Epigenetic Alterations in Age-Related Macular Degeneration: Mechanisms and Implications
Abstract
1. Introduction
2. The Pathophysiology of Age-Related Macular Degeneration
3. Genetic Risk Factors in Age-Related Macular Degeneration
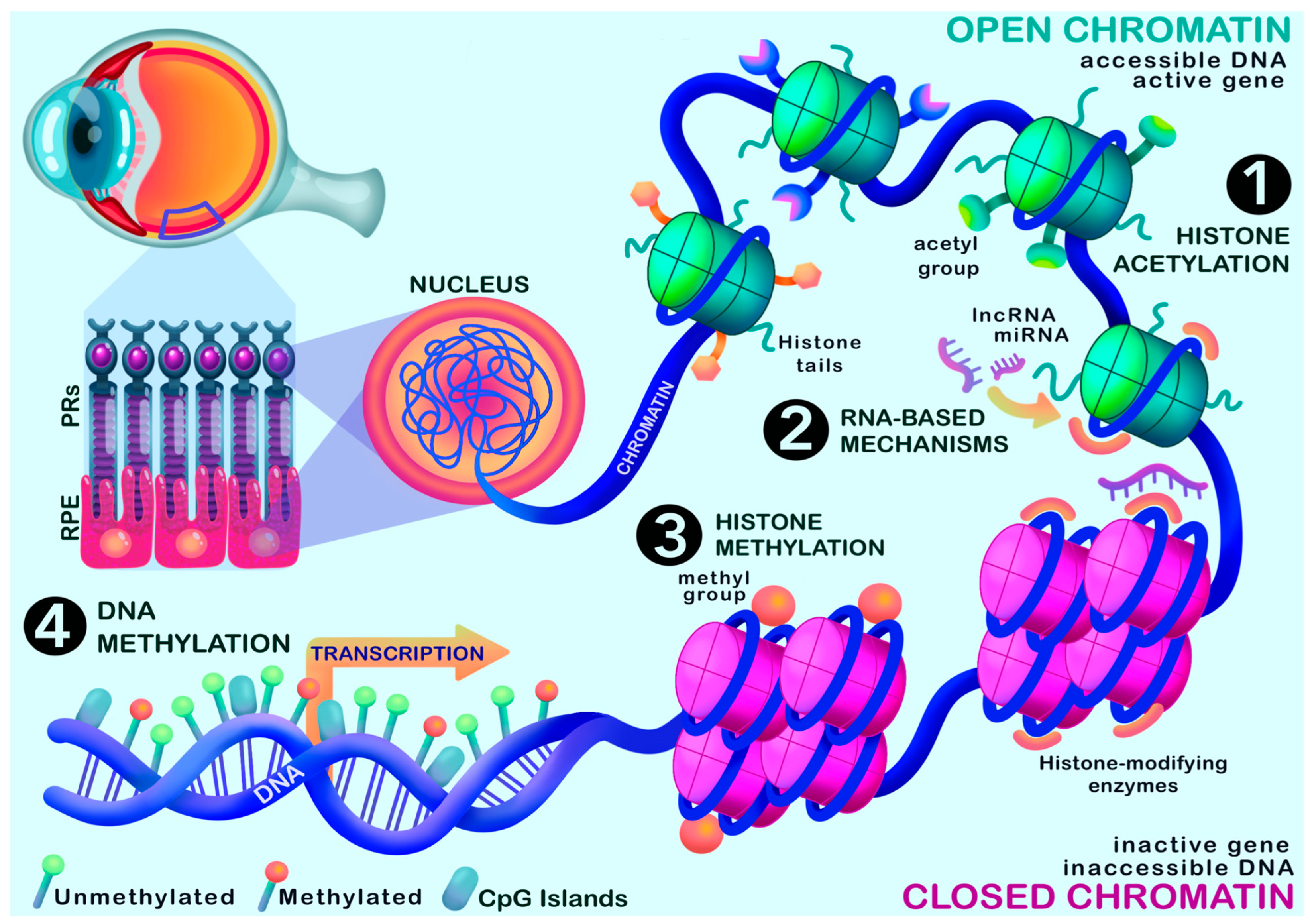
4. Evidence for Epigenetic Regulation in Age-Related Macular Degeneration
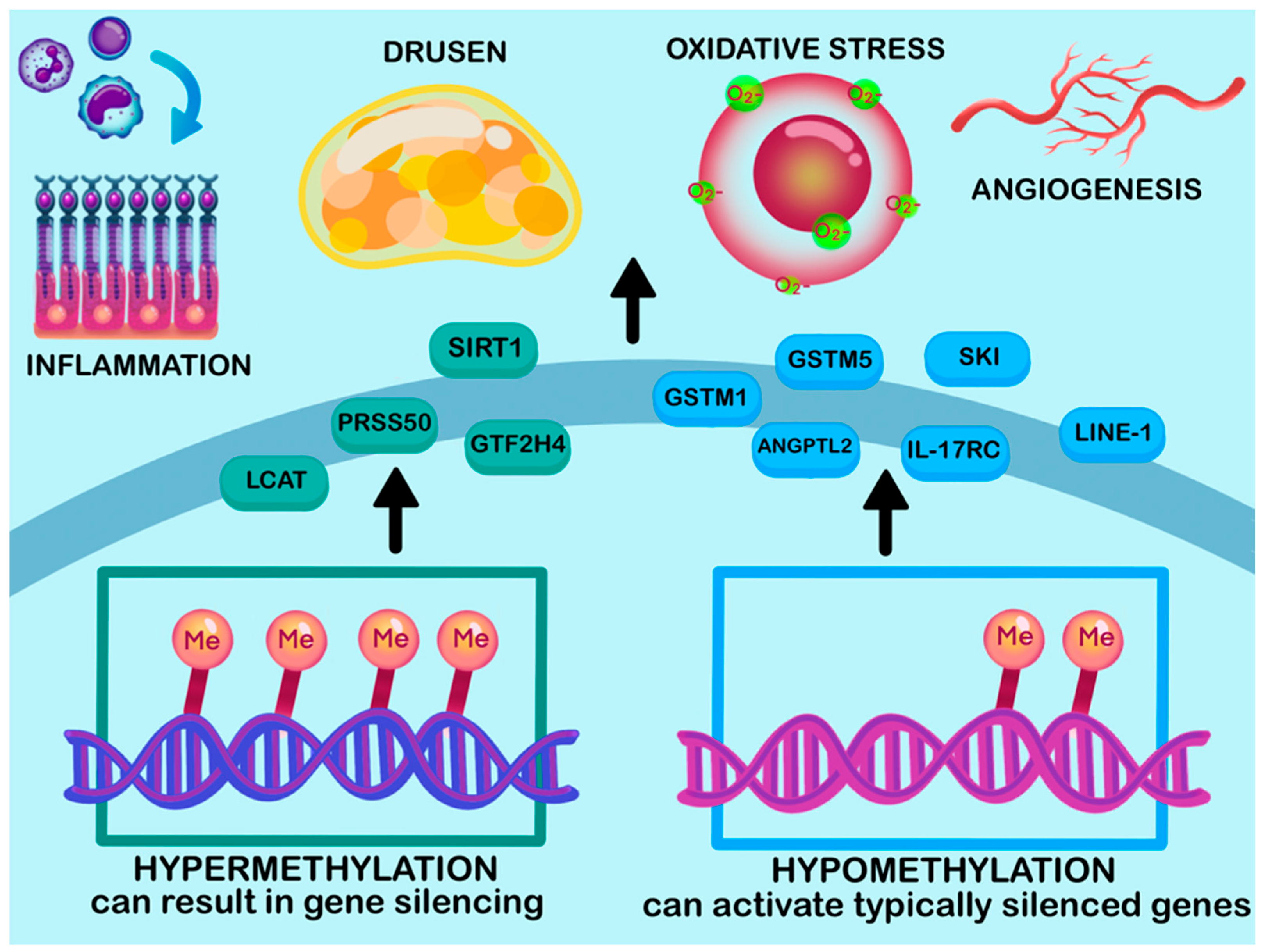
4.1. DNA Methylation
| Name of Gene | Methylation Status | Tissue or Source | Regulation in AMD | Proposed Function | Reference |
|---|---|---|---|---|---|
| GSTM1 and GSTM5 | Hypomethylated | RPE/choroid | Downregulated | Reduces antioxidant defense. Involved in increasing RPE vulnerability Involved in oxidative stress response | [129,130] |
| IL17RC | Hypomethylated | Blood and Retina | Upregulated | Enhances chronic inflammation | [131,144] |
| ANGPTL2 | Hypomethylated | AMD retina | Upregulated | Involved in Angiogenesis Increases risk of CNV | [138,145] |
| SKI | Hypomethylated | AMD RPE | Upregulated | Impacts oxidative stress pathway Associated with TGF-β signaling | [129,137] |
| GTF2H4 | Hypermethylated | AMD RPE | Downregulated | Impaired DNA repair and transcription, affecting degeneration | [137] |
| LINE-1 | Hypomethylated | Peripheral Blood | Downregulated | Increased transcription and genomic instability | [132,139,140,141] |
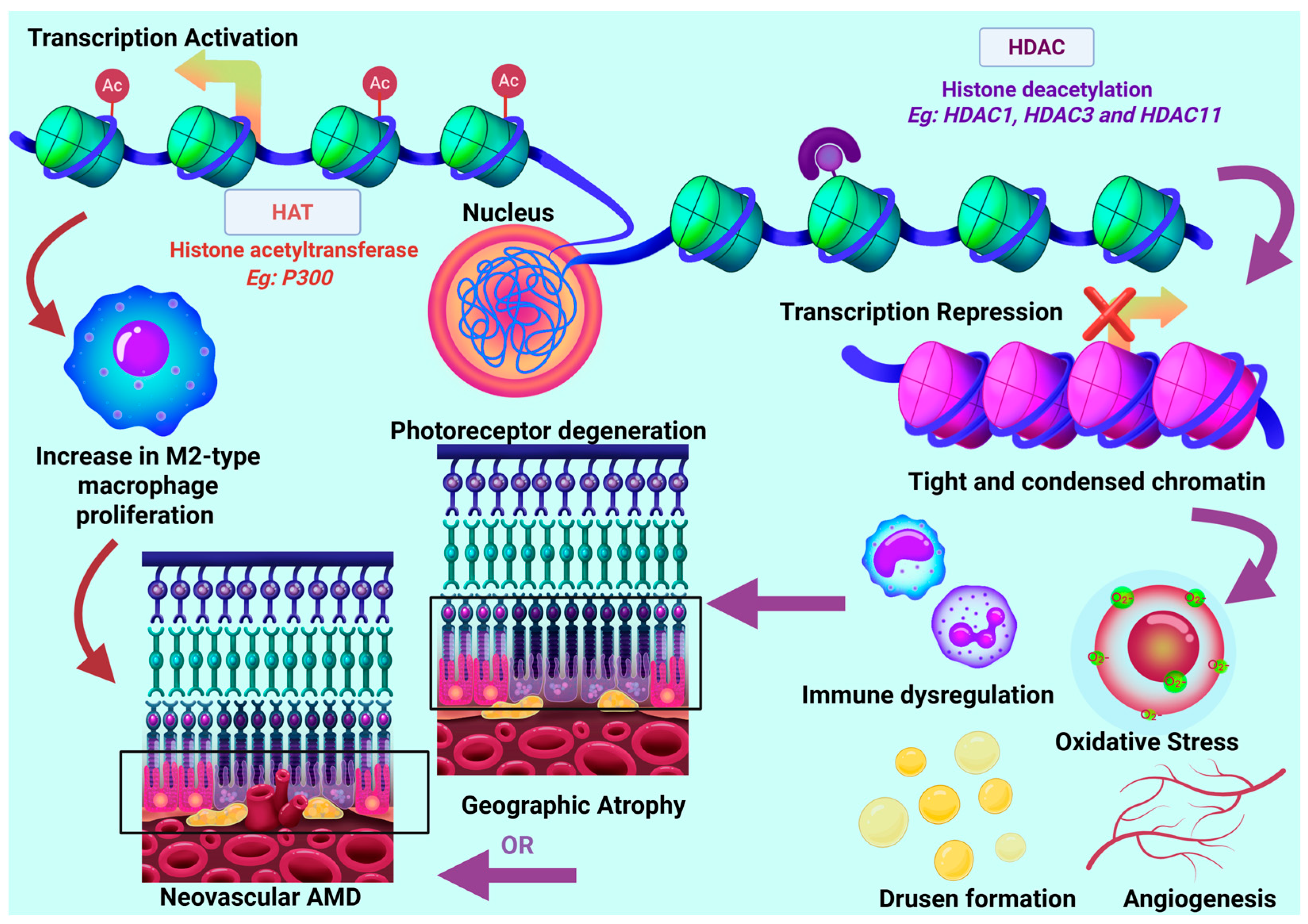
4.2. Histone Variants and Modifiers
| HDAC Isoform | Expression Changes | Proposed Role/Function | Reference |
|---|---|---|---|
| HDAC1 and HDAC2 | Downregulated in retinal cells with advanced GA. Upregulated in cybrid model | Chromatin compaction transcription repression DNA damage response. Represses Inflammation | [19,27,157] |
| HDAC 3 | Downregulated in retinal models Upregulated in cybrid models | Modulates oxidative stress and immune response | [19,157,158] |
| HDAC9 | Downregulated in cybrid AMD models | Regulation of angiogenesis, apoptosis, inflammation | [19] |
| HDAC10 | Downregulated in cybrid AMD models | Regulation of metabolic and cellular stress response. | [19] |
| HDAC11 | Upregulated in retinal and cybrid AMD models | Regulates Inflammation. Prompts photoreceptor degeneration. | [19,161,162] |
| SIRT1 | Downregulated in retinal AMD models | Regulates expression of VEGF. Protect against oxidative stress Regulates inflammation | [132,164,165,166] |
4.3. Chromatin Accessibility
4.4. Long Noncoding RNA
| Name of lncRNA | Expression Changes | Proposed Role | Tissue/Model | Reference |
|---|---|---|---|---|
| RP11-234O6.2 | Downregulated | Downregulated in RPE/choroid of AMD patients, implicated in protecting RPE cells from oxidative damage-induced cell death. | AMD RPE/choroid Human RPE culture model | [207] |
| PWRN2 | Upregulated | Involved in promoting RPE cell death and stress-related mitochondrial damage. | Human RPE culture model | [206] |
| MEG3 | Upregulated | Implicated in promoting apoptosis via association with p53 transactivation. | Mouse photooxidative damage model | [209] |
| Vax2os1, Vax2os2 | Upregulated | Enriched in aqueous humor of nAMD patients. Predicted interaction with NFκB, involved in inflammation and angiogenesis. | nAMD aqueous humor | [198,199] |
| MALAT1 | Upregulated | Increased in experimental CNV, suppression reduces CNV lesion size. Implicated in promoting choroidal neovascularisation via modulation of VEGF-A expression. | Mouse CNV model | [212] |
| ZNF503-AS1 | Downregulated | Decreased in RPE/choroid of GA patient specimens. Implicated RPE protection by suppressing dedifferentiation and pathology, in vitro. | AMD RPE/choroid Human RPE culture model | [208,213] |
| LINC00167 | Downregulated | Decreased in RPE/choroid of AMD patient specimens. Suppression promotes RPE dedifferentiation and mitochondrial/phagocytic dysfunction in vitro, indicative of a protective role. | AMD RPE/choroid Human RPE culture model | [204,205] |
