Formation of the Vasculogenic Mimicry Phenotype in Melanoma Mel Z Cells Is Coupled with Changes in Inter-Chromosomal Contacts of Developmental Genes with rDNA Clusters
Abstract
1. Introduction
2. Results
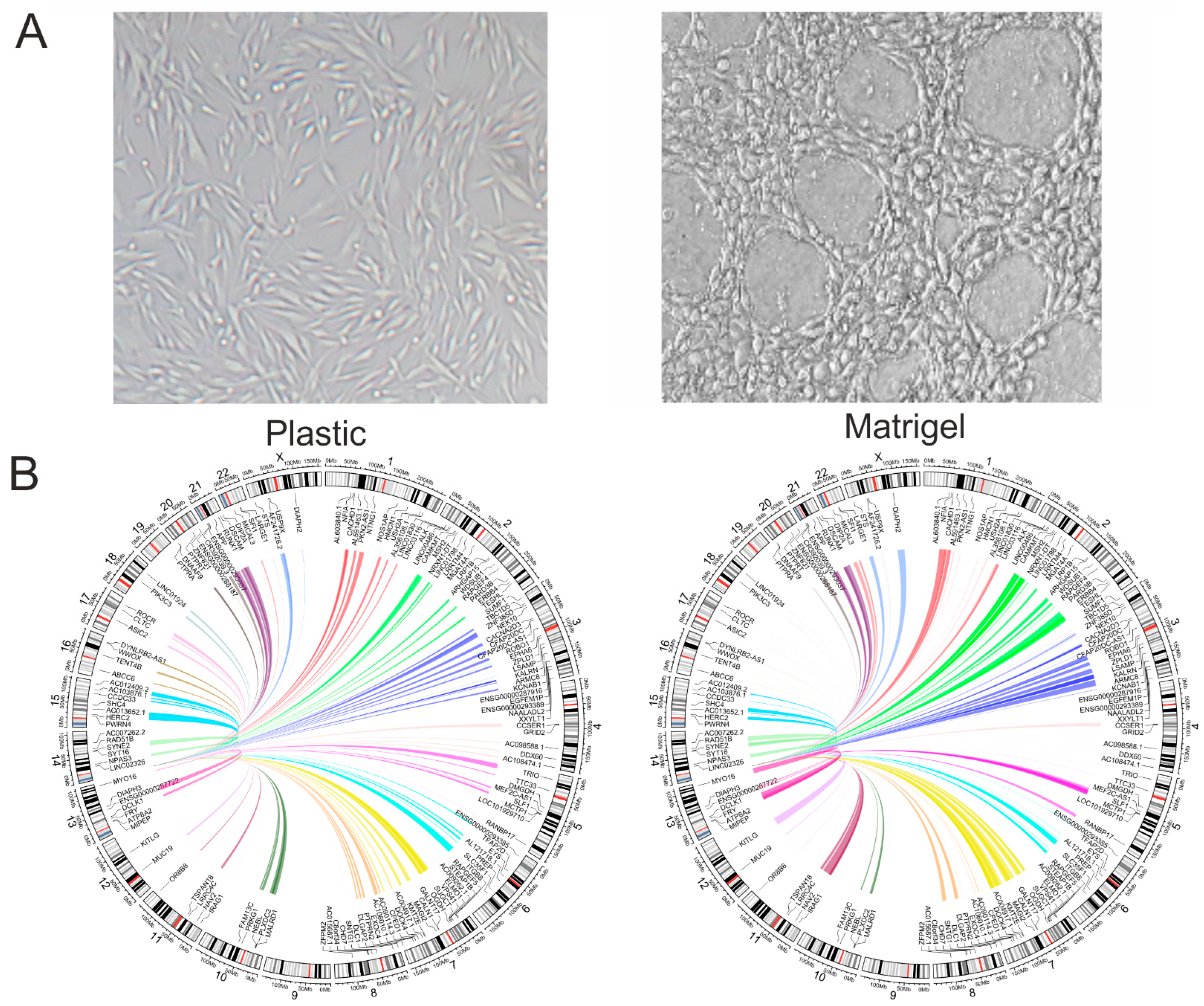
2.1. Transferring Melanoma Cells from a Plastic Surface to Matrigel Leads to the Formation of the VM Phenotype and Changes in rDNA Inter-Chromosomal Contacts
2.2. Analysis of Biological Processes in Which rDNA-Contacting Genes in Mel Z Cells Are Involved
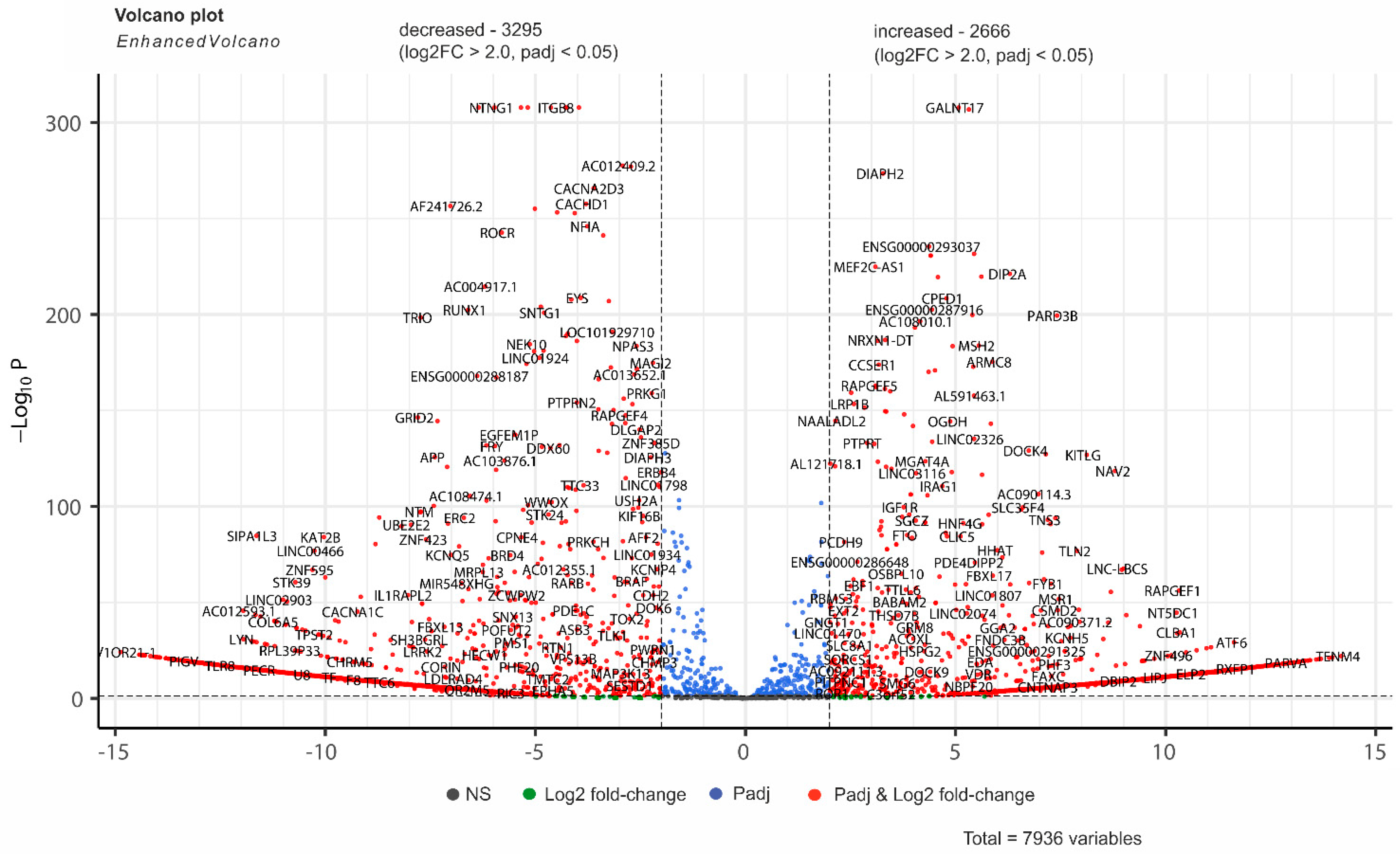
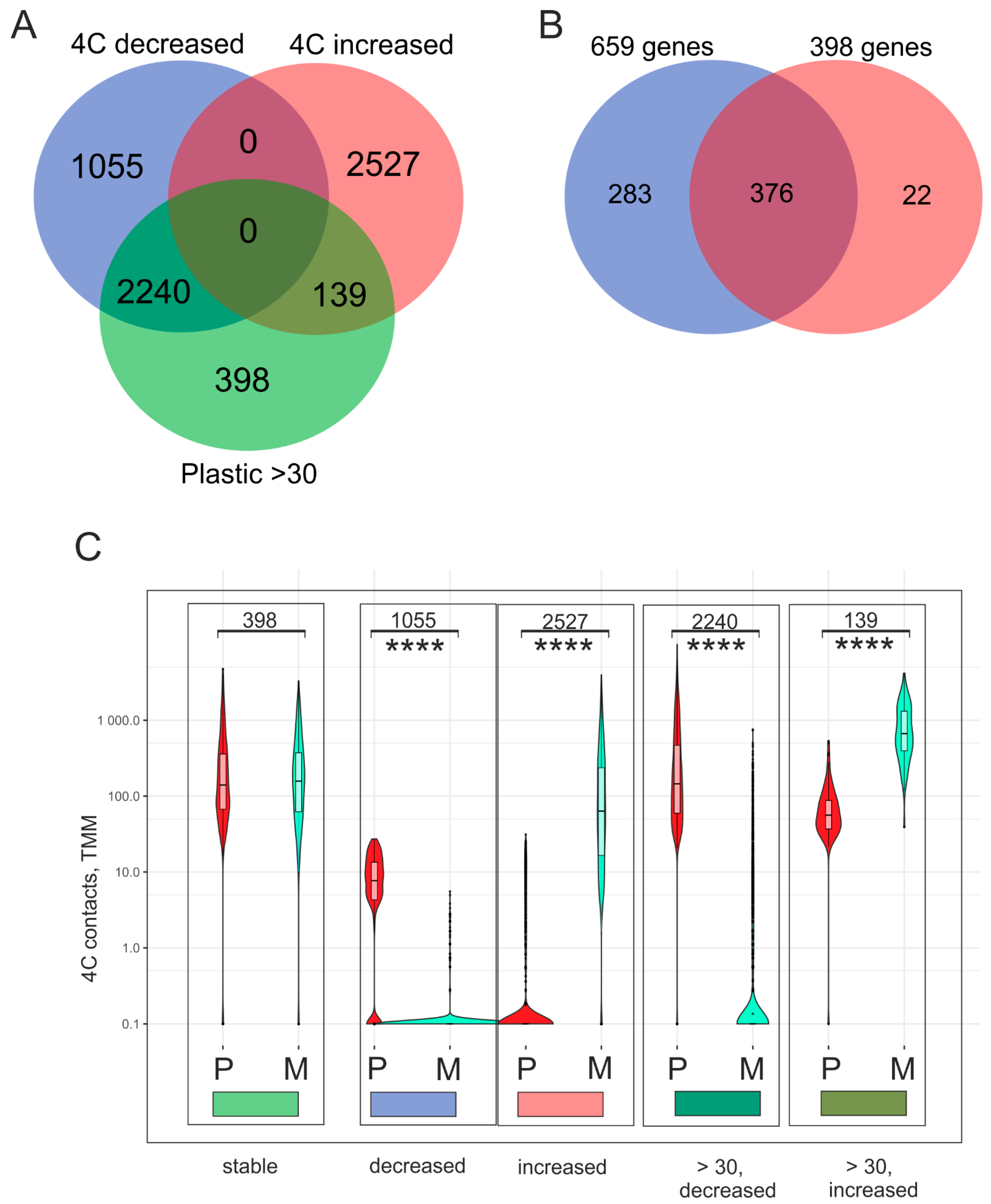
2.3. The Amount of Contacts Between Genes and rDNA Clusters in Mel Z Cells Changes Significantly
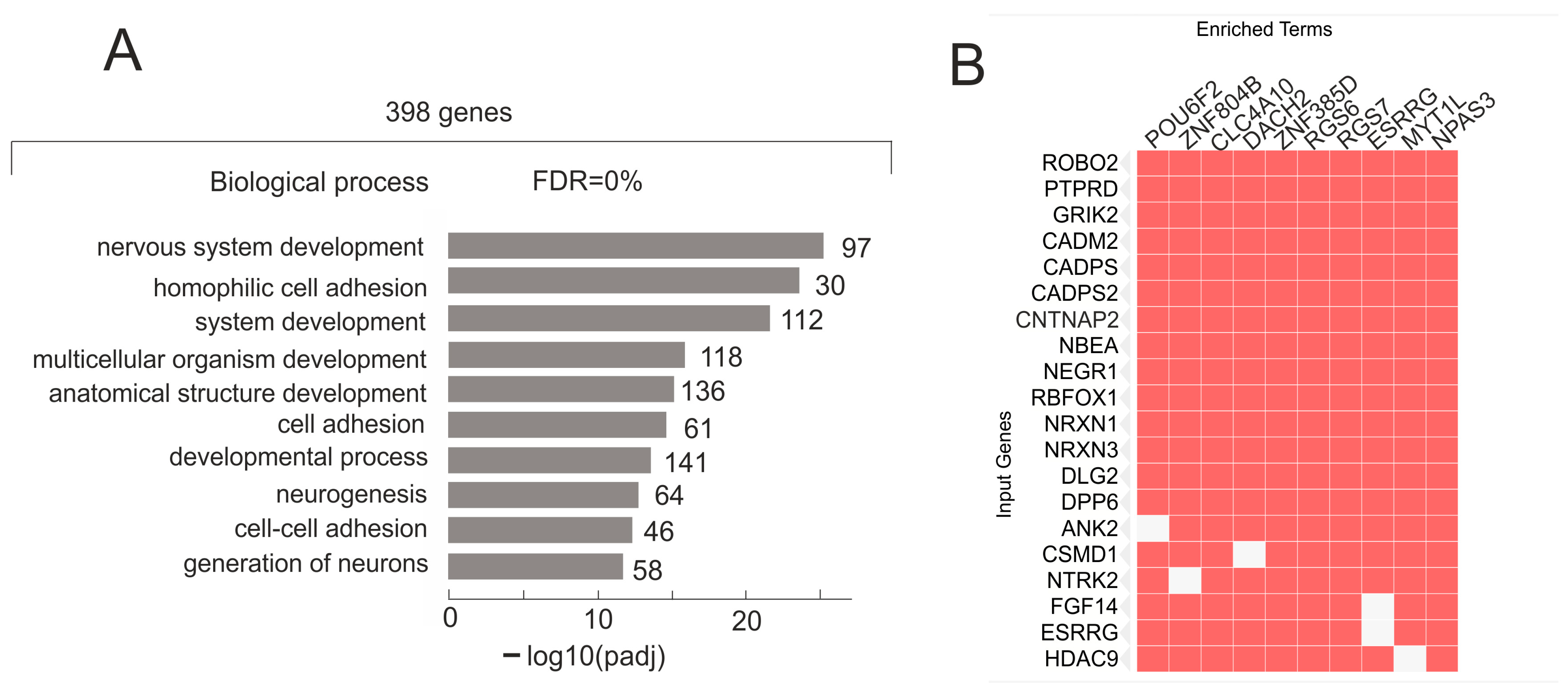
2.4. Only 398 Genes That Stably Retain the Numbers of Their Contacts with rDNA Clusters Are Co-Expressed with Numerous lincRNAs and Are Highly Associated with H3K27me3 Marks
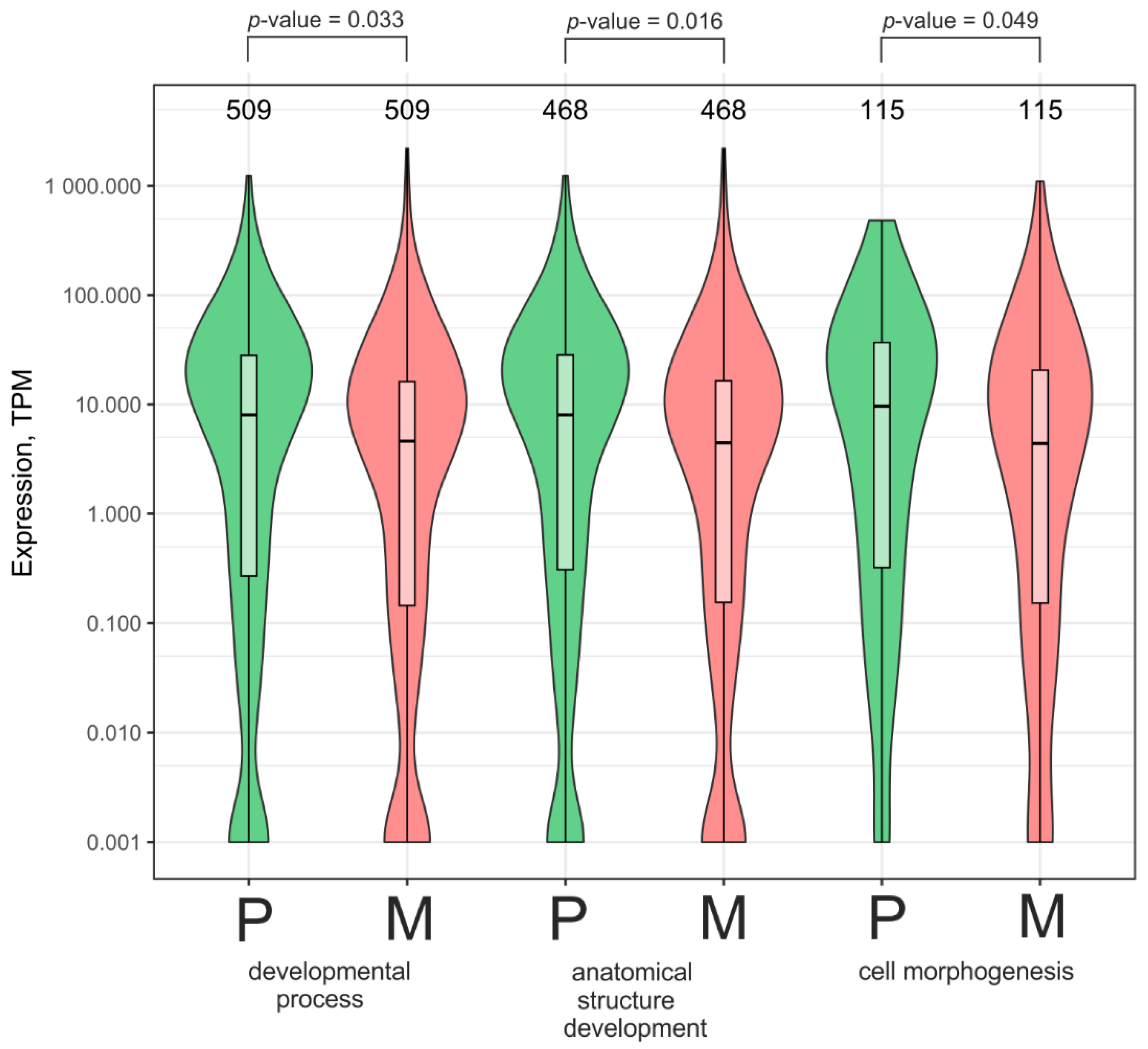
