Analysis of Selective Pressure on Ancient Human Mitochondrial Genomes Reveals the Presence of Widespread Sequencing Artefacts
Abstract
1. Introduction
2. Results
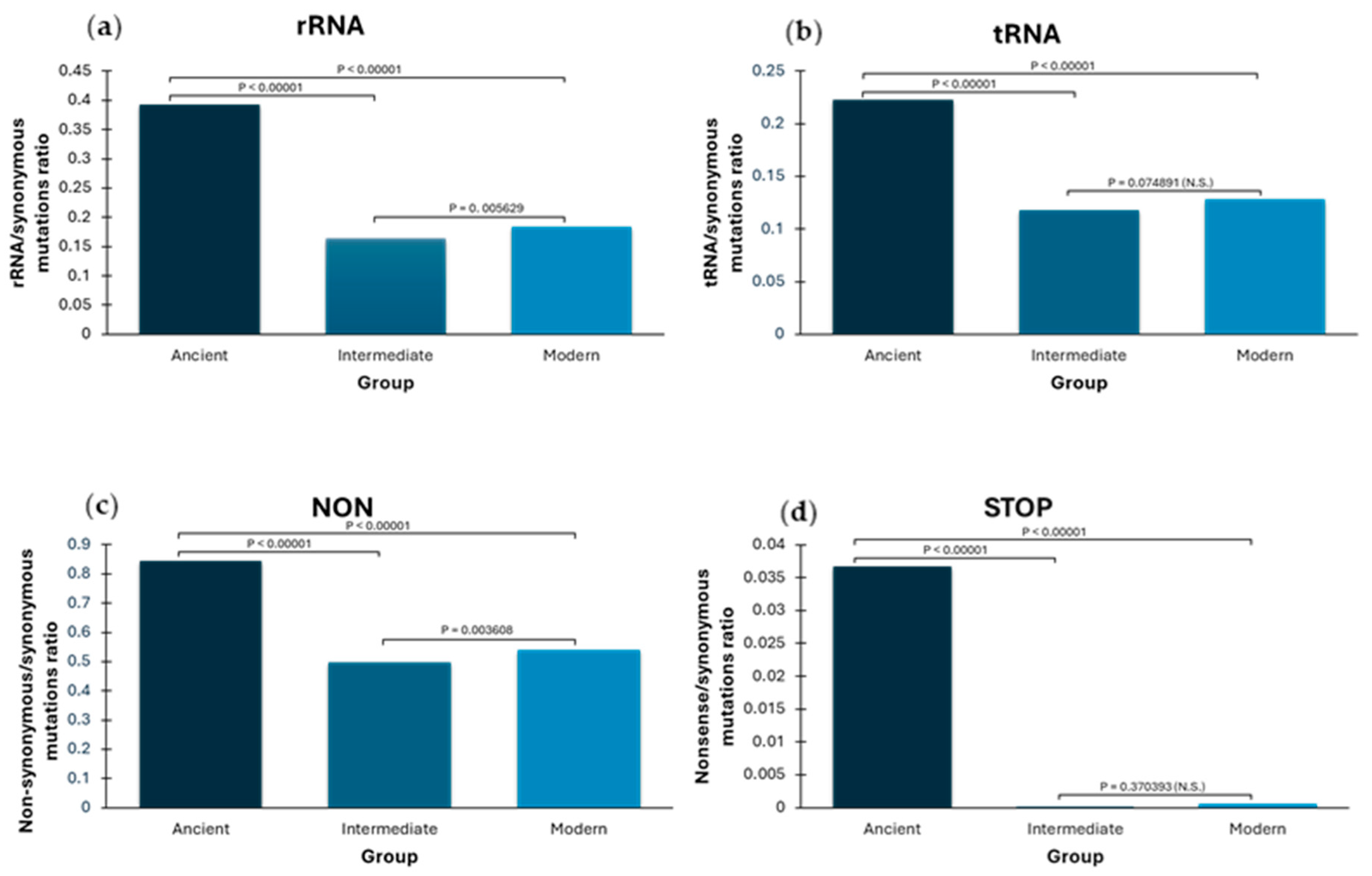
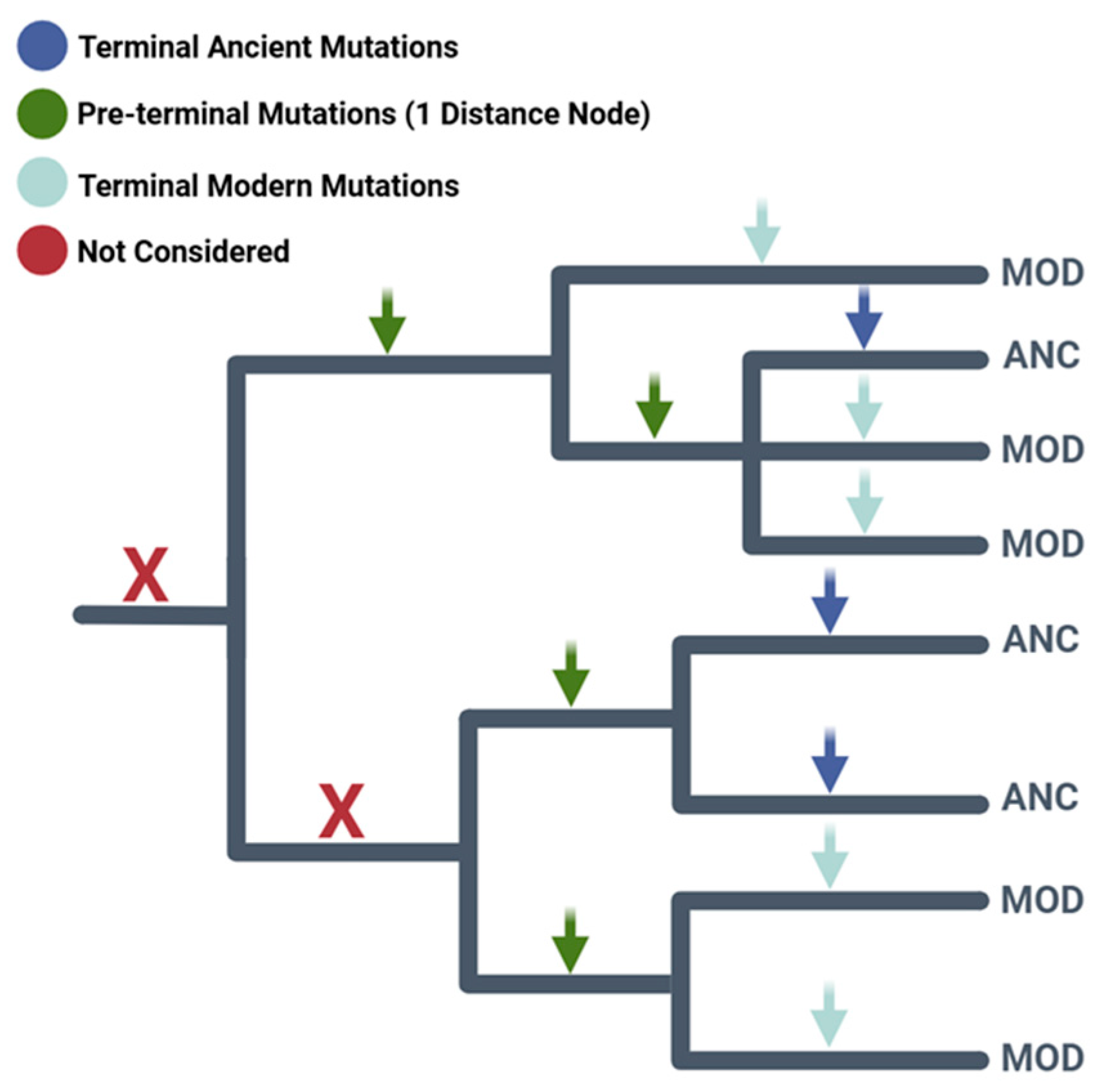
2.1. Higher Ratios of Ancient Terminal Variants: Mutation Load Characterized by “Phantom Mutations”
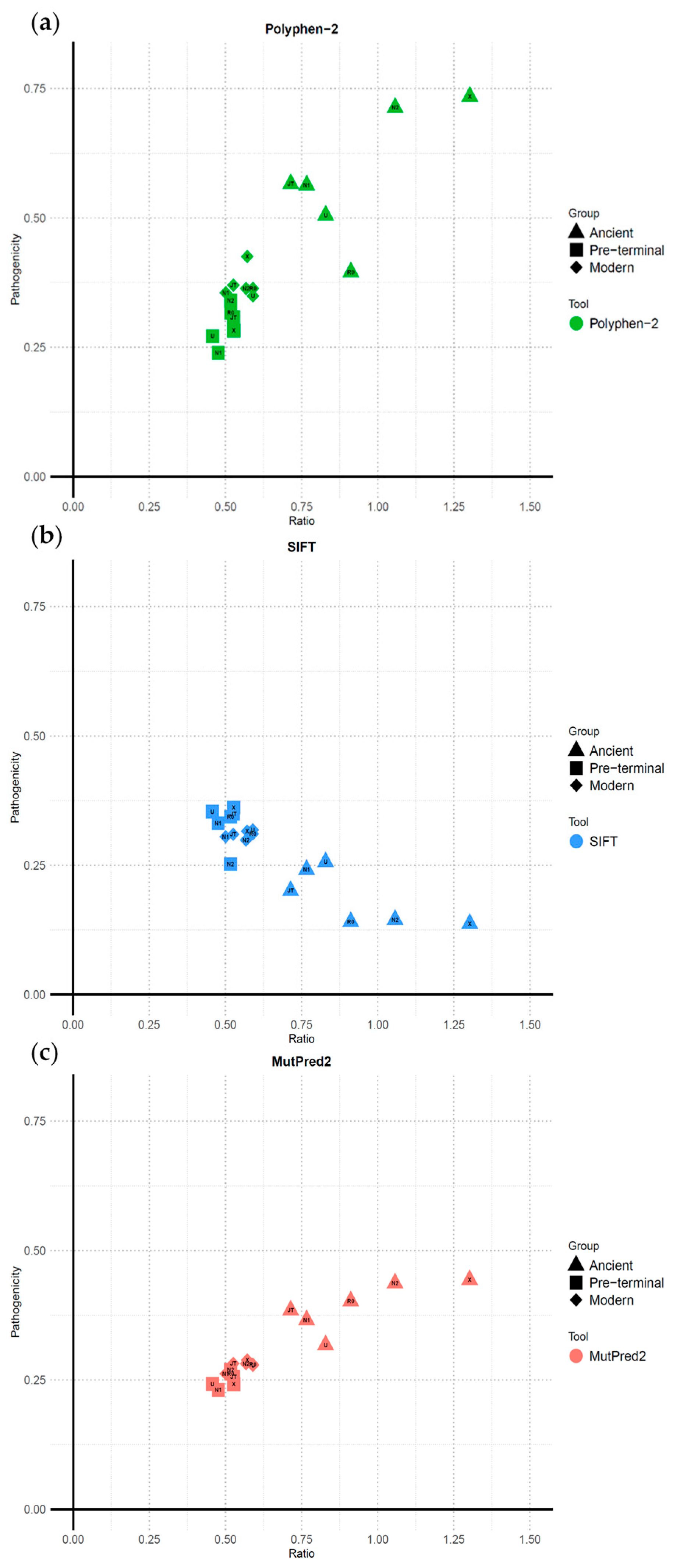
2.2. Greater Pathogenicity Scores in Ancient Terminal Mutations
2.3. Ancient Terminal Variants Exhibit Consistently Higher Ratios and Pathogenicity Across Haplogroups
3. Discussions
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AADR | Allen Ancient DNA Resource |
| aDNA | Ancient DNA |
| NGS | Next-Generation Sequencing |
| mtDNA | Mitochondrial DNA |
References
- Dabney, J.; Meyer, M.; Paabo, S. Ancient DNA Damage. Cold Spring Harb. Perspect. Biol. 2013, 5, a012567. [Google Scholar] [CrossRef] [PubMed]
- Dalal, V.; Pasupuleti, N.; Chaubey, G.; Rai, N.; Shinde, V. Advancements and Challenges in Ancient DNA Research: Bridging the Global North–South Divide. Genes 2023, 14, 479. [Google Scholar] [CrossRef]
- Allentoft, M.E.; Collins, M.; Harker, D.; Haile, J.; Oskam, C.L.; Hale, M.L.; Campos, P.F.; Samaniego, J.A.; Gilbert, M.T.P.; Willerslev, E.; et al. The Half-Life of DNA in Bone: Measuring Decay Kinetics in 158 Dated Fossils. Proc. R. Soc. B Biol. Sci. 2012, 279, 4724–4733. [Google Scholar] [CrossRef]
- Olivieri, C.; Ermini, L.; Rizzi, E.; Corti, G.; Bonnal, R.; Luciani, S.; Marota, I.; De Bellis, G.; Rollo, F. Characterization of Nucleotide Misincorporation Patterns in the Iceman’s Mitochondrial DNA. PLoS ONE 2010, 5, e8629. [Google Scholar] [CrossRef]
- Briggs, A.W.; Stenzel, U.; Meyer, M.; Krause, J.; Kircher, M.; Pääbo, S. Removal of Deaminated Cytosines and Detection of in Vivo Methylation in Ancient DNA. Nucleic Acids Res. 2010, 38, e87. [Google Scholar] [CrossRef]
