Regulatory Mechanism of the GmMYB14 Transcription Factor on Auxin-Related Proteins in Soybean
Abstract
1. Introduction
2. Results
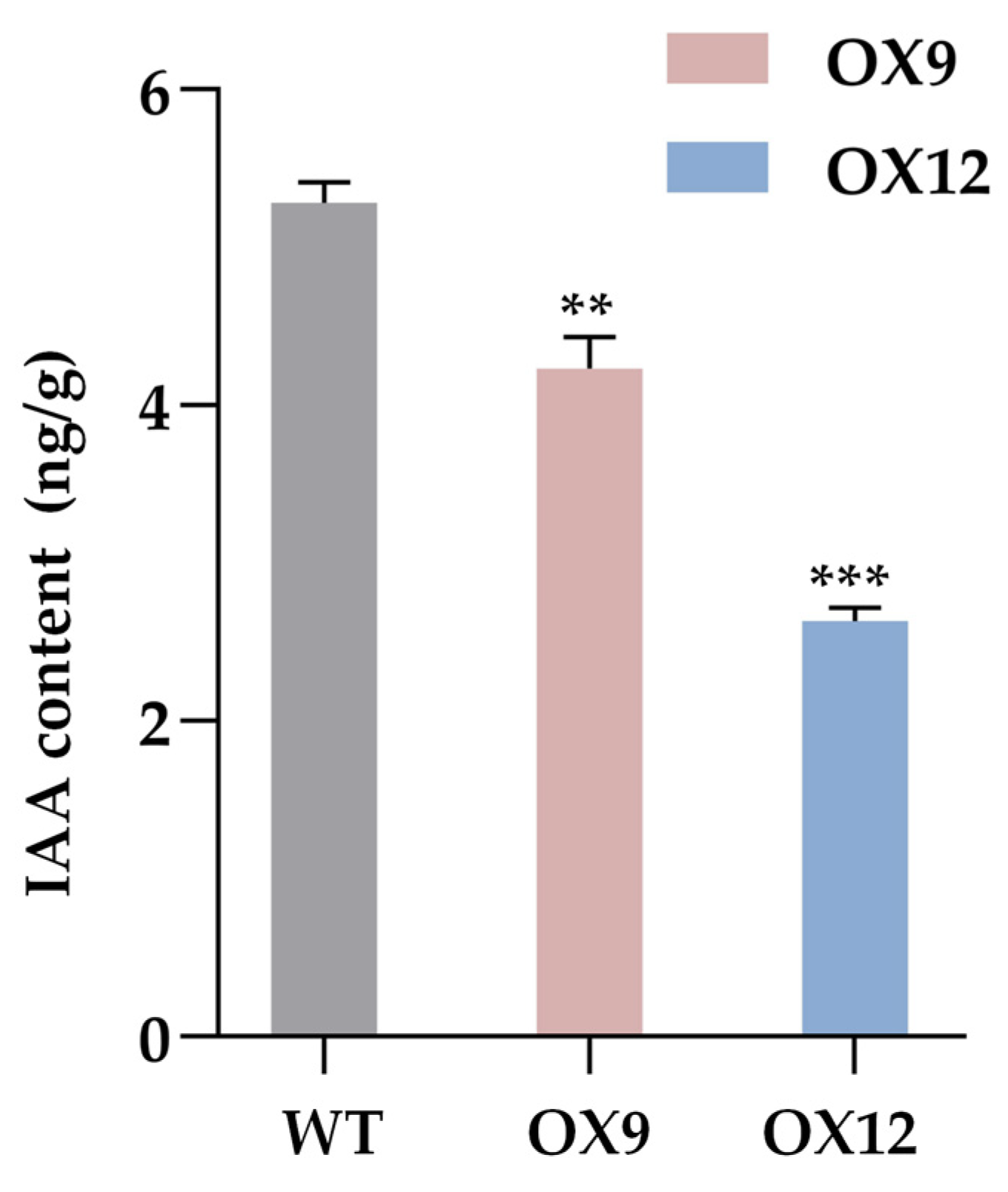
2.1. Endogenous IAA Content of GmMYB14-OX Plants Decreased
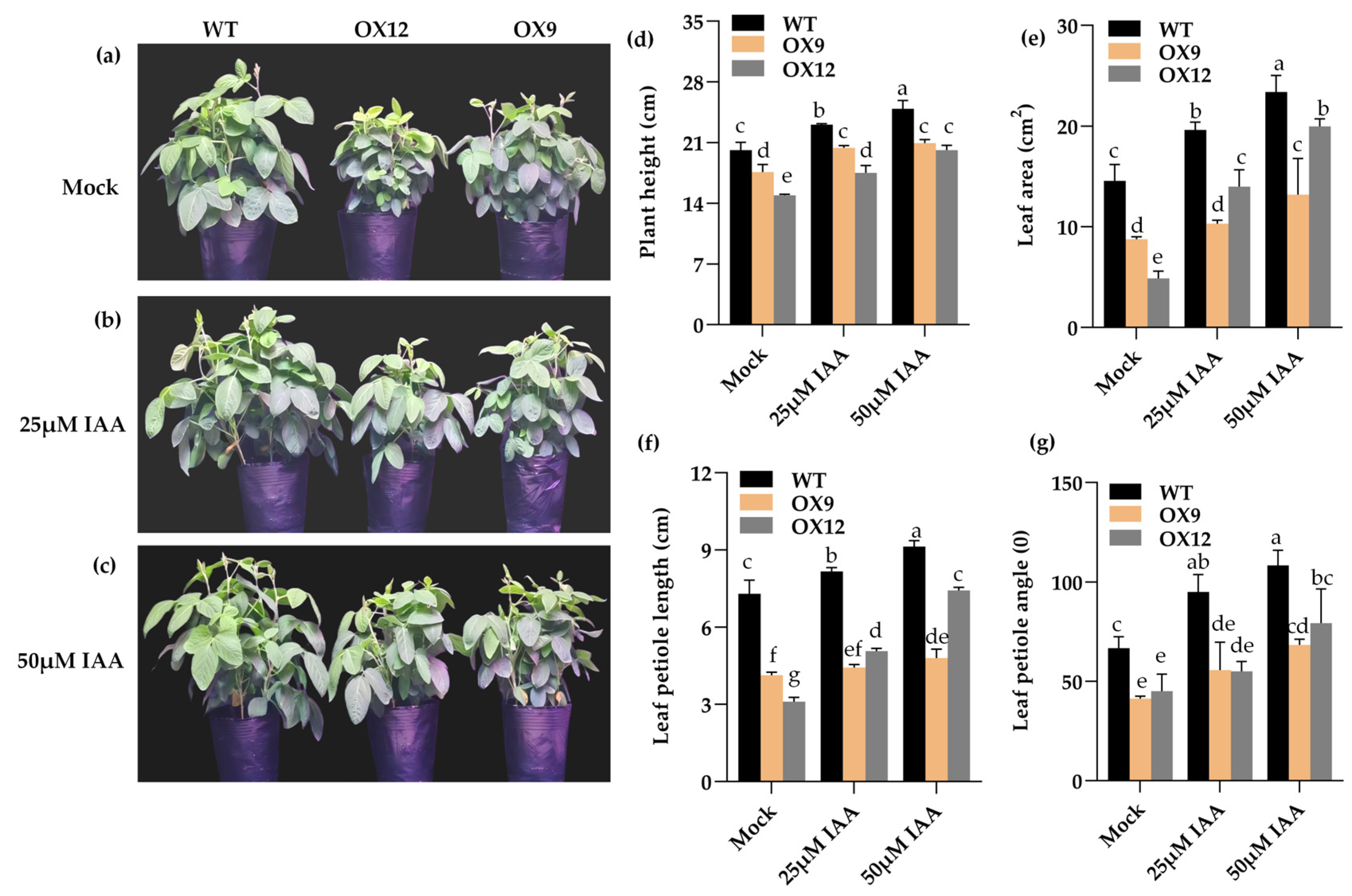
2.2. Phenotype of GmMYB14-OX Plants Partially Recovered After Treatment with IAA
2.3. Differentially Expressed Genes (DEGs) in GmMYB14-OX and WT Plants
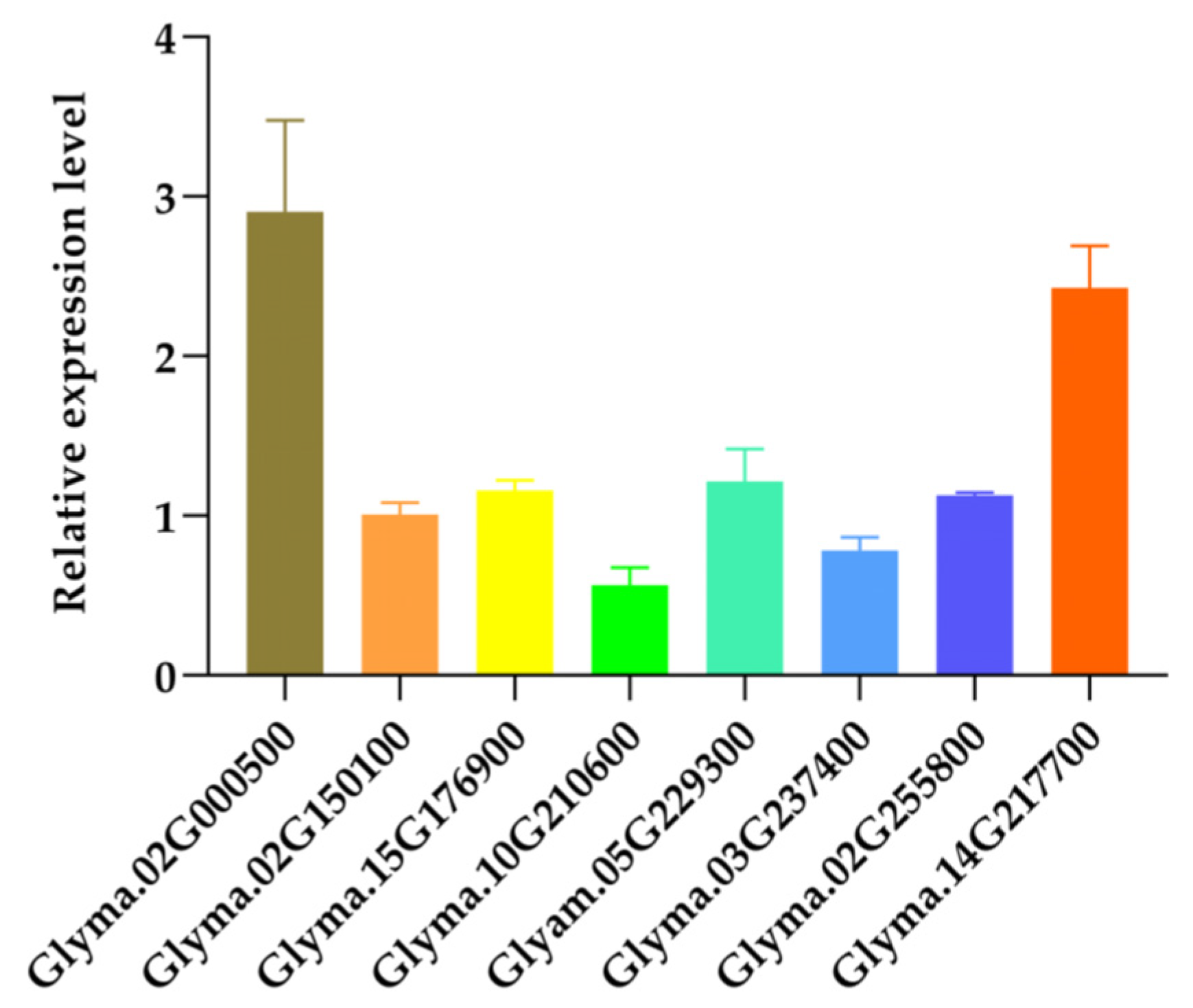
2.4. High Expression of GmIAA1 in OX12 Leaves
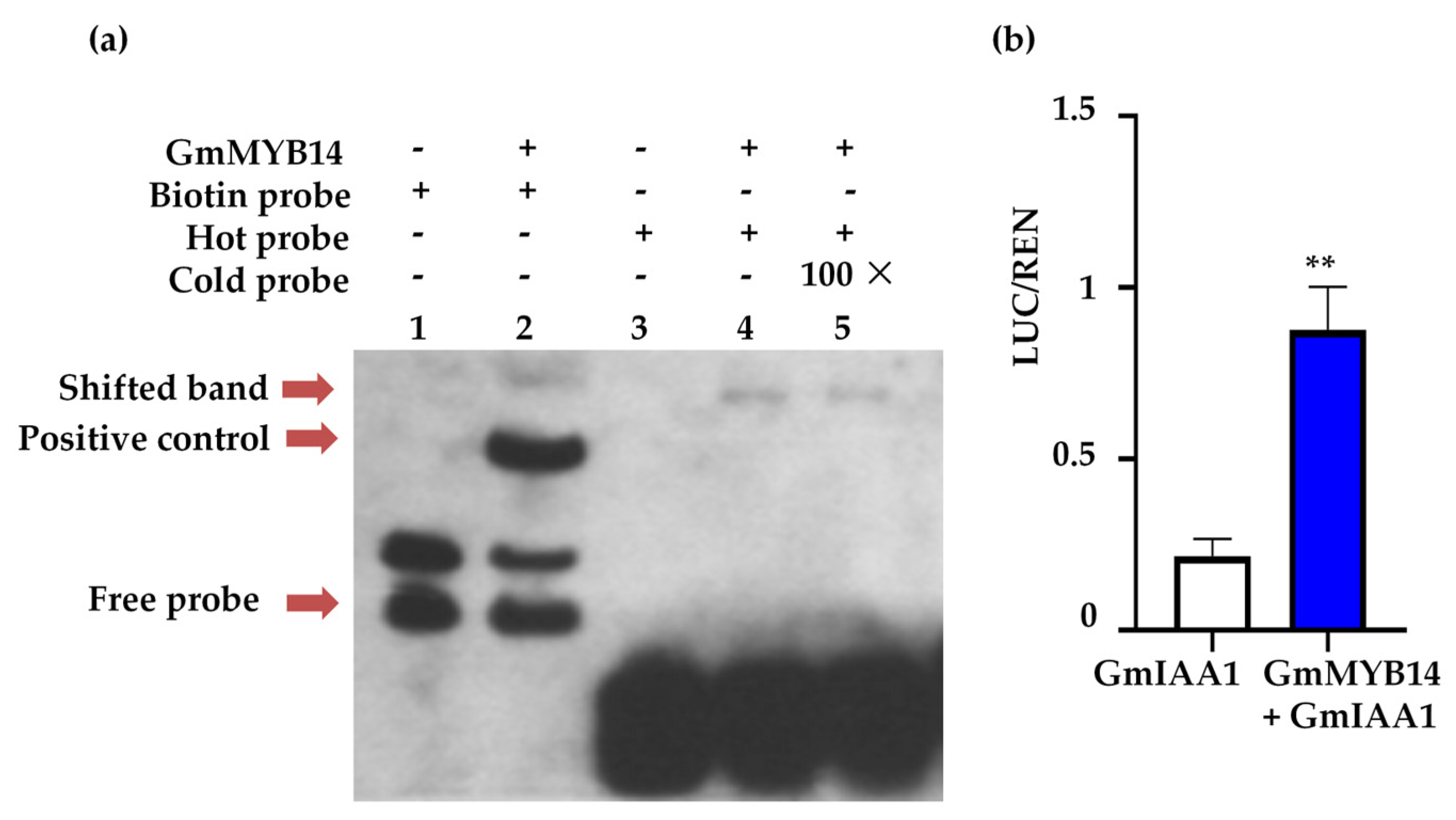
2.5. GmMYB14 Directly Regulated the Expression of GmIAA1
3. Discussion
4. Materials and Methods
4.1. Plants Materials
4.2. Endogenous IAA Content Measurement
4.3. Application of Exogenous IAA
4.4. Transcriptome Analysis
4.5. Quantitative Reverse Transcription PCR (qRT-PCR)
4.6. Electrophoretic Mobility Shift Assay (EMSA)
4.7. Dual-Luciferase Assay
4.8. Data Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kumar, M.; Suhag, R.; Hasan, M.; Dhumal, S.; Radha; Pandiselvam, R.; Senapathy, M.; Sampathrajan, V.; Punia, S.; Sayed, A.; et al. Black soybean (Glycine max (L.) Merr.): Paving the way toward new nutraceutical. Crit. Rev. Food Sci. Nutr. 2023, 63, 6208–6234. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X.; Hao, L.; Zhang, G.; Jin, Z.; Li, C.; Yang, Y.; Rao, J.; Chen, B. Traditional fermented soybean products: Processing, flavor formation, nutritional and biological activities. Crit. Rev. Food Sci. Nutr. 2022, 62, 1971–1989. [Google Scholar] [CrossRef]
- Kim, I.S.; Yang, W.S.; Kim, C.H. Beneficial effects of soybean-derived bioactive peptides. Int. J. Mol. Sci. 2021, 22, 8570. [Google Scholar] [CrossRef]
- He, Y.; Wang, Y.J.; Zhang, S.Z.; Tao, W. Investigation and analysis of soybean meal supply in 2020. Feed Rev. 2021, 7, 48–50. [Google Scholar]
- Hou, Z.; Huang, H.; Wang, Y.; Chen, L.; Yue, L.; Liu, B.; Kong, F.; Yang, H. Molecular regulation of shoot architecture in soybean. Plant Cell Environ. 2024. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.B.; Ma, J. The genetic basis of shoot architecture in soybean. Mol. Breed. 2023, 43, 55. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, D.; Mao, Y.; Zhou, Y.; Zhao, L.; Zhang, C.; Liu, Y.; Chen, J. The organ size and morphological change during the domestication process of soybean. Front. Plant Sci. 2022, 13, 913238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Song, Y.; Ou, R.; Liu, Y.; Li, S.; Lu, X.; Xu, S.; Su, Y.; Jiang, D.; Ding, Y.; et al. SCAG: A stratified, clustered, and growing-based algorithm for soybean branch angle extraction and ideal plant architecture evaluation. Plant Phenomics 2024, 6, 190. [Google Scholar] [CrossRef]
- Clark, C.B.; Wang, W.; Wang, Y.; Fear, G.J.; Wen, Z.; Wang, D.; Ren, B.; Ma, J. Identification and molecular mapping of a major quantitative trait locus underlying branch angle in soybean. Theor. Appl. Genet. 2022, 135, 777–784. [Google Scholar] [CrossRef]
- Liu, W.; Wang, T.; Wang, Y.; Liang, X.; Han, J.; Han, D. MbMYBC1, a M. baccata MYB transcription factor, contribute to cold and drought stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 2023, 14, 1141446. [Google Scholar] [CrossRef]
- Li, P.H. Study on Transcriptional Regulation Mechanism of Flavonoid and Isoflavonoid Biosynthesis in Legume Plants. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2017. [Google Scholar]
- Chen, L.; Yang, H.; Fang, Y.; Guo, W.; Chen, H.; Zhang, X.; Dai, W.; Chen, S.; Hao, Q.; Yuan, S.; et al. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 2021, 19, 702–716. [Google Scholar] [CrossRef]
- Wang, T.; Jin, Y.; Deng, L.; Li, F.; Wang, Z.; Zhu, Y.; Wu, Y.; Qu, H.; Zhang, S.; Liu, Y.; et al. The transcription factor MYB110 regulates plant height, lodging resistance, and grain yield in rice. Plant Cell 2024, 36, 298–323. [Google Scholar] [CrossRef]
- Li, D.; Lu, X.; Qian, D.; Wang, P.; Tang, D.; Zhong, Y.; Shang, Y.; Guo, H.; Wang, Z.; Zhu, G.; et al. Dissected leaf 1 encodes an MYB transcription factor that controls leaf morphology in potato. Theor. Appl. Genet. 2023, 136, 183. [Google Scholar] [CrossRef]
- Han, Y.; Qu, M.; Liu, Z.; Kang, C. Transcription factor FveMYB117a inhibits axillary bud outgrowth by regulating cytokinin homeostasis in woodland strawberry. Plant Cell 2024, 36, 2427–2446. [Google Scholar] [CrossRef]
