Integrated Analysis of Physiological Responses and Transcriptome of Cotton Seedlings Under Drought Stress
Abstract
1. Introduction
2. Results
2.1. Evaluation and Validation of RNA-Seq
2.1.1. Sequencing Data Analysis
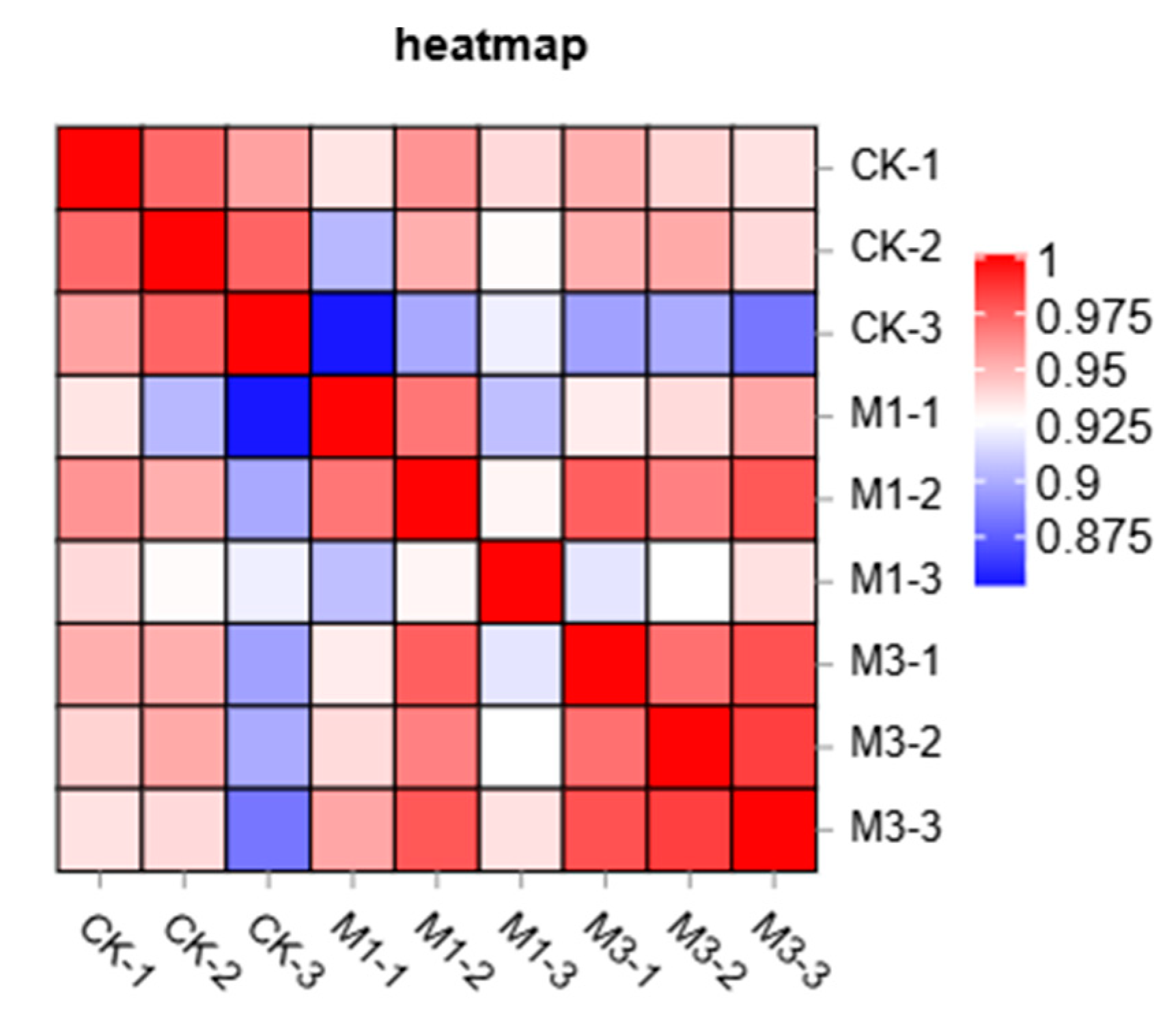
2.1.2. Correlation Analysis Between Samples
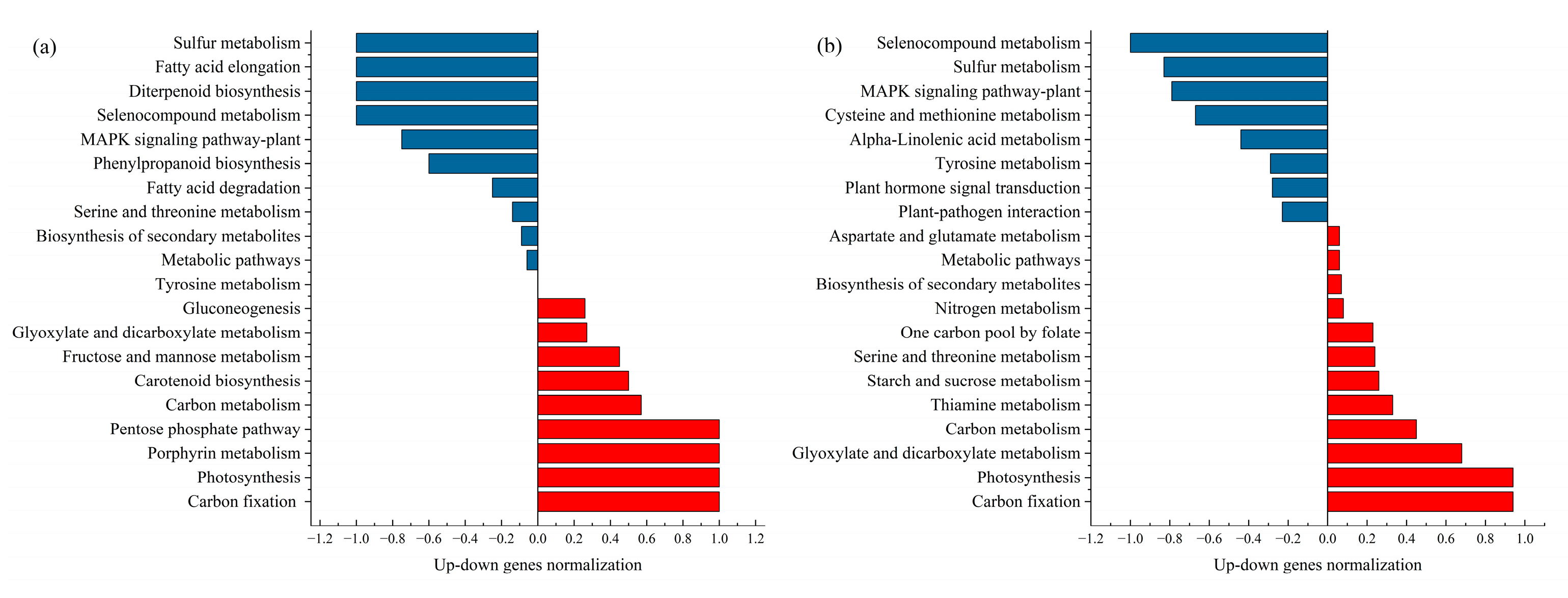
2.2. KEGG (Kyoto Encyclopedia of Genes and Genomes) Metabolic Pathway Analysis of DEGs in Cotton Seedings Under Drought Stress
2.3. An Analysis of the Effects of Drought Stress on the Physiological Indicators of Cotton Seedlings and the DEGs in Related Metabolic Pathways
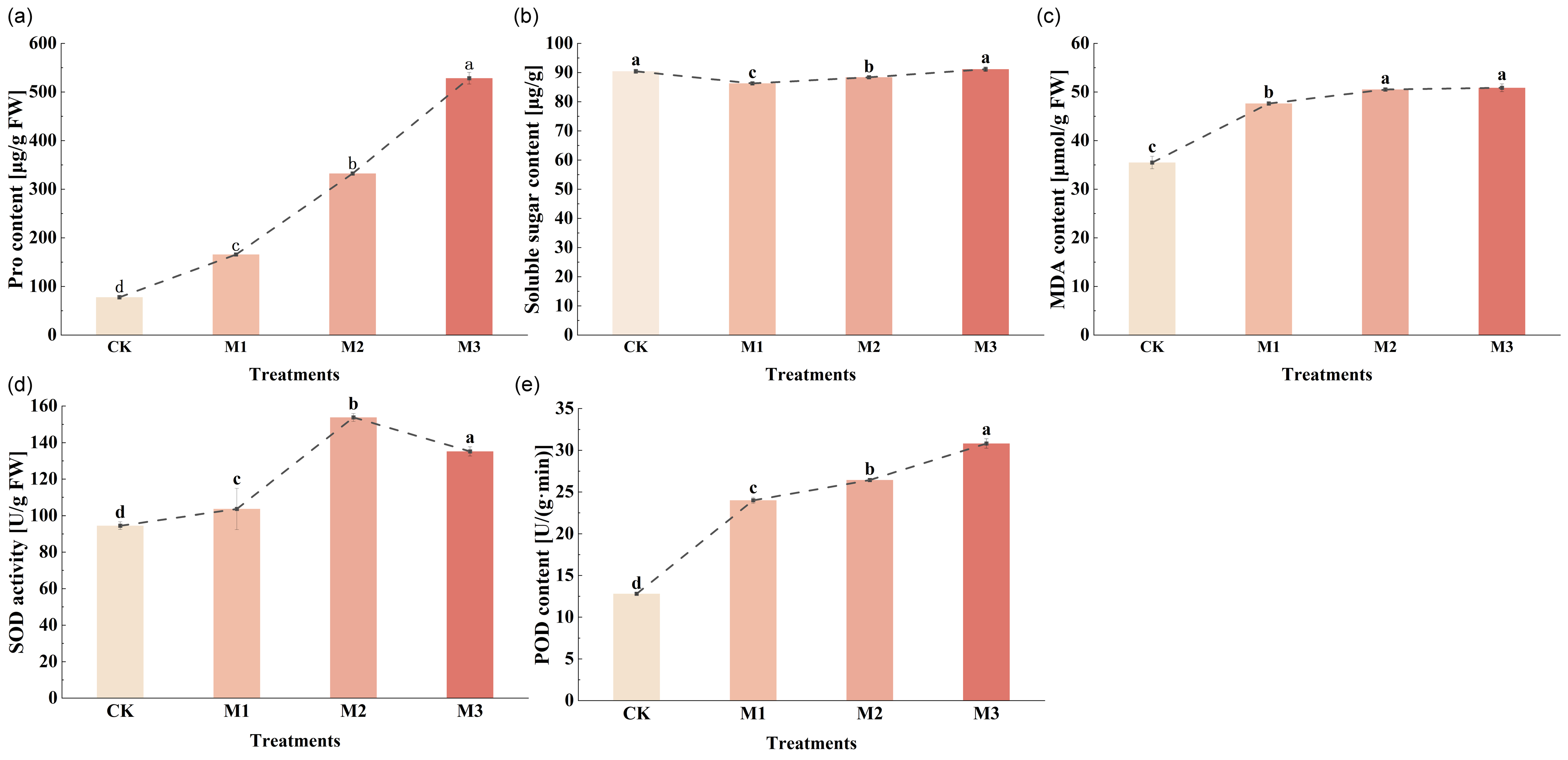
2.3.1. The Physiological Indicators of Cotton Seedlings Under Drought Stress
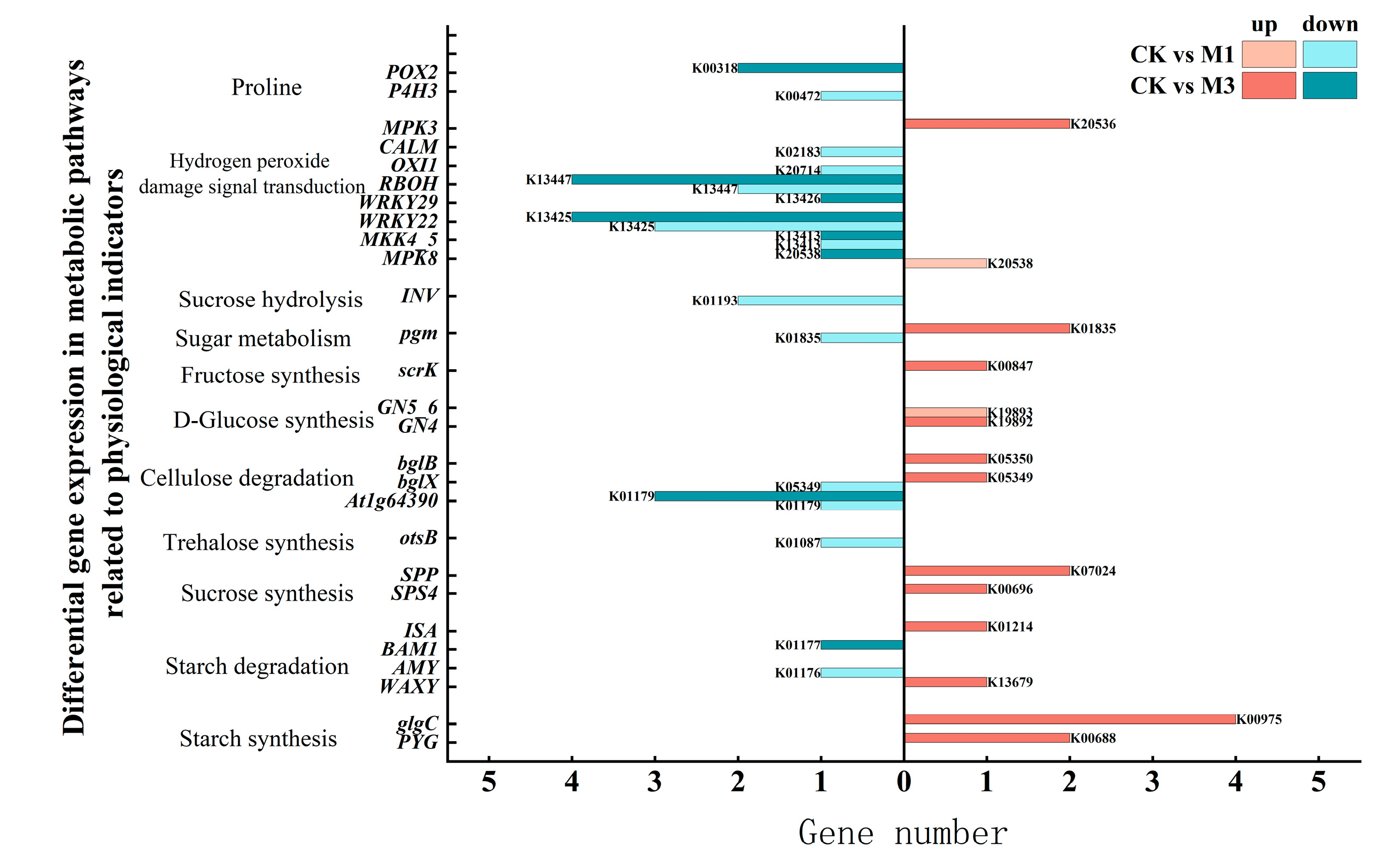
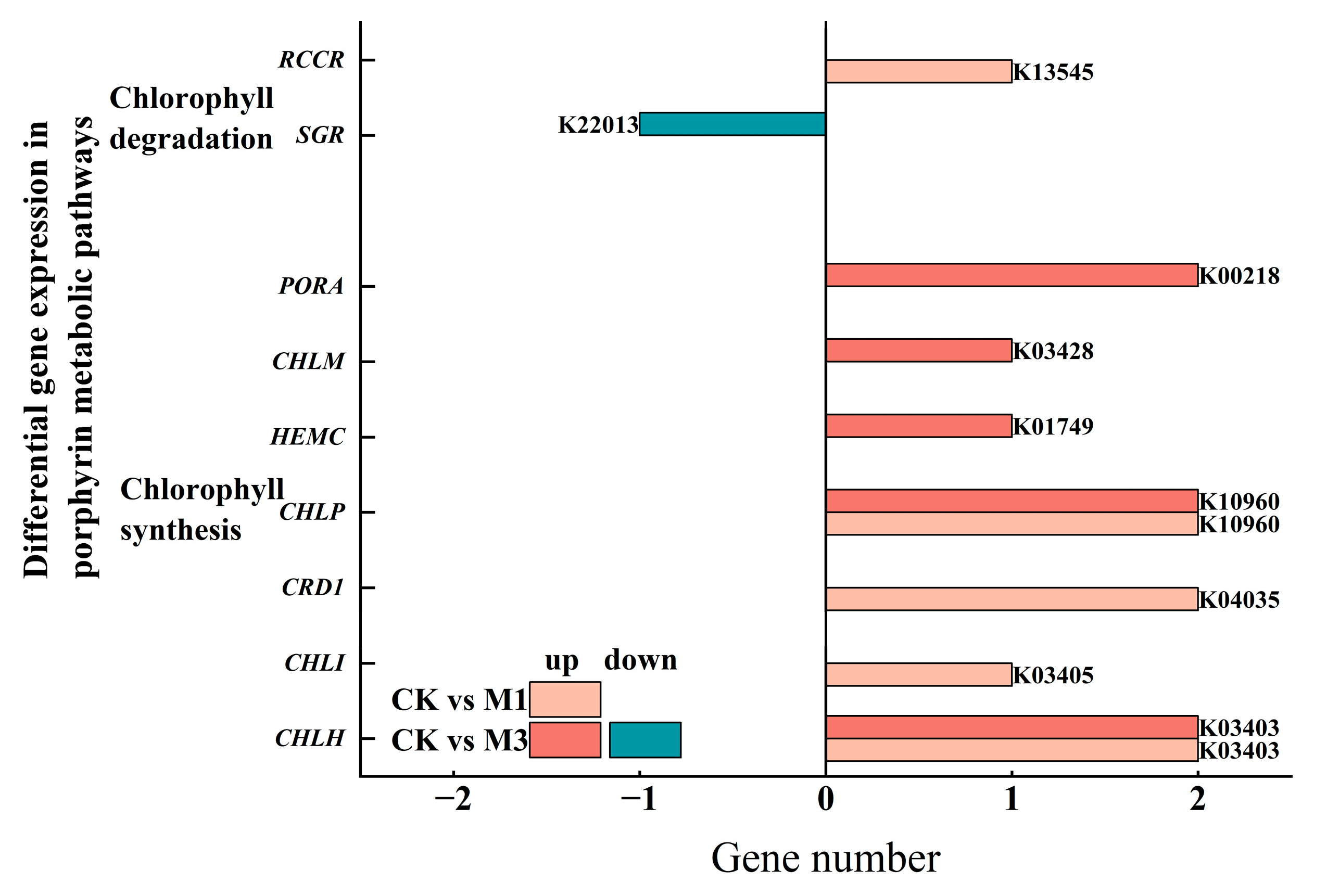
2.3.2. Analysis of DEGs in Pathways Related to Physiological Indicators of Cotton Seedlings Under Drought Stress
2.4. An Analysis of the Effects of Drought Stress on the Photosynthesis of Cotton Seedlings and the DEGs in Related Metabolic Pathways
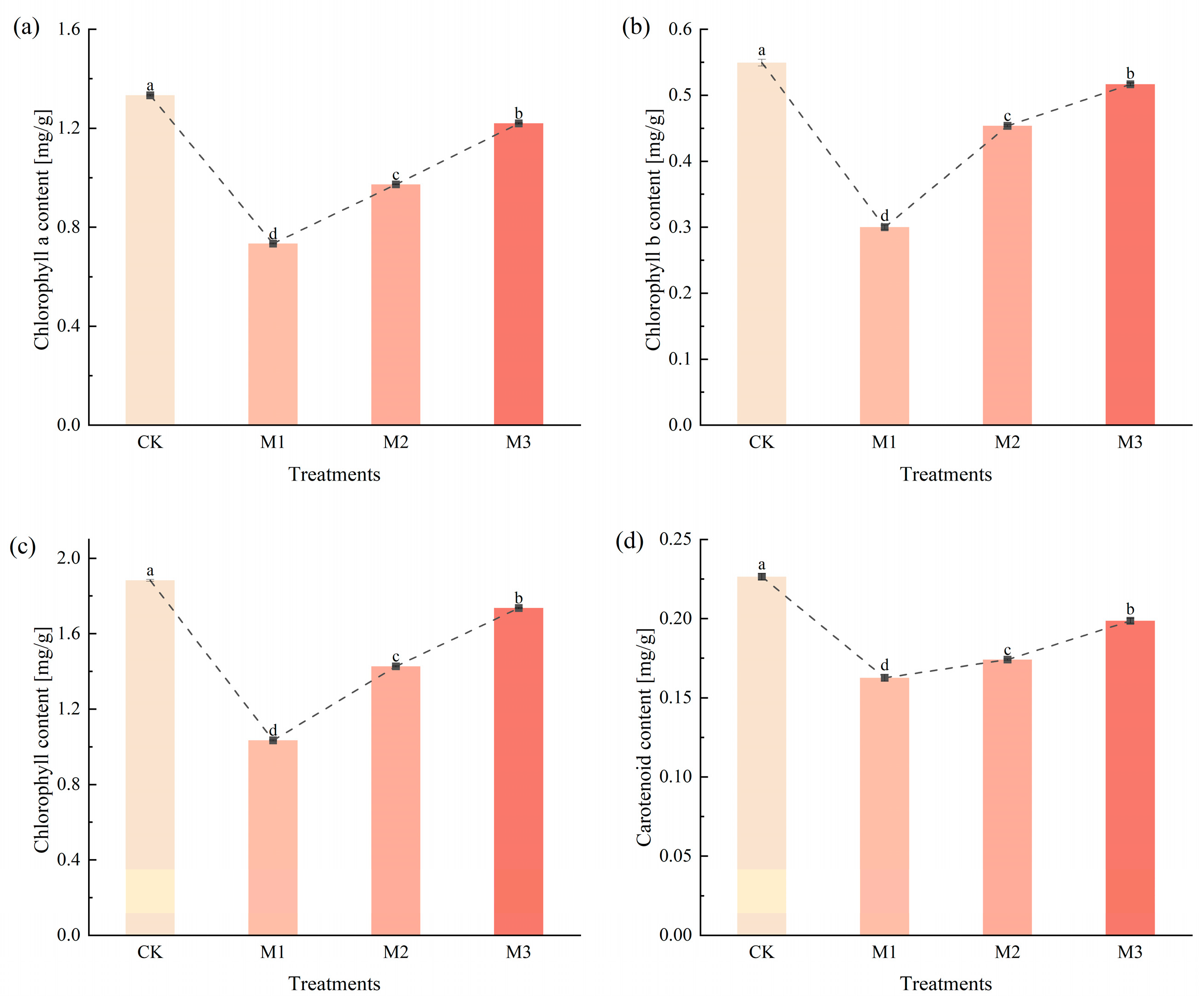
2.4.1. Changes in Chlorophyll Content and Analysis of DEGs in Cotton Seedlings Under Drought Stress
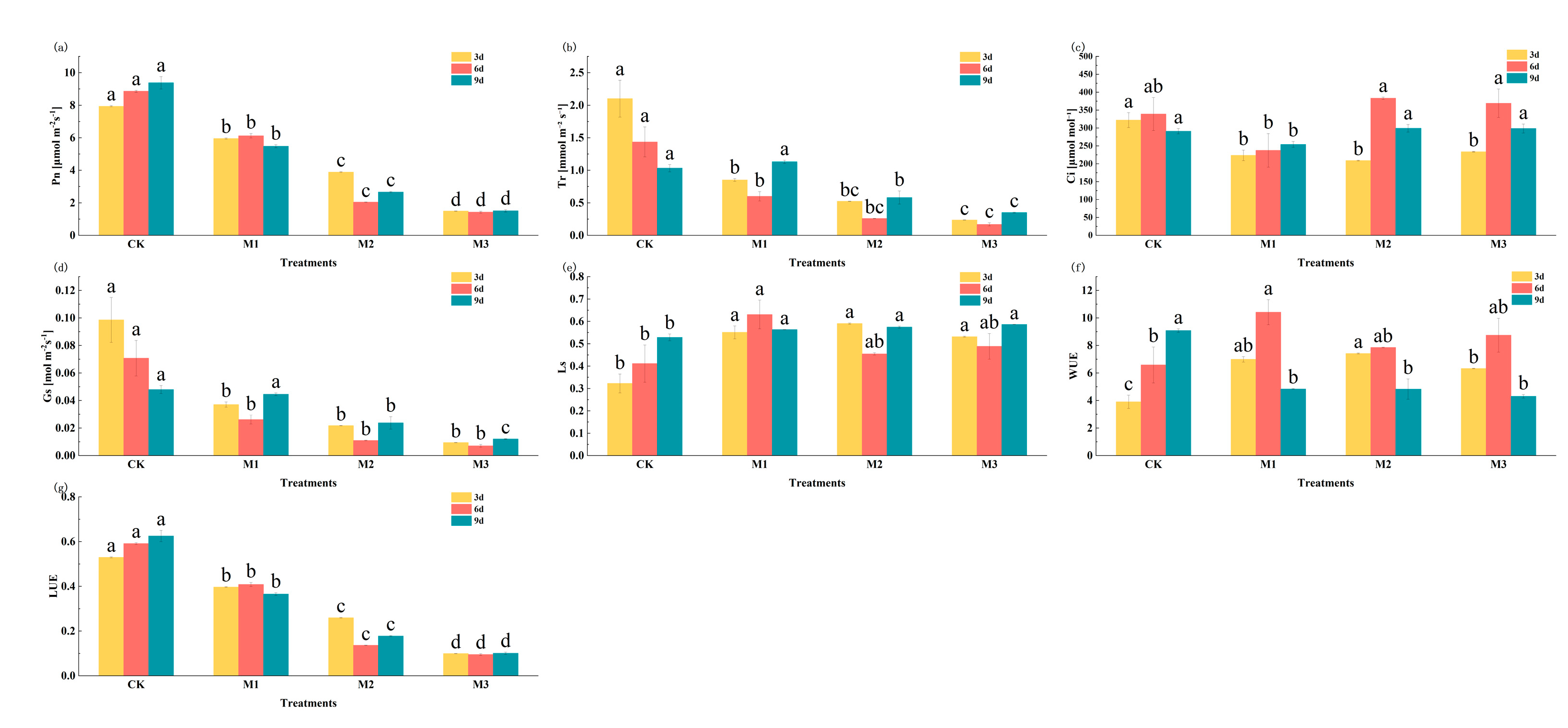
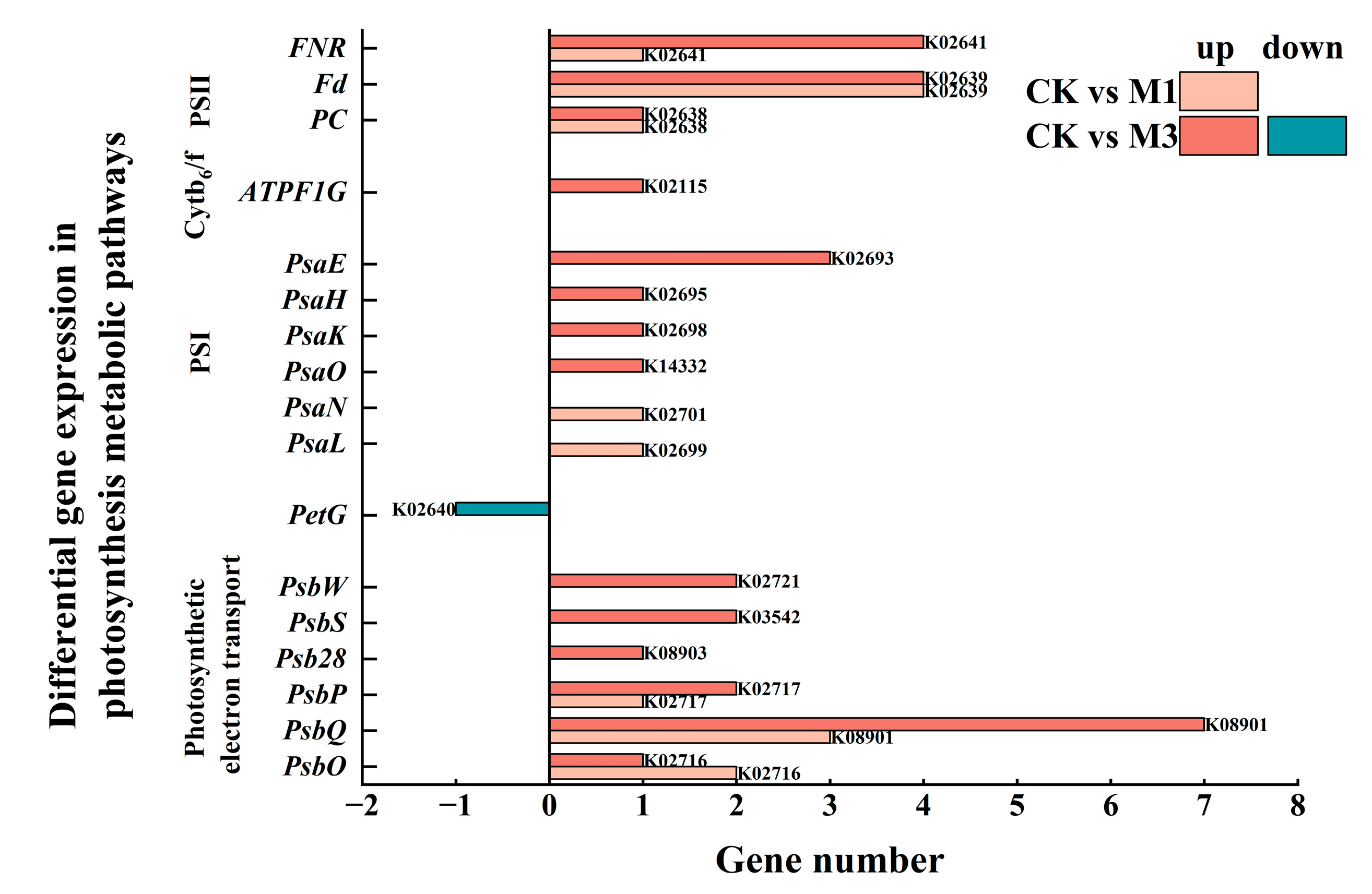
2.4.2. Changes in Gas Exchange Parameters and Analysis of DEGs in Photosynthesis Metabolic Pathway in Cotton Seedlings Under Drought Stress
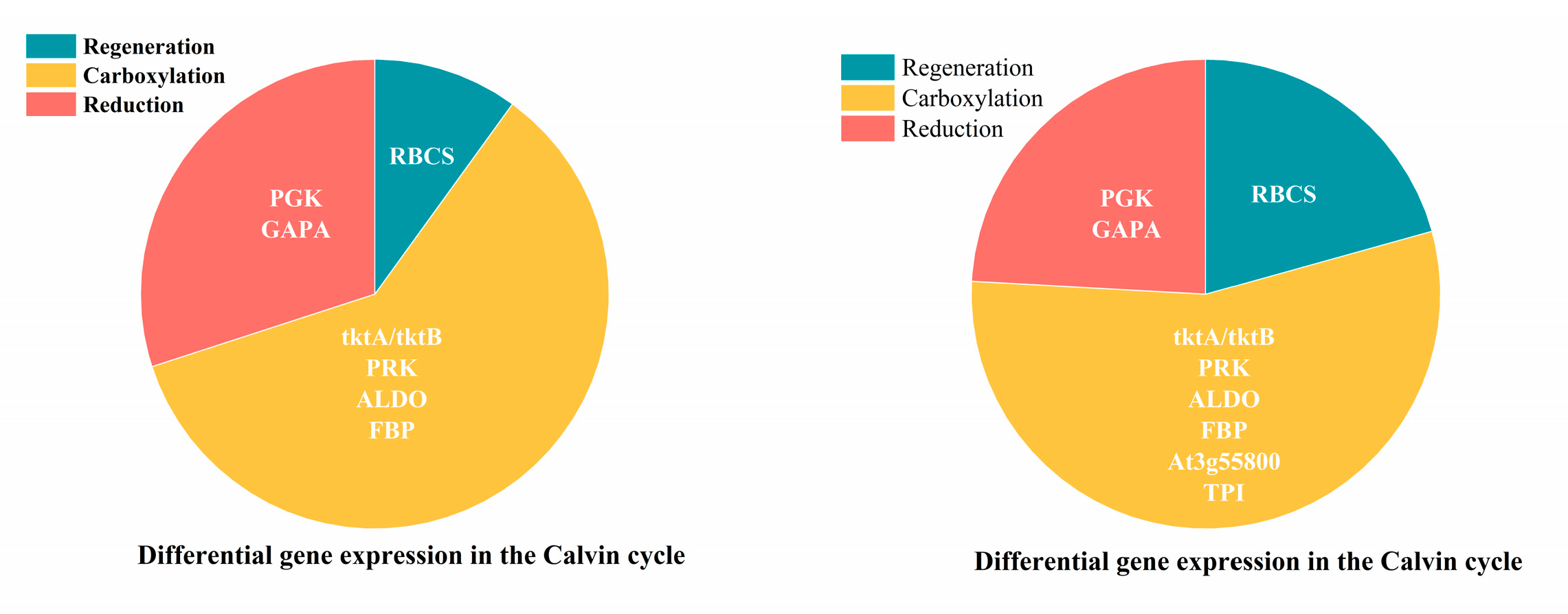
2.4.3. Analysis of DEGs in Carbon Fixation Metabolic Pathway in Cotton Seedlings Under Drought Stress
2.5. An Analysis of the Effects of Drought Stress on Hormones in Cotton Seedlings and the DEGs in Related Metabolic Pathways
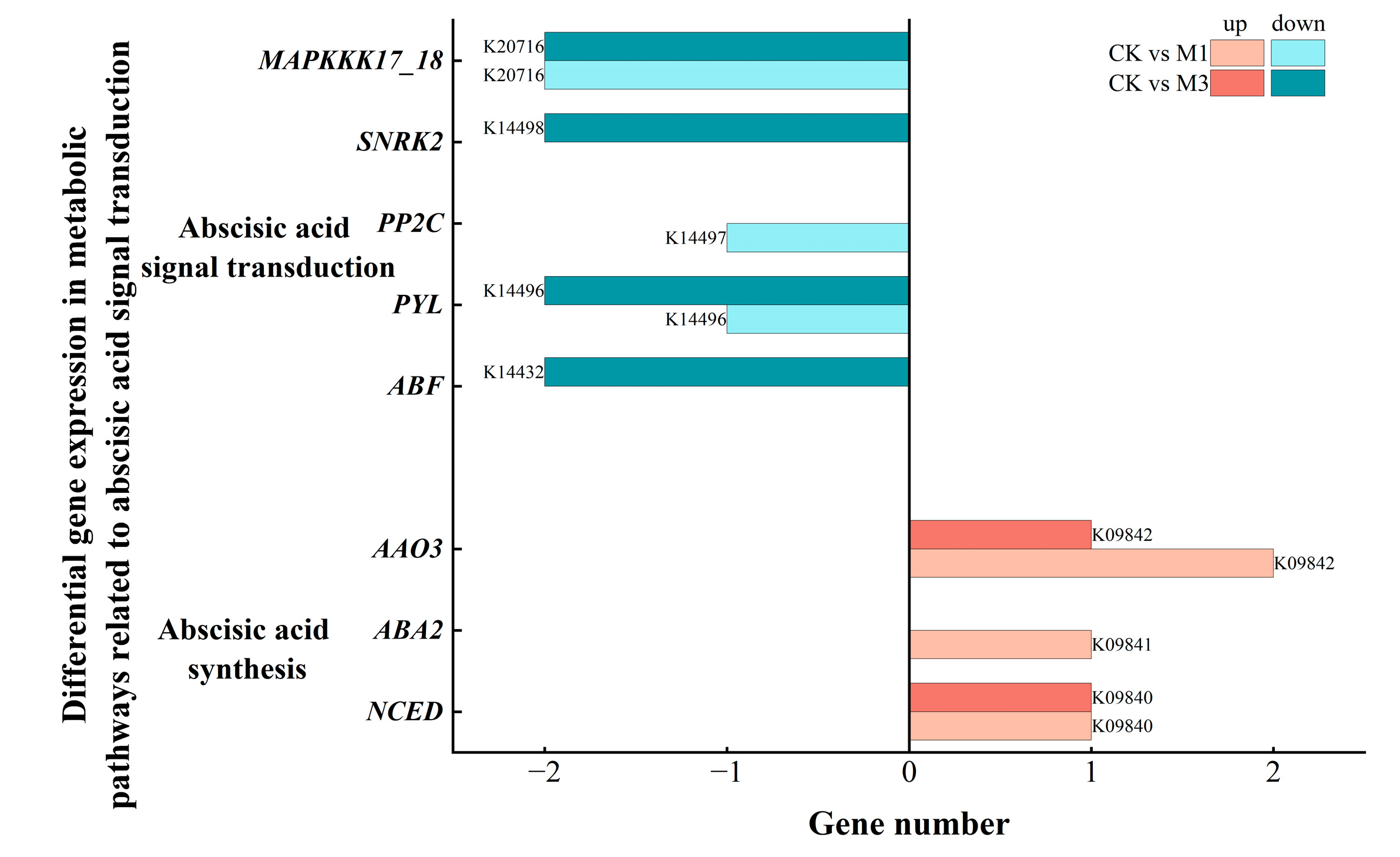
2.5.1. Analysis of DEGs in Abscisic Acid-Related Metabolic Pathways in Cotton Seedlings Under Drought Stress
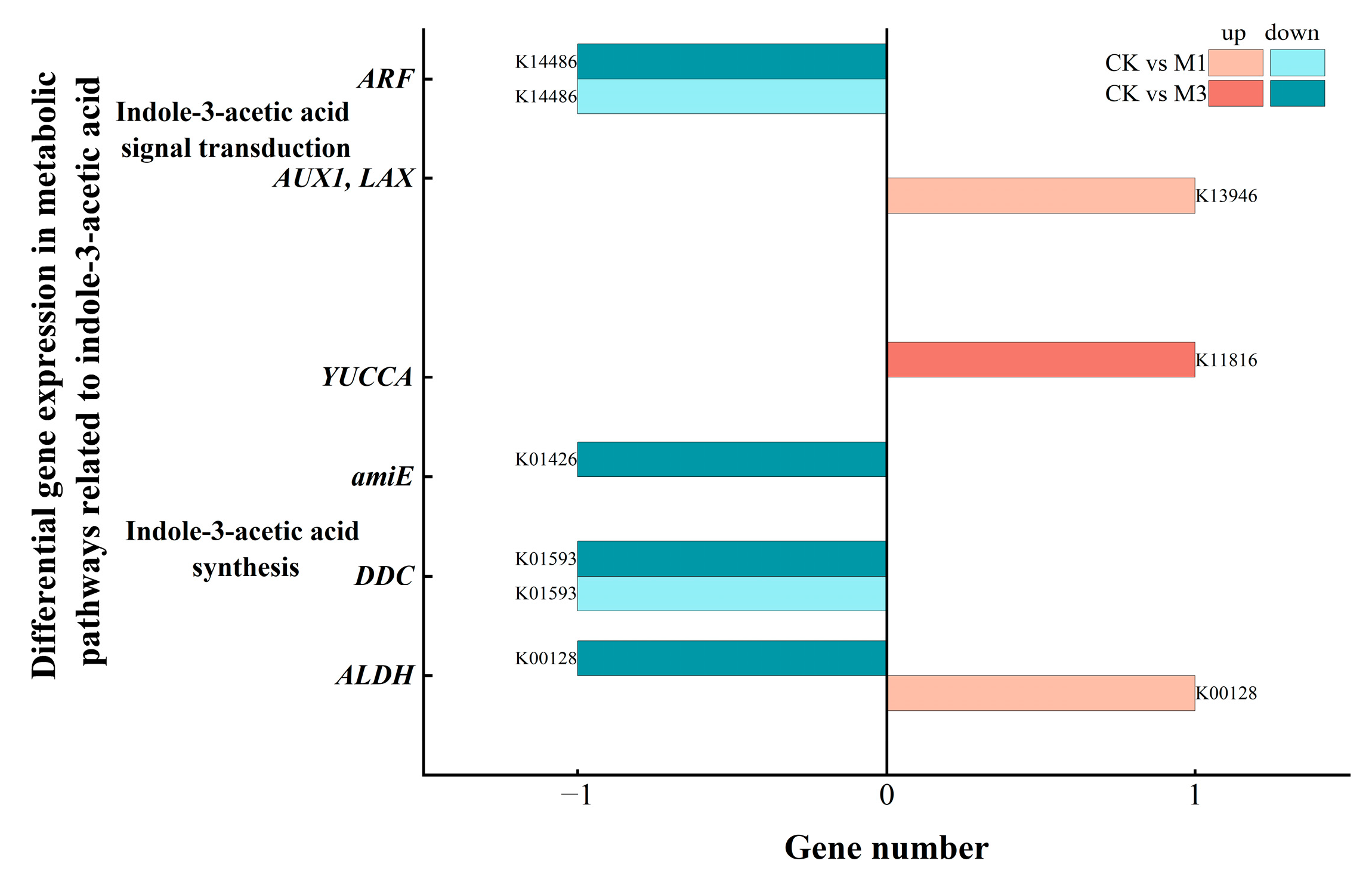
2.5.2. Analysis of DEGs in Indole-3-Acetic Acid-Related Metabolic Pathways in Cotton Seedlings Under Drought Stress
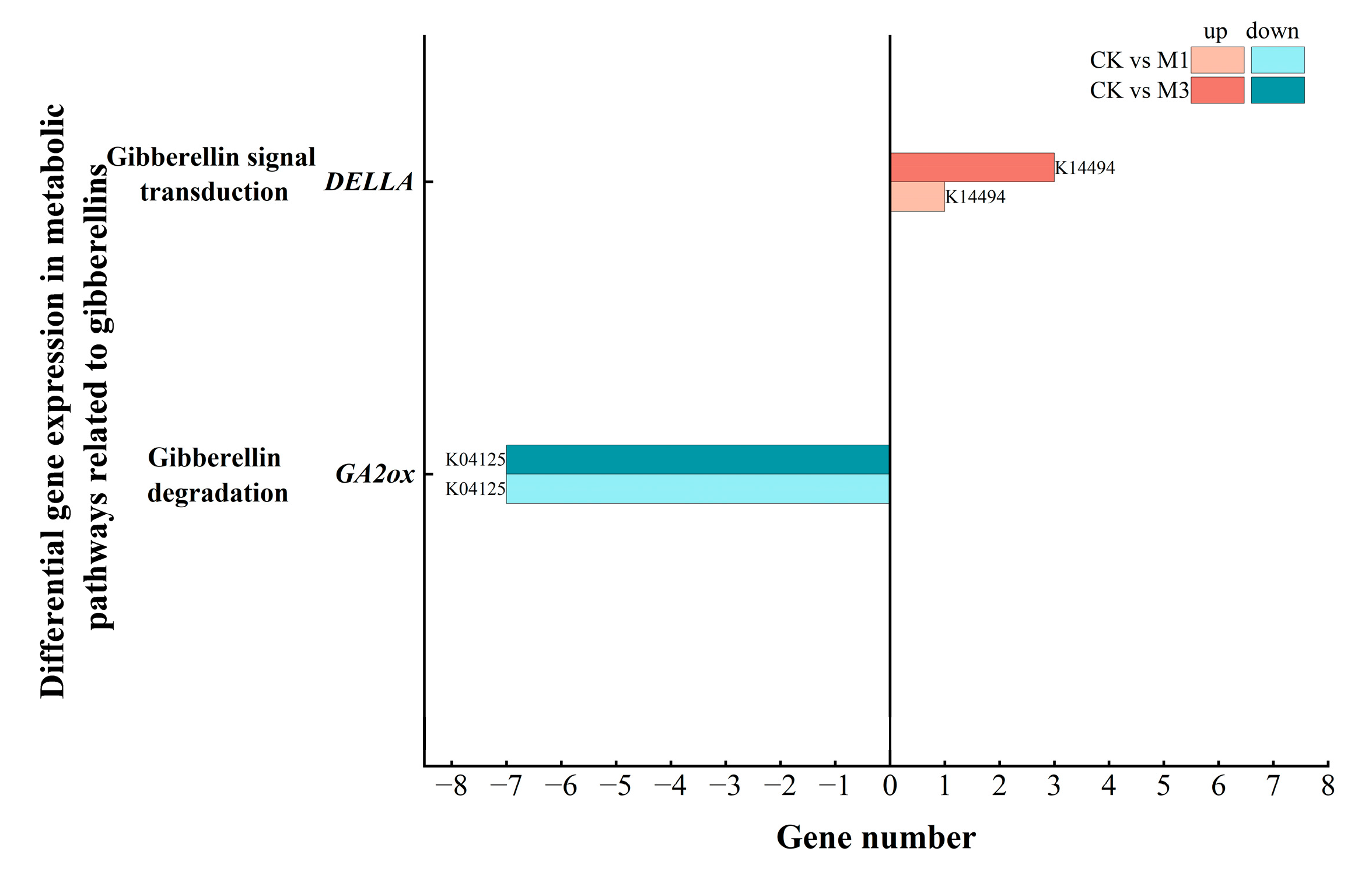
2.5.3. Analysis of DEGs in Gibberellin-Related Metabolic Pathways in Cotton Seedlings Under Drought Stress
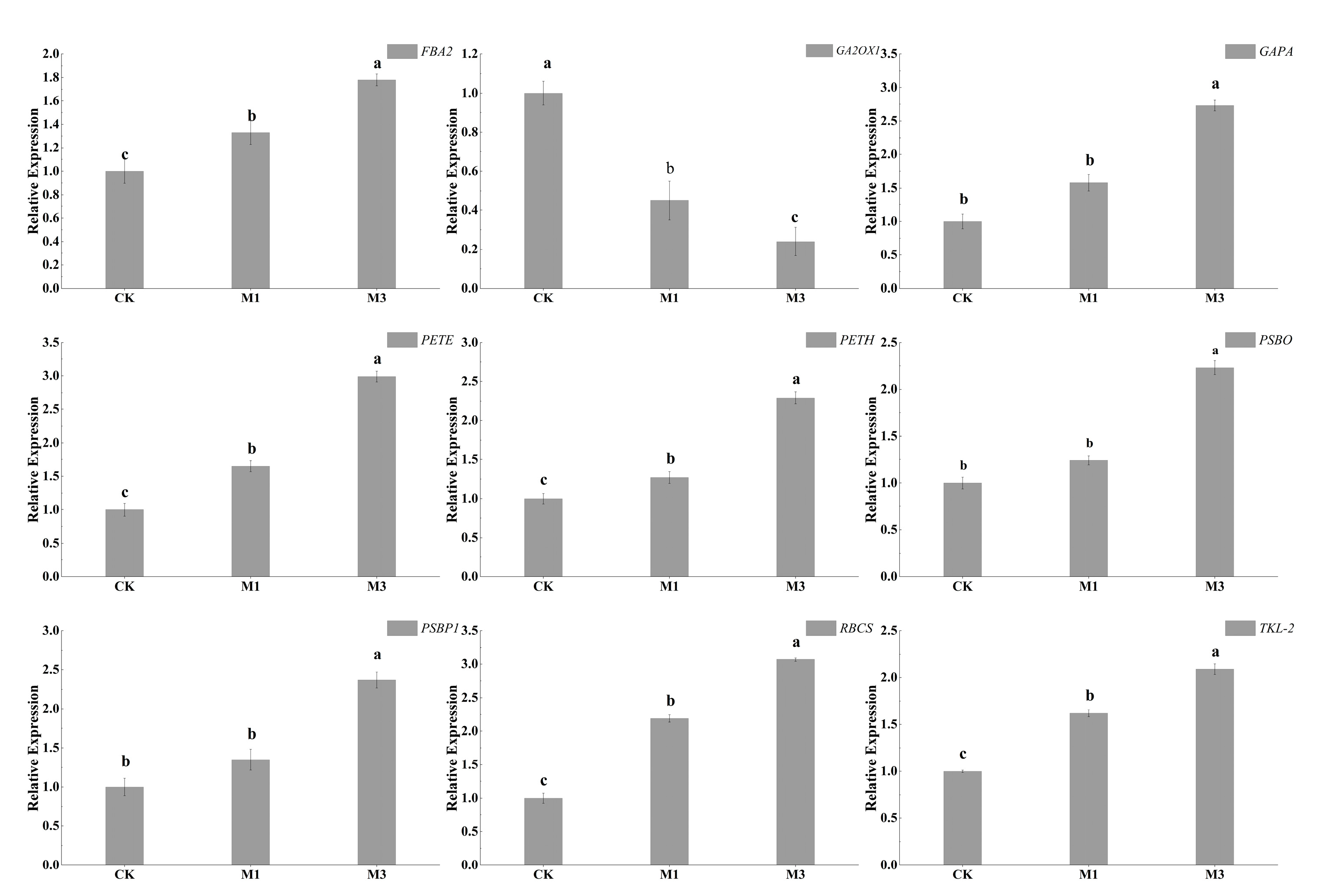
2.6. Verification of DEGs by RT-qPCR
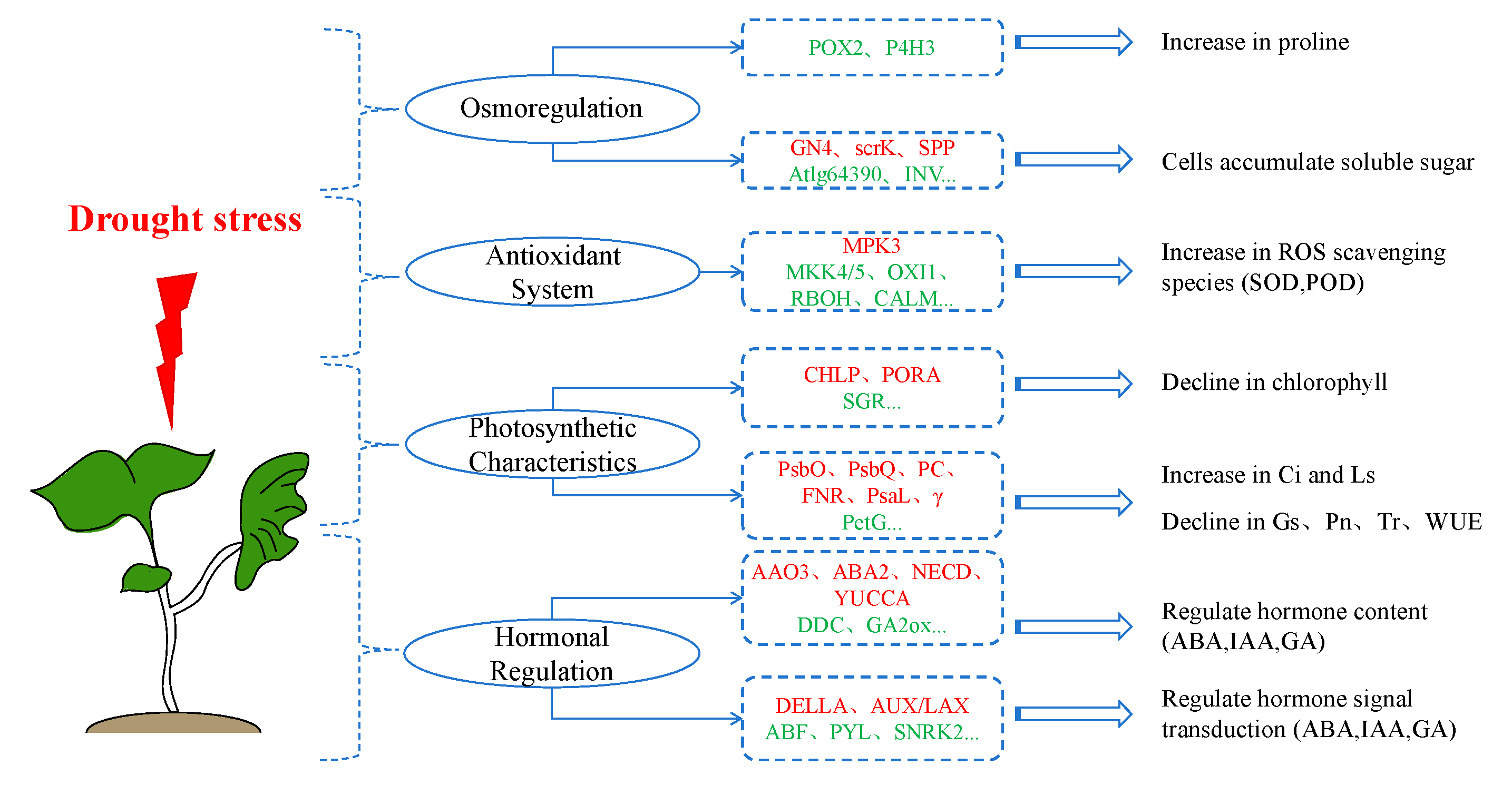
3. Discussion
4. Materials and Methods
4.1. Plant Cultivation
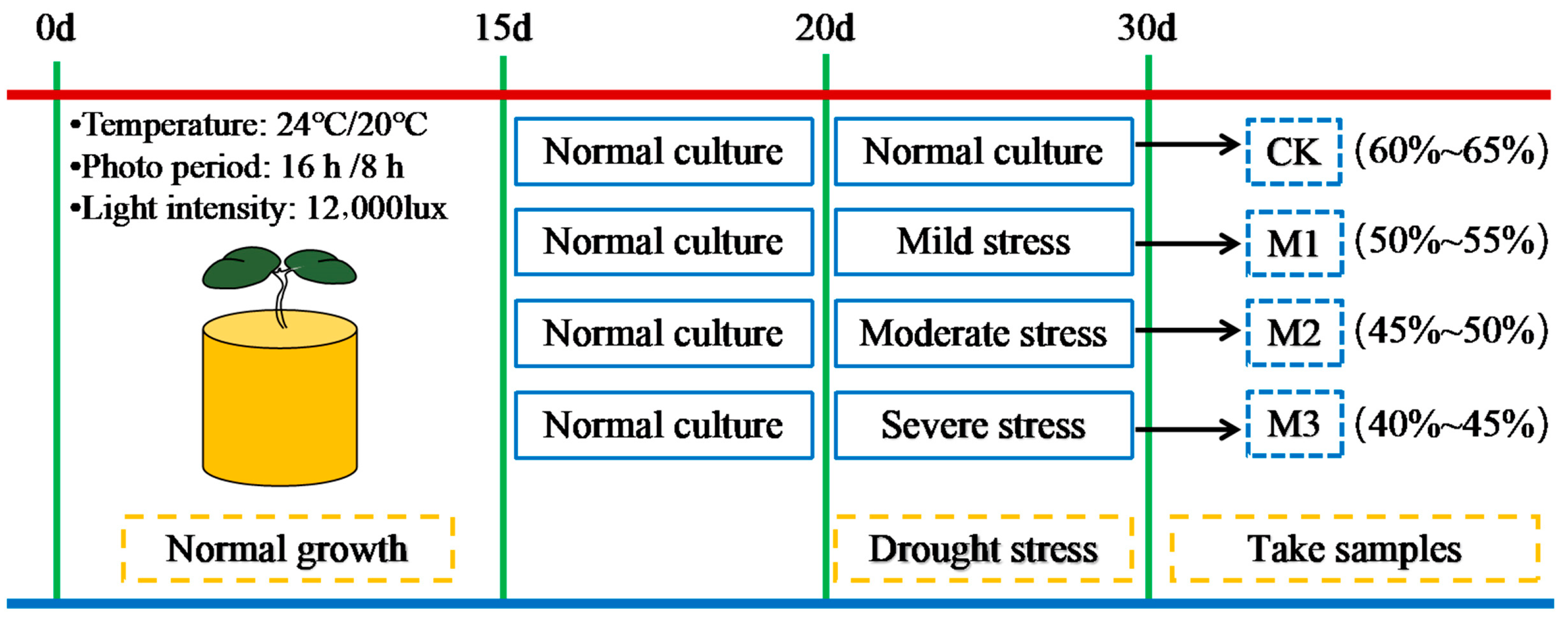
4.2. Experimental Design
4.3. Determination of Physiological Indicators
4.3.1. Determination of Malondialdehyde Content
4.3.2. Determination of Proline Content
4.3.3. Determination of Superoxide Dismutase Activity
4.3.4. Determination of Peroxidase Content
4.4. Determination of Chlorophyll Content and Hormone
4.5. Measurement of Gas Exchange Parameters
4.6. RNA Sequencing and Data Analysis
4.7. Real-Time Quantitative PCR
4.8. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef]
- Dubey, A.; Kumar, A.; Malla, M.A.; Chowdhary, K.; Singh, G.; Gudasalamani Ravikanth, H.; Sharma, S.; Saati-Santamaria, Z.; Menéndez, E.; Dames, J.F. Approaches for the Amelioration of Adverse Effects of Drought Stress on Crop Plants. Front. Biosci. 2021, 26, 928–947. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The Physiology of Plant Responses to Drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Darko, R.O.; Yuan, S.; Hong, L.; Liu, J.; Yan, H. Irrigation, a Productive Tool for Food Security—A Review. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2016, 66, 191–206. [Google Scholar] [CrossRef]
- Haghpanah, M.; Hashemipetroudi, S.; Arzani, A.; Araniti, F. Drought Tolerance in Plants: Physiological and Molecular Responses. Plants 2024, 13, 2962. [Google Scholar] [CrossRef]