- Briggs, A.W.; Stenzel, U.; Johnson, P.L.F.; Green, R.E.; Kelso, J.; Prüfer, K.; Meyer, M.; Krause, J.; Ronan, M.T.; Lachmann, M.; et al. Patterns of Damage in Genomic DNA Sequences from a Neandertal. Proc. Natl. Acad. Sci. USA 2007, 104, 14616–14621. [Google Scholar] [CrossRef]
- Richards, M.B.; Sykes, B.C.; Hedges, R.E.M. Authenticating DNA Extracted from Ancient Skeletal Remains. J. Archaeol. Sci. 1995, 22, 291–299. [Google Scholar] [CrossRef]
- Ehler, E.; Novotný, J.; Juras, A.; Chyleński, M.; Moravčík, O.; Pačes, J. AmtDB: A Database of Ancient Human Mitochondrial Genomes. Nucleic Acids Res. 2019, 47, D29–D32. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, U.; Hadly, E.A. Using Phylochronology to Reveal Cryptic Population Histories: Review and Synthesis of 29 Ancient DNA Studies. Mol. Ecol. 2009, 18, 1310–1330. [Google Scholar] [CrossRef] [PubMed]
- Amorim, A.; Fernandes, T.; Taveira, N. Mitochondrial DNA in Human Identification: A Review. PeerJ 2019, 7, e7314. [Google Scholar] [CrossRef]
- Pakendorf, B.; Stoneking, M. Mitochondrial DNA and Human Evolution. Annu. Rev. Genom. Hum. Genet. 2005, 6, 165–186. [Google Scholar] [CrossRef]
- Silva, M.; Oliveira, M.; Vieira, D.; Brandão, A.; Rito, T.; Pereira, J.B.; Fraser, R.M.; Hudson, B.; Gandini, F.; Edwards, C.; et al. A Genetic Chronology for the Indian Subcontinent Points to Heavily Sex-Biased Dispersals. BMC Evol. Biol. 2017, 17, 88. [Google Scholar] [CrossRef] [PubMed]
- Soares, P.; Ermini, L.; Thomson, N.; Mormina, M.; Rito, T.; Röhl, A.; Salas, A.; Oppenheimer, S.; Macaulay, V.; Richards, M.B. Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock. Am. J. Hum. Genet. 2009, 84, 740–759. [Google Scholar] [CrossRef]
- Richards, M.B.; Torroni, A.; Soares, P. Phylogeography. In The International Encyclopedia of Biological Anthropology; Trevathan, W., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 1–3. ISBN 10: 1118584422. [Google Scholar]
- Destro-Bisol, G.; Jobling, M.A.; Rocha, J.; Novembre, J.; Richards, M.B.; Mulligan, C.; Batini, C.; Manni, F. Molecular Anthropology in the Genomic Era. J. Anthropol. Sci. 2010, 88, 93–112. [Google Scholar] [PubMed]