- Kim, J.S.; Chae, S.; Jo, J.E.; Kim, K.D.; Song, S.; Park, S.H.; Choi, S.; Jun, K.M.; Shim, S.; Jeon, J.; et al. OsMYB14, an R2R3-MYB transcription factor, regulates plant height through the control of hormone metabolism in rice. Mol. Cells 2024, 47, 100093. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, X.; Thompson, A.; Guo, M.; Yoshida, S.; Asami, T.; Chory, J.; Yin, Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009, 58, 275–286. [Google Scholar] [CrossRef]
- Li, Y.Y.; Qi, Y.H. Research advances in biological functions of plant Aux/IAA gene family. Chin. Bull. Bot. 2022, 57, 30–41. [Google Scholar]
- Luo, J.; Zhou, J.; Zhang, J. Aux/IAA gene family in plants: Molecular structure, regulation, and function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef] [PubMed]
- Cancé, C.; Martin-Arevalillo, R.; Boubekeur, K.; Dumas, R. Auxin response factors are keys to the many auxin doors. New Phytol. 2022, 235, 402–419. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, L.; Yu, B.; Guo, J.; Zhao, Y.; Zhu, Y. Expression Analysis of AUX/IAA family genes in apple under salt stress. Biochem. Genet. 2022, 60, 1205–1221. [Google Scholar] [CrossRef]
- Wang, F.; Niu, H.; Xin, D.; Long, Y.; Wang, G.; Liu, Z.; Li, G.; Zhang, F.; Qi, M.; Ye, Y.; et al. OsIAA18, an Aux/IAA transcription factor gene, is involved in salt and drought tolerance in rice. Front. Plant Sci. 2021, 12, 738660. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Tanaka, A.; Inahashi, H.; Nishizawa, N.K.; Tsutsumi, N.; Inukai, Y.; Nakazono, M. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 20770–20775. [Google Scholar] [CrossRef]
- Cao, M.; Chen, R.; Li, P.; Yu, Y.; Zheng, R.; Ge, D.; Zheng, W.; Wang, X.; Gu, Y.; Gelová, Z.; et al. TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 2019, 568, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Si, C.; Zeng, D.; Da Silva, J.A.T.; Qiu, S.; Duan, J.; Bai, S.; He, C. Genome-wide identification of Aux/IAA and ARF gene families reveals their potential roles in flower opening of dendrobium officinale. BMC Genom. 2023, 24, 199. [Google Scholar] [CrossRef]
- Lin, N.; Wang, M.; Jiang, J.; Zhou, Q.; Yin, J.; Li, J.; Lian, J.; Xue, Y.; Chai, Y. Down regulation of Brassica napus MYB69 (BnMYB69) increases biomass growth and disease susceptibility via remodeling phytohormone, chlorophyll, shikimate and lignin levels. Front. Plant Sci. 2023, 14, 1157836. [Google Scholar] [CrossRef]