2.5. Numerous Genes Whose Number of Contacts with rDNA Clusters Increased After Growth on Matrigel Are Involved in Development and Morphogenesis and Are Subject to Silencing
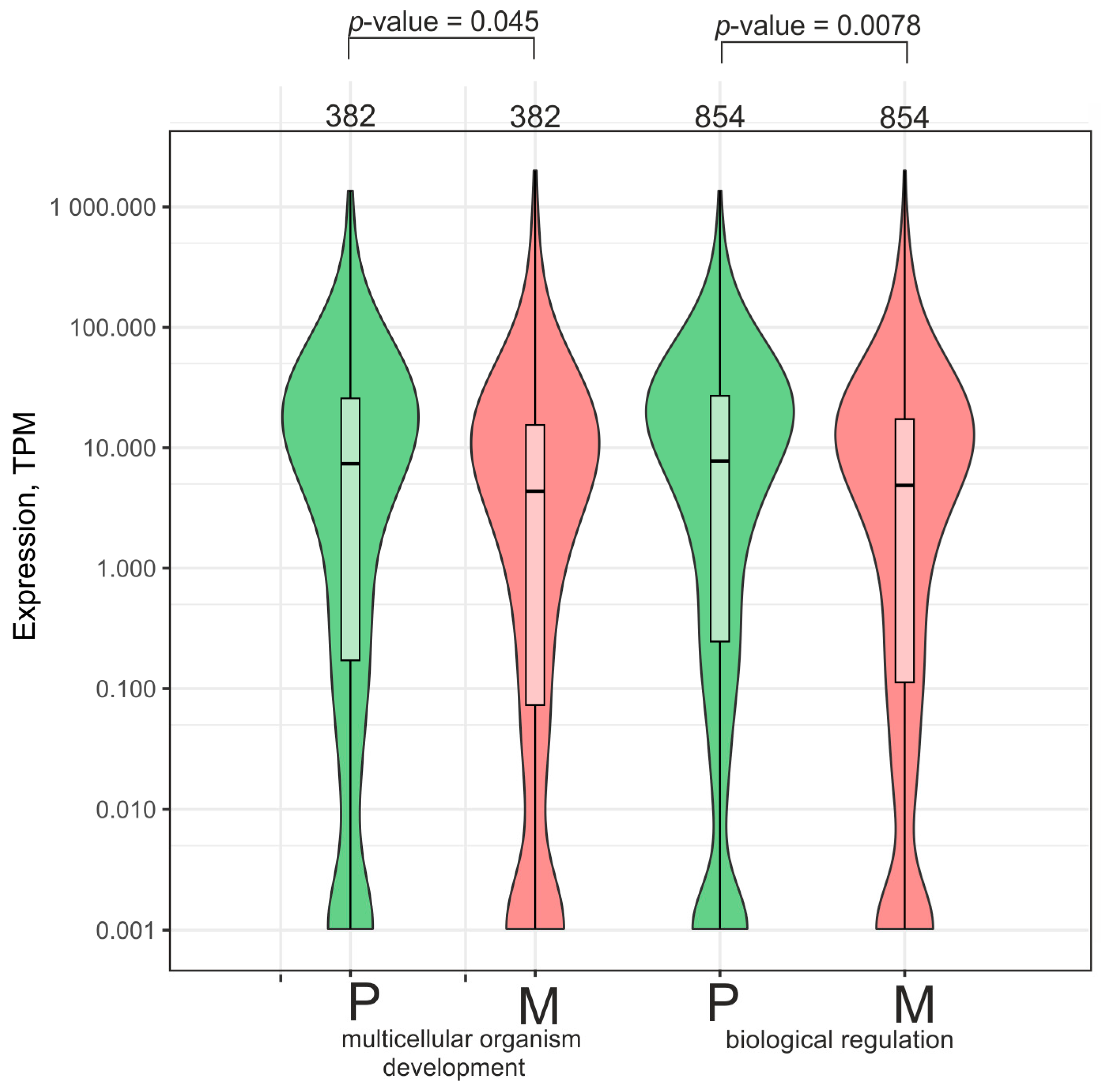
2.6. Numerous Genes Whose Number of Contacts with rDNA Clusters Decreased While Growing on Matrigel Are Involved in Development and Biological Regulation
3. Discussion
3.1. Development of Vasculogenic Mimicry Is Connected with the Functions of rDNA-Contacting Genes
3.2. Genes Forming Stable Contacts with Nucleoli Are Strongly Associated with lincRNAs and H3K27ac Marks
3.3. Possible Role of lincRNAs and ZNF Genes in Inter-Chromosomal Interactions
3.4. How Nucleoli May Be Involved in Both the Activation and Repression of rDNA-Contacting Genes
4. Materials and Methods
4.1. Cell Culture
4.2. 4C-rDNA Procedure
4.3. 4C Mapping and Processing
