Data Mining and Biochemical Profiling Reveal Novel Biomarker Candidates in Alzheimer’s Disease
Abstract
1. Introduction
2. Results
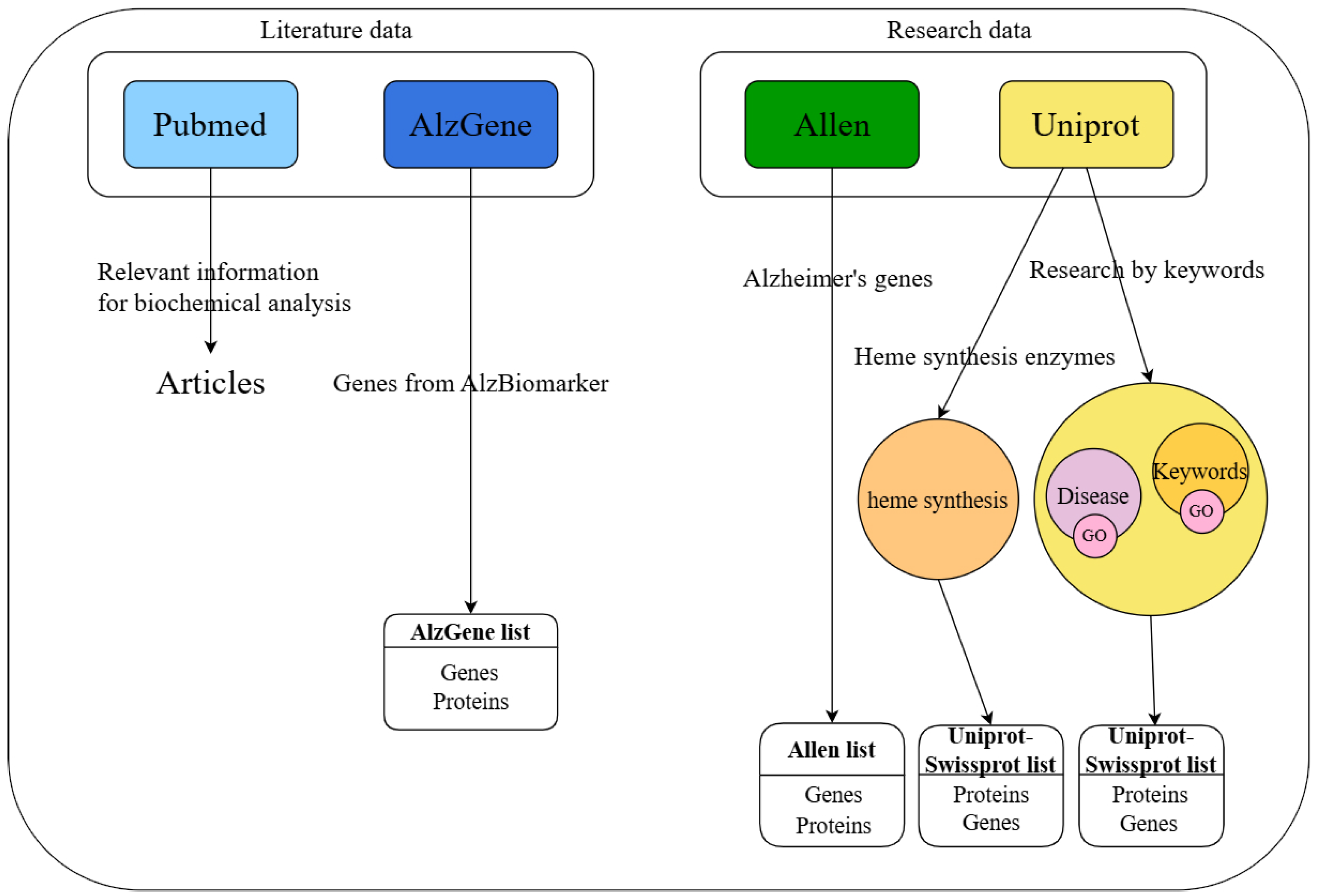
2.1. Dataset Collection
2.1.1. UniProt Investigation Results
- Phosphatidylinositol-binding clathrin assembly protein (PICALM, gene: PICALM) acts as a modulator of the internalization of the transferrin receptor, thus modulating the entry of iron into cells [24];
- Appoptosin, also called mitochondrial glycine transporter (gene: SLC25A38), is important for heme synthesis, as it transports the glycine necessary for the first reaction of heme biosynthesis into the mitochondria [25];
- Humanin (gene: MT-RNR2) is a very small peptide encoded by the mitochondrial genome. It is associated with a longer life, and has antioxidant and mitochondria protective functions [26]. Because its transcription and amino acid content differ from the rest of the nuclear encoded proteins, we did not include this peptide in further analysis.