- Shan, B.H. Functional Study and Molecular Mechanisms of GmMYB133 in Regulating Plant Height in Soybean (Glycine max). Master’s Thesis, Jilin University, Changchun, China, 2022. [Google Scholar]
- Yuan, Y.J. Auxin Signaling Pathway Dependent on R2R3 MYB Regulates Tomato Trichome Formation. Ph.D. Thesis, Chongqing University, Chongqing, China, 2020. [Google Scholar]
- Ma, G.; Li, M.; Wu, Y.; Jiang, C.; Chen, Y.; Xing, D.; Zhao, Y.; Liu, Y.; Jiang, X.; Xia, T.; et al. Camellia sinensis CsMYB4a participates in regulation of stamen growth by interaction with auxin signaling transduction repressor CsAUX/IAA4. Crop. J. 2024, 12, 188–201. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Strader, L.C.; Bartel, B. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 2011, 4, 477–486. [Google Scholar] [CrossRef]
- Casanova-Sáez, R.; Mateo-Bonmatí, E.; Ljung, K. Auxin metabolism in plants. Cold Spring Harb. Perspect. Biol. 2021, 13, a039867. [Google Scholar] [CrossRef]
- Wang, Y.; Jiao, Y. Auxin and above-ground meristems. J. Exp. Bot. 2018, 69, 147–154. [Google Scholar] [CrossRef]
- Dong, J.; Huang, H. Auxin polar transport flanking incipient primordium initiates leaf adaxial-abaxial polarity patterning. J. Integr. Plant Biol. 2018, 60, 455–464. [Google Scholar] [CrossRef]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertova, D.; Jurgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef]
- Wang, Q.; Marconi, M.; Guan, C.; Wabnik, K.; Jiao, Y. Polar auxin transport modulates early leaf flattening. Proc. Natl. Acad. Sci. USA 2022, 119, e2079398177. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yoon, J.; Kim, H.; Lee, S.; Kim, T.; Kang, K.; Paek, N. OsMYB7 determines leaf angle at the late developmental stage of lamina joints in rice. Front. Plant Sci. 2023, 14, 1167202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, S.; Lin, Y.; Qiao, J.; Han, N.; Li, Y.; Feng, Y.; Li, D.; Qi, Y. Auxin transporter OsPIN1b, a novel regulator of leaf inclination in rice (Oryza sativa L.). Plants 2023, 12, 409. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, L.; Ke, M.; Gao, Z.; Tu, T.; Huang, L.; Chen, J.; Guan, Y.; Huang, X.; Chen, X. GmPIN1-mediated auxin asymmetry regulates leaf petiole angle and plant architecture in soybean. J. Integr. Plant Biol. 2022, 64, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Chen, Z.; Cui, Y.; Ke, M.; Xu, H.; Xu, Q.; Chen, J.; Li, Y.; Huang, L.; Zhao, H.; et al. GmPIN-dependent polar auxin transport is involved in soybean nodule development. Plant Cell 2021, 33, 2981–3003. [Google Scholar] [CrossRef]