- Javed, Q. Impacts of Salt Stress on The Physiology of Plants and Opportunity to Rewater The Stressed Plants With Diluted Water: A Review. Appl. Ecol. Environ. Res. 2019, 17, 12583–12604. [Google Scholar] [CrossRef]
- Agurla, S.; Gahir, S.; Munemasa, S.; Murata, Y.; Raghavendra, A.S. Mechanism of Stomatal Closure in Plants Exposed to Drought and Cold Stress. In Survival Strategies in Extreme Cold and Desiccation: Adaptation Mechanisms and Their Applications; Springer: Singapore, 2018; pp. 215–232. [Google Scholar]
- Liu, Z.; Fu, W.G. Physiological response of lettuce leaves from the same plant to different air humidity under drought stress. Jiangsu Agric. Sci. 2025, 53, 180–185. [Google Scholar]
- Ghosh, U.K.; Islam, M.N.; Siddiqui, M.N.; Khan, M.A.R. Understanding the Roles of Osmolytes for Acclimatizing Plants to Changing Environment: A Review of Potential Mechanism. Plant Signal. Behav. 2021, 16, 1913306. [Google Scholar] [CrossRef]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S. Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef]
- Van Oosten, M.J.; Costa, A.; Punzo, P.; Landi, S.; Ruggiero, A.; Batelli, G.; Grillo, S. Genetics of Drought Stress Tolerance in Crop Plants. In Drought Stress Tolerance in Plants: Molecular and Genetic Perspectives; Springer: Cham, Switzerland, 2016; Volume 2, pp. 39–70. [Google Scholar]
- Mahmood, T.; Khalid, S.; Abdullah, M.; Ahmed, Z.; Shah, M.K.N.; Ghafoor, A.; Du, X. Insights into Drought Stress Signaling in Plants and the Molecular Genetic Basis of Cotton Drought Tolerance. Cells 2019, 9, 105. [Google Scholar] [CrossRef]
- Wang, X.; Li, N.; Li, W.; Gao, X.; Cha, M.; Qin, L.; Liu, L. Advances in Transcriptomics in the Response to Stress in Plants. Glob. Med. Genet. 2020, 7, 030–034. [Google Scholar] [CrossRef]
- Picelli, S. Single-Cell RNA-Sequencing: The Future of Genome Biology Is Now. RNA Biol. 2017, 14, 637–650. [Google Scholar] [CrossRef]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription Factors as Key Molecular Target to Strengthen the Drought Stress Tolerance in Plants. Physiol. Plant. 2021, 172, 847–868. [Google Scholar] [CrossRef]
- Iqbal, S.; Wang, X.; Mubeen, I.; Kamran, M.; Kanwal, I.; Díaz, G.A.; Abbas, A.; Parveen, A.; Atiq, M.N.; Alshaya, H. Phytohormones Trigger Drought Tolerance in Crop Plants: Outlook and Future Perspectives. Front. Plant Sci. 2022, 12, 799318. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; He, S.; Sun, G.; Pan, Z.; Sun, J.; Geng, X.; Peng, Z.; Gong, W.; Wang, L.; Pang, B. Favorable Pleiotropic Loci for Fiber Yield and Quality in Upland Cotton (Gossypium hirsutum). Sci. Rep. 2021, 11, 15935. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, Z.; Liang, Y.; Gong, Z.; Zhang, N.; Ditta, A.; Sang, Z.; Wang, J.; Li, X. Whole Transcriptome Sequencing Reveals Drought Resistance-Related Genes in Upland Cotton. Genes 2022, 13, 1159. [Google Scholar] [CrossRef]
- Han, B.; Wang, F.; Liu, Z.; Chen, L.; Yue, D.; Sun, W.; Lin, Z.; Zhang, X.; Zhou, X.; Yang, X. Transcriptome and Metabolome Profiling of Interspecific CSSLs Reveals General and Specific Mechanisms of Drought Resistance in Cotton. Theor. Appl. Genet. 2022, 135, 3375–3391. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, C.; Zhu, W.; Yan, G.; Chen, X.; Qiu, P.; Ruzimurod, B.; Ye, W.; Qaraevna, B.Z.; Yin, Z. Molecular Mechanism That Underlies Cotton Response to Salt and Drought Stress Revealed by Complementary Transcriptomic and iTRAQ Analyses. Environ. Exp. Bot. 2023, 209, 105288. [Google Scholar] [CrossRef]