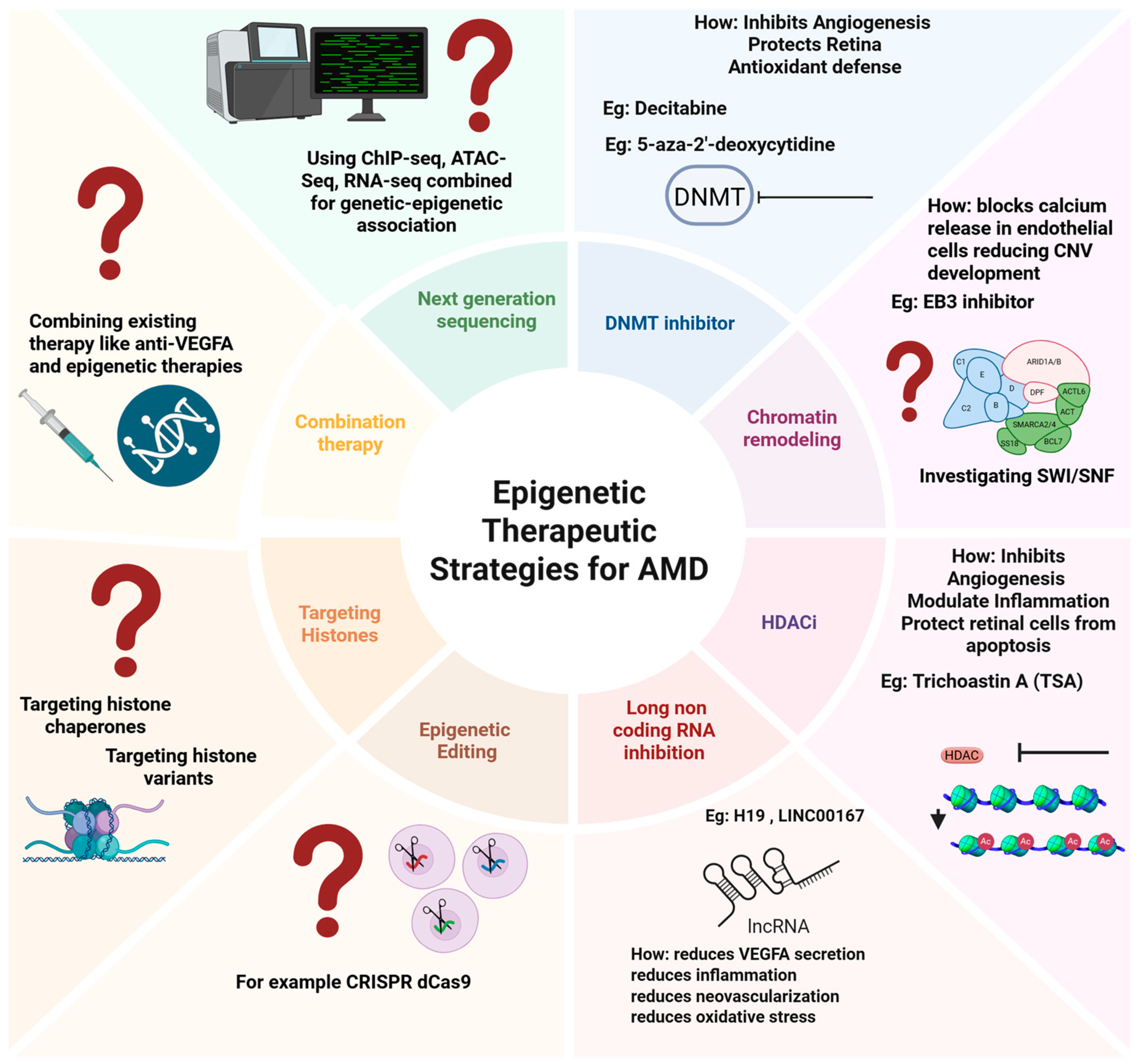
5. Epigenetics as a Potential Therapeutic Target?
6. Conclusions
Funding
Conflicts of Interest
Abbreviations
| 5-AZA-dc | 5-aza-2′-deoxycytidine |
| ABCA4 | ATP-binding cassette subfamily A member 4 |
| AMD | Age-related macular degeneration |
| ANGPTL2 | Angiopoietin-like 2 |
| ANRIL | Anti-sense noncoding RNA |
| APOE | Apolipoprotein E |
| ARMS2 | Age-related maculopathy susceptibility 2 |
| ATAC-seq | Assay for transposase accessible chromatin sequencing |
| BrM | Bruch’s membrane |
| C3 | Complement 3 |
| C5 | Complement 5 |
| C7 | Complement factor 7 |
| CD46 | Cluster of differentiation 46 |
| CETP | Cholesteryl ester transfer protein |
| CF1 | Complement factor I |
| CFH | Complement factor H |
| ChIP-seq | Chromatin immunoprecipitation sequencing |
| CLU | Clusterin |
| CNV | Choroidal neovascularization |
| CRISPR-Dcas9 | Clustered regularly interspaced short palindromic repeats—dead cas 9 |
| CYTOR | Cytoskeleton regulator RNA |
| DNA | Deoxyribonucleic acid |
| DNMT1 | DNA methyltransferase 1 |
| DNMT3 | DNA methyltransferase 3 |
| EBIN | End-binding protein 3 |
| ECM | Extracellular matrix |
| EMT | Epithelial to mesenchymal transition |
| eQTL | Expression quantitative trait locus |
| FRZB | Frizzled-related protein |
| GA | Geographic atrophy |
| GSTM1 | Glutathione S-transferase isoform mu1 |
| GSTM5 | Glutathione S-transferase isoform mu1 |
| GTFH4 | General transcription factor IIH subunit 4. |
| GWAS | Genome-wide association studies |
| H3K27ac | Histone 3 lysine 27 acetylation |
| H3K27me3 | Histone 3 lysine 27 trimethylation |
| H3K4me1 | Histone 3 lysine 4 monomethylation |
| H3K4me3 | Histone 3 lysine trimethylation |
| HAT | Histone acetyltransferase |
| HDAC | Histone Deacetylase |
| HDACi | Histone deacetylase inhibitors |
| HIF1A | Hypoxia inducible factor 1-alpha |
| HTRA1 | High temperature requirement A serine peptidase 1 |
| IL | Interleukin |
| IL17RC | Interleukin 17 receptor C |
| iPSC | Induced pluripotent stem cells |
| LIPC | hepatic lipase |
| lncRNA | Long noncoding RNA |
| LINE1 | Long interspersed nuclear element-1 |
| MALAT1 | Metastasis-associated lung adenocarcinoma transcript 1 |
| MAPK | Mitogen-activated protein kinase signaling |
| MEG3 | Maternally expressed gene 3 |
| miRNA | MicroRNA |
| MTND-2 | Mitochondrially encoded NADH dehydrogenase 2 |
| nAMD | Neovascular age-related macular degeneration |
| ncRNA | Noncoding RNA |
| NF-κB | Nuclear factor kappa B |
| NGFR | Nerve growth factor receptor |
| PRS | Polygenic risk scores |
| PRSS50 | Protease serine 50 |
| PTM | Post-translational modifications |
| PWRN2 | Prader–Willi region nonprotein-coding RNA |
| QC | Quality control |
| ROS | Reactive oxygen species |
| RPE | Retinal pigment epithelium |
| scATAC-seq | Single-cell assay for transposase accessible chromatin sequencing |
| scRNA-seq | Single-cell RNA sequencing |
| SHARE-seq | Simultaneous high-throughput ATAC and RNA expression with sequencing |
| siRNA | Small interfering RNA |
| SIRT1 | Silent mating type information regulation 2 homolog 1 |
| SMAD2 | Mothers against decapentaplegic homolog 2 |
| SNARE-seq | Single-nucleus chromatin accessibility and mRNA expression sequencing |
| SNP | Single-nucleotide polymorphism |
| snRNA-seq | Single-nucleus RNA-seq |
| SOC3 | Suppressor of cytokine signaling 3 |
| SOD2 | Superoxide dismutase 2 |
| SW/SNF | SWItch/sucrose non-fermentable |
| TF | Transcription factor |
| TGFβ1 | transforming growth factor beta-1 |
| TLE2 | Transducing-like enhancer protein 2 |
| TNF | Tumor necrosis factor |
| TSA | Trichoastin A |
| VEGF | Vascular endothelial growth factor |
| VEGFA | Vascular endothelial growth factor A |
| XBP1 | Spliced X-box binding protein 1 |
| ZNF503-AS1 | Zinc finger protein 503 antisense RNA |
References
- Smith, W.; Assink, J.; Klein, R.; Mitchell, P.; Klaver, C.C.; Klein, B.E.; Hofman, A.; Jensen, S.; Wang, J.J.; de Jong, P.T.V.M.; et al. Risk factors for age-related macular degeneration: Pooled findings from three continents. Ophthalmology 2001, 108, 697–704. [Google Scholar] [CrossRef]
- Vyawahare, H.; Shinde, P. Age-Related Macular Degeneration: Epidemiology, Pathophysiology, Diagnosis, and Treatment. Cureus 2022, 14, e29583. [Google Scholar] [CrossRef]
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.G.; Klein, R.; Cheng, C.-Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Fleckenstein, M.; Keenan, T.D.; Guymer, R.H.; Chakravarthy, U.; Schmitz-Valckenberg, S.; Klaver, C.C.; Wong, W.T.; Chew, E.Y. Age-related macular degeneration. Nat. Rev. Dis. Primers 2021, 7, 31. [Google Scholar] [CrossRef]
- Provis, J.M.; Penfold, P.L.; Cornish, E.E.; Sandercoe, T.M.; Madigan, M.C. Anatomy and development of the macula: Specialisation and the vulnerability to macular degeneration. Clin. Exp. Optom. 2005, 88, 269–281. [Google Scholar] [CrossRef]
- Hageman, G.S.; Anderson, D.H.; Johnson, L.V.; Hancox, L.S.; Taiber, A.J.; Hardisty, L.I.; Hageman, J.L.; Stockman, H.A.; Borchardt, J.D.; Gehrs, K.M. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2005, 102, 7227–7232. [Google Scholar] [CrossRef]
- Klein, R.J.; Zeiss, C.; Chew, E.Y.; Tsai, J.-Y.; Sackler, R.S.; Haynes, C.; Henning, A.K.; SanGiovanni, J.P.; Mane, S.M.; Mayne, S.T.; et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005, 308, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Haines, J.L.; Hauser, M.A.; Schmidt, S.; Scott, W.K.; Olson, L.M.; Gallins, P.; Spencer, K.L.; Kwan, S.Y.; Noureddine, M.; Gilbert, J.R.; et al. Complement factor H variant increases the risk of age-related macular degeneration. Science 2005, 308, 419–421. [Google Scholar] [CrossRef] [PubMed]
- Lorés-Motta, L.; Paun, C.C.; Corominas, J.; Pauper, M.; Geerlings, M.J.; Altay, L.; Schick, T.; Daha, M.R.; Fauser, S.; Hoyng, C.B. Genome-wide association study reveals variants in CFH and CFHR4 associated with systemic complement activation: Implications in age-related macular degeneration. Ophthalmology 2018, 125, 1064–1074. [Google Scholar] [CrossRef] [PubMed]
- Karimi, S.; Nouri, H.; Mahmoudinejad-Azar, S.; Abtahi, S.-H. Smoking and environmental tobacco smoke exposure: Implications in ocular disorders. Cutan. Ocul. Toxicol. 2023, 42, 1–7. [Google Scholar] [CrossRef]
- West, S.K. Smoking and the risk of eye diseases. In Nutritional and Environmental Influences on the Eye; CRC Press: Boca Raton, FL, USA, 2021; pp. 151–164. [Google Scholar]
- Seddon, J.M.; Reynolds, R.; Shah, H.R.; Rosner, B. Smoking, dietary betaine, methionine, and vitamin D in monozygotic twins with discordant macular degeneration: Epigenetic implications. Ophthalmology 2011, 118, 1386–1394. [Google Scholar] [CrossRef]
- Keeling, E.; Lynn, S.A.; Koh, Y.M.; Scott, J.A.; Kendall, A.; Gatherer, M.; Page, A.; Cagampang, F.R.; Lotery, A.J.; Ratnayaka, J.A. A High Fat “Western-style” Diet Induces AMD-like Features in Wildtype Mice. Mol. Nutr. Food Res. 2022, 66, 2100823. [Google Scholar] [CrossRef]
- Figueiredo, I.; Farinha, C.; Barreto, P.; Coimbra, R.; Pereira, P.; Marques, J.P.; Pires, I.; Cachulo, M.L.; Silva, R. Nutritional Genomics: Implications for Age-Related Macular Degeneration. Nutrients 2024, 16, 4124. [Google Scholar] [CrossRef]
- Seddon, J.M.; Cote, J.; Page, W.F.; Aggen, S.H.; Neale, M.C. The US twin study of age-related macular degeneration: Relative roles of genetic and environmental influences. Arch. Ophthalmol. 2005, 123, 321–327. [Google Scholar] [CrossRef]
- Millen, A.E.; Meyers, K.J.; Liu, Z.; Engelman, C.D.; Wallace, R.B.; LeBlanc, E.S.; Tinker, L.F.; Iyengar, S.K.; Robinson, J.G.; Sarto, G.E.; et al. Association between vitamin D status and age-related macular degeneration by genetic risk. JAMA Ophthalmol. 2015, 133, 1171–1179. [Google Scholar] [CrossRef]
- Gemenetzi, M.; Lotery, A.J. The role of epigenetics in age-related macular degeneration. Eye 2014, 28, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Desmettre, T. Epigenetics in age-related macular degeneration (AMD). J. Fr. D’ophtalmologie 2018, 41, e407–e415. [Google Scholar] [CrossRef]
- Nashine, S.; Nesburn, A.B.; Kuppermann, B.D.; Kenney, M.C. Age-related macular degeneration (AMD) mitochondria modulate epigenetic mechanisms in retinal pigment epithelial cells. Exp. Eye Res. 2019, 189, 107701. [Google Scholar] [CrossRef] [PubMed]