- Wiśniewska, J.; Kęsy, J.; Mucha, N.; Tyburski, J. Auxin resistant 1 gene (AUX1) mediates auxin effect on Arabidopsis thaliana callus growth by regulating its content and distribution pattern. J. Plant Physiol. 2024, 293, 154168. [Google Scholar] [CrossRef]
- Yang, Z.; Wei, H.; Gan, Y.; Liu, H.; Cao, Y.; An, H.; Que, X.; Gao, Y.; Zhu, L.; Tan, S.; et al. Structural insights into auxin influx mediated by the Arabidopsis AUX1. Cell 2025, 188, 3960–3973.e15. [Google Scholar] [CrossRef]
- Mellor, N.; Bennett, M.J.; King, J.R. GH3-mediated auxin conjugation can result in either transient or oscillatory transcriptional auxin responses. Bull. Math. Biol. 2016, 78, 210–234. [Google Scholar] [CrossRef] [PubMed]
- Péret, B.; Middleton, A.M.; French, A.P.; Larrieu, A.; Bishopp, A.; Njo, M.; Wells, D.M.; Porco, S.; Mellor, N.; Band, L.R.; et al. Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Mol. Syst. Biol. 2013, 9, 699. [Google Scholar] [CrossRef]
- Tognacca, R.S.; Ljung, K.; Botto, J.F. Unveiling molecular signatures in light-induced seed germination: Insights from PIN3, PIN7, and AUX1 in Arabidopsis thaliana. Plants. 2024, 13, 408. [Google Scholar] [CrossRef] [PubMed]
- Seifu, Y.W.; Pukyšová, V.; Rýdza, N.; Bilanovičová, V.; Zwiewka, M.; Sedláček, M.; Nodzyński, T. Mapping the membrane orientation of auxin homeostasis regulators PIN5 and PIN8 in Arabidopsis thaliana root cells reveals their divergent topology. Plant Methods 2024, 20, 84. [Google Scholar] [CrossRef]
- Mravec, J.; Skůpa, P.; Bailly, A.; Hoyerová, K.; Křeček, P.; Bielach, A.; Petrášek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.; et al. Subcellular homeostasis of phytohormoneauxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef]
- Hayashi, K.; Arai, K.; Aoi, Y.; Tanaka, Y.; Hira, H.; Guo, R.; Hu, Y.; Ge, C.; Zhao, Y.; Kasahara, H.; et al. The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 2021, 12, 6752. [Google Scholar] [CrossRef]
