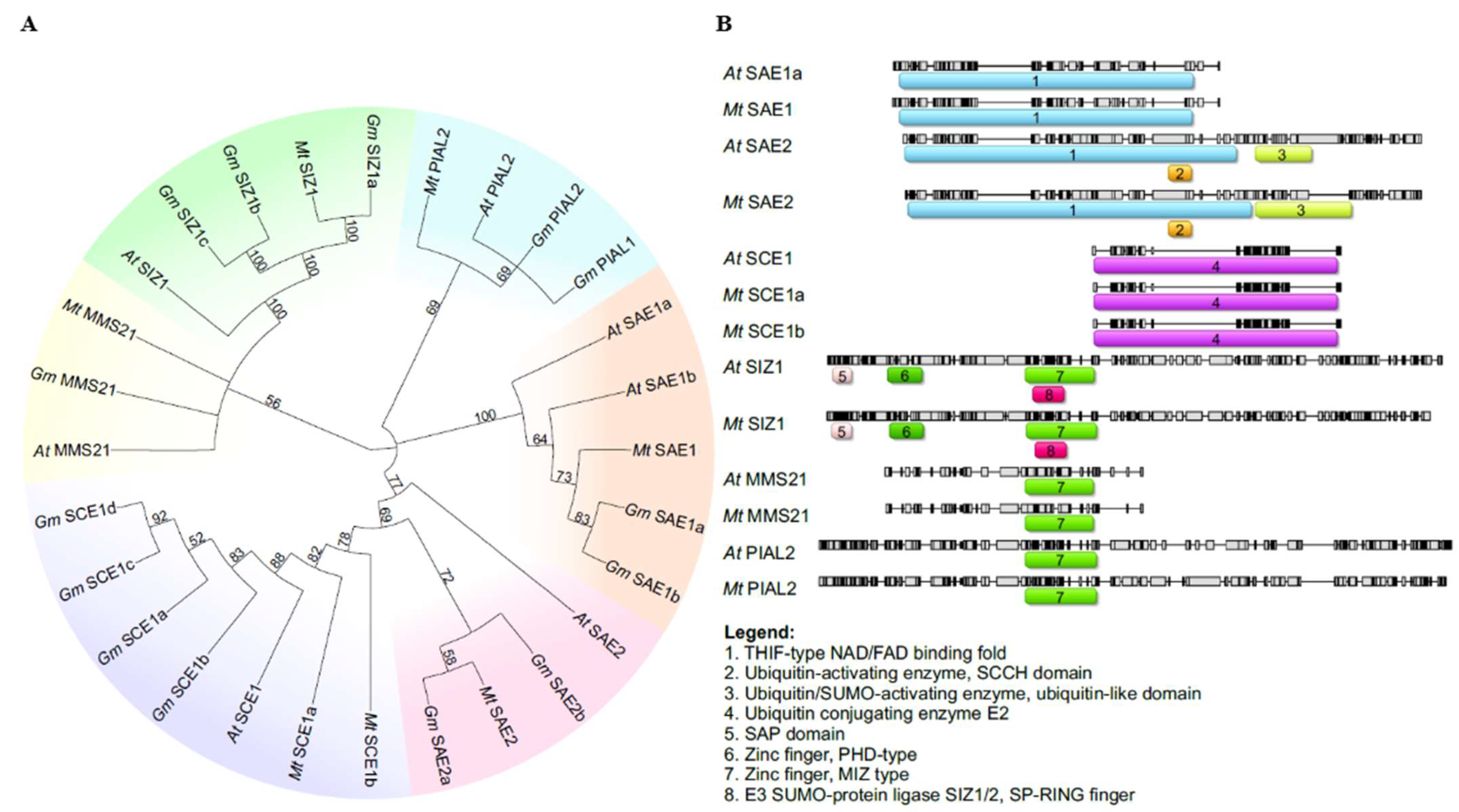

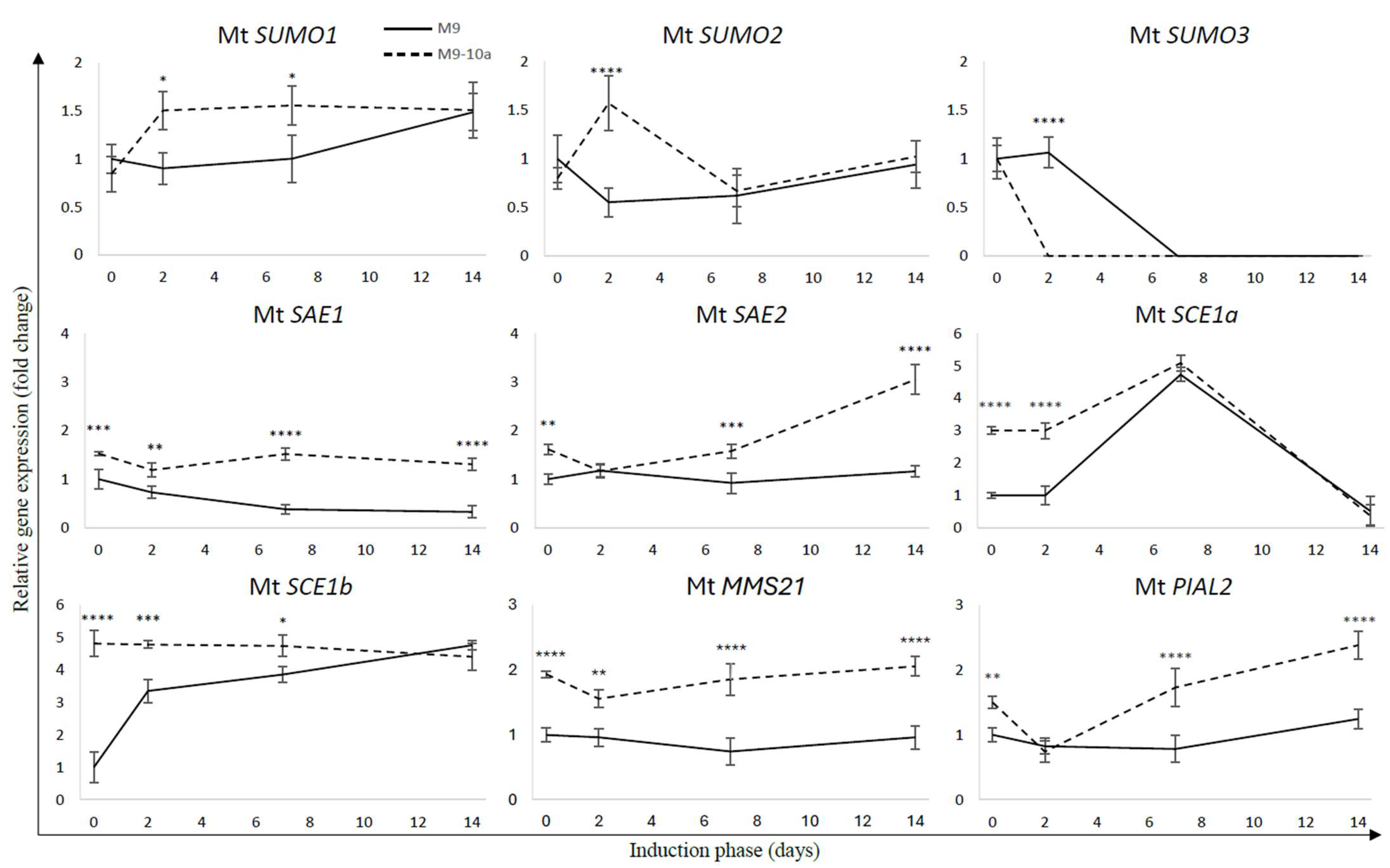
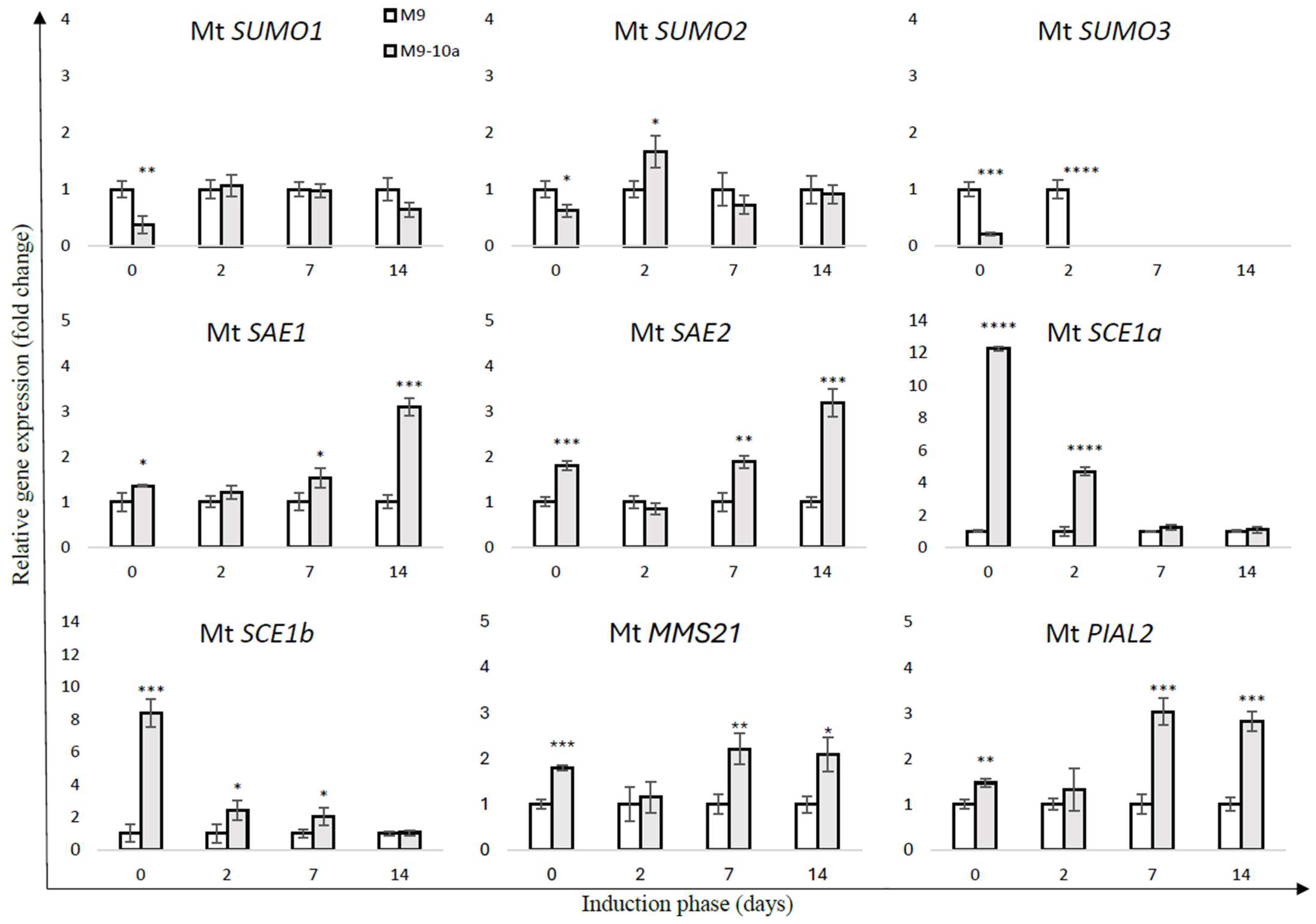
| Group | Gene Name | Locus ID | Chromosome | Gene Accession Number | Exon No. | Protein Accession Number | Protein Length (aa) |
|---|---|---|---|---|---|---|---|
| SUMO | Mt SUMO1 | MtrunA17_Chr3g0099761 | 3 | XM_013604411.3 | 3 | XP_013459865.1 | 101 |
| Mt SUMO2 | MtrunA17_Chr3g0099771 | 3 | XM_013604412.3 | 3 | XP_013459866.1 | 101 | |
| Mt SUMO3 | - | 8 | XM_039829461.1 | 4 | XP_039685395.1 | 110 | |
| E1 | Mt SAE1 | MtrunA17_Chr8g0386251 | 8 | XM_013591535.3 | 11 | XP_013446989.2 | 325 |
| Mt SAE2 | MtrunA17_Chr8g0352371 | 8 | XM_013589621.3 | 11 | XP_013445075.1 | 635 | |
