Overexpression of the β-Glucosidase Gene SpBGLU25 from the Desert Pioneer Plant Stipagrostis pennata Enhances the Drought Tolerance in Arabidopsis
Abstract
1. Introduction
2. Results
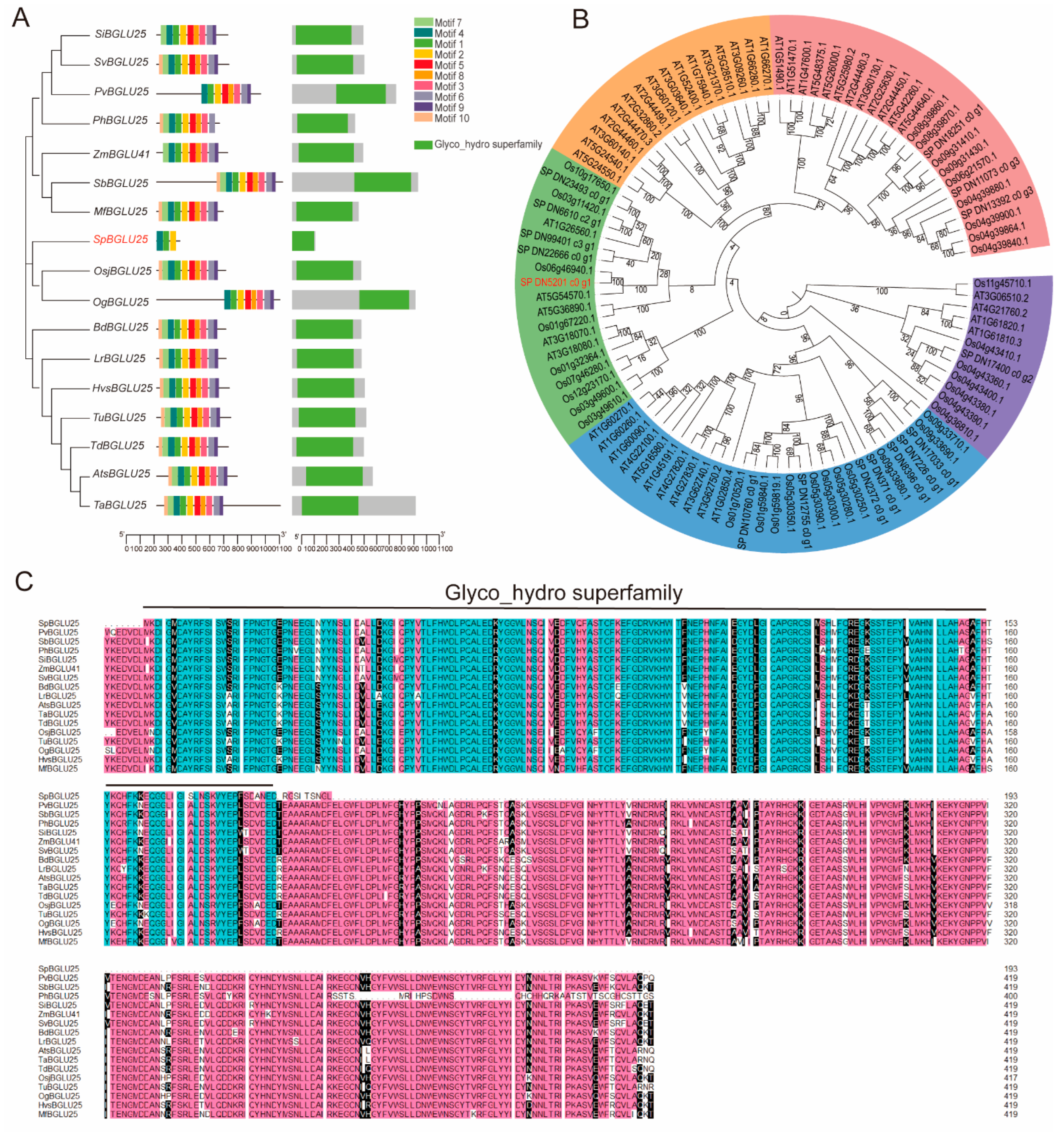
2.1. Phylogenetic Analysis of the SpBGLU25 Gene Sequence in Different Closely Related Species
2.2. Expression Analysis of SpBGLU25
2.3. SpBGLU25 Involvement in Plant Response to Drought Stress
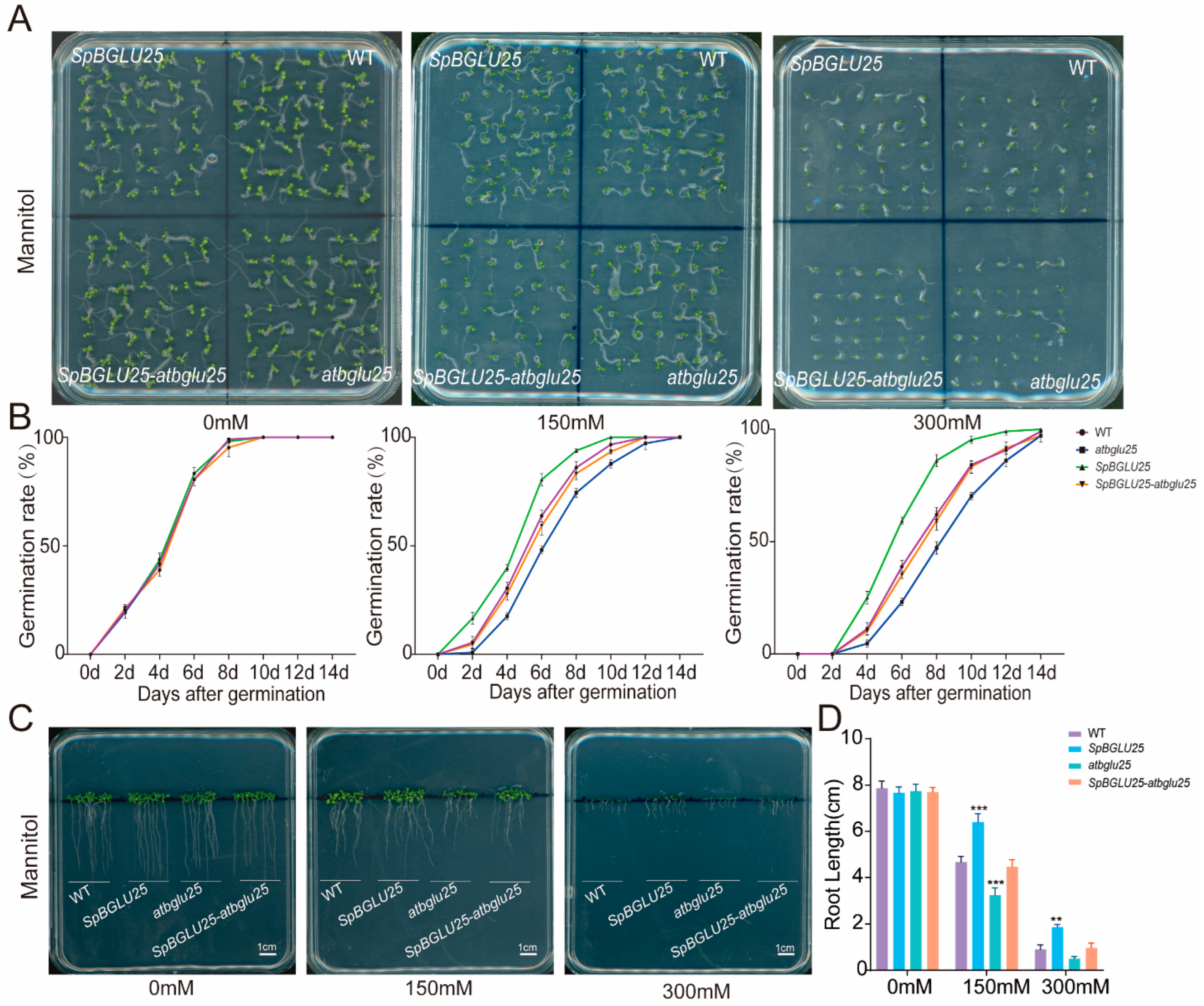
2.3.1. SpBGLU25 Enhances Seed Germination Vitality and Root Growth of Arabidopsis Under Drought Stress
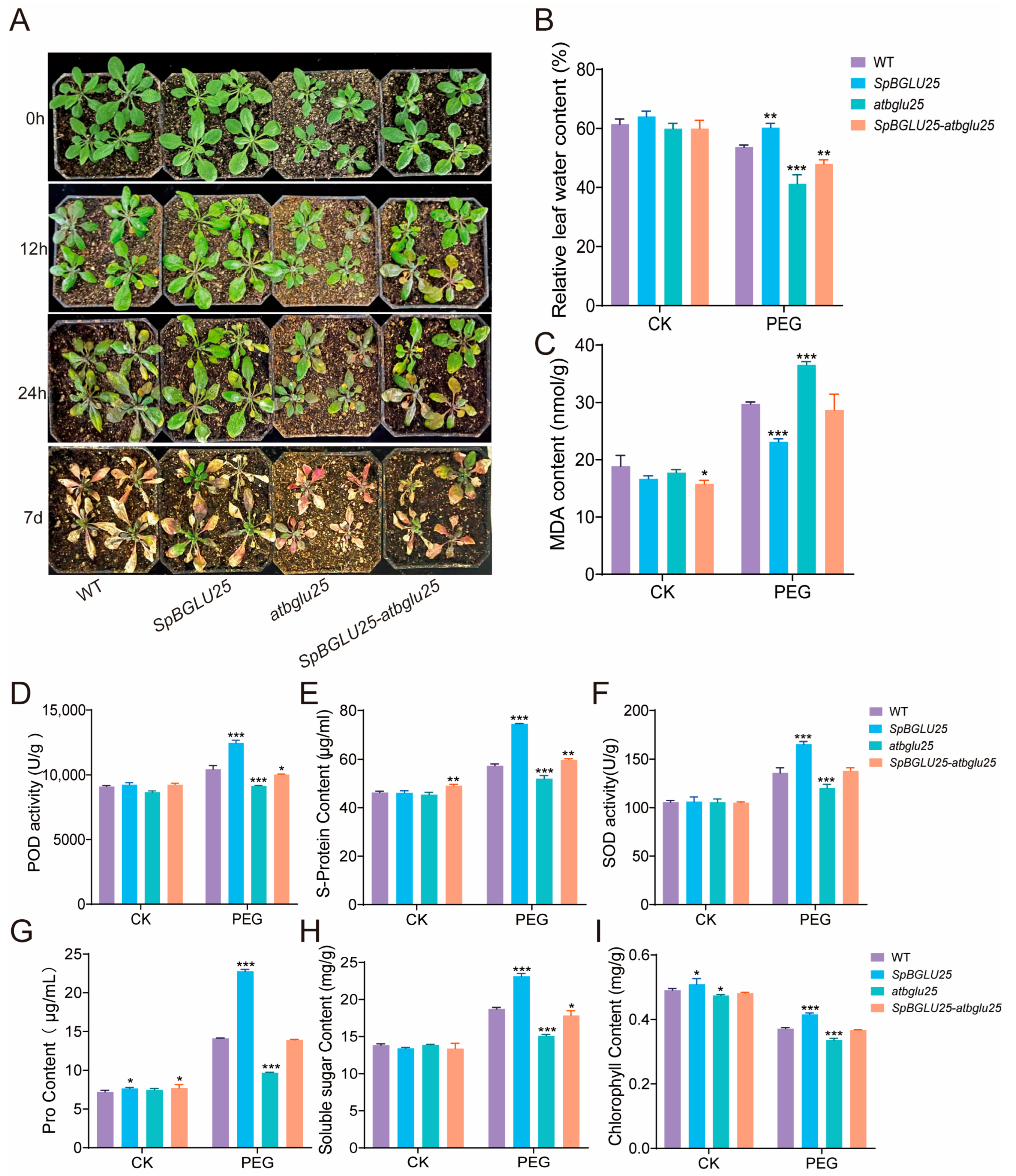
2.3.2. SpBGLU25 Significantly Enhances Drought Resistance in Arabidopsis by Strengthening Antioxidant Defense and Osmotic Regulation
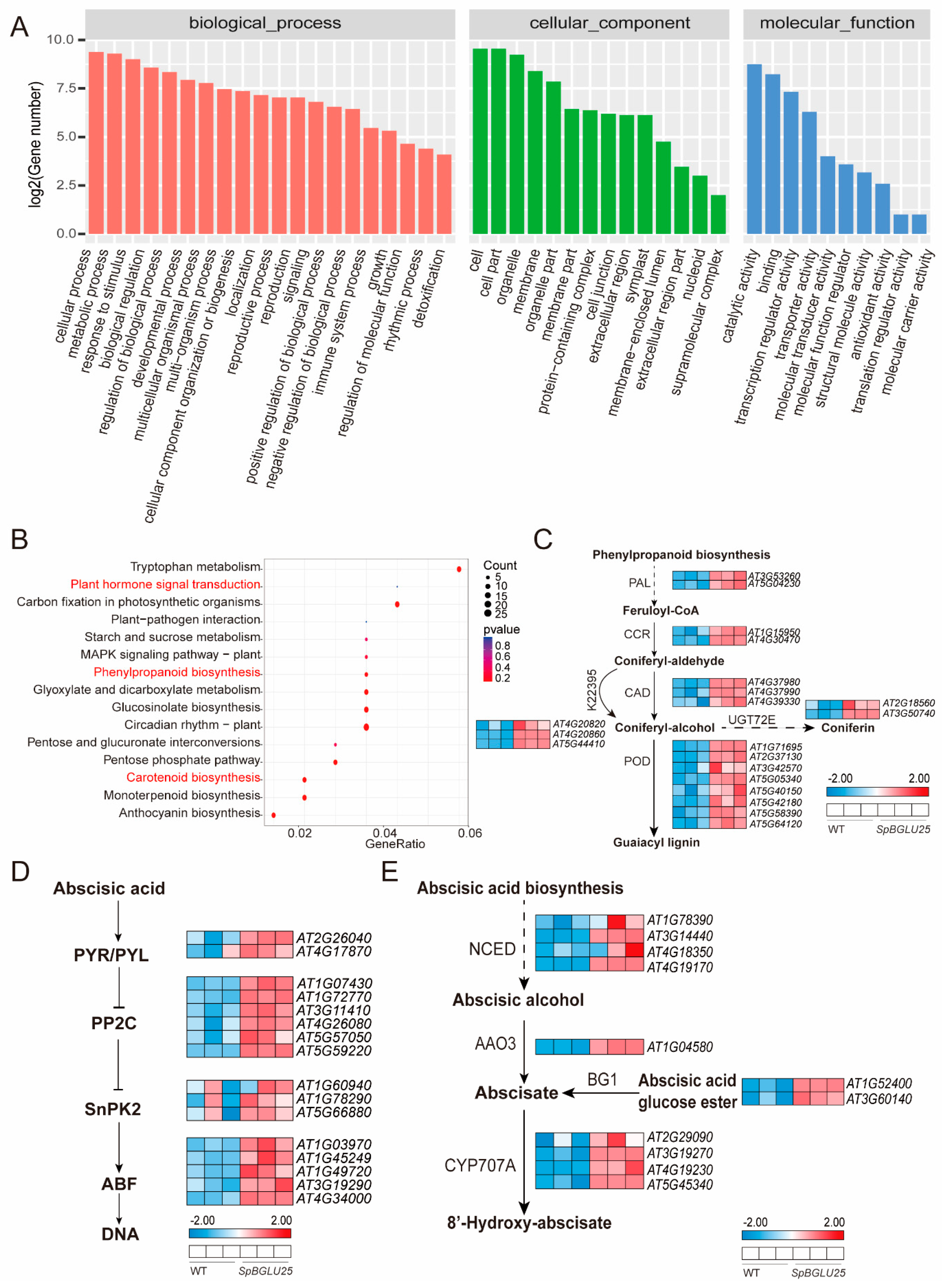
2.4. SpBGLU25 Enhances Drought Resistance in Arabidopsis by Activating Phenylpropanoid Metabolism and ABA Signaling Pathways
2.5. Detection of Germination Rate and Root Length of Four Arabidopsis Types Seeds Under ABA Treatment
3. Discussion
3.1. The Transgenic Arabidopsis SpBGLU25 Adapts to Drought Stress by Increasing Phenolic Compounds and Cell Wall Strength
3.2. Transgenic Arabidopsis SpBGLU25 Adapts to Drought Stress Through Plant Hormone Signaling
4. Materials and Methods
4.1. Materials
4.1.1. Plant Materials
4.1.2. Experimental Reagents
4.2. Methods
4.2.1. Planting of S. pennata Seeds
4.2.2. RNA Extraction and cDNA Synthesis of S. pennata
4.2.3. Cloning of the SpBGLU25 Gene
4.2.4. Gene Sequence Analysis
4.2.5. Construction of Plant Expression Vectors
4.2.6. Subcellular Localization
4.2.7. Gene Expression Characterization Analysis
4.2.8. Obtaining and Identifying SpBGLU25 Gene Overexpression in Arabidopsis
4.2.9. The Effect of Drought Stress on the Phenotype of Arabidopsis Overexpressing the SpBGLU25 Gene