- 4C full-length adapters (both direct and reverse complement (RC)):>A1D TTCACTTCTGACATCCCAGATTTGATCTCCCTACAGAATGCTGTACAGAACTGGCGAGTTGATTTCTGGACTT>A1RC AAGTCCAGAAATCAACTCGCCAGTTCTGTACAGCATTCTGTAGGGAGATCAAATCTGGGATGTCAGAAGTGAA>A2D TCTTTGAAAAAAATCCCAGAAGTGGTTTTGGCTTTTTGGCTAGGAGGCCTAAGCCTGCTGAGAACTTTCCTGCCCAGGATCCT>A2RC AGGATCCTGGGCAGGAAAGTTCTCAGCAGGCTTAGGCCTCCTAGCCAAAAAGCCAAAACCACTTCTGGGATTTTTTTCAAAGAwere removed at 5′ ends with the options -O 10 (minimal overlap adapter with the read), --trim-n (omit Ns at the ends of reads), --times = 4 (search for the adapter up to 4 times in the read consequently), --minimum-length 20 (minimum acceptable read length after trimming), and -q 24 (minimal acceptable quality). All untrimmed reads were separated into a particular file for further processing.
- Illumina 3′ adapter arrays from AGATCGGAAGAGC to AGATCGGAAGAGCNNNNNNNNNN and from GATCGGAAGAGC to GATCGGAAGAGCNNNNNNNNNN anchored to the 3′ end of reads were deleted using cutadapt with the following options: -O 10 (minimal overlap adapter with the read), --times = 4 (search