- Zhang, B.; Long, Y.; Pei, L.; Huang, X.; Li, B.; Han, B.; Zhang, M.; Lindsey, K.; Zhang, X.; Wang, M. Drought Response Revealed by Chromatin Organization Variation and Transcriptional Regulation in Cotton. BMC Biol. 2024, 22, 114. [Google Scholar] [CrossRef]
- Peng, Z.; He, S.; Gong, W.; Sun, J.; Pan, Z.; Xu, F.; Lu, Y.; Du, X. Comprehensive Analysis of Differentially Expressed Genes and Transcriptional Regulation Induced by Salt Stress in Two Contrasting Cotton Genotypes. BMC Genom. 2014, 15, 760. [Google Scholar] [CrossRef]
- Zhu, Y.-N.; Shi, D.-Q.; Ruan, M.-B.; Zhang, L.-L.; Meng, Z.-H.; Liu, J.; Yang, W.-C. Transcriptome Analysis Reveals Crosstalk of Responsive Genes to Multiple Abiotic Stresses in Cotton (Gossypium hirsutum L.). PLoS ONE 2013, 8, e80218. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Chao, D.; Zhao, Y. How Plants Sense and Respond to Osmotic Stress. J. Integr. Plant Biol. 2024, 66, 394–423. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, M.; Turkyilmaz Unal, B.; García-Caparrós, P.; Khursheed, A.; Gul, A.; Hasanuzzaman, M. Osmoregulation and Its Actions during the Drought Stress in Plants. Physiol. Plant. 2021, 172, 1321–1335. [Google Scholar] [CrossRef]
- Zuo, Z.Y.; Guo, J.H.; Xin, C.Y.; Liu, S.Q.; Mao, H.P.; Wang, Y.J.; Li, X.N. Salt acclimation induced salt tolerance in wild-type and abscisic acid-deficient mutant barley. Plant Soil Environ. 2019, 65, 516–521. [Google Scholar] [CrossRef]
- Furlan, A.L.; Bianucci, E.; Giordano, W.; Castro, S.; Becker, D.F. Proline Metabolic Dynamics and Implications in Drought Tolerance of Peanut Plants. Plant Physiol. Biochem. 2020, 151, 566–578. [Google Scholar] [CrossRef]
- Tiainen, P.; Myllyharju, J.; Koivunen, P. Characterization of a Second Arabidopsis Thaliana Prolyl 4-Hydroxylase with Distinct Substrate Specificity. J. Biol. Chem. 2005, 280, 1142–1148. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, Y.; Yadav, V.; Zhao, W.; He, Y.; Zhang, X.; Wei, C. Drought-Induced Proline Is Mainly Synthesized in Leaves and Transported to Roots in Watermelon under Water Deficit. Hortic. Plant J. 2022, 8, 615–626. [Google Scholar] [CrossRef]
- Dai, M.; Zhang, Y.; Lu, X.; Wang, D.; Zhang, Y.; Zhou, N.; Wu, Z.; Liu, L.; Rui, C.; Wang, X. Starch Granules of the Sugar-Pathway Were Eliminated under the Stress of PEG-Drought Compared with Soil-Drought. Ind. Crops Prod. 2023, 193, 116158. [Google Scholar] [CrossRef]
- Wu, F.; Fu, J.; Chen, Y.; Huang, J.; Su, Q.; Zheng, J.; Chen, Z.; Zhu, H.; Zheng, Y.; Huang, J. Effect of Salt Stress on Anti-adversity Physiology of Baccaurea ramiflora Seedling. J. Trop. Subtrop. Bot. 2025, 33, 207–212. [Google Scholar]
- Li, S.; Li, H.; Yang, W.; Yang, Z.; Wang, X.; Chai, H. Physiological and molecular mechanisms of response to drought stress in alfalfa. Pratacult. Sci. 2018, 35, 331–340. [Google Scholar]
- Boriboonkaset, T.; Theerawitaya, C.; Yamada, N.; Pichakum, A.; Supaibulwatana, K.; Cha-Um, S.; Takabe, T.; Kirdmanee, C. Regulation of Some Carbohydrate Metabolism-Related Genes, Starch and Soluble Sugar Contents, Photosynthetic Activities and Yield Attributes of Two Contrasting Rice Genotypes Subjected to Salt Stress. Protoplasma 2013, 250, 1157–1167. [Google Scholar] [CrossRef]
- Mansoor, S.; Ali Wani, O.; Lone, J.K.; Manhas, S.; Kour, N.; Alam, P.; Ahmad, A.; Ahmad, P. Reactive Oxygen Species in Plants: From Source to Sink. Antioxidants 2022, 11, 225. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Reddy, A.R.; Chaitanya, K.V.; Vivekanandan, M. Drought-Induced Responses of Photosynthesis and Antioxidant Metabolism in Higher Plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar] [CrossRef]