4.2.10. Determination of Germination Rate and Root Length of SpBGLU25 Gene Overexpressing Arabidopsis Seeds
4.2.11. Transcriptome Analysis
4.2.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sun, B.; Yang, D.; Wang, F.; Li, R.; Li, H. Cloning and Expression of the Sugar Transporter Gene SpSWEET3 from the Desert Plant Stipagrostis pennata. Chin. J. Desert Res. 2023, 43, 129–138. [Google Scholar]
- Yin, S.; Tang, R.; Yin, Q.; Wang, F.; Li, R.; Li, H. Cloning and Expression of the Arabinofuranosidase Gene SpARAF1 from Stipagrostis pennata (Stipagrostis pennata). Chin. J. Desert Res. 2024, 44, 48–57. [Google Scholar]
- Zhu, L.-J.; Wang, S.-M.; Xia, J.; Zhu, H.-W. Clonal Configuration and Ramet Population Characteristics of Stipagrostis pennata in Different Habitats. J. Arid Land 2012, 29, 770–775. [Google Scholar]
- Liu, Y.; Xu, H.; Yan, P. A Taxonomic Study on Polygonaceae from Pamirs of China. J. Shihezi Univ. Nat. Sci. Ed. 2009, 27, 162–168. [Google Scholar]
- Ren, M.; Wang, S.; Zhang, X.; Wang, Z.; Yang, M. Rhizosheath soil microbial functional diversity of two typical Gramineae plants in the southern margin of the Junggar basin. J. Ecol. 2017, 37, 5630–5639. [Google Scholar][Green Version]
- Jiao, S.; Li, X.; Zhang, T.; Ma, L. Isolation, Identification and Phylogenetic Tree Analysis of Endophytic Bacteria from Stipagrostis pennata. N. Hortic. 2020, 77–84. [Google Scholar]
- Jiehua, C. Study on the cellular characteristics of different populations of Stipagrostis pennata in the Junggar Basin. Ph.D. Thesis, Shihezi University, Shihezi, China, 2013. [Google Scholar]
- Baohua, G. Preliminary study on the drought resistance physiological mechanisms of Stipagrostis pennata. Ph.D. Thesis, Shihezi University, Shihezi, China, 2009. [Google Scholar]