- Macaulay, V.; Soares, P.; Richards, M.B. Rectifying Long-Standing Misconceptions about the ρ Statistic for Molecular Dating. PLoS ONE 2019, 14, e0212311. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Mittnik, A.; Johnson, P.L.F.; Bos, K.; Lari, M.; Bollongino, R.; Sun, C.; Giemsch, L.; Schmitz, R.; Burger, J.; et al. A Revised Timescale for Human Evolution Based on Ancient Mitochondrial Genomes. Curr. Biol. 2013, 23, 553–559. [Google Scholar] [CrossRef]
- Duchêne, S.; Lanfear, R.; Ho, S.Y.W. The Impact of Calibration and Clock-Model Choice on Molecular Estimates of Divergence Times. Mol. Phylogenet. Evol. 2014, 78, 277–289. [Google Scholar] [CrossRef]
- Li, W.H.; Tanimura, M. The Molecular Clock Runs More Slowly in Man than in Apes and Monkeys. Nature 1987, 326, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Elson, J.L.; Turnbull, D.M.; Howell, N. Comparative Genomics and the Evolution of Human Mitochondrial DNA: Assessing the Effects of Selection. Am. J. Hum. Genet. 2004, 74, 229–238. [Google Scholar] [CrossRef]
- Kivisild, T.; Shen, P.; Wall, D.P.; Do, B.; Sung, R.; Davis, K.; Passarino, G.; Underhill, P.A.; Scharfe, C.; Torroni, A.; et al. The Role of Selection in the Evolution of Human Mitochondrial Genomes. Genetics 2006, 172, 373–387. [Google Scholar] [CrossRef]
- Lipson, M.; Loh, P.-R.; Sankararaman, S.; Patterson, N.; Berger, B.; Reich, D. Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes. PLoS Genet. 2015, 11, e1005550. [Google Scholar] [CrossRef]
- Fu, Q.; Li, H.; Moorjani, P.; Jay, F.; Slepchenko, S.M.; Bondarev, A.A.; Johnson, P.L.F.; Aximu-Petri, A.; Prüfer, K.; De Filippo, C.; et al. Genome Sequence of a 45,000-Year-Old Modern Human from Western Siberia. Nature 2014, 514, 445–449. [Google Scholar] [CrossRef]
- Rieux, A.; Balloux, F. Inferences from Tip-calibrated Phylogenies: A Review and a Practical Guide. Mol. Ecol. 2016, 25, 1911–1924. [Google Scholar] [CrossRef]