- Zhang, X.B.; Ni, Z.; Zhuang, Y.Q. Effects of exogenous trehalose on soybean seedlings under NaCl stress. Jiangsu Agricultural Sciences. 2024, 52, 116–122. [Google Scholar]
- Luo, J.; Zhao, L.-L.; Gong, S.-Y.; Sun, X.; Li, P.; Qin, L.-X.; Zhou, Y.; Xu, W.-L.; Li, X.-B. A Cotton Mitogen-Activated Protein Kinase (GhMPK6) Is Involved in ABA-Induced CAT1 Expression and H2O2 Production. J. Genet. Genom. 2011, 38, 557–565. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, S. Mitogen-activated Protein Kinase Cascades in Plant Signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef]
- Zhang, J.-B.; Wang, X.-P.; Wang, Y.-C.; Chen, Y.-H.; Luo, J.-W.; Li, D.-D.; Li, X.-B. Genome-Wide Identification and Functional Characterization of Cotton (Gossypium hirsutum) MAPKKK Gene Family in Response to Drought Stress. BMC Plant Biol. 2020, 20, 217. [Google Scholar] [CrossRef]
- Hong, C.; Guo, L.N.; Zhang, Z.Y.; Ouyang, N.N.; Wu, P.; Ma, H.L. Transcriptomics Analysis of the Mechanism Underlying the Synthesis of Phenolic Compounds in Fresh-Cut Red Cabbage under Ultrasound Stress. Food Sci. 2025, 46, 37–48. [Google Scholar]
- Impa, S.; Nadaradjan, S.; Jagadish, S. Drought Stress Induced Reactive Oxygen Species and Anti-Oxidants in Plants. In Abiotic Stress Responses in Plants: Metabolism, Productivity and Sustainability; Springer: New York, NY, USA, 2012; pp. 131–147. [Google Scholar]
- Eggink, L.L.; Park, H.; Hoober, J.K. The Role of Chlorophyll b in Photosynthesis: Hypothesis. BMC Plant Biol. 2001, 1, 2. [Google Scholar] [CrossRef]
- Hu, F.; Zhang, Y.; Guo, J. Effects of Drought Stress on Photosynthetic Physiological Characteristics, Leaf Microstructure, and Related Gene Expression of Yellow Horn. Plant Signal. Behav. 2023, 18, 2215025. [Google Scholar] [CrossRef] [PubMed]
- Reinbothe, C.; El Bakkouri, M.; Buhr, F.; Muraki, N.; Nomata, J.; Kurisu, G.; Fujita, Y.; Reinbothe, S. Chlorophyll Biosynthesis: Spotlight on Protochlorophyllide Reduction. Trends Plant Sci. 2010, 15, 614–624. [Google Scholar] [CrossRef]
- Blankenship, R.E. Molecular Mechanisms of Photosynthesis; John Wiley & Sons: Hoboken, NJ, USA, 2021; Volume 117–257, ISBN 1-119-80011-0. [Google Scholar]
- Zhang, Z.-F.; Li, Y.-Y.; Xiao, B.-Z. Comparative Transcriptome Analysis Highlights the Crucial Roles of Photosynthetic System in Drought Stress Adaptation in Upland Rice. Sci. Rep. 2016, 6, 19349. [Google Scholar] [CrossRef]
- Yamaoka, C.; Suzuki, Y.; Makino, A. Differential Expression of Genes of the Calvin–Benson Cycle and Its Related Genes During Leaf Development in Rice. Plant Cell Physiol. 2016, 57, 115–124. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Yi, J.; Yang, Y.; Lei, C.; Gong, M. Transcriptome Response to Drought, Rehydration and Re-Dehydration in Potato. Int. J. Mol. Sci. 2019, 21, 159. [Google Scholar] [CrossRef]
- Khan, R.; Zhou, P.; Ma, X.; Zhou, L.; Wu, Y.; Ullah, Z.; Wang, S. Transcriptome Profiling, Biochemical and Physiological Analyses Provide New Insights towards Drought Tolerance in Nicotiana tabacum L. Genes 2019, 10, 1041. Genes 2019, 10, 1041. [Google Scholar] [CrossRef]
- Zhi, Q.-Q.; Chen, Y.; Hu, H.; Huang, W.-Q.; Bao, G.-G.; Wan, X.-R. Physiological and Transcriptome Analyses Reveal Tissue-Specific Responses of Leucaena Plants to Drought Stress. Plant Physiol. Biochem. 2024, 214, 108926. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, C.; Zhang, J.; Li, Z.; Zeng, Q.; Sun, Q.; Wang, X.; Zhao, L.; Zhang, L.; Li, B. Physiological and Transcriptome Analyses of Chinese Cabbage in Response to Drought Stress1. J. Integr. Agric. 2024, 23, 2255–2269. [Google Scholar] [CrossRef]