- Devi, S.M.; Mahalaxmi, I.; Kaavya, J.; Chinnkulandhai, V.; Balachandar, V. Does epigenetics have a role in age related macular degeneration and diabetic retinopathy? Genes. Dis. 2021, 8, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. In Seminars in Reproductive Medicine; Thieme Medical Publishers: Stuttgart, Germany, 2009; pp. 351–357. [Google Scholar]
- Jin, B.; Li, Y.; Robertson, K.D. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes. Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef]
- Henikoff, S.; Smith, M.M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 2015, 7, a019364. [Google Scholar] [CrossRef]
- Sadakierska-Chudy, A.; Filip, M. A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotox. Res. 2015, 27, 172–197. [Google Scholar] [CrossRef]
- Wei, J.-W.; Huang, K.; Yang, C.; Kang, C.-S. Non-coding RNAs as regulators in epigenetics. Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef]
- Al Aboud, N.M.; Tupper, C.; Jialal, I. Genetics, epigenetic mechanism. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Dubey, S.K.; Dubey, R.; Prajapati, S.C.; Jung, K.; Mohan, K.; Liu, X.; Roney, J.; Tian, W.; Abney, J.; Giarmarco, M.M.; et al. Histone deficiency and hypoacetylation in the aging retinal pigment epithelium. Aging Cell 2024, 23, e14108. [Google Scholar] [CrossRef]
- Villota-Salazar, N.A.; Mendoza-Mendoza, A.; González-Prieto, J.M. Epigenetics: From the past to the present. Front. Life Sci. 2016, 9, 347–370. [Google Scholar] [CrossRef]
- Yu, X.; Zhao, H.; Wang, R.; Chen, Y.; Ouyang, X.; Li, W.; Sun, Y.; Peng, A. Cancer epigenetics: From laboratory studies and clinical trials to precision medicine. Cell Death Discov. 2024, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Ge, T.; Gu, X.; Jia, R.; Ge, S.; Chai, P.; Zhuang, A.; Fan, X. Crosstalk between metabolic reprogramming and epigenetics in cancer: Updates on mechanisms and therapeutic opportunities. Cancer Commun. 2022, 42, 1049–1082. [Google Scholar] [CrossRef]
- Park, Y.J.; Han, S.M.; Huh, J.Y.; Kim, J.B. Emerging roles of epigenetic regulation in obesity and metabolic disease. J. Biol. Chem. 2021, 297, 101296. [Google Scholar] [CrossRef]
- Basavarajappa, B.S.; Subbanna, S. Histone methylation regulation in neurodegenerative disorders. Int. J. Mol. Sci. 2021, 22, 4654. [Google Scholar] [CrossRef] [PubMed]
- Mohd Murshid, N.; Aminullah Lubis, F.; Makpol, S. Epigenetic changes and its intervention in age-related neurodegenerative diseases. Cell. Mol. Neurobiol. 2022, 42, 577–595. [Google Scholar] [CrossRef]
- Wang, K.; Liu, H.; Hu, Q.; Wang, L.; Liu, J.; Zheng, Z.; Zhang, W.; Ren, J.; Zhu, F.; Liu, G.-H. Epigenetic regulation of aging: Implications for interventions of aging and diseases. Signal Transduct. Target. Ther. 2022, 7, 374. [Google Scholar] [CrossRef]
- Saul, D.; Kosinsky, R.L. Epigenetics of Aging and Aging-Associated Diseases. Int. J. Mol. Sci. 2021, 22, 401. [Google Scholar] [CrossRef]
- Amini, M.A.; Karbasi, A.; Vahabirad, M.; Khanaghaei, M.; Alizamir, A. Mechanistic Insight into Age-Related Macular Degeneration (AMD): Anatomy, Epidemiology, Genetics, Pathogenesis, Prevention, Implications, and Treatment Strategies to Pace AMD Management. Chonnam Med. J. 2023, 59, 143–159. [Google Scholar] [CrossRef]
- Flores, R.; Carneiro, Â.; Vieira, M.; Tenreiro, S.; Seabra, M.C. Age-related macular degeneration: Pathophysiology, management, and future perspectives. Ophthalmologica 2021, 244, 495–511. [Google Scholar] [CrossRef]
- Bhutto, I.; Lutty, G. Understanding age-related macular degeneration (AMD): Relationships between the photoreceptor/retinal pigment epithelium/Bruch’s membrane/choriocapillaris complex. Mol. Asp. Med. 2012, 33, 295–317. [Google Scholar] [CrossRef]
- García-Layana, A.; Cabrera-López, F.; García-Arumí, J.; Arias-Barquet, L.; Ruiz-Moreno, J.M. Early and intermediate age-related macular degeneration: Update and clinical review. Clin. Interv. Aging 2017, 12, 1579–1587. [Google Scholar] [CrossRef]
- Khan, K.N.; Mahroo, O.A.; Khan, R.S.; Mohamed, M.D.; McKibbin, M.; Bird, A.; Michaelides, M.; Tufail, A.; Moore, A.T. Differentiating drusen: Drusen and drusen-like appearances associated with ageing, age-related macular degeneration, inherited eye disease and other pathological processes. Prog. Retin. Eye Res. 2016, 53, 70–106. [Google Scholar] [CrossRef]
- Schlanitz, F.G.; Baumann, B.; Kundi, M.; Sacu, S.; Baratsits, M.; Scheschy, U.; Shahlaee, A.; Mittermüller, T.J.; Montuoro, A.; Roberts, P.; et al. Drusen volume development over time and its relevance to the course of age-related macular degeneration. Br. J. Ophthalmol. 2017, 101, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Abdelsalam, A.; Del Priore, L.; Zarbin, M.A. Drusen in age-related macular degeneration: Pathogenesis, natural course, and laser photocoagulation–induced regression. Surv. Ophthalmol. 1999, 44, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Murali, A.; Krishnakumar, S.; Subramanian, A.; Parameswaran, S. Bruch’s membrane pathology: A mechanistic perspective. Eur. J. Ophthalmol. 2020, 30, 1195–1206. [Google Scholar] [CrossRef] [PubMed]
- Tisi, A.; Feligioni, M.; Passacantando, M.; Ciancaglini, M.; Maccarone, R. The impact of oxidative stress on blood-retinal barrier physiology in age-related macular degeneration. Cells 2021, 10, 64. [Google Scholar] [CrossRef]
- Haddad, S.; Chen, C.A.; Santangelo, S.L.; Seddon, J.M. The Genetics of Age-Related Macular Degeneration: A Review of Progress to Date. Surv. Ophthalmol. 2006, 51, 316–363. [Google Scholar] [CrossRef]
- Schultz, N.M.; Bhardwaj, S.; Barclay, C.; Gaspar, L.; Schwartz, J. Global Burden of Dry Age-Related Macular Degeneration: A Targeted Literature Review. Clin. Ther. 2021, 43, 1792–1818. [Google Scholar] [CrossRef]
- Ferris, F.L.; Fine, S.L.; Hyman, L. Age-related macular degeneration and blindness due to neovascular maculopathy. Arch. Ophthalmol. 1984, 102, 1640–1642. [Google Scholar] [CrossRef]
- Ratnayaka, J.A.; Lotery, A.J. Challenges in studying geographic atrophy (GA) age-related macular degeneration: The potential of a new mouse model with GA-like features. Neural Regen. Res. 2020, 15, 863–864. [Google Scholar] [CrossRef]
- Arya, M.; Sabrosa, A.S.; Duker, J.S.; Waheed, N.K. Choriocapillaris changes in dry age-related macular degeneration and geographic atrophy: A review. Eye Vis 2018, 5, 22. [Google Scholar] [CrossRef]
- Bakri, S.J.; Bektas, M.; Sharp, D.; Luo, R.; Sarda, S.P.; Khan, S. Geographic atrophy: Mechanism of disease, pathophysiology, and role of the complement system. J. Manag. Care Spec. Pharm. 2023, 29 (Suppl. S5-a), S2–S11. [Google Scholar] [CrossRef] [PubMed]
- Madheswaran, G.; Ramesh, S.V.; Pardhan, S.; Sapkota, R.; Raman, R. Impact of living with a bilateral central vision loss due to geographic atrophy—Qualitative study. BMJ Open 2021, 11, e047861. [Google Scholar] [CrossRef] [PubMed]
- Sivaprasad, S.; Tschosik, E.A.; Guymer, R.H.; Kapre, A.; Suñer, I.J.; Joussen, A.M.; Lanzetta, P.; Ferrara, D. Living with geographic atrophy: An ethnographic study. Ophthalmol. Ther. 2019, 8, 115–124. [Google Scholar] [CrossRef]
- Sacconi, R.; Corbelli, E.; Querques, L.; Bandello, F.; Querques, G. A review of current and future management of geographic atrophy. Ophthalmol. Ther. 2017, 6, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Danis, R.P.; Lavine, J.A.; Domalpally, A. Geographic atrophy in patients with advanced dry age-related macular degeneration: Current challenges and future prospects. Clin. Ophthalmol. 2015, 9, 2159–2174. [Google Scholar] [CrossRef]
- Borchert, G.A.; Shamsnajafabadi, H.; Hu, M.L.; De Silva, S.R.; Downes, S.M.; MacLaren, R.E.; Xue, K.; Cehajic-Kapetanovic, J. The Role of Inflammation in Age-Related Macular Degeneration—Therapeutic Landscapes in Geographic Atrophy. Cells 2023, 12, 2092. [Google Scholar] [CrossRef]
- Almony, A.; Keyloun, K.R.; Shah-Manek, B.; Multani, J.K.; McGuiness, C.B.; Chen, C.-C.; Campbell, J.H. Clinical and economic burden of neovascular age-related macular degeneration by disease status: A US claims-based analysis. J. Manag. Care Spec. Pharm. 2021, 27, 1260–1272. [Google Scholar] [CrossRef]
- Schmid, A.; Bucher, F.; Liczenczias, E.; Maslanka Figueroa, S.; Müller, B.; Agostini, H. nAMD: Optimization of patient care and patient-oriented information with the help of an internet-based survey. Graefe’s Arch. Clin. Exp. Ophthalmol. 2022, 260, 3241–3253. [Google Scholar] [CrossRef]