- Rieux, A.; Eriksson, A.; Li, M.; Sobkowiak, B.; Weinert, L.A.; Warmuth, V.; Ruiz-Linares, A.; Manica, A.; Balloux, F. Improved Calibration of the Human Mitochondrial Clock Using Ancient Genomes. Mol. Biol. Evol. 2014, 31, 2780–2792. [Google Scholar] [CrossRef]
- Soares, P.; Abrantes, D.; Rito, T.; Thomson, N.; Radivojac, P.; Li, B.; Macaulay, V.; Samuels, D.C.; Pereira, L. Evaluating Purifying Selection in the Mitochondrial DNA of Various Mammalian Species. PLoS ONE 2013, 8, e58993. [Google Scholar] [CrossRef]
- Pereira, L.; Soares, P.; Radivojac, P.; Li, B.; Samuels, D.C. Comparing Phylogeny and the Predicted Pathogenicity of Protein Variations Reveals Equal Purifying Selection across the Global Human mtDNA Diversity. Am. J. Hum. Genet. 2011, 88, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R. Synonymous and Nonsynonymous Rate Variation in Nuclear Genes of Mammals. J. Mol. Evol. 1998, 46, 409–418. [Google Scholar] [CrossRef]
- Bandelt, H.-J.; Quintana-Murci, L.; Salas, A.; Macaulay, V. The Fingerprint of Phantom Mutations in Mitochondrial DNA Data. Am. J. Hum. Genet. 2002, 71, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Levin, L.; Zhidkov, I.; Gurman, Y.; Hawlena, H.; Mishmar, D. Functional Recurrent Mutations in the Human Mitochondrial Phylogeny: Dual Roles in Evolution and Disease. Genome Biol. Evol. 2013, 5, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.B.; Freyer, C.; Elson, J.L.; Wredenberg, A.; Cansu, Z.; Trifunovic, A.; Larsson, N.-G. Strong Purifying Selection in Transmission of Mammalian Mitochondrial DNA. PLoS Biol. 2008, 6, e10. [Google Scholar] [CrossRef]
- Gunning, A.C.; Fryer, V.; Fasham, J.; Crosby, A.H.; Ellard, S.; Baple, E.L.; Wright, C.F. Assessing Performance of Pathogenicity Predictors Using Clinically Relevant Variant Datasets. J. Med. Genet. 2021, 58, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wen, S.; Lu, C.; Zhou, B.; Curnoe, D.; Lu, H.; Li, H.; Wang, W.; Cheng, H.; Yi, S.; et al. Ancient DNA and Multimethod Dating Confirm the Late Arrival of Anatomically Modern Humans in Southern China. Proc. Natl. Acad. Sci. USA 2021, 118, e2019158118. [Google Scholar] [CrossRef] [PubMed]