- Qiao, J.; Zhang, Y.; Han, S.; Chang, S.; Gao, Z.; Qi, Y.; Qian, Q. OsARF4 regulates leaf inclination via auxin and brassinosteroid pathways in rice. Front. Plant Sci. 2022, 13, 979033. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, S.; Xu, Y.; Yu, C.; Shen, C.; Qian, Q.; Geisler, M.; Jiang, D.A.; Qi, Y. Auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015, 38, 638–654. [Google Scholar] [CrossRef] [PubMed]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef]
- Bian, S.; Jin, D.; Sun, G.; Shan, B.; Zhou, H.; Wang, J.; Zhai, L.; Li, X. Characterization of the soybean R2R3-MYB transcription factor GmMYB81 and its functional roles under abiotic stresses. Gene 2020, 753, 144803. [Google Scholar] [CrossRef]
- Song, Y.; You, J.; Xiong, L. Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol. Biol. 2009, 70, 297–309. [Google Scholar] [CrossRef]
- Su, B.; Wu, H.; Guo, Y.; Gao, H.; Wei, Z.; Zhao, Y.; Qiu, L. GmIAA27 encodes an AUX/IAA protein involved in dwarfing and multi-branching in soybean. Int. J. Mol. Sci. 2022, 23, 8643. [Google Scholar] [CrossRef]
- Bar, M.; Israeli, A.; Levy, M.; Gera, H.B.; Jiménez-Gómez, J.M.; Kouril, S.; Tarkowski, P.; Ori, N. CLAUSA is a MYB transcription factor that promotes leaf differentiation by attenuating cytokinin signaling. Plant Cell 2016, 28, 1602–1615. [Google Scholar] [CrossRef]
- Lee, K.W.; Chen, J.J.W.; Wu, C.S.; Chang, H.C.; Chen, H.Y.; Kuo, H.H.; Lee, Y.S.; Chang, Y.L.; Chang, H.C.; Shiue, S.Y.; et al. Auxin plays a role in the adaptation of rice to anaerobic germination and seedling establishment. Plant Cell Environ. 2023, 46, 1157–1175. [Google Scholar] [CrossRef] [PubMed]
- Gören-Sağlam, N.; Harrison, E.; Breeze, E.; Öz, G.; Buchanan-Wollaston, V. Analysis of the impact of indole-3-acetic acid (IAA) on gene expression during leaf senescence in Arabidopsis thaliana. Physiol. Mol. Biol. Plants 2020, 26, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.M.; Zhou, X.A.; Li, W.B.; Chang, W.; Zhou, R.; Wang, C.; Sha, A.H.; Shan, Z.H.; Zhang, C.J.; Qiu, D.Z.; et al. Genome-wide transcriptional analysis of two soybean genotypes under dehydration and rehydration conditions. BMC Genomics. 2013, 14, 687. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Motulsky, H.J.; Brown, R.E. Detecting outliers when fitting data with nonlinear regression—A new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform. 2006, 7, 123. [Google Scholar] [CrossRef]