for the adapter up to 4 times in the read), --minimum-length 20 (minimum acceptable read length after trimming), and -q 24 (minimal acceptable quality).
- Incomplete 5′ 4C full-length adapter arrays from TTCACTTCTGACATCCCAGATTTGATCTCCCTACAGAATGCTGTACAGAACTGGCGAGTTGATTTCTGGACTT to TTCACTTCTGACATCCCAGA (minimal length = 20) and from TCTTTGAAAAAAATCCCAGAAGTGGTTTTGGCTTTTTGGCTAGGAGGCCTAAGCCTGCTGAGAACTTTCCTGCCCAGGATCCT to TCTTTGAAAAAAATCCCAGA (minimal length = 20), both direct and reverse complement, anchored to the 5′ end of reads, were filtered out using cutadapt with the same options as above.
- Illumina adapter GATCGGAAGAGC and IlluminaPE adapter AGATCGGAAGAGC were removed using cutadapt from the 3′ ends of reads with the following options: -O 5 (minimal overlap adapter with the read), --times = 4 (search for the adapter up to 4 times in the read), --minimum-length 20 (minimum acceptable read length after trimming), and -q 24 (minimal acceptable quality).
- Incomplete 3′ 4C full-length adapter arrays from TTCACTTCTGACATCCCAGATTTGATCTCCCTACAGAATGCTGTACAGAACTGGCGAGTTGATTTCTGGACTT to GCGAGTTGATTTCTGGACTT (minimal length = 20) and from TCTTTGAAAAAAATCCCAGAAGTGGTTTTGGCTTTTTGGCTAGGAGGCCTAAGCCTGCTGAGAACTTTCCTGCCCAGGATCCT to ACTTTCCTGCCCAGGATCCT (minimal length = 20), both direct and reverse complement, were deleted using cutadapt from the 3′ end of reads with the same options as in points 2 and 3 of this protocol.
- All untrimmed reads separated at the first step of the procedure (i.e., the reads where no adapters were found) were trimmed again by the described procedure (points 1–5) with the following differences: at the first step, the changed set of cutadapt options was applied: -e 0.2 (this option raises the error rate to 0.2, thus enabling researchers to find adapters that were read with more errors) and -O 15 (instead of -O 10, thus requiring longer overlap of adapters with the reads).