- Mitoferrin-1 (gene: SLC25A37) is a mitochondrial iron transporter that specifically mediates iron uptake in developing erythroid cells, thereby playing an essential role in heme biosynthesis [27];
- Mitoferrin-2 (gene: SLC25A28) is a mitochondrial iron transporter ubiquitously expressed that mediates iron uptake in all tissues [28];
- Frataxin (gene FXN), is a high-affinity iron-binding partner for ferrochelatase that is capable of both delivering iron to ferrochelatase and mediating the terminal step in mitochondrial heme biosynthesis [29].
- The proteins most sensitive to oxygen, based on the E/Q ratio, were mitoferrin-2 (E/Q = 1062) and frataxin (E/Q = 1429). Because they require a well oxygenated tissue for their synthesis, these proteins are scarcely produced in the hypoxic brain and their downregulation can impair neuronal function, leading to neurodegenerative diseases. Next, to fully investigate the whole heme synthesis pathway, we searched the information available in UniProt about the eight single biosynthetic enzymes: ALA synthase, ALA dehydratase, porphobilinogen synthase, uroporphyrinogen-III synthase, uroporphyrinogen decarboxylase, coproporphyrinogen-III oxidase, protoporphyrinogen oxidase, and ferrochelatase (heme synthase). Their amino acid composition was downloaded from UniProt, and the E/Q ratio was calculated to discover which proteins were most sensitive to oxygen. The E/Q value was always greater than 1 for all enzymes, indicating that they all should be synthesized in an environment that needs to be well oxygenated. However, none of these enzymes were linked to AD; therefore, we excluded these proteins from our final analysis.
2.1.2. AlzGene Investigation Results
2.1.3. AHBA Investigation Results
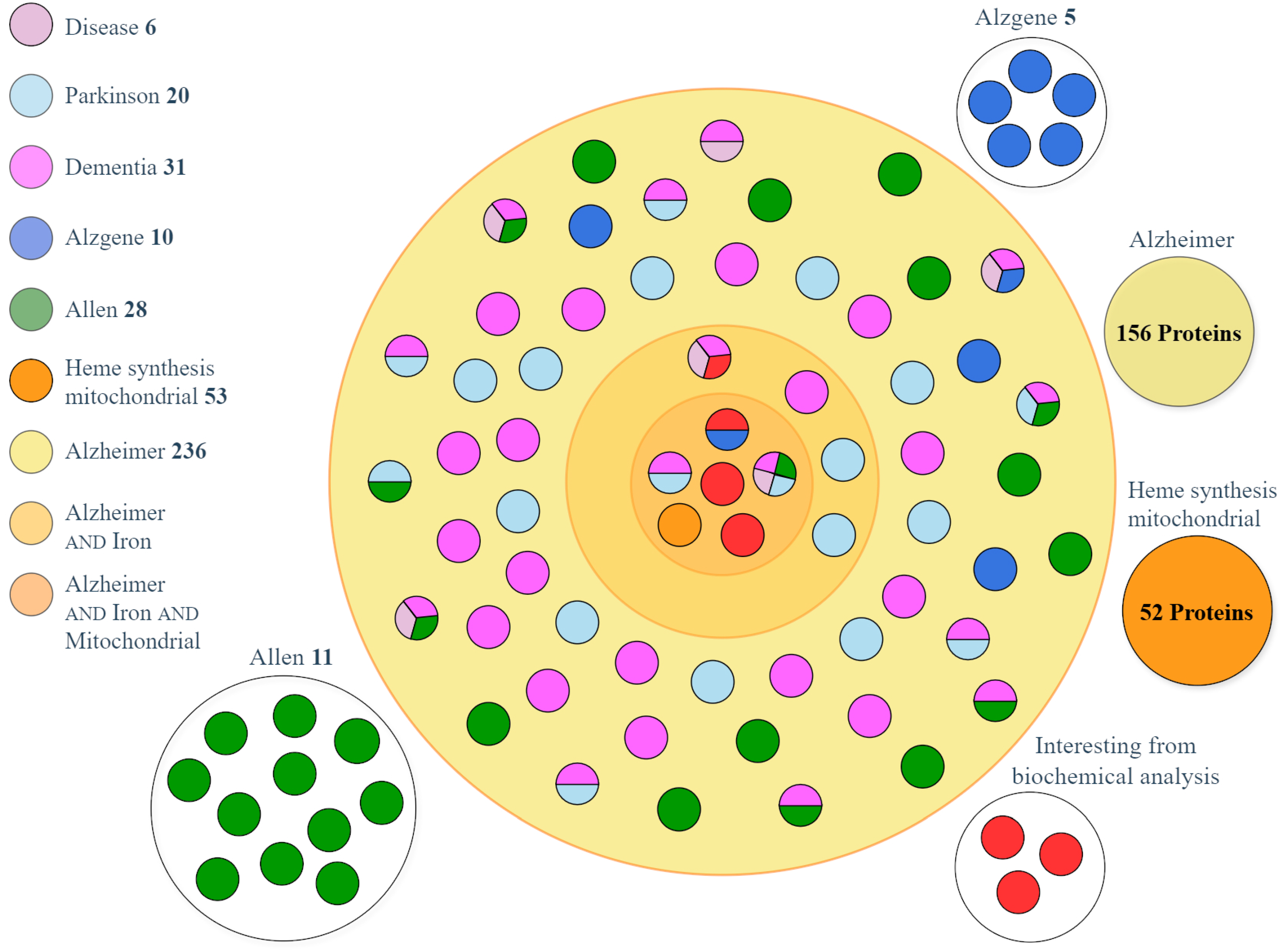
2.2. Dataset Matching and Analysis
- Amyloid-beta precursor protein (APP, gene: APP) is a precursor that, upon proteolytic processing, generates the amyloid-beta (Aβ) peptide, which is the main constituent of amyloid plaques implicated in the etiology of AD [31];
- Alpha-synuclein (gene: SNCA) is a protein involved in the regulation of pre-synaptic function and the release of neurotransmitters, which in the phosphorylated form has a higher propensity for aggregation and formation of neurotoxic fibrils not only in Parkinson’s disease but also in AD [32];
- Prostaglandin G/H synthase 1 (COX-1, gene: PTGS1) is an enzyme that transforms arachidonate to pro-inflammatory prostanoids; its expression is increased in AD, and the nonsteroidal anti-inflammatory drugs (NSAIDs) that inhibit COX slow the progression and delay the onset of AD [33].