- Long, L.; Wang, H.; Ma, X.; Xu, H.; Wang, S. Seed Rain of Stipagrostis pennata in the Gurbantonggut Desert. Arid Zone Res. 2014, 31, 516–522. [Google Scholar][Green Version]
- Wang, C.; Li, J. Research progress on plant β-glucosidase. Biol. Res. 2021, 43, 101–109. [Google Scholar][Green Version]
- Ketudat, C.J.; Esen, A. beta-Glucosidases. Cell. Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Yang, J.; Ma, L.; Jiang, W.; Yao, Y.; Tang, Y.; Pang, Y. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula. Plant Physiol. Biochem. 2021, 158, 21–33. [Google Scholar] [CrossRef]
- Kong, H.; Song, J.; Ma, S.; Yang, J.; Shao, Z.; Li, Q.; Li, Z.; Xie, Z.; Yang, P.; Cao, Y. Genome-wide identification and expression analysis of the glycosyl hydrolase family 1 genes in Medicago sativa revealed their potential roles in response to multiple abiotic stresses. BMC Genom. 2024, 25, 20. [Google Scholar] [CrossRef] [PubMed]
- Baiya, S.; Mahong, B.; Lee, S.K.; Jeon, J.S.; Cairns, J.R.K. Demonstration of monolignol beta-glucosidase activity of rice Os4BGlu14, Os4BGlu16 and Os4BGlu18 in Arabidopsis thaliana bglu45 mutant. Plant Physiol. Biochem. 2018, 127, 223–230. [Google Scholar] [CrossRef]
- Xu, M.; Li, H.; Luo, H.; Liu, J.; Li, K.; Li, Q.; Yang, N.; Xu, D. Unveiling the Role of beta-Glucosidase Genes in Bletilla striata’s Secondary Metabolism: A Genome-Wide Analysis. Int. J. Mol. Sci. 2024, 25, 13191. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.X.; Miao, L.L.; Liu, Y.; Liu, H.C.; Liu, Z.P. Gene cloning and characterization of a cold-adapted beta-glucosidase belonging to glycosyl hydrolase family 1 from a psychrotolerant bacterium Micrococcus antarcticus. Enzyme Microb. Technol. 2011, 49, 94–99. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Feng, X.; Peng, F.; Mazoor, M.A.; Zhang, Y.; Zhao, Y.; Han, W.L.; Lu, L.; Cao, Y.; et al. Analysis of the beta-Glucosidase Family Reveals Genes Involved in the Lignification of Stone Cells in Chinese White Pear (Pyrus bretschneideri Rehd.). J. Front. Plant Sci. 2022, 13, 852001. [Google Scholar] [CrossRef]
- Han, Y.; Watanabe, S.; Shimada, H.; Sakamoto, A. Dynamics of the leaf endoplasmic reticulum modulate beta-glucosidase-mediated stress-activated ABA production from its glucosyl ester. J. Exp. Bot. 2020, 71, 2058–2071. [Google Scholar] [CrossRef] [PubMed]
- Brzobohatý, B.; Moore, I.; Kristoffersen, P.; Bako, L.; Campos, N.; Schell, J.; Palme, K. Release of active cytokinin by a beta-glucosidase localized to the maize root meristem. Science 1993, 262, 1051–1054. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Lee, K.H.; Dong, T.; Jeong, J.C.; Jin, J.B.; Kanno, Y.; Kim, D.H.; Kim, S.Y.; Seo, M.; Bressan, R.A.; et al. A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell. 2012, 24, 2184–2199. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.A.; Morais, M.A.B.; Terrett, O.M.; Lyczakowski, J.J.; Zanphorlin, L.M.; Ferreira-Filho, J.A.; Tonoli, C.C.C.; Murakami, M.T.; Dupree, P.; Souza, A.P. An engineered GH1 beta-glucosidase displays enhanced glucose tolerance and increased sugar release from lignocellulosic materials. Sci. Rep. 2019, 9, 4903. [Google Scholar] [CrossRef]
- Lee, K.H.; Piao, H.L.; Kim, H.Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.J.; Hwang, I. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef]