- Wong, T.; Chakravarthy, U.; Klein, R.; Mitchell, P.; Zlateva, G.; Buggage, R.; Fahrbach, K.; Probst, C.; Sledge, I. The natural history and prognosis of neovascular age-related macular degeneration: A systematic review of the literature and meta-analysis. Ophthalmology 2008, 115, 116–126.e1. [Google Scholar] [CrossRef]
- Chappelow, A.V.; Kaiser, P.K. Neovascular age-related macular degeneration: Potential therapies. Drugs 2008, 68, 1029–1036. [Google Scholar] [CrossRef]
- Banister, K.; Cook, J.A.; Scotland, G.; Azuara-Blanco, A.; Goulao, B.; Heimann, H.; Hernandez, R.; Hogg, R.; Kennedy, C.; Sivaprasad, S.; et al. Non-invasive testing for early detection of neovascular macular degeneration in unaffected second eyes of older adults: EDNA diagnostic accuracy study. Health Technol. Assess. 2022, 26, 1–142. [Google Scholar] [CrossRef]
- Pugazhendhi, A.; Hubbell, M.; Jairam, P.; Ambati, B. Neovascular macular degeneration: A review of etiology, risk factors, and recent advances in research and therapy. Int. J. Mol. Sci. 2021, 22, 1170. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Nawaz, M.I. Molecular mechanism of VEGF and its role in pathological angiogenesis. J. Cell. Biochem. 2022, 123, 1938–1965. [Google Scholar] [CrossRef] [PubMed]
- Curry, B.; Bylsma, G.; Hewitt, A.W.; Verma, N. The VEGF treatment of AMD switch study (The vTAS Study). Asia-Pac. J. Ophthalmol. 2017, 6, 481–487. [Google Scholar]
- Amoaku, W.M.; Chakravarthy, U.; Gale, R.; Gavin, M.; Ghanchi, F.; Gibson, J.; Harding, S.; Johnston, R.; Kelly, S.; Lotery, A.; et al. Defining response to anti-VEGF therapies in neovascular AMD. Eye 2015, 29, 721–731. [Google Scholar] [CrossRef]
- Boyle, J.; Vukicevic, M.; Koklanis, K.; Itsiopoulos, C. Experiences of patients undergoing anti-VEGF treatment for neovascular age-related macular degeneration: A systematic review. Psychol. Health Med. 2015, 20, 296–310. [Google Scholar] [CrossRef]
- Bressler, N.M.; Chang, T.S.; Suñer, I.J.; Fine, J.T.; Dolan, C.M.; Ward, J.; Ianchulev, T. Vision-related function after ranibizumab treatment by better-or worse-seeing eye: Clinical trial results from MARINA and ANCHOR. Ophthalmology 2010, 117, 747–756.e744. [Google Scholar] [CrossRef]
- Heier, J.S.; Lad, E.M.; Holz, F.G.; Rosenfeld, P.J.; Guymer, R.H.; Boyer, D.; Grossi, F.; Baumal, C.R.; Korobelnik, J.-F.; Slakter, J.S.; et al. Pegcetacoplan for the treatment of geographic atrophy secondary to age-related macular degeneration (OAKS and DERBY): Two multicentre, randomised, double-masked, sham-controlled, phase 3 trials. Lancet 2023, 402, 1434–1448. [Google Scholar] [CrossRef]
- Wykoff, C.C.; Ou, W.C.; Brown, D.M.; Croft, D.E.; Wang, R.; Payne, J.F.; Clark, W.L.; Abdelfattah, N.S.; Sadda, S.R.; Group, T.-A.S. Randomized trial of treat-and-extend versus monthly dosing for neovascular age-related macular degeneration: 2-year results of the TREX-AMD study. Ophthalmol. Retin. 2017, 1, 314–321. [Google Scholar] [CrossRef]
- Nadeem, A.; Malik, I.A.; Shariq, F.; Afridi, E.K.; Taha, M.; Raufi, N.; Naveed, A.K.; Iqbal, J.; Habte, A. Advancements in the treatment of geographic atrophy: Focus on pegcetacoplan in age-related macular degeneration. Ann. Med. Surg. 2023, 85, 6067–6077. [Google Scholar] [CrossRef] [PubMed]
- Schachar, I.H. Concerning Syfovre Approval for Geographic Atrophy. JAMA Ophthalmol. 2024, 142, 85–86. [Google Scholar] [CrossRef] [PubMed]
- Danzig, C.J.; Khanani, A.M.; Loewenstein, A. C5 inhibitor avacincaptad pegol treatment for geographic atrophy: A comprehensive review. Immunotherapy 2024, 16, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, G.J.; Westby, K.; Csaky, K.G.; Monés, J.; Pearlman, J.A.; Patel, S.S.; Joondeph, B.C.; Randolph, J.; Masonson, H.; Rezaei, K.A. C5 inhibitor avacincaptad pegol for geographic atrophy due to age-related macular degeneration: A randomized pivotal phase 2/3 trial. Ophthalmology 2021, 128, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.S.; Lally, D.R.; Hsu, J.; Wykoff, C.C.; Eichenbaum, D.; Heier, J.S.; Jaffe, G.J.; Westby, K.; Desai, D.; Zhu, L.; et al. Avacincaptad pegol for geographic atrophy secondary to age-related macular degeneration: 18-month findings from the GATHER1 trial. Eye 2023, 37, 3551–3557. [Google Scholar] [CrossRef]
- Mousavi, M.; Armstrong, R.A. Genetic risk factors and age-related macular degeneration (AMD). J. Optom. 2013, 6, 176–184. [Google Scholar] [CrossRef]
- Chen, Y.; Bedell, M.; Zhang, K. Age-related macular degeneration: Genetic and environmental factors of disease. Mol. Interv. 2010, 10, 271–281. [Google Scholar] [CrossRef]
- Deng, Y.; Qiao, L.; Du, M.; Qu, C.; Wan, L.; Li, J.; Huang, L. Age-related macular degeneration: Epidemiology, genetics, pathophysiology, diagnosis, and targeted therapy. Genes. Dis. 2022, 9, 62–79. [Google Scholar] [CrossRef]
- Klein, M.L.; Schultz, D.W.; Edwards, A.; Matise, T.C.; Rust, K.; Berselli, C.B.; Trzupek, K.; Weleber, R.G.; Ott, J.; Wirtz, M.K.; et al. Age-related macular degeneration: Clinical features in a large family and linkage to chromosome 1q. Arch. Ophthalmol. 1998, 116, 1082–1088. [Google Scholar] [CrossRef]
- Shahid, H.; Khan, J.C.; Cipriani, V.; Sepp, T.; Matharu, B.K.; Bunce, C.; Harding, S.P.; Clayton, D.G.; Moore, A.T.; Yates, J.R.; et al. Age-related macular degeneration: The importance of family history as a risk factor. Br. J. Ophthalmol. 2012, 96, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, S.K.; Song, D.; Klein, B.E.; Klein, R.; Schick, J.H.; Humphrey, J.; Millard, C.; Liptak, R.; Russo, K.; Jun, G.; et al. Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am. J. Hum. Genet. 2004, 74, 20–39. [Google Scholar] [CrossRef] [PubMed]
- Majewski, J.; Schultz, D.W.; Weleber, R.G.; Schain, M.B.; Edwards, A.O.; Matise, T.C.; Acott, T.S.; Ott, J.; Klein, M.L. Age-related macular degeneration—A genome scan in extended families. Am. J. Hum. Genet. 2003, 73, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Bhumika; Bora, N.S.; Bora, P.S. Genetic insights into age-related macular degeneration. Biomedicines 2024, 12, 1479. [Google Scholar] [CrossRef]
- Shughoury, A.; Sevgi, D.D.; Ciulla, T.A. Molecular genetic mechanisms in age-related macular degeneration. Genes. 2022, 13, 1233. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 48, 134–143. [Google Scholar] [CrossRef]
- Schick, T.; Lorés-Motta, L.; Altay, L.; Fritsche, L.G.; den Hollander, A.I.; Fauser, S. The effect of genetic variants associated with age-related macular degeneration varies with age. Investig. Ophthalmol. Vis. Sci. 2020, 61, 17. [Google Scholar] [CrossRef]
- Black, J.R.M.; Clark, S.J. Age-related macular degeneration: Genome-wide association studies to translation. Genet. Med. 2016, 18, 283–289. [Google Scholar] [CrossRef]
- Piri, N.; Kaplan, H.J. Role of complement in the onset of age-related macular degeneration. Biomolecules 2023, 13, 832. [Google Scholar] [CrossRef]
- Nischler, C.; Oberkofler, H.; Ortner, C.; Paikl, D.; Riha, W.; Lang, N.; Patsch, W.; Egger, S.F. Complement factor H Y402H gene polymorphism and response to intravitreal bevacizumab in exudative age-related macular degeneration. Acta Ophthalmol. 2011, 89, e344–e349. [Google Scholar] [CrossRef]
- Clark, S.J.; Higman, V.A.; Mulloy, B.; Perkins, S.J.; Lea, S.M.; Sim, R.B.; Day, A.J. His-384 allotypic variant of factor H associated with age-related macular degeneration has different heparin binding properties from the non-disease-associated form. J. Biol. Chem. 2006, 281, 24713–24720. [Google Scholar] [CrossRef] [PubMed]
- Seddon, J.M.; Francis, P.J.; George, S.; Schultz, D.W.; Rosner, B.; Klein, M.L. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA 2007, 297, 1793–1800. [Google Scholar] [CrossRef]
- Laine, M.; Jarva, H.; Seitsonen, S.; Haapasalo, K.; Lehtinen, M.J.; Lindeman, N.; Anderson, D.H.; Johnson, P.T.; JärVelä, I.; Jokiranta, T.S.; et al. Y402H polymorphism of complement factor H affects binding affinity to C-reactive protein. J. Immunol. 2007, 178, 3831–3836. [Google Scholar] [CrossRef] [PubMed]
- Kubicka-Trząska, A.; Żuber-Łaskawiec, K.; Dziedzina, S.; Sanak, M.; Romanowska-Dixon, B.; Karska-Basta, I. Genetic variants of complement factor H Y402H (rs1061170), C2 R102G (rs2230199), and C3 E318D (rs9332739) and response to intravitreal anti-VEGF treatment in patients with exudative age-related macular degeneration. Medicina 2022, 58, 658. [Google Scholar] [CrossRef]