- Posth, C.; Renaud, G.; Mittnik, A.; Drucker, D.G.; Rougier, H.; Cupillard, C.; Valentin, F.; Thevenet, C.; Furtwängler, A.; Wißing, C.; et al. Pleistocene Mitochondrial Genomes Suggest a Single Major Dispersal of Non-Africans and a Late Glacial Population Turnover in Europe. Curr. Biol. 2016, 26, 827–833. [Google Scholar] [CrossRef]
- The 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Schönherr, S.; Weissensteiner, H.; Kronenberg, F.; Forer, L. Haplogrep 3—An Interactive Haplogroup Classification and Analysis Platform. Nucleic Acids Res. 2023, 51, W263–W268. [Google Scholar] [CrossRef]
- Mallick, S.; Micco, A.; Mah, M.; Ringbauer, H.; Lazaridis, I.; Olalde, I.; Patterson, N.; Reich, D. The Allen Ancient DNA Resource (AADR) a Curated Compendium of Ancient Human Genomes. Sci. Data 2024, 11, 182. [Google Scholar] [CrossRef]
- Van Oven, M. PhyloTree Build 17: Growing the Human Mitochondrial DNA Tree. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e392–e394. [Google Scholar] [CrossRef]
- Vieira, D.; Almeida, M.; Richards, M.B.; Soares, P. An Efficient and User-Friendly Implementation of the Founder Analysis Methodology. In Practical Applications of Computational Biology and Bioinformatics, 13th International Conference; Fdez-Riverola, F., Rocha, M., Mohamad, M.S., Zaki, N., Castellanos-Garzón, J.A., Eds.; Advances in Intelligent Systems and Computing; Springer International Publishing: Cham, Switzerland, 2020; Volume 1005, pp. 121–128. ISBN 978-3-030-23872-8. [Google Scholar]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A Method and Server for Predicting Damaging Missense Mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting Amino Acid Changes That Affect Protein Function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT Web Server: Predicting Effects of Amino Acid Substitutions on Proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Pejaver, V.; Urresti, J.; Lugo-Martinez, J.; Pagel, K.A.; Lin, G.N.; Nam, H.