- Wang, C.; Chen, S.; Dong, Y.; Ren, R.; Chen, D.; Chen, X. Chloroplastic Os3BGlu6 contributes significantly to cellular ABA pools and impacts drought tolerance and photosynthesis in rice. New Phytol. 2020, 226, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Yuan, X.; Zhao, Y.; Wang, X.; Lu, L.; Wang, H.; Li, Y.; Gao, J.; Wang, L.; Zhang, H. Identification of ARF Genes and Elucidation of the Regulatory Effects of PsARF16a on the Dormancy of Tree Peony Plantlets. Genes 2024, 15, 666. [Google Scholar] [CrossRef]
- Wan, Q.; Yao, R.; Zhao, Y.; Xu, Y. JA and ABA signaling pathways converge to protect plant regeneration in stress conditions. Cell. Rep. 2025, 44, 115423. [Google Scholar] [CrossRef] [PubMed]
- Baiya, S.; Hua, Y.; Ekkhara, W.; Cairns, J.R.K. Expression and enzymatic properties of rice (Oryza sativa L.) monolignol beta-glucosidases. Plant Sci. 2014, 227, 101–109. [Google Scholar] [CrossRef]
- Pereira, A. Plant Abiotic Stress Challenges from the Changing Environment. Front. Plant Sci. 2016, 7, 1123. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Yang, J.; Wang, X.; Zhang, N.; Si, H. Potato Stu-miR398b-3p Negatively Regulates Cu/Zn-SOD Response to Drought Tolerance. Int. J. Mol. Sci. 2023, 24, 2525. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Wang, W.; Yi, H.; Zhang, R.; Han, Y.; Wang, Y.; Zhou, F. Physiological and Ecological Changes of Ginkgo biloba Leaves During Yellowing in Semi-arid Areas. J. Anhui Agric. Sci. 2025, 53, 118–123. [Google Scholar]
- Wang, K.; Nan, L.; Li, J.; Liang, P.; Chen, J.; Wei, S.; Liu, X. Effects of drought stress on endogenous hormonecontents of different root-type alfalfa. China Agric. Res. Arid. Areas 2022, 40, 30–36. [Google Scholar]
- Nong, Y.; Wu, H.; Weng, X.; Wei, J.; Luo, Y.; Yang, J.; Chen, X.; Chen, Q. Effects of Drought Stress on the Physiological Characteristics of Seedlings of Different Tea Varieties. China J. Anhui Agric. Sci. 2024, 52, 96–100. [Google Scholar]