- Gold, B.; Merriam, J.E.; Zernant, J.; Hancox, L.S.; Taiber, A.J.; Gehrs, K.; Cramer, K.; Neel, J.; Bergeron, J.; Barile, G.R. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat. Genet. 2006, 38, 458–462. [Google Scholar] [CrossRef]
- Pilotti, C.; Greenwood, J.; Moss, S.E. Functional evaluation of AMD-associated risk variants of complement factor B. Investig. Ophthalmol. Vis. Sci. 2020, 61, 19. [Google Scholar] [CrossRef] [PubMed]
- Armento, A.; Ueffing, M.; Clark, S.J. The complement system in age-related macular degeneration. Cell. Mol. Life Sci. 2021, 78, 4487–4505. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Faber, C.; Falk, M.; Nissen, M.H.; Hviid, T.V.; Sørensen, T.L. Altered expression of CD46 and CD59 on leukocytes in neovascular age-related macular degeneration. Am. J. Ophthalmol. 2012, 154, 193–199.e2. [Google Scholar] [CrossRef]
- Kavanagh, D.; Yu, Y.; Schramm, E.C.; Triebwasser, M.; Wagner, E.K.; Raychaudhuri, S.; Daly, M.J.; Atkinson, J.P.; Seddon, J.M. Rare genetic variants in the CFI gene are associated with advanced age-related macular degeneration and commonly result in reduced serum factor I levels. Hum. Mol. Genet. 2015, 24, 3861–3870. [Google Scholar] [CrossRef]
- Hallam, T.M.; Marchbank, K.J.; Harris, C.L.; Osmond, C.; Shuttleworth, V.G.; Griffiths, H.; Cree, A.J.; Kavanagh, D.; Lotery, A.J. Rare genetic variants in complement factor I lead to low FI plasma levels resulting in increased risk of age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2020, 61, 18. [Google Scholar] [CrossRef]
- Lu, Z.-G.; May, A.; Dinh, B.; Lin, V.; Su, F.; Tran, C.; Adivikolanu, H.; Ehlen, R.; Che, B.; Wang, Z.-H.; et al. The interplay of oxidative stress and ARMS2-HTRA1 genetic risk in neovascular AMD. Vessel. Plus 2021, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Tamura, H.; Tsujikawa, A.; Yamashiro, K.; Akagi-Kurashige, Y.; Nakata, I.; Nakanishi, H.; Hayashi, H.; Ooto, S.; Otani, A.; Yoshimura, N. Association of ARMS2 genotype with bilateral involvement of exudative age-related macular degeneration. Am. J. Ophthalmol. 2012, 154, 542–548.e1. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Camp, N.J.; Sun, H.; Tong, Z.; Gibbs, D.; Cameron, D.J.; Chen, H.; Zhao, Y.; Pearson, E.; Li, X.; et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science 2006, 314, 992–993. [Google Scholar] [CrossRef]
- Boulton, M.E.; Qi, X.; Kanda, A.; Nellissery, J.; Mitter, S.K.; Grant, M.B.; Swaroop, A. ARMS2 association with the mitochondrial outer membrane is reduced in RPE cells exposed to oxidative stress and in AMD. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5301. [Google Scholar]
- Chang, Y.-J.; Jenny, L.A.; Li, Y.-S.; Cui, X.; Kong, Y.; Li, Y.; Sparrow, J.R.; Tsang, S.H. CRISPR editing demonstrates rs10490924 raised oxidative stress in iPSC-derived retinal cells from patients with ARMS2/HTRA1-related AMD. Proc. Natl. Acad. Sci. USA 2023, 120, e2215005120. [Google Scholar] [CrossRef]
- Dewan, A.; Liu, M.; Hartman, S.; Zhang, S.S.; Liu, D.T.; Zhao, C.; Tam, P.O.; Chan, W.M.; Lam, D.S.; Snyder, M.; et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 2006, 314, 989–992. [Google Scholar] [CrossRef]
- Williams, B.L.; Seager, N.A.; Gardiner, J.D.; Pappas, C.M.; Cronin, M.C.; Amat di San Filippo, C.; Anstadt, R.A.; Liu, J.; Toso, M.A.; Nichols, L.; et al. Chromosome 10q26-driven age-related macular degeneration is associated with reduced levels of HTRA1 in human retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 2021, 118, e2103617118. [Google Scholar] [CrossRef]
- Fan, D.; Kassiri, Z. Biology of Tissue Inhibitor of Metalloproteinase 3 (TIMP3), and Its Therapeutic Implications in Cardiovascular Pathology. Front. Physiol. 2020, 11, 661. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, R.; Zhao, X.; Ma, Z.; Xin, J.; Xu, S.; Guo, D. The role of oxidative stress in the pathogenesis of ocular diseases: An overview. Mol. Biol. Rep. 2024, 51, 454. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Jiang, S.; Gericke, A. Age-related macular degeneration: Role of oxidative stress and blood vessels. Int. J. Mol. Sci. 2021, 22, 1296. [Google Scholar] [CrossRef] [PubMed]
- Gabrielle, P.-H. Lipid Metabolism and Retinal Diseases; Wiley Online Library: Hoboken, NJ, USA, 2022. [Google Scholar]
- Wang, Y.-F.; Han, Y.; Zhang, R.; Qin, L.; Wang, M.-X.; Ma, L. CETP/LPL/LIPC gene polymorphisms and susceptibility to age-related macular degeneration. Sci. Rep. 2015, 5, 15711. [Google Scholar] [CrossRef]
- Li, B.; Chang, F.-Y.; Arunkumar, R.; Wan, Z.; Addo, E.K.; Bernstein, P.S. Hepatic Lipase (LIPC) Knockdown Increases Macular Carotenoid Influx and Cholesterol Efflux in ARPE-19 Cells. Investig. Ophthalmol. Vis. Sci. 2023, 64, 5202. [Google Scholar]
- Jacobo-Albavera, L.; Domínguez-Pérez, M.; Medina-Leyte, D.J.; González-Garrido, A.; Villarreal-Molina, T. The role of the ATP-binding cassette A1 (ABCA1) in human disease. Int. J. Mol. Sci. 2021, 22, 1593. [Google Scholar] [CrossRef]
- Peters, F.; Ebner, L.J.; Atac, D.; Maggi, J.; Berger, W.; den Hollander, A.I.; Grimm, C. Regulation of ABCA1 by AMD-associated genetic variants and hypoxia in iPSC-RPE. Int. J. Mol. Sci. 2022, 23, 3194. [Google Scholar] [CrossRef]
- Bao, X.; Zhang, Z.; Guo, Y.; Buser, C.; Kochounian, H.; Wu, N.; Li, X.; He, S.; Sun, B.; Ross-Cisneros, F.N.; et al. Human RGR gene and associated features of age-related macular degeneration in models of retina-choriocapillaris atrophy. Am. J. Pathol. 2021, 191, 1454–1473. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Cui, H.; Bao, X.; Huang, L.; He, S.; Fong, H.K.; Zhao, M. Proteopathy Linked to Exon-Skipping Isoform of RGR-Opsin Contributes to the Pathogenesis of Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2023, 64, 41. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, S.; Guan, W.; Xu, N.; Zhu, L.; Du, W.; Liu, Z.; Fong, H.K.; Huang, L.; Zhao, M. Retinal G-protein-coupled receptor deletion exacerbates AMD-like changes via the PINK1–parkin pathway under oxidative stress. FASEB J. 2024, 38, e70135. [Google Scholar] [CrossRef]
- Yu, C.; Robman, L.; He, W.; Woods, R.L.; Thao, L.T.P.; Wolfe, R.; Phung, J.; Makeyeva, G.A.; Hodgson, L.A.; McNeil, J.J.; et al. Predictive performance of an updated polygenic risk score for age-related macular degeneration. Ophthalmology 2024, 131, 880–891. [Google Scholar] [CrossRef]
- Barnstable, C.J. Epigenetics and degenerative retinal diseases: Prospects for new therapeutic approaches. Asia-Pac. J. Ophthalmol. 2022, 11, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Farkas, M.H.; DeAngelis, M.M. Age-related macular degeneration: From epigenetics to therapeutic implications. Age-Relat. Macular Degener. Clin. Genes. Back. Patient Manag. 2021, 1256, 221–235. [Google Scholar]
- Caputo, V.; Strafella, C.; Termine, A.; Fabrizio, C.; Ruffo, P.; Cusumano, A.; Giardina, E.; Ricci, F.; Cascella, R. Epigenomic signatures in age-related macular degeneration: Focus on their role as disease modifiers and therapeutic targets. Eur. J. Ophthalmol. 2021, 31, 2856–2867. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wu, J.; Guo, H.; Yao, W.; Li, S.; Lu, Y.; Jia, Y.; Liang, X.; Tang, J.; Zhang, H. Post-translational modifications of histones: Mechanisms, biological functions, and therapeutic targets. MedComm 2023, 4, e292. [Google Scholar] [CrossRef]
- Quina, A.; Buschbeck, M.; Di Croce, L. Chromatin structure and epigenetics. Biochem. Pharmacol. 2006, 72, 1563–1569. [Google Scholar] [CrossRef]
- Jiang, C.; Pugh, B.F. Nucleosome positioning and gene regulation: Advances through genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Cao, Y.; Qin, J.; Song, X.; Zhang, Q.; Shi, Y.; Cao, L. DNA methylation, its mediators and genome integrity. Int. J. Biol. Sci. 2015, 11, 604. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Singal, R.; Ginder, G.D. DNA methylation. Blood J. Am. Soc. Hematol. 1999, 93, 4059–4070. [Google Scholar]
- Rajanala, K.; Upadhyay, A. Epigenetic switches in retinal homeostasis and target for drug development. Int. J. Mol. Sci. 2024, 25, 2840. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Meng, C.; He, S.; Gu, C.; Lhamo, T.; Draga, D.; Luo, D.; Qiu, Q. DNA methylation in diabetic retinopathy: Pathogenetic role and potential therapeutic targets. Cell Biosci. 2022, 12, 186. [Google Scholar] [CrossRef] [PubMed]