| Class | Gene ID | Fold Changes | A. thaliana | Annotations | |
|---|---|---|---|---|---|
| Up | GmARF | Glyma.14G217700 | 4.71 | AT1G19850 | Auxin response factor 5 |
| Glyma.10G210600 | 3.48 | AT4G30080 | Auxin response factor 10 related | ||
| GmAUX/IAA | Glyma.02G000500 | 2.49 | AT5G43700 | Auxin responsive protein IAA 1 related | |
| Glyma.05G229300 | 3.28 | AT4G29080 | Auxin induced protein | ||
| GmABP | Glyma.15G176900 | 3.91 | AT5G20630 | Germin like protein subfamily 3 member 3 | |
| Glyma.02G150100 | 4.58 | AT4G02980 | Auxin binding protein 1 | ||
| GmAUX1/LAX | Glyma.02G255800 | 4.65 | AT2G38120 | Amino acid transporter | |
| GmAXR4 | Glyma.03G237400 | 2.09 | AT1G54990 | Protein auxin response 4 like | |
| Down | GmARF | Glyma.11G145500 | −2.73 | AT2G28350 | Auxin response factor 10 related |
| Glyma.09G072200 | −3.37 | AT5G20730 | Auxin response factor 19 related | ||
| Glyma.01G002100 | −3.53 | AT5G20730 | Auxin response factor | ||
| Glyma.13G112600 | −4.41 | AT5G20730 | Auxin response factor 19 related | ||
| GmAUX/IAA | Glyma.02G142500 | −2.23 | AT3G23050 | Auxin responsive protein IAA16 | |
| Glyma.03G158700 | −2.24 | AT4G14550 | Auxin induced protein AUX28 | ||
| Glyma.20G210500 | −2.14 | AT5G43700 | Auxin responsive protein IAA 1 related | ||
| Glyma.10G031900 | −2.33 | AT3G23050 | Auxin responsive protein IAA16 | ||
| GmAUX1/LAX | Glyma.06G110200 | −2.09 | AT1G77690 | Auxin transporter protein 2 related | |
| Glyma.03G063600 | −2.64 | AT2G38120 | Auxin transporter protein 1 related | ||
| GmPIN | Glyma.18G241000 | −3.84 | AT5G16530 | Auxin efflux carrier component 8 related | |
| Glyma.07G164600 | −4.12 | AT1G70940 | Auxin efflux carrier component 3 related | ||
| GmSAUR | Glyma.08G004100 | −2.33 | AT2G46690 | Small auxin up RNA | |
| Glyma.09G221900 | −3.70 | AT4G38840 | Small auxin up RNA | ||
| Glyma.09G221800 | −4.78 | AT4G38840 | Small auxin up RNA | ||
| Glyma.16G020700 | −2.26 | AT2G46690 | Small auxin up RNA | ||
| GmILR1 | Glyma.06G115100 | −6.10 | AT1G44350 | IAA amino acid hydrolase ILR1 like 6 |
| Name | Primer Sequences (5’-3’) | Application |
|---|---|---|
| Glyma.02G255800-F | TTGACACATTTGGACTCTTGC | qRT-PCR |
| Glyma.02G255800-R | TCAATGATGCAGCTTTGTGTTG | |