3. Discussion
- ApoE (gene: APOE): the APOE4 gene is the strongest genetic risk factor for the development of LOAD [54].
- ATP-binding cassette transporter A7 (gene: ABCA7): a susceptibility factor of LOAD. The reduction in ABCA7 expression or loss of function increases amyloid production [37].
- Disintegrin and metalloproteinase domain-containing protein 10 (gene: ADAM10): transmembrane metalloproteinase responsible for alpha-secretase cleavage of APP. The non-amyloidogenic APP processing by ADAM10 is decreased in AD [39].
- Drebrin (gene: DBN1): is involved in memory-related synaptic plasticity in the hippocampus. AD brains show remarkable reductions in drebrin immunoreactivity [40].
- Ubiquitin carboxyl-terminal hydrolase isozyme L1 (Gene: UCHL1): regulates APP processing by promoting BACE1 degradation, its downregulation increases APP [41].
- NADH-ubiquinone oxidoreductase chain 1 (gene: MT-ND1): core subunit of the mitochondrial membrane respiratory chain Complex I. Mitochondrial gene transcript is altered in AD [42].
- Sortilin-related receptor (gene: SORL1): because it is a sorting receptor for APP, regulating its intracellular trafficking and processing into amyloidogenic-beta peptides, SorL1 deficiency is a genetic predisposition to AD. SorL1 depletion leads to the disturbance of iron homeostasis in the rat hippocampus, mitochondrial oxidative stress, hippocampal degeneration, and impaired spatial memory [43].
- Kallikrein-6 (gene: KLK6): serine protease that degrades alpha-synuclein and prevents its polymerization. The dysregulation of kallikreins has been linked to several neurological disorders, including AD [44].
- RE1-silencing transcription factor (gene: REST): a transcriptional repressor of neuronal genes in neural stem cells. The deletion of REST in mouse models accelerates neurodegeneration and cognitive decline by increasing Aβ deposition and the accumulation of misfolded and phosphorylated tau [45].
- Sequestosome-1 (gene: SQSTM1): a multifunctional scaffolding protein that plays a central role in autophagy. Genetic association studies have reported that it may play an important role in the progression of AD via associations with Aβ levels in cerebrospinal fluid and Aβ deposition in the brain of patients with AD [46].
Limitations
4. Materials and Methods
4.1. Data Sources
4.2. Database Investigation
4.2.1. UniProt Investigation from Disease Section
4.2.2. UniProt Investigation with Keywords
4.2.3. UniProt Investigation by Keywords Relative to Similar Cognitive Pathologies
4.2.4. UniProt Investigation of the Heme Synthesis Pathway
4.2.5. PubMed Investigation
4.2.6. AlzGene Investigation
4.2.7. AHBA Investigation
4.3. Analysis of Amino Acid Content
4.4. Data Matching
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| E/Q | Glutamate/glutamine |
| GO | Gene Ontology |
References
- Xiaopeng, Z.; Jing, Y.; Xia, L.; Xingsheng, W.; Juan, D.; Yan, L.; Baoshan, L. Global Burden of Alzheimer’s Disease and Other Dementias in Adults Aged 65 Years and Older, 1991–2021: Population-Based Study. Front. Public Health 2025, 13, 1585711. [Google Scholar] [CrossRef] [PubMed]
- Ziaunys, M.; Sakalauskas, A.; Mikalauskaite, K.; Smirnovas, V. Polymorphism of Alpha-Synuclein Amyloid Fibrils Depends on Ionic Strength and Protein Concentration. Int. J. Mol. Sci. 2021, 22, 12