- Abokyi, S.; To, C.-H.; Lam, T.T.; Tse, D.Y. Central role of oxidative stress in age-related macular degeneration: Evidence from a review of the molecular mechanisms and animal models. Oxidative Med. Cell. Longev. 2020, 2020, 7901270. [Google Scholar] [CrossRef]
- Advani, J.; Mehta, P.A.; Hamel, A.R.; Mehrotra, S.; Kiel, C.; Strunz, T.; Corso-Díaz, X.; Kwicklis, M.; van Asten, F.; Ratnapriya, R.; et al. QTL mapping of human retina DNA methylation identifies 87 gene-epigenome interactions in age-related macular degeneration. Nat. Commun. 2024, 15, 1972. [Google Scholar] [CrossRef]
- Hunter, A.; Spechler, P.A.; Cwanger, A.; Song, Y.; Zhang, Z.; Ying, G.-s.; Hunter, A.K.; Dezoeten, E.; Dunaief, J.L. DNA methylation is associated with altered gene expression in AMD. Investig. Ophthalmol. Vis. Sci. 2012, 53, 2089–2105. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, D.; Thombre, R.; Dhar, A.; Anant, S. DNA methyltransferases: A novel target for prevention and therapy. Front. Oncol. 2014, 4, 80. [Google Scholar] [CrossRef]
- Maugeri, A.; Barchitta, M.; Fallico, M.; Castellino, N.; Reibaldi, M.; Agodi, A. Characterization of SIRT1/DNMTs functions and LINE-1 methylation in patients with age-related macular degeneration. J. Clin. Med. 2019, 8, 159. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Liu, B.; Tuo, J.; Shen, D.; Chen, P.; Li, Z.; Liu, X.; Ni, J.; Dagur, P.; Sen, H.N. Hypomethylation of the IL17RC promoter associates with age-related macular degeneration. Cell Rep. 2012, 2, 1151–1158. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, Y.; Chu, F.; Liao, K.; Cui, Z.; Chen, J.; Tang, S. Integrated Analysis of DNA methylation and transcriptome profile to identify key features of age-related macular degeneration. Bioengineered 2021, 12, 7061–7078. [Google Scholar] [CrossRef] [PubMed]
- Oliver, V.F.; Franchina, M.; Jaffe, A.E.; Branham, K.E.; Othman, M.; Heckenlively, J.R.; Swaroop, A.; Campochiaro, B.; Vote, B.J.; Craig, J.E.; et al. Hypomethylation of the IL17RC promoter in peripheral blood leukocytes is not a hallmark of age-related macular degeneration. Cell Rep. 2013, 5, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Kenney, M.C.; Nashine, S. Further understanding of epigenetic dysfunction of the retinal pigment epithelium in AMD. Expert. Rev. Ophthalmol. 2020, 15, 221–231. [Google Scholar] [CrossRef]
- Porter, L.F.; Saptarshi, N.; Fang, Y.; Rathi, S.; Den Hollander, A.I.; De Jong, E.K.; Clark, S.J.; Bishop, P.N.; Olsen, T.W.; Liloglou, T.; et al. Whole-genome methylation profiling of the retinal pigment epithelium of individuals with age-related macular degeneration reveals differential methylation of the SKI, GTF2H4, and TNXB genes. Clin. Epigenetics 2019, 11, 6. [Google Scholar] [CrossRef]
- Hirasawa, M.; Takubo, K.; Osada, H.; Miyake, S.; Toda, E.; Endo, M.; Umezawa, K.; Tsubota, K.; Oike, Y.; Ozawa, Y. Angiopoietin-like protein 2 is a multistep regulator of inflammatory neovascularization in a murine model of age-related macular degeneration. J. Biol. Chem. 2016, 291, 7373–7385. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Hou, Y.; Fan, Y.; Jiang, H.; Li, B.; Zhu, H.; Liu, Y.; Zhang, L.; Zhang, J.; et al. Association of Plasma Vitamins and Carotenoids, DNA Methylation of LCAT, and Risk of Age-Related Macular Degeneration. Nutrients 2023, 15, 2985. [Google Scholar] [CrossRef]
- Khan, M.; Shah, S.; Lv, B.; Lv, Z.; Ji, N.; Song, Z.; Wu, P.; Wang, X.; Mehmood, A. Molecular mechanisms of Alu and LINE-1 interspersed repetitive sequences reveal diseases of visual system dysfunction. Ocul. Immunol. Inflamm. 2023, 31, 1848–1858. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Barchitta, M.; Mazzone, M.G.; Giuliano, F.; Basile, G.; Agodi, A. Resveratrol Restores LINE-1 Methylation Levels by Modulating SIRT1 and DNMTs Functions in Cellular Models of Age-Related Macular Degeneration. Preprints 2018. [Google Scholar] [CrossRef]
- Merbs, S.L.; Khan, M.A.; Hackler, L., Jr.; Oliver, V.F.; Wan, J.; Qian, J.; Zack, D.J. Cell-Specific DNA Methylation Patterns of Retina-Specific Genes. PLoS ONE 2012, 7, e32602. [Google Scholar] [CrossRef]
- Karemaker, I.D.; Vermeulen, M. Single-cell DNA methylation profiling: Technologies and biological applications. Trends Biotechnol. 2018, 36, 952–965. [Google Scholar] [CrossRef]
- Camacho, P.; Ribeiro, E.; Pereira, B.; Varandas, T.; Nascimento, J.; Henriques, J.; Dutra-Medeiros, M.; Delgadinho, M.; Oliveira, K.; Silva, C.; et al. DNA methyltransferase expression (DNMT1, DNMT3a and DNMT3b) as a potential biomarker for anti-VEGF diabetic macular edema response. Eur. J. Ophthalmol. 2023, 33, 2267–2274. [Google Scholar] [CrossRef]
- Cui, X.; Zhao, Q.; Mahata, B.; Wen, D.; Yu-Wai-Man, P.; Li, X. Multiomic Screening Unravels the Immunometabolic Signatures and Drug Targets of Age-Related Macular Degeneration. bioRxiv 2024. [Google Scholar] [CrossRef]
- McGinty, R.K.; Tan, S. Histone, nucleosome, and chromatin structure. In Fundamentals of Chromatin; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–28. [Google Scholar]
- Peterson, C.L.; Laniel, M.-A. Histones and histone modifications. Curr. Biol. 2004, 14, R546–R551. [Google Scholar] [CrossRef]
- Campos, E.I.; Reinberg, D. Histones: Annotating chromatin. Annu. Rev. Genet. 2009, 43, 559–599. [Google Scholar] [CrossRef]
- Khorasanizadeh, S. The nucleosome: From genomic organization to genomic regulation. Cell 2004, 116, 259–272. [Google Scholar] [CrossRef]
- Kimura, H. Histone modifications for human epigenome analysis. J. Hum. Genet. 2013, 58, 439–445. [Google Scholar] [CrossRef]
- Turner, B.M. Histone acetylation and control of gene expression. J. Cell Sci. 1991, 99, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Struhl, K. Histone acetylation and transcriptional regulatory mechanisms. Genes. Dev. 1998, 12, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002, 12, 198–209. [Google Scholar] [CrossRef]
- Park, P.J. ChIP–seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009, 10, 669–680. [Google Scholar] [CrossRef]
- Ruijter, A.J.d.; GENNIP, A.H.v.; Caron, H.N.; Kemp, S.; KUILENBURG, A.B.v. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749. [Google Scholar] [CrossRef]
- Husain, S.; Obert, E.; Singh, S.; Schnabolk, G. Inhibition of HDAC1 and 3 in the Presence of Systemic Inflammation Reduces Retinal Degeneration in a Model of Dry Age-Related Macular Degeneration. J. Ocul. Pharmacol. Ther. 2024, 40, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Schnabolk, G.; Obert, E.; Singh, S.; Guzman, W.; Husain, S. Effect of HDAC Inhibition on a Model of Dry AMD in the Presence of Systemic Inflammation. Investig. Ophthalmol. Vis. Sci. 2023, 64, 3909. [Google Scholar]
- Jun, J.H.; Kim, J.-S.; Palomera, L.F.; Jo, D.-G. Dysregulation of histone deacetylases in ocular diseases. Arch. Pharmacal Res. 2024, 47, 20–39. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Feng, S.; Zhang, Q.; Qin, H.; Xu, C.; Fu, X.; Yan, L.; Zhao, Y.; Yao, K. Roles of histone acetyltransferases and deacetylases in the retinal development and diseases. Mol. Neurobiol. 2023, 60, 2330–2354. [Google Scholar] [CrossRef]
- Wang, J.; Zibetti, C.; Shang, P.; Sripathi, S.R.; Zhang, P.; Cano, M.; Hoang, T.; Xia, S.; Ji, H.; Merbs, S.L.; et al. ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nat. Commun. 2018, 9, 1364. [Google Scholar] [CrossRef]
- Luu, J.; Kallestad, L.; Hoang, T.; Lewandowski, D.; Dong, Z.; Blackshaw, S.; Palczewski, K. Epigenetic hallmarks of age-related macular degeneration are recapitulated in a photosensitive mouse model. Hum. Mol. Genet. 2020, 29, 2611–2624. [Google Scholar] [CrossRef]
- Mimura, T.; Kaji, Y.; Noma, H.; Funatsu, H.; Okamoto, S. The role of SIRT1 in ocular aging. Exp. Eye Res. 2013, 116, 17–26. [Google Scholar] [CrossRef]
- Zhou, M.; Luo, J.; Zhang, H. Role of Sirtuin 1 in the pathogenesis of ocular disease. Int. J. Mol. Med. 2018, 42, 13–20. [Google Scholar] [CrossRef]
- Karbasforooshan, H.; Karimi, G. The role of SIRT1 in diabetic retinopathy. Biomed. Pharmacother. 2018, 97, 190–194. [Google Scholar] [CrossRef]