4.4. Differential 4C Analysis
4.5. Violin Plots for 4C-rDNA Data
4.6. Violin Plots for Gene Expression Data
4.7. Code Accessibility
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hendrix, M.J.; Seftor, E.A.; Seftor, R.E.; Chao, J.-T.; Chien, D.-S.; Chu, Y.-W. “Tumor cell vascular mimicry: Novel targeting opportunity in melanoma”. Pharmacol. Ther. 2016, 159, 83–92. [Google Scholar] [CrossRef]
- Wei, X.; Chen, Y.; Jiang, X.; Peng, M.; Liu, Y.; Mo, Y.; Ren, D.; Hua, Y.; Yu, B.; Zhou, Y.; et al. Mechanisms of vasculogenic mimicry in hypoxic tumor microenvironments. Mol. Cancer 2021, 20, 7. [Google Scholar] [CrossRef] [PubMed]
- Maniotis, A.J.; Folberg, R.; Hess, A.; Seftor, E.A.; Gardner, L.M.; Pe’er, J.; Trent, J.M.; Meltzer, P.S.; Hendrix, M.J. Vascular channel formation by human melanoma cells in vivo and in vitro: Vasculogenic mimicry. Am. J. Pathol. 1999, 155, 739–752. [Google Scholar] [CrossRef]
- Cao, Z.; Bao, M.; Miele, L.; Sarkar, F.H.; Wang, Z.; Zhou, Q. Tumour vasculogenic mimicry is associated with poor prognosis of human cancer patients: A systemic review and meta-analysis. Eur. J. Cancer 2013, 49, 3914–3923. [Google Scholar] [CrossRef]
- Liu, H.; Wang, H.; Chen, D.; Gu, C.; Huang, J.; Mi, K. Endoplasmic reticulum stress inhibits 3D Matrigel-induced vasculogenic mimicry of breast cancer cells via TGF-β1/Smad2/3 and β-catenin signaling. FEBS Open Bio 2021, 11, 2607–2618. [Google Scholar] [CrossRef]
- Pathan, S.; Ali, T.; Vincent, S.; Nanjappa, Y.; David, R.M.; Kumar, O.P. A Dermoscopic Inspired System for Localization and Malignancy Classification of Melanocytic Lesions. Appl. Sci. 2022, 12, 4243. [Google Scholar] [CrossRef]
- Vartanian, A.; Stepanova, E.; Grigorieva, I.; Solomko, E.; Belkin, V.; Baryshnikov, A.; Lichinitser, M. Melanoma vasculogenic mimicry capillary-like structure formation depends on integrin and calcium signaling. Microcirculation 2011, 18, 390–399. [Google Scholar] [CrossRef]
- Diesch, J.; Bywater, M.J.; Sanij, E.; Cameron, D.P.; Schierding, W.; Brajanovski, N.; Son, J.; Sornkom, J.; Hein, N.; Evers, M.; et al. Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II. gene programs during malignant transformation. Commun. Biol. 2019, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- Tchurikov, N.A.; Fedoseeva, D.M.; Klushevskaya, E.S.; Slovohotov, I.Y.; Chechetkin, V.R.; Kravatsky, Y.V.; Kretova, O.V. rDNA Clusters Make Contact with Genes that Are Involved in Differentiation and Cancer and Change Contacts after Heat Shock Treatment. Cells 2019, 8, 1393. [Google Scholar] [CrossRef]
- Tchurikov, N.A.; Klushevskaya, E.S.; Alembekov, I.R.; Kretova, A.N.; Chechetkin, V.R.; Kravatskaya, G.I.; Kravatsky, Y.V. Induction of the Erythroid Differentiation of K562 Cells Is Coupled with Changes in the Inter-Chromosomal Contacts of rDNA Clusters. Int. J. Mol. Sci. 2023, 24, 9842. [Google Scholar] [CrossRef] [PubMed]
- Tchurikov, N.A.; Vartanian, A.A.; Klushevskaya, E.S.; Alembekov, I.R.; Kretova, A.N.; Lukicheva, V.N.; Chechetkin, V.R.; Kravatskaya, G.I.; Kosorukov, V.S.; Kravatsky, Y.V. Strong Activation of ID1, ID2, and ID3 Genes Is Coupled with the Formation of Vasculogenic Mimicry Phenotype in Melanoma Cells. Int. J. Mol. Sci. 2024, 25, 9291. [Google Scholar] [CrossRef]
- Vartanian, A.A.; Khochenkova, Y.A.; Kosobokova, E.N.; Baryshnikova, M.A.; Kosorukov, V.S. CD437 Reduces Metastatic Potential of Melanoma Cells. Mosc. Univ. Chem. Bull. 2021, 76, 208–214. [Google Scholar] [CrossRef]
- Price, K.J.; Tsykin, A.; Giles, K.M.; Sladic, R.T.; Epis, M.R.; Ganss, R.; Goodall, G.J.; Leedman, P.J. Matrigel basement membrane matrix influences expression of microRNAs in cancer cell lines. Biochem. Biophys. Res. Commun. 2012, 427, 343–348. [Google Scholar] [CrossRef]
- Vartanian, A.; Karshieva, S.; Dombrovsky, V.; Belyavsky, A. Melanoma educates mesenchymal stromal cells towards vasculogenic mimicry. Oncol. Lett. 2016, 11, 4264–4268. [Google Scholar] [CrossRef]
- Vartanian, A.; Baryshnikova, M.; Burova, O.; Afanasyeva, D.; Misyurin, V.; Belyavsky, A.; Shprakh, Z. Inhibitor of vasculogenic mimicry restores sensitivity of resistant melanoma cells to DNA-damaging agents. Melanoma Res. 2017, 27, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Jögi, A.; Vaapil, M.; Johansson, M.; Påhlman, S. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Upsala J. Med. Sci. 2012, 117, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Seipel, K.; Medley, Q.G.; Kedersha, N.L.; Zhang, X.A.; O’bRien, S.P.; Serra-Pages, C.; Hemler, M.E.; Streuli, M. Trio amino-terminal guanine nucleotide exchange factor domain expression promotes actin cytoskeleton reorganization, cell migration and anchorage-independent cell growth. J. Cell Sci. 1999, 112 Pt 12, 1825–1834. [Google Scholar] [CrossRef] [PubMed]