| Glyma.14G217700-F | ATGAAGACCACACTTGCAGCAG | |
| Glyma.14G217700-R | TATCAATTTTGCCAAGAGGGTG | |
| Glyma.02G150100-F | GGTAGAACGTTGTACGAGTCCG | |
| Glyma.02G150100-R | AGTCATGTGAGAGAGACCAGCC | |
| Glyma.15G176900-F | GCCTCCAGATTCTGGACTTTTC | |
| Glyma.15G176900-R | TTAACCTGACCCTCCAAGAACA | |
| Glyma.10G210600-F | CACAAGACATCTAATGCTTCCGAT | |
| Glyma.10G210600-R | TTGTCAGTTCCAAGATCAGACTTG | |
| Glyam.05G229300-F | ATGTCTGAGGCATTGGAAGATGT | |
| Glyam.05G229300-R | ATCCACCATTCCAGTTTTCATCA | |
| Glyma.03G237400-F | AGGAAAGTTGAACAGGAGTCCAC | |
| Glyma.03G237400-R | GCGTCCATATAATTAGCTCCATG | |
| Glyma.02G255800-F | TTGACACATTTGGACTCTTTGC | |
| Glyma.02G255800-R | TCAATGATGCAGCTTTGTGTTG | |
| GmActin-F | ATCTTGACTGAGCGTGGTTATTCC | |
| GmActin-R | GCTGGTCCTGGCTGTCTCC | |
| GmIAA1-F | AGCTGTTTAACGGTATGTTTAG | EMSA |
| GmIAA-LUC-F | ACGGTATCGATAAGCTTCGCATTTAAGCTGTTTAACGG | Dual-luciferase assay |
| GmIAA-LUC-R | AGAACTAGTGGATCCGTGACCAGATTATCAGAATCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, L.; Liu, Y.; Yang, H.; Guo, W.; Hao, Q.; Chen, S.; Yuan, S.; Zhang, C.; Yang, Z.; Han, B.; et al. Regulatory Mechanism of the GmMYB14 Transcription Factor on Auxin-Related Proteins in Soybean. Int. J. Mol. Sci. 2025, 26, 7763. https://doi.org/10.3390/ijms26167763
Peng L, Liu Y, Yang H, Guo W, Hao Q, Chen S, Yuan S, Zhang C, Yang Z, Han B, et al. Regulatory Mechanism of the GmMYB14 Transcription Factor on Auxin-Related Proteins in Soybean. International Journal of Molecular Sciences. 2025; 26(16):7763. https://doi.org/10.3390/ijms26167763
Chicago/Turabian StylePeng, Lihua, Yangyan Liu, Hongli Yang, Wei Guo, Qingnan Hao, Shuilian Chen, Songli Yuan, Chanjuan Zhang, Zhonglu Yang, Bei Han, and et al. 2025. "Regulatory Mechanism of the GmMYB14 Transcription Factor on Auxin-Related Proteins in Soybean" International Journal of Molecular Sciences 26, no. 16: 7763. https://doi.org/10.3390/ijms26167763
APA StylePeng, L., Liu, Y., Yang, H., Guo, W., Hao, Q., Chen, S., Yuan, S., Zhang, C., Yang, Z., Han, B., Huang, Y., Shan, Z., Chen, L., & Chen, H. (2025). Regulatory Mechanism of the GmMYB14 Transcription Factor on Auxin-Related Proteins in Soybean. International Journal of Molecular Sciences, 26(16), 7763. https://doi.org/10.3390/ijms26167763