- Zhang, H.; He, S.; Spee, C.; Ishikawa, K.; Hinton, D.R. SIRT1 mediated inhibition of VEGF/VEGFR2 signaling by Resveratrol and its relevance to choroidal neovascularization. Cytokine 2015, 76, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.; Gong, Z. The beneficial roles of SIRT1 in neuroinflammation-related diseases. Oxidative Med. Cell. Longev. 2020, 2020, 6782872. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, Y.; Wang, Y.; Chao, Y.; Zhang, J.; Jia, Y.; Tie, J.; Hu, D. Regulation of SIRT1 and its roles in inflammation. Front. Immunol. 2022, 13, 831168. [Google Scholar] [CrossRef]
- Maugeri, A.; Barchitta, M.; Mazzone, M.G.; Giuliano, F.; Basile, G.; Agodi, A. Resveratrol modulates SIRT1 and DNMT functions and restores LINE-1 methylation levels in ARPE-19 cells under oxidative stress and inflammation. Int. J. Mol. Sci. 2018, 19, 2118. [Google Scholar] [CrossRef]
- Sun, H.; Li, D.; Wei, C.; Liu, L.; Xin, Z.; Gao, H.; Gao, R. The relationship between SIRT1 and inflammation: A systematic review and meta-analysis. Front. Immunol. 2024, 15, 1465849. [Google Scholar] [CrossRef]
- Donmez, G.; Outeiro, T.F. SIRT1 and SIRT2: Emerging targets in neurodegeneration. EMBO Mol. Med. 2013, 5, 344–352. [Google Scholar] [CrossRef]
- Martin, A.; Tegla, C.A.; Cudrici, C.D.; Kruszewski, A.M.; Azimzadeh, P.; Boodhoo, D.; Mekala, A.P.; Rus, V.; Rus, H. Role of SIRT1 in autoimmune demyelination and neurodegeneration. Immunol. Res. 2015, 61, 187–197. [Google Scholar] [CrossRef]
- Pallàs, M.; Casadesús, G.; Smith, M.A.; Coto-Montes, A.; Pelegri, C.; Vilaplana, J.; Camins, A. Resveratrol and neurodegenerative diseases: Activation of SIRT1 as the potential pathway towards neuroprotection. Curr. Neurovascular Res. 2009, 6, 70–81. [Google Scholar] [CrossRef]
- Li, W.; Wang, Y.; Zhu, L.; Du, S.; Mao, J.; Wang, Y.; Wang, S.; Bo, Q.; Tu, Y.; Yi, Q. The P300/XBP1s/Herpud1 axis promotes macrophage M2 polarization and the development of choroidal neovascularization. J. Cell. Mol. Med. 2021, 25, 6709–6720. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.; Zhao, M. An updated review of the epigenetic mechanism underlying the pathogenesis of age-related macular degeneration. Aging Dis. 2020, 11, 1219. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015, 109, 21.29.1–21.29.9. [Google Scholar] [CrossRef] [PubMed]
- Shang, P.; Daley, R.; Mahally, E.R.; Stepicheva, N.A.; Ghosh, S.; Liu, H.; Strizhakova, A.; Chowdhury, O.; Koontz, V.; Hose, S.L. HDAC11 is a crucial regulator for visual cycle genes and retinal function. Investig. Ophthalmol. Vis. Sci. 2022, 63, 4115-F0352. [Google Scholar]
- Hamid, M.A.; Moustafa, M.T.; Càceres-del-Carpio, J.; Kuppermann, B.D.; Kenney, M.C. Effects of antiangiogenic drugs on expression patterns of epigenetic pathway genes. Ophthalmic Surg. Lasers Imaging Retin. 2018, 49, S29–S33. [Google Scholar] [CrossRef]
- Gemenetzi, M.; Lotery, A. Epigenetics in age-related macular degeneration: New discoveries and future perspectives. Cell. Mol. Life Sci. 2020, 77, 807–818. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Razin, S.V.; Iarovaia, O.V.; Sjakste, N.; Sjakste, T.; Bagdoniene, L.; Rynditch, A.V.; Eivazova, E.R.; Lipinski, M.; Vassetzky, Y.S. Chromatin domains and regulation of transcription. J. Mol. Biol. 2007, 369, 597–607. [Google Scholar] [CrossRef]
- Grandi, F.C.; Modi, H.; Kampman, L.; Corces, M.R. Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 2022, 17, 1518–1552. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.N.; D’Antonio-Chronowska, A.; Greenwald, W.W.; Borja, V.; Aguiar, L.R.; Pogue, R.; Matsui, H.; Benaglio, P.; Borooah, S.; D’Antonio, M.; et al. Human iPSC-Derived Retinal Pigment Epithelium: A Model System for Prioritizing and Functionally Characterizing Causal Variants at AMD Risk Loci. Stem Cell Rep. 2019, 12, 1342–1353. [Google Scholar] [CrossRef]
- Baek, S.; Lee, I. Single-cell ATAC sequencing analysis: From data preprocessing to hypothesis generation. Comput. Struct. Biotechnol. J. 2020, 18, 1429–1439. [Google Scholar] [CrossRef]
- Hansen, K.H.; Bracken, A.P.; Pasini, D.; Dietrich, N.; Gehani, S.S.; Monrad, A.; Rappsilber, J.; Lerdrup, M.; Helin, K. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 2008, 10, 1291–1300. [Google Scholar] [CrossRef]
- Bae, S.; Lesch, B.J. H3K4me1 distribution predicts transcription state and poising at promoters. Front. Cell Dev. Biol. 2020, 8, 289. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lake, B.B.; Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 2019, 37, 1452–1457. [Google Scholar] [CrossRef]
- Ma, S.; Zhang, B.; LaFave, L.M.; Earl, A.S.; Chiang, Z.; Hu, Y.; Ding, J.; Brack, A.; Kartha, V.K.; Tay, T. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 2020, 183, 1103–1116.e20. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15 (Suppl. S1), R17–R29. [Google Scholar] [CrossRef]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef] [PubMed]
- Koffler-Brill, T.; Noy, Y.; Avraham, K.B. The long and short: Non-coding RNAs in the mammalian inner ear. Hear. Res. 2023, 428, 108666. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Kaiser, P.K.; Symons, R.A.; Shah, S.M.; Quinlan, E.J.; Tabandeh, H.; Do, D.V.; Reisen, G.; Lockridge, J.A.; Short, B.; Guerciolini, R.; et al. RNAi-based treatment for neovascular age-related macular degeneration by Sirna-027. Am. J. Ophthalmol. 2010, 150, 33–39.e2. [Google Scholar] [CrossRef]
- Zhang, C.; Owen, L.A.; Lillvis, J.H.; Zhang, S.X.; Kim, I.K.; DeAngelis, M.M. AMD genomics: Non-coding RNAs as biomarkers and therapeutic targets. J. Clin. Med. 2022, 11, 1484. [Google Scholar] [CrossRef] [PubMed]
- Berber, P.; Grassmann, F.; Kiel, C.; Weber, B.H. An eye on age-related macular degeneration: The role of microRNAs in disease pathology. Mol. Diagn. Ther. 2017, 21, 31–43. [Google Scholar] [CrossRef]
- Wang, L.; Lee, A.Y.W.; Wigg, J.P.; Peshavariya, H.; Liu, P.; Zhang, H. miRNA involvement in angiogenesis in age-related macular degeneration. J. Physiol. Biochem. 2016, 72, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Gemayel, M.C.; Bhatwadekar, A.D.; Ciulla, T. RNA therapeutics for retinal diseases. Expert. Opin. Biol. Ther. 2021, 21, 603–613. [Google Scholar] [CrossRef]
- Meola, N.; Pizzo, M.; Alfano, G.; Surace, E.M.; Banfi, S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. Rna 2012, 18, 111–123. [Google Scholar] [CrossRef]
- Xu, X.-D.; Li, K.-R.; Li, X.-M.; Yao, J.; Qin, J.; Yan, B. Long non-coding RNAs: New players in ocular neovascularization. Mol. Biol. Rep. 2014, 41, 4493–4505. [Google Scholar] [CrossRef]
- Almalki, W.H.; Almujri, S.S. The impact of NF-κB on inflammatory and angiogenic processes in age-related macular degeneration. Exp. Eye Res. 2024, 248, 110111. [Google Scholar] [CrossRef]
- Zhang, P.; Lu, B.; Xu, F.; Wang, C.; Zhang, R.; Liu, Y.; Wei, C.; Mei, L. Analysis of long noncoding RNAs in choroid neovascularization. Curr. Eye Res. 2020, 45, 1403–1414. [Google Scholar] [CrossRef]
- Zhang, X.; Du, S.; Yang, D.; Jin, X.; Zhang, Y.; Wang, D.; Wang, H.; Zhang, Y.; Zhu, M. LncRNA MALAT1 knockdown inhibits the development of choroidal neovascularization. Heliyon 2023, 9, e19503. [Google Scholar] [CrossRef]
- Zhou, M.; Geathers, J.S.; Grillo, S.L.; Weber, S.R.; Wang, W.; Zhao, Y.; Sundstrom, J.M. Role of Epithelial-Mesenchymal Transition in Retinal Pigment Epithelium Dysfunction. Front. Cell Dev. Biol. 2020, 8, 501. [Google Scholar] [CrossRef]
- Chen, X.; Sun, R.; Yang, D.; Jiang, C.; Liu, Q. LINC00167 regulates RPE differentiation by targeting the miR-203a-3p/SOCS3 Axis. Mol. Ther. Nucleic Acids 2020, 19, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Zuo, C.; Liao, N.; Yao, L.; Yang, R.; Chen, H.; Wen, F. Identification of key lncRNAs in age-related macular degeneration through integrated bioinformatics and experimental validation. Aging 2024, 16, 5435. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Luo, Y.; Chen, G.; Liu, H.; Tian, N.; Zen, X.; Huang, Y. Long non-coding RNA PWRN2 regulates cytotoxicity in an in vitro model of age-related macular degeneration. Biochem. Biophys. Res. Commun. 2021, 535, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Meng, Y.-F.; Xing, Q.; Tao, J.-J.; Lu, J.; Wu, Y. Identification of lncRNAs involved in biological regulation in early age-related macular degeneration. Int. J. Nanomed. 2017, 12, 7589–7602. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, C.; Qin, B.; Liu, G.; Ji, J.; Sun, X.; Xu, M.; Ding, S.; Zhu, M.; Huang, G.; et al. LncRNA ZNF503-AS1 promotes RPE differentiation by downregulating ZNF503 expression. Cell Death Dis. 2017, 8, e3046. [Google Scholar] [CrossRef]