- Li, M.; Li, H.; Sun, A.; Wang, L.; Ren, C.; Liu, J.; Gao, X. Transcriptome Analysis Reveals Key Drought-Stress-Responsive Genes in Soybean. Front. Genet. 2022, 13, 1060529. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kang, K.; Lee, K.; Back, K. Characterization of Rice Tryptophan Decarboxylases and Their Direct Involvement in Serotonin Biosynthesis in Transgenic Rice. Planta 2007, 227, 263–272. [Google Scholar] [CrossRef]
- Shah, G.; Fiaz, S.; Attia, K.A.; Khan, N.; Jamil, M.; Abbas, A.; Yang, S.H.; Jumin, T. Indole Pyruvate Decarboxylase Gene Regulates the Auxin Synthesis Pathway in Rice by Interacting with the Indole-3-Acetic Acid–Amido Synthetase Gene, Promoting Root Hair Development under Cadmium Stress. Front. Plant Sci. 2022, 13, 1023723. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, J.; Xu, Y.; Abbas, F.; Xu, D.; Tao, S.; Xie, X.; Song, F.; Huang, Q.; Sharma, A. Genome-Wide Identification and Expression Analysis of AUX/LAX Family Genes in Chinese Hickory (Carya cathayensis Sarg.) Under Various Abiotic Stresses and Grafting. Front. Plant Sci. 2023, 13, 1060965. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shu, D.; Tan, Z.; Ma, M.; Guo, N.; Gao, S.; Duan, G.; Kuai, B.; Hu, Y.; Li, S. The Arabidopsis IDD14 Transcription Factor Interacts with bZIP-type ABFs/AREBs and Cooperatively Regulates ABA-mediated Drought Tolerance. New Phytol. 2022, 236, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Ptošková, K.; Szecówka, M.; Jaworek, P.; Tarkowská, D.; Petřík, I.; Pavlović, I.; Novák, O.; Thomas, S.G.; Phillips, A.L.; Hedden, P. Changes in the Concentrations and Transcripts for Gibberellins and Other Hormones in a Growing Leaf and Roots of Wheat Seedlings in Response to Water Restriction. BMC Plant Biol. 2022, 22, 284. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Wu, W.; Abrams, S.R.; Cutler, A.J. The Relationship of Drought-Related Gene Expression in Arabidopsis Thaliana to Hormonal and Environmental Factors. J. Exp. Bot. 2008, 59, 2991–3007. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Dong, W.; Zhang, Y.; He, Z. Gibberellins Modulate Abiotic Stress Tolerance in Plants. Sci. Sin. Vitae 2013, 43, 1119–1126. [Google Scholar] [CrossRef]
- Lin, Q.; Wu, F.; Sheng, P.; Zhang, Z.; Zhang, X.; Guo, X.; Wang, J.; Cheng, Z.; Wang, J.; Wang, H.; et al. The SnRK2-APC/CTE Regulatory Module Mediates the Antagonistic Action of Gibberellic Acid and Abscisic Acid Pathways. Nat. Commun. 2015, 6, 7981. [Google Scholar] [CrossRef]
- Sarwar, R.; Zhu, K.; Jiang, T.; Ding, P.; Gao, Y.; Tan, X. DELLAs Directed Gibberellins Responses Orchestrate Crop Development: A Brief Review. Crop Sci. 2023, 63, 1–28. [Google Scholar] [CrossRef]
- Gao, J.-F. Experimental Guidance for Plant Physiology; Higer Education Press: Beijing, China, 2006. [Google Scholar]
- Zhang, S.Q. Plant Physiology Experimental Techniques Tutorial; Science Press: Beijing, China, 2011. [Google Scholar]
- IBM Corp. IBM SPSS Statistics, version 25.0; IBM Corp.: Armonk, NY, USA, 2017.
- OriginLab Corporation. Origin, version 2021; OriginLab Corporation: Northampton, MA, USA, 2021.

| Samples | Clean Reads | CLean_Bases | GC Content | % ≥ Q30 |
|---|---|---|---|---|
| CK (normal culture)-1 | 43,657,062 | 6,514,911,687 | 43.49% | 91.81% |
| CK-2 | 41,758,086 | 6,232,287,901 | 43.71% | 91.28% |
| CK-3 | 38,925,918 | 5,813,473,089 | 43.75% | 92.10% |
| M1-1 | 37,720,786 | 5,616,674,520 | 43.30% | 90.53% |
| M1-2 | 54,369,244 | 8,111,458,084 | 43.81% | 92.04% |
| M1-3 | 43,414,136 | 6,481,368,190 | 43.90% | 92.07% |
| M3-1 | 44,289,592 | 6,605,707,682 | 43.61% | 91.77% |
| M3-2 | 35,961,940 | 5,367,231,914 | 43.72% | 91.34% |