-J.; Mort, M.; Cooper, D.N.; Sebat, J.; Iakoucheva, L.M.; et al. Inferring the Molecular and Phenotypic Impact of Amino Acid Variants with MutPred2. Nat. Commun. 2020, 11, 5918. [Google Scholar] [CrossRef] [PubMed]
- Kakar, M.U.; Matloob, M.; Dai, R.; Deng, Y.; Ullah, K.; Kakar, I.U.; Khaliq, G.; Umer, M.; Bhutto, Z.A.; Fazlani, S.A.; et al. In Silico Screening and Identification of Deleterious Missense SNPs along with Their Effects on CD-209 Gene: An Insight to CD-209 Related-Diseases. PLoS ONE 2021, 16, e0247249. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, P.; Pinho, B.; Miguéis, B.; Almeida, J.B.; Rito, T.; Soares, P. Analysis of Selective Pressure on Ancient Human Mitochondrial Genomes Reveals the Presence of Widespread Sequencing Artefacts. Int. J. Mol. Sci. 2025, 26, 7739. https://doi.org/10.3390/ijms26167739
Fernandes P, Pinho B, Miguéis B, Almeida JB, Rito T, Soares P. Analysis of Selective Pressure on Ancient Human Mitochondrial Genomes Reveals the Presence of Widespread Sequencing Artefacts. International Journal of Molecular Sciences. 2025; 26(16):7739. https://doi.org/10.3390/ijms26167739
Chicago/Turabian StyleFernandes, Pedro, Bernardo Pinho, Bárbara Miguéis, João B. Almeida, Teresa Rito, and Pedro Soares. 2025. "Analysis of Selective Pressure on Ancient Human Mitochondrial Genomes Reveals the Presence of Widespread Sequencing Artefacts" International Journal of Molecular Sciences 26, no. 16: 7739. https://doi.org/10.3390/ijms26167739
APA StyleFernandes, P., Pinho, B., Miguéis, B., Almeida, J. B., Rito, T., & Soares, P. (2025). Analysis of Selective Pressure on Ancient Human Mitochondrial Genomes Reveals the Presence of Widespread Sequencing Artefacts. International Journal of Molecular Sciences, 26(16), 7739. https://doi.org/10.3390/ijms26167739