- Räschle, M.; Smeenk, G.; Hansen, R.K.; Temu, T.; Oka, Y.; Hein, M.Y.; Nagaraj, N.; Long, D.T.; Walter, J.C.; Hofmann, K.; et al. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 2015, 348, 1253671. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, Q.; Maniatis, T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 1999, 97, 779–790. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, W.; Tan, Y.; Su, G.; Xu, L.; Jiang, M.L.; Li, S.; Meir, Y.-J.J.; Wang, Y.; Li, G.; et al. Limbal Niche Cells and Three-Dimensional Matrigel-Induced Dedifferentiation of Mature Corneal Epithelial Cells. Investig. Ophthalmol. Vis. Sci. 2022, 63, 1. [Google Scholar] [CrossRef]
- Zheng, Y.-L.; Sun, Y.-P.; Zhang, H.; Liu, W.-J.; Jiang, R.; Li, W.-Y.; Zheng, Y.-H.; Zhang, Z.-G.; Ivanovic, Z. Mesenchymal Stem Cells Obtained from Synovial Fluid Mesenchymal Stem Cell-Derived Induced Pluripotent Stem Cells on a Matrigel Coating Exhibited Enhanced Proliferation and Differentiation Potential. PLoS ONE 2015, 10, e0144226. [Google Scholar] [CrossRef]
- Lang, S.H.; Sharrard, R.M.; Stark, M.; Villette, J.M.; Maitland, N.J. Prostate epithelial cell lines form spheroids with evidence of glandular differentiation in three-dimensional Matrigel cultures. Br. J. Cancer 2001, 85, 590–599. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Tchurikov, N.A.; Kravatsky, Y.V. The Role of rDNA Clusters in Global Epigenetic Gene Regulation. Front. Genet. 2021, 12, 730633. [Google Scholar] [CrossRef] [PubMed]
- Latonen, L. Phase-to-Phase With Nucleoli—Stress Responses, Protein Aggregation and Novel Roles of RNA. Front. Cell Neurosci. 2019, 13, 151. [Google Scholar] [CrossRef]
- Gupta, S.; Santoro, R. Regulation and Roles of the Nucleolus in Embryonic Stem Cells: From Ribosome Biogenesis to Genome Organization. Stem Cell Rep. 2020, 15, 1206–1219, ISSN 2213-6711. [Google Scholar] [CrossRef] [PubMed]
- Ananiev, E.V.; Barsky, V.E.; Ilyin, Y.V.; Churikov, N.A. Localization of nucleoli in Drosophila melanogaster polytene chromosomes. Chromosoma 1981, 81, 619–628. [Google Scholar] [CrossRef]
- Yu, S.; Lemos, B. The long-range interaction map of ribosomal DNA arrays. PLoS Genet. 2018, 14, e1007258. [Google Scholar] [CrossRef]
- Li, X.; Tsai, P.; Wieder, E.D.; Kribben, A.; Van Putten, V.; Schrier, R.W.; Nemenoff, R.A. Vascular smooth muscle cells grown on Matrigel. A model of the contractile phenotype with decreased activation of mitogen-activated protein kinase. J. Biol. Chem. 1994, 269, 19653–19658. [Google Scholar] [PubMed]
- Wang, J.; Edeen, K.; Manzer, R.; Chang, Y.; Wang, S.; Chen, X.; Funk, C.J.; Cosgrove, G.P.; Fang, X.; Mason, R.J. Differentiated human alveolar epithelial cells and reversibility of their phenotype in vitro. Am. J. Respir. Cell Mol. Biol. 2007, 36, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Tchurikov, N.A.; Alembekov, I.R.; Klushevskaya, E.S.; Kretova, A.N.; Lukicheva, V.N.; Chechetkin, V.R.; Kravatskaya, G.I.; Kravatsky, Y.V. Preferential Co-Expression and Colocalization of rDNA-Contacting Genes with LincRNAs Suggest Their Involvement in Shaping Inter-Chromosomal Interactions with Nucleoli. Int. J. Mol. Sci. 2024, 25, 6333. [Google Scholar] [CrossRef]
- Wang, C.; Wang, L.; Ding, Y.; Lu, X.; Zhang, G.; Yang, J.; Zheng, H.; Wang, H.; Jiang, Y.; Xu, L. LncRNA Structural Characteristics in Epigenetic Regulation. Int. J. Mol. Sci. 2017, 18, 2659. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Deniz, E.; Erman, B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct. Integr. Genom. 2017, 17, 135–143. [Google Scholar] [CrossRef]
- Liang, Y.; Liu, M.; Liu, B.; Ziman, B.; Peng, G.; Mao, Q.; Wang, X.; Jiang, L.; Lin, D.-C.; Zheng, Y. Comprehensive analysis of H3K27me3 LOCKs under different DNA methylation contexts reveal epigenetic redistribution in tumorigenesis. Epigenet. Chromatin 2025, 18, 6. [Google Scholar] [CrossRef]
- Tchurikov, N.A.; Klushevskaya, E.S.; Kravatsky, Y.V.; Kravatskaya, G.I.; Fedoseeva, D.M. Interchromosomal Contacts of rDNA Clusters in Three Human Cell Lines Are Associated with Silencing of Genes Controlling Morphogenesis. Dokl. Biochem. Biophys. 2021, 496, 22–26. [Google Scholar] [CrossRef]
- Begnis, M.; Duc, J.; Offner, S.; Grun, D.; Sheppard, S.; Rosspopoff, O.; Trono, D. Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments. Sci. Adv. 2024, 10, eado1662. [Google Scholar] [CrossRef]
- Fedoseeva, D.M.; Kretova, O.V.; Gorbacheva, M.A.; Tchurikov, N.A. Individual effects of the copia and gypsy enhancer and insulator on chromatin marks, eRNA synthesis, and binding of insulator proteins in transfected genetic constructs. Gene 2018, 641, 151–160. [Google Scholar] [CrossRef]
- Tchurikov, N.A.; Fedoseeva, D.M.; Sosin, D.V.; Snezhkina, A.V.; Melnikova, N.V.; Kudryavtseva, A.V.; Kravatsky, Y.V.; Kretova, O.V. Hot spots of DNA double-strand breaks and genomic contacts of human rDNA units are involved in epigenetic regulation. J. Mol. Cell Biol. 2015, 7, 366–382. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-enhancers in the control of cell identity and disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator Condensation at Super-Enhancers Links Phase Separation and Gene Control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bushnell, B. BBTools: A Suite of Fast, Multithreaded Bioinformatics Tools Designed for Analysis of DNA and RNA Sequence Data. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 1 August 2025).