| M3-3 | 53,940,862 | 8,050,791,622 | 43.82% | 92.80% |
| Samples | Clean Read Number | Total Mapped | Unique Mapped | Multiple Mapped |
|---|---|---|---|---|
| CK-1 | 43,507,538 | 41,842,721 (96.17%) | 40,106,591 (92.18%) | 1,736,130 (3.99%) |
| CK-2 | 41,512,474 | 39,801,003 (95.88%) | 38,059,123 (91.68%) | 1,741,880 (4.20%) |
| CK-3 | 38,740,216 | 37,245,056 (96.14%) | 35,681,502 (92.10%) | 1,563,554 (4.04%) |
| M1-1 | 37,515,964 | 35,178,416 (93.77%) | 33,588,795 (89.53%) | 1,589,621 (4.24%) |
| M1-2 | 54,016,576 | 51,767,192 (95.84%) | 49,485,423 (91.61%) | 2,281,769 (4.22%) |
| M1-3 | 43,182,614 | 41,068,568 (95.10%) | 39,307,967 (91.03%) | 1,760,601 (4.08%) |
| M3-1 | 44,085,140 | 41,683,531 (94.55%) | 39,873,331 (90.45%) | 1,810,200 (4.11%) |
| M3-2 | 35,766,806 | 33,894,337 (94.76%) | 32,427,872 (90.66%) | 1,466,465 (4.10%) |
| M3-3 | 53,671,742 | 51,261,221 (95.51%) | 49,041,525 (91.37%) | 2,219,696 (4.14%) |
| Pathway | K_ID | Gene | Gene Type |
|---|---|---|---|
| Sucrose and starch metabolism (ko00500) | K00688 | PYG | PYG, glgP; glycogen phosphorylase [EC: 2.4.1.1] |
| K00975 | glgC | glgC; glucose-1-phosphate adenylyltransferase [EC: 2.7.7.27] | |
| K13679 | WAXY | WAXY; granule-bound starch synthase [EC: 2.4.1.242] | |
| K01176 | AMY | AMY, amyA, malS; alpha-amylase [EC: 3.2.1.1] | |
| K01177 | BAM1 | E3.2.1.2; beta-amylase [EC: 3.2.1.2] | |
| K01214 | ISA | ISA, treX; isoamylase [EC: 3.2.1.68] | |
| K00696 | SPS4 | E2.4.1.14; sucrose-phosphate synthase [EC: 2.4.1.14] | |
| K07024 | SPP | SPP; sucrose-6-phosphatase [EC: 3.1.3.24] | |
| K01193 | INV | INV, sacA; beta-fructofuranosidase [EC: 3.2.1.26] | |
| K01087 | otsB | otsB; trehalose 6-phosphate phosphatase [EC: 3.1.3.12] | |
| K01179 | At1g64390 | E3.2.1.4; endoglucanase [EC: 3.2.1.4] | |
| K05349 | bglX | bglX; beta-glucosidase [EC: 3.2.1.21] | |
| K05350 | bglB | bglB; beta-glucosidase [EC: 3.2.1.21] | |
| K00847 | scrK | E2.7.1.4, scrK; fructokinase [EC: 2.7.1.4] | |
| K01835 | pgm | pgm; phosphoglucomutase [EC: 5.4.2.2] | |
| K19892 | GN4 | GN4; glucan endo-1,3-beta-glucosidase 4 [EC: 3.2.1.39] | |
| K19893 | GN5/6 | GN5/6; glucan endo-1,3-beta-glucosidase 5/6 [EC: 3.2.1.39] | |
| MAPK signaling pathway—plant (ko04016) | K20538 | MPK8 | MPK8; mitogen-activated protein kinase 8 [EC: 2.7.11.24] |
| K13413 | MKK4/5 | MKK4/5; mitogen-activated protein kinase kinase 4/5 [EC: 2.7.12.2] | |
| K13425 | WRKY22 | WRKY22; WRKY transcription factor 22 | |
| K13426 | WRKY29 | WRKY29; WRKY transcription factor 29 | |
| K13447 | RBOH | RBOH; respiratory burst oxidase [EC: 1.6.3.- 1.11.1.-] | |
| K20714 | OXI1 | OXI1; serine/threonine-protein kinase OXI1 [EC: 2.7.11.1] | |
| K02183 | CALM | CALM; calmodulin | |
| K20536 | MPK3 | MPK3; mitogen-activated protein kinase 3 [EC: 2.7.11.24] | |
| Arginine and proline metabolism (ko00330) | K00472 | P4H3 | P4HA; prolyl 4-hydroxylase [EC: 1.14.11.2] |
| K00318 | POX2 | PRODH, fadM, putB; proline dehydrogenase [EC: 1.5.5.2] |
| Pathway | K_ID | Gene | Gene Type |
|---|---|---|---|
| Porphyrin metabolism (ko00860) | K03403 | CHLH | chlH, bchH; magnesium chelatase subunit H [EC: 6.6.1.1] |
| K03405 | CHLI | chlI, bchI; magnesium chelatase subunit I [EC: 6.6.1.1] | |
| K04035 | CRD1 | E1.14.13.81, acsF, chlE; magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase [EC: 1.14.13.81] | |
| K10960 | CHLP | chlP, bchP; geranylgeranyl diphosphate/geranylgeranyl-bacteriochlorophyllide a reductase [EC: 1.3.1.83 1.3.1.111] | |
| K01749 | HEMC | hemC, HMBS; hydroxymethylbilane synthase [EC: 2.5.1.61] | |