| E2 | Mt SCE1a | MtrunA17_Chr4g0053191 | 4 | XM_013601981.3 | 5 | XP_013457435.1 | 159 |
| Mt SCE1b | MtrunA17_Chr5g0401831 | 5 | XM_003611630.4 | 5 | XP_003611678.1 | 159 | |
| E3 | Mt MMS21 | MtrunA17_Chr2g0316861 | 2 | XM_013609188.3 | 7 | XP_013464642.1 | 244 |
| Mt PIAL2 | MtrunA17_Chr8g0356491 | 8 | XM_039828859.1 | 17 | XP_039684793.1 | 822 | |
| Mt SIZ1 | MtrunA17_Chr4g0029701 | 4 | XM_039833913.1 | 17 | XP_003606454.1 | 882 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kujawska, A.; Król, P. Identification of SUMO Proteins and Their Expression Profile During Induction of Somatic Embryogenesis in Medicago truncatula Gaertn. Int. J. Mol. Sci. 2025, 26, 8133. https://doi.org/10.3390/ijms26178133
Kujawska A, Król P. Identification of SUMO Proteins and Their Expression Profile During Induction of Somatic Embryogenesis in Medicago truncatula Gaertn. International Journal of Molecular Sciences. 2025; 26(17):8133. https://doi.org/10.3390/ijms26178133
Chicago/Turabian StyleKujawska, Anna, and Paulina Król. 2025. "Identification of SUMO Proteins and Their Expression Profile During Induction of Somatic Embryogenesis in Medicago truncatula Gaertn." International Journal of Molecular Sciences 26, no. 17: 8133. https://doi.org/10.3390/ijms26178133
APA StyleKujawska, A., & Król, P. (2025). Identification of SUMO Proteins and Their Expression Profile During Induction of Somatic Embryogenesis in Medicago truncatula Gaertn. International Journal of Molecular Sciences, 26(17), 8133. https://doi.org/10.3390/ijms26178133