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar]
- Storer, J.; Hubley, R.; Rosen, J.; Wheeler, T.J.; Smit, A.F. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob. DNA 2021, 12, 2. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef]
- Barrett, T.; Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Stetsenko, P. data.table: Extension of “data.frame.”. The Comprehensive R Archive Network, 2024. [Google Scholar] [CrossRef]
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation. The Comprehensive R Archive Network, 2023. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Blighe, K.; Lewis, M.; Rana, S. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 1 August 2025).
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant graphics for data analysis. In Use R! 2nd ed.; Springer: Cham, Switzerland, 2016; p. 1. [Google Scholar]
- Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Lee, S.; Cook, D.; Lawrence, M. Plyranges: A grammar of genomic data transformation. Genome Biol. 2019, 20, 4. [Google Scholar] [CrossRef] [PubMed]
- Coombes, K.R.; Brock, G.; Abrams, Z.B.; Abruzzo, L.V. Polychrome: Creating and assessing qualitative palettes with many colors. J. Stat. Softw. 2019, 90, 1–23. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Neph, S.; Kuehn, M.S.; Reynolds, A.P.; Haugen, E.; Thurman, R.E.; Johnson, A.K.; Rynes, E.; Maurano, M.T.; Vierstra, J.; Thomas, S.; et al. BEDOPS: High-performance genomic feature operations. Bioinformatics 2012, 28, 1919–1920. [Google Scholar] [CrossRef]
- Wickham, H.; Vaughan, D.; Girlich, M. tidyr: Tidy Messy Data. The Comprehensive R Archive Network, 2024. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tchurikov, N.A.; Klushevskaya, E.S.; Lukicheva, V.N.; Kretova, A.N.; Poperekova, E.N.; Chechetkin, V.R.; Kravatskaya, G.I.; Vartanian, A.A.; Kosorukov, V.S.; Alembekov, I.R.; et al. Formation of the Vasculogenic Mimicry Phenotype in Melanoma Mel Z Cells Is Coupled with Changes in Inter-Chromosomal Contacts of Developmental Genes with rDNA Clusters. Int. J. Mol. Sci. 2025, 26, 8085. https://doi.org/10.3390/ijms26168085
Tchurikov NA, Klushevskaya ES, Lukicheva VN, Kretova AN, Poperekova EN, Chechetkin VR, Kravatskaya GI, Vartanian AA, Kosorukov VS, Alembekov IR, et al. Formation of the Vasculogenic Mimicry Phenotype in Melanoma Mel Z Cells Is Coupled with Changes in Inter-Chromosomal Contacts of Developmental Genes with rDNA Clusters. International Journal of Molecular Sciences. 2025; 26(16):8085. https://doi.org/10.3390/ijms26168085
Chicago/Turabian StyleTchurikov, Nickolai A., Elena S. Klushevskaya, Viktoriya N. Lukicheva, Antonina N. Kretova, Elizaveta N. Poperekova, Vladimir R. Chechetkin, Galina I. Kravatskaya, Amalia A. Vartanian, Vyacheslav S. Kosorukov, Ildar R. Alembekov, and et al. 2025. "Formation of the Vasculogenic Mimicry Phenotype in Melanoma Mel Z Cells Is Coupled with Changes in Inter-Chromosomal Contacts of Developmental Genes with rDNA Clusters" International Journal of Molecular Sciences 26, no. 16: 8085. https://doi.org/10.3390/ijms26168085
APA StyleTchurikov, N. A., Klushevskaya, E. S., Lukicheva, V. N., Kretova, A. N., Poperekova, E. N., Chechetkin, V. R., Kravatskaya, G. I., Vartanian, A. A., Kosorukov, V. S., Alembekov, I. R., & Kravatsky, Y. V. (2025). Formation of the Vasculogenic Mimicry Phenotype in Melanoma Mel Z Cells Is Coupled with Changes in Inter-Chromosomal Contacts of Developmental Genes with rDNA Clusters. International Journal of Molecular Sciences, 26(16), 8085. https://doi.org/10.3390/ijms26168085