| K03428 | CHLM | bchM, chlM; magnesium-protoporphyrin O-methyltransferase [EC: 2.1.1.11] | |
| K00218 | PORA | por; protochlorophyllide reductase [EC: 1.3.1.33] | |
| K22013 | SGR | SGR, SGRL; magnesium dechelatase [EC: 4.99.1.10] | |
| K13545 | RCCR | RCCR, ACD2; red chlorophyll catabolite reductase [EC: 1.3.7.12] |
| Category | Protein Subunit | K_ID | Gene Type |
|---|---|---|---|
| PSII | PsbO | K02716 | psbO; photosystem II oxygen-evolving enhancer protein 1 |
| PsbQ | K08901 | psbQ; photosystem II oxygen-evolving enhancer protein 3 | |
| PsbP | K02717 | psbP; photosystem II oxygen-evolving enhancer protein 2 | |
| Psb28 | K08903 | psb28; photosystem II 13kDa protein | |
| PsbS | K03542 | psbS; photosystem II 22kDa protein | |
| PsbW | K02721 | psbW; photosystem II PsbW protein | |
| Cytb6/f | PetG | K02640 | petG; cytochrome b6-f complex subunit 5 |
| ATPase | γ | K02115 | ATPF1G, atpG; F-type H+-transporting ATPase subunit gamma |
| PSⅠ | PsaL | K02699 | psaL; photosystem I subunit XI |
| PsaN | K02701 | psaN; photosystem I subunit PsaN | |
| PsaO | K14332 | psaO; photosystem I subunit PsaO | |
| PsaK | K02698 | psaK; photosystem I subunit X | |
| PsaH | K02695 | psaH; photosystem I subunit VI | |
| PsaE | K02693 | psaE; photosystem I subunit IV | |
| Photosynthetic electron transport | PC | K02638 | petE; plastocyanin |
| Fd | K02639 | petF; ferredoxin | |
| FNR | K02641 | petH; ferredoxin--NADP+ reductase |
| Gene ID | Symbol | K_ID | Description |
|---|---|---|---|
| GH_D13G1739 | GA2OX1 | K04125 | PREDICTED: GA2ox2 |
| GH_A11G1964 | PSBO | K02716 | PREDICTED: oxygen-evolving enhancer protein 1, chloroplastic-like |
| GH_A05G4010 | PSBP1 | K02717 | PREDICTED: oxygen-evolving enhancer protein 2, chloroplastic-like |
| GH_D05G1861 | PETE | K02638 | PREDICTED: plastocyanin |
| GH_D11G1863 | PETH | K02641 | PREDICTED: ferredoxin—NADP reductase, leaf isozyme, chloroplastic |
| GH_A07G1943 | RBCS | K01602 | PREDICTED: ribulose bisphosphate carboxylase small chain, chloroplastic |
| GH_A13G0295 | FBA2 | K01623 | PREDICTED: fructose-bisphosphate aldolase 1, chloroplastic-like |
| GH_A10G0699 | GAPA | K05298 | PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic-like |
| GH_D07G1598 | TKL-2 | K00615 | PREDICTED: transketolase, chloroplastic |
| Pathway | K_ID | Gene | Gene Name |
|---|---|---|---|
| Carbon fixation in photosynthetic organisms (ko00710) | K00615 | tktA/tktB | E2.2.1.1, tktA, tktB; transketolase [EC: 2.2.1.1] |
| K00855 | PRK | PRK, prkB; phosphoribulokinase [EC: 2.7.1.19] | |
| K00927 | PGK | PGK, pgk; phosphoglycerate kinase [EC: 2.7.2.3] | |
| K01602 | rbcS | rbcS; ribulose-bisphosphate carboxylase small chain [EC: 4.1.1.39] | |
| K01623 | ALDO | ALDO; fructose-bisphosphate aldolase, class I [EC: 4.1.2.13] | |
| K03841 | FBP | FBP, fbp; fructose-1,6-bisphosphatase I [EC: 3.1.3.11] | |
| K05298 | GAPA | GAPA; glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) [EC: 1.2.1.13] | |
| K01100 | At3g55800 | E3.1.3.37; sedoheptulose-bisphosphatase [EC: 3.1.3.37] | |
| K01803 | TPI | TPI, tpiA; triosephosphate isomerase (TIM) [EC: 5.3.1.1] | |
| K00026 | MDH2 | MDH2; malate dehydrogenase [EC: 1.1.1.37] | |
| K01006 | ppdK | ppdK; pyruvate, orthophosphate dikinase [EC: 2.7.9.1] | |
| K01595 | ppc | ppc; phosphoenolpyruvate carboxylase [EC: 4.1.1.31] | |
| K14272 | GGAT | GGAT; glutamate—glyoxylate aminotransferase [EC: 2.6.1.4 2.6.1.2 2.6.1.44] |
| Pathway | K_ID | Gene | Gene Type |
|---|---|---|---|
| Carotenoid biosynthesis (ko00906) | K09840 | NCED | NCED; 9-cis-epoxycarotenoid dioxygenase [EC: 1.13.11.51] |
| K09841 | ABA2 | ABA2; xanthoxin dehydrogenase [EC: 1.1.1.288] | |