- Zhu, Y.-X.; Yao, J.; Liu, C.; Hu, H.-T.; Li, X.-M.; Ge, H.-M.; Zhou, Y.-F.; Shan, K.; Jiang, Q.; Yan, B. Long non-coding RNA MEG3 silencing protects against light-induced retinal degeneration. Biochem. Biophys. Res. Commun. 2018, 496, 1236–1242. [Google Scholar] [CrossRef]
- Acar, I.E.; Galesloot, T.E.; Luhmann, U.F.O.; Fauser, S.; Gayán, J.; den Hollander, A.I.; Nogoceke, E. Whole Genome Sequencing Identifies Novel Common and Low-Frequency Variants Associated With Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2023, 64, 24. [Google Scholar] [CrossRef] [PubMed]
- Farashi, S.; Abbott, C.J.; Ansell, B.R.; Wu, Z.; Altay, L.; Arnon, E.; Arnould, L.; Bagdasarova, Y.; Balaskas, K.; Chen, F.K.; et al. Genetic Risk of Reticular Pseudodrusen in Age-Related Macular Degeneration: HTRA1/lncRNA BX842242.1 dominates, with no evidence for Complement Cascade involvement. medRxiv 2024. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, L.; Li, Y.; Gui, C.; Pei, Y.; Zhou, G. Roles and mechanisms of long non-coding RNAs in age-related macular degeneration. Heliyon 2023, 9, e22307. [Google Scholar] [CrossRef]
- Sharma, A.; Singh, N.K. Long non-coding RNAs and proliferative retinal diseases. Pharmaceutics 2023, 15, 1454. [Google Scholar] [CrossRef]
- ElSheikh, R.H.; Chauhan, M.Z.; Sallam, A.B. Current and Novel Therapeutic Approaches for Treatment of Neovascular Age-Related Macular Degeneration. Biomolecules 2022, 12, 1629. [Google Scholar] [CrossRef] [PubMed]
- Lanza, M.; Benincasa, G.; Costa, D.; Napoli, C. Clinical role of epigenetics and network analysis in eye diseases: A translational science review. J. Ophthalmol. 2019, 2019, 2424956. [Google Scholar] [CrossRef]
- Camacho, P.; Ribeiro, E.; Pereira, B.; Nascimento, J.; Rosa, P.C.; Henriques, J.; Barrão, S.; Sadio, S.; Quendera, B.; Delgadinho, M.; et al. DNA Methyltransferase Expression (DNMT1, DNMT3a, and DNMT3b) as a Potential Biomarker in Age-Related Macular Degeneration. J. Clin. Med. 2025, 14, 559. [Google Scholar] [CrossRef]
- Oliver, V.F.; Jaffe, A.E.; Song, J.; Wang, G.; Zhang, P.; Branham, K.E.; Swaroop, A.; Eberhart, C.G.; Zack, D.J.; Qian, J. Differential DNA methylation identified in the blood and retina of AMD patients. Epigenetics 2015, 10, 698–707. [Google Scholar] [CrossRef]
- Jin, Y.; LI, A.; Zheng, X.; WU, H.; Sun, G.; Zhang, X. Effects of 5-Aza-dC combined with chemotherapy regimens on the apoptosis of lung adenocarcinoma cells. Clin. Med. China 2022, 12, 129–134. [Google Scholar]
- Samardzija, M.; Corna, A.; Gomez-Sintes, R.; Jarboui, M.A.; Armento, A.; Roger, J.E.; Petridou, E.; Haq, W.; Paquet-Durand, F.; Zrenner, E.; et al. HDAC inhibition ameliorates cone survival in retinitis pigmentosa mice. Cell Death Differ. 2021, 28, 1317–1332. [Google Scholar] [CrossRef]
- Trifunović, D.; Arango-Gonzalez, B.; Comitato, A.; Barth, M.; Del Amo, E.M.; Kulkarni, M.; Sahaboglu, A.; Hauck, S.M.; Urtti, A.; Arsenijevic, Y. HDAC inhibition in the cpfl1 mouse protects degenerating cone photoreceptors in vivo. Hum. Mol. Genet. 2016, 25, 4462–4472. [Google Scholar]
- Schnichels, S.; Schultheiß, M.; Hofmann, J.; Szurman, P.; Bartz-Schmidt, K.U.; Spitzer, M.S. Trichostatin A induces cell death at the concentration recommended to differentiate the RGC-5 cell line. Neurochem. Int. 2012, 60, 581–591. [Google Scholar] [CrossRef]
- Desjardins, D.; Liu, Y.; Crosson, C.E.; Ablonczy, Z. Histone deacetylase inhibition restores retinal pigment epithelium function in hyperglycemia. PLoS ONE 2016, 11, e0162596. [Google Scholar] [CrossRef]
- Tokarz, P.; Kaarniranta, K.; Blasiak, J. Inhibition of DNA methyltransferase or histone deacetylase protects retinal pigment epithelial cells from DNA damage induced by oxidative stress by the stimulation of antioxidant enzymes. Eur. J. Pharmacol. 2016, 776, 167–175. [Google Scholar] [CrossRef]
- Yang, C.; Ge, L.; Yu, X.; Lazarovici, P.; Zheng, W. Artemisinin Confers Cytoprotection toward Hydrogen Peroxide-Induced Cell Apoptosis in Retinal Pigment Epithelial Cells in Correlation with the Increased Acetylation of Histone H4 at Lysine 8. Molecules 2024, 29, 1789. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-S.; Wu, F.; Jin, Y.-M.; Chang, W.-Q.; Xu, T.-M. HDAC11: A rising star in epigenetics. Biomed. Pharmacother. 2020, 131, 110607. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.S.; Dubey, R.; Dubey, S.K.; Ashley, N.F.; Tian, W.V.; Kleinman, M.E. HDAC1/2 inhibition induces cell-type dependent effects on viability and histone H3 and H4 acetylation. Investig. Ophthalmol. Vis. Sci. 2023, 64, 2108. [Google Scholar]
- Kleinman, M.E.; Dubey, R.; Davila, A.; Dubey, S.K. Acetyl-histone profiling of degenerating RPE after selective HDAC1/2 inhibition. Investig. Ophthalmol. Vis. Sci. 2022, 63, 799-F0358. [Google Scholar]
- Lee, Q.; Chan, W.C.; Qu, X.; Sun, Y.; Abdelkarim, H.; Le, J.; Saqib, U.; Sun, M.Y.; Kruse, K.; Banerjee, A.; et al. End binding-3 inhibitor activates regenerative program in age-related macular degeneration. Cell Rep. Med. 2023, 4, 101223. [Google Scholar] [CrossRef]
- Chan, M.; Le, J.; Maienschein-Cline, M.; Komarova, Y. EB3 inhibitor prevents AMD via widespread opening of chromatin. Investig. Ophthalmol. Vis. Sci. 2022, 63, 367-F0198. [Google Scholar]
- Sun, S.; Chen, Y.; Ouyang, Y.; Tang, Z. Regulatory Roles of SWI/SNF Chromatin Remodeling Complexes in Immune Response and Inflammatory Diseases. Clin. Rev. Allergy Immunol. 2024, 68, 2. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, X.; Xin, J.; Ding, Z.; Liu, S.; Fang, Q.; Yang, N.; Xu, R.-m.; Cai, G. Architecture of SWI/SNF chromatin remodeling complex. Protein Cell 2018, 9, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Chmykhalo, V.K.; Deev, R.V.; Tokarev, A.T.; Polunina, Y.A.; Xue, L.; Shidlovskii, Y.V. SWI/SNF complex connects signaling and epigenetic state in cells of nervous system. Mol. Neurobiol. 2025, 62, 1536–1557. [Google Scholar] [CrossRef]
- Gatchalian, J.; Liao, J.; Maxwell, M.B.; Hargreaves, D.C. Control of stimulus-dependent responses in macrophages by SWI/SNF chromatin remodeling complexes. Trends Immunol. 2020, 41, 126–140. [Google Scholar] [CrossRef]
- Wang, D.; Wang, J.; Zhou, D.; Wu, Z.; Liu, W.; Chen, Y.; Chen, G.; Zhang, J. SWI/SNF complex genomic alterations as a predictive biomarker for response to immune checkpoint inhibitors in multiple cancers. Cancer Immunol. Res. 2023, 11, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.A.; Biswas, S.; Feng, B.; Chen, S.; Gonder, J.; Chakrabarti, S. lncRNA H19 prevents endothelial–mesenchymal transition in diabetic retinopathy. Diabetologia 2019, 62, 517–530. [Google Scholar] [CrossRef]
- Sun, B.; Ding, Y.; Jin, X.; Xu, S.; Zhang, H. Long non-coding RNA H19 promotes corneal neovascularization by targeting microRNA-29c. Biosci. Rep. 2019, 39, BSR20182394. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Lv, R.; Shi, X.e.; Yang, G.; Jin, J. CRISPR/dCas9 tools: Epigenetic mechanism and application in gene transcriptional regulation. Int. J. Mol. Sci. 2023, 24, 14865. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, P.; Kuppe, C. Spatial multi-omics: Novel tools to study the complexity of cardiovascular diseases. Genome Medicine 2024, 16, 14. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kisswani, D.; Carroll, C.; Valdes-Mora, F.; Rutar, M. Epigenetic Alterations in Age-Related Macular Degeneration: Mechanisms and Implications. Int. J. Mol. Sci. 2025, 26, 7601. https://doi.org/10.3390/ijms26157601
Kisswani D, Carroll C, Valdes-Mora F, Rutar M. Epigenetic Alterations in Age-Related Macular Degeneration: Mechanisms and Implications. International Journal of Molecular Sciences. 2025; 26(15):7601. https://doi.org/10.3390/ijms26157601
Chicago/Turabian StyleKisswani, Dana, Christina Carroll, Fatima Valdes-Mora, and Matt Rutar. 2025. "Epigenetic Alterations in Age-Related Macular Degeneration: Mechanisms and Implications" International Journal of Molecular Sciences 26, no. 15: 7601. https://doi.org/10.3390/ijms26157601
APA StyleKisswani, D., Carroll, C., Valdes-Mora, F., & Rutar, M. (2025). Epigenetic Alterations in Age-Related Macular Degeneration: Mechanisms and Implications. International Journal of Molecular Sciences, 26(15), 7601. https://doi.org/10.3390/ijms26157601