- Chu, F.; Liu, Y.; Wang, W.; Hu, Q.; Yang, A. Effects of Drought Stress on Active Oxygen Metabolism, Osmotic Regulators, SPAD and Chlorophyll Fluorescence Characteristics of Sweet Potato. Chin. Agric. Bull. 2019, 35, 29–34. [Google Scholar]
- Shao, Z.; Yang, B.; Zhu, C.; Li, D.; Guo, J. Phenolic Content and Antioxidant Activity in Leaves of Cerasus humilis at Different Growth Stages. China Acta Bot. Northwestica 2022, 42, 1720–1727. [Google Scholar]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Qi, X.; Zhu, R.; Ye, D.; Shou, M.; Peng, L.; Qiu, M.; Shi, M.; Kai, G. Transcriptome Analysis of Salvia miltiorrhiza under Drought Stress. Plants 2024, 13, 161. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Yoon, S.; Kim, Y.S.; Kim, J.J. Rice OsERF71-mediated root modification affects shoot drought tolerance. Plant Signal. Behav. 2017, 12, e1268311. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, Z.; Ni, H.; Yuan, F.; Zhang, S. Identification and Functional Analysis of CAD Gene Family in Pomegranate (Punica granatum). Genes 2022, 14, 26. [Google Scholar] [CrossRef]
- Moura, J.C.M.S.; Bonine, C.A.V.; De Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Wang, Y. Transgenic Study on the Relationship between Poplar Glycosyltransferase and Lignin Synthesis. Shandong University, 2012. [Google Scholar]
- Lanot, A.; Hodge, D.; Lim, E.K.; Vaistij, F.E.; Bowles, D.J. Redirection of flux through the phenylpropanoid pathway by increased glucosylation of soluble intermediates. Planta 2008, 228, 609–616. [Google Scholar] [CrossRef]
- Yuan, H.; Guo, W.; Zhao, L.; Yu, Y.; Wu, J.; Cheng, L.; Zhao, D.; Kang, Q.; Huang, W. Cloning and Expression Analysis of the Glycosyltransferase Gene LuUGT72E1 in Flax. China J. Crops 2016, 62–67. [Google Scholar]