| K09842 | AAO3 | AAO3; abscisic-aldehyde oxidase [EC: 1.2.3.14] | |
| Diterpenoid biosynthesis (ko00904) | K04125 | GA2ox | E1.14.11.13; gibberellin 2beta-dioxygenase [EC: 1.14.11.13] |
| Tryptophan metabolism (ko00380) | K00128 | ALDH | ALDH; aldehyde dehydrogenase (NAD+) [EC: 1.2.1.3] |
| K01593 | DDC | DDC, TDC; aromatic-L-amino-acid/L-tryptophan decarboxylase [EC: 4.1.1.28 4.1.1.105] | |
| K01426 | amiE | E3.5.1.4, amiE; amidase [EC: 3.5.1.4] | |
| K11816 | YUCCA | YUCCA; indole-3-pyruvate monooxygenase [EC: 1.14.13.168] | |
| Signal transduction (ko04075, ko04016) | K14432 | ABF | ABF; ABA responsive element binding factor |
| K14496 | PYL | PYL; abscisic acid receptor PYR/PYL family | |
| K14497 | PP2C | PP2C; protein phosphatase 2C [EC: 3.1.3.16] | |
| K14498 | SNRK2 | SNRK2; serine/threonine-protein kinase SRK2 [EC: 2.7.11.1] | |
| K20716 | MAPKKK17/18 | MAPKKK17_18; mitogen-activated protein kinase kinase kinase 17/18 | |
| K14494 | DELLA | DELLA; DELLA protein | |
| K13946 | AUX1/LAX | AUX1, LAX; auxin influx carrier (AUX1 LAX family) | |
| K14486 | ARF | K14486, ARF; auxin response factor |
| Gene Name | Primer Name | Primer Sequence (5′–3′) | Amplicon Size (bp) |
|---|---|---|---|
| GA2OX1 | GA2OX1-F | TTGTTCCCTCCTCTTATCCC | 233 |
| GA2OX1-R | GGTCCAATCCTTTTATTACCAT | 233 | |
| PSBO | PSBO-F | TCGCTCTTGCTACATCTGC | 115 |
| PSBO-R | CCTGTTCCTTTGACTTCCAT | 115 | |
| PSBP1 | PSBP1-F | CCACCACGCACTCACAAC | 121 |
| PSBP1-R | TCCATCATCTTCCTGCTTTT | 121 | |
| PETE | PETE-F | GGGGTCTGGCTTTCATTC | 115 |
| PETE-R | CTTGGGATTTCGTCCTCAT | 115 | |
| PETH | PETH-F | ATGCTCGCTACTGGAACTG | 133 |
| PETH-R | GCAATGAACTACTCGTGGG | 133 | |
| RBCS | RBCS-F | ACTACGATGGACGCTACTGG | 108 |
| RBCS-R | CATTGGGGTATTCCTTCTTG | 108 | |
| FBA2 | FBA2-F | ACGGTCTTTCATCCCGCACAG | 132 |
| FBA2-R | AGCGAGCAAGTCCCCAAGC | 132 | |
| GAPA | GAPA-F | ACATCGTCCCGACTTCAA | 168 |
| GAPA-R | CAGCGTTCACCTCTTCAGC | 168 | |
| TKL-2 | TKL-2-F | CCGTTTCATTCTGTCCGC | 149 |
| TKL-2-R | CTCCAGGTGTTTCAAAGTTCTC | 149 | |
| UBC28 | UBC28-F | AGCGGATTTTGAAGGAACT | 127 |
| UBC28-R | GCATAAGGGCTATCTGAGGG | 127 |
| Reagent | Volume (μL) | Step | Time | Cycles |
|---|---|---|---|---|
| 2xqPCRmix | 5.0 | 95 °C | 30 s | |
| F Primer (10 pmol/μL) | 0.25 | 95 °C | 10 s | 40 cycles |
| R Primer (10 pmol/μL) | 0.25 | 60 °C | 30 s | 92.10% |
| DNA template | 2.0 | 95 °C | 15 s | |
| ddH2O | 2.5 | 60 °C | 60 s | fluorescence detection in 0.5 °C steps |
| total | 10.0 | 95 °C | 15 s | 92.07% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhao, Y.; Gao, C.; Li, X.; Wu, K.; Lin, M.; Sun, W. Integrated Analysis of Physiological Responses and Transcriptome of Cotton Seedlings Under Drought Stress. Int. J. Mol. Sci. 2025, 26, 7824. https://doi.org/10.3390/ijms26167824
Li X, Zhao Y, Gao C, Li X, Wu K, Lin M, Sun W. Integrated Analysis of Physiological Responses and Transcriptome of Cotton Seedlings Under Drought Stress. International Journal of Molecular Sciences. 2025; 26(16):7824. https://doi.org/10.3390/ijms26167824
Chicago/Turabian StyleLi, Xin, Yuhao Zhao, Chen Gao, Xiaoya Li, Kunkun Wu, Meiwei Lin, and Weihong Sun. 2025. "Integrated Analysis of Physiological Responses and Transcriptome of Cotton Seedlings Under Drought Stress" International Journal of Molecular Sciences 26, no. 16: 7824. https://doi.org/10.3390/ijms26167824
APA StyleLi, X., Zhao, Y., Gao, C., Li, X., Wu, K., Lin, M., & Sun, W. (2025). Integrated Analysis of Physiological Responses and Transcriptome of Cotton Seedlings Under Drought Stress. International Journal of Molecular Sciences, 26(16), 7824. https://doi.org/10.3390/ijms26167824