- Yanqing, J. Study on the Mechanism of Peroxidase Gene TaPrx109-B1 in Enhancing Drought Resistance in Wheat. Ph.D. Thesis, Henan Agricultural University, Henan, China, 2024. [Google Scholar]
- Zhang, L.; Dai, Y.; Yue, L.; Chen, G.; Yuan, L.; Zhang, S.; Li, F.; Zhang, H.; Li, G.; Zhu, S.; et al. Heat stress response in Chinese cabbage (Brassica rapa L.) revealed by transcriptome and physiological analysis. PeerJ 2022, 10, e13427. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, F.; Tang, M. Transcriptome Analysis of Arbuscular Mycorrhizal Casuarina glauca in Damage Mitigation of Roots on NaCl Stress. Microorganisms 2021, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Yong, Q.; Li, Z.; Qu, Y. Response of Endogenous Hormones in Camellia oleifera to Drought Stress and Rehydration Based on Transcriptome Analysis. Southwest Agric. J. 2024, 37, 913–924. [Google Scholar]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012, 35, 53–60. [Google Scholar] [CrossRef]
- Fatima, O.A. A Comprehensive Analysis of the 9-Cis Epoxy Carotenoid Dioxygenase Gene Family and Their Responses to Salt Stress in Hordeum vulgare L. Plants 2024, 13, 3327. [Google Scholar] [CrossRef]
- Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P.W.; Kayastha, A.M.; Singh, V.K. MFPPI—Multi FASTA ProtParam Interface. Bioinformation 2016, 12, 74–77. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50(D1), D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]


| Primer Name | Primer Sequence (from 5′ to 3′) | Primer Function |
|---|---|---|
| SpBGLU25-F | ATGGAAGTACTTTGTGAAACC | Gene cloning |
| SpBGLU25-R | CCTGCGGTCATTGTCAT | |
| SpBGLU25-tong-F | atttggagaggacagggtaccATGAAGGACATTGGCATGGATG | Gene vector construction |
| SpBGLU25-tong-R | ggtactagtgtcgactctagaAAGTCCATTGCTCGTGATGCTG | |
| q-GAPDH-F | AGTCCGTCGCCATCGTCA | The reference gene |
| q-GAPDH-R | CGTGCCCATGCCTTCTGT | |
| q-SpBGL25-F | AGCTCATGCTGGTGCTTTTC | Gene expression analysis |
| q-SpBGL25-R | GTCCATTGCTCGTGATGCTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, J.; Wang, J.; Zhu, F.; Li, X.; Feng, J.; Fan, J.; Chen, M.; Li, X.; Hu, M.; Song, Z.; et al. Overexpression of the β-Glucosidase Gene SpBGLU25 from the Desert Pioneer Plant Stipagrostis pennata Enhances the Drought Tolerance in Arabidopsis. Int. J. Mol. Sci. 2025, 26, 6663. https://doi.org/10.3390/ijms26146663
Niu J, Wang J, Zhu F, Li X, Feng J, Fan J, Chen M, Li X, Hu M, Song Z, et al. Overexpression of the β-Glucosidase Gene SpBGLU25 from the Desert Pioneer Plant Stipagrostis pennata Enhances the Drought Tolerance in Arabidopsis. International Journal of Molecular Sciences. 2025; 26(14):6663. https://doi.org/10.3390/ijms26146663
Chicago/Turabian StyleNiu, Jiahuan, Jingru Wang, Faren Zhu, Xuechi Li, Jianting Feng, Jiliang Fan, Mingsu Chen, Xiaoying Li, Ming Hu, Zhangqi Song, and et al. 2025. "Overexpression of the β-Glucosidase Gene SpBGLU25 from the Desert Pioneer Plant Stipagrostis pennata Enhances the Drought Tolerance in Arabidopsis" International Journal of Molecular Sciences 26, no. 14: 6663. https://doi.org/10.3390/ijms26146663
APA StyleNiu, J., Wang, J., Zhu, F., Li, X., Feng, J., Fan, J., Chen, M., Li, X., Hu, M., Song, Z., Li, Z., Wang, F., Li, R., & Li, H. (2025). Overexpression of the β-Glucosidase Gene SpBGLU25 from the Desert Pioneer Plant Stipagrostis pennata Enhances the Drought Tolerance in Arabidopsis. International Journal of Molecular Sciences, 26(14), 6663. https://doi.org/10.3390/ijms26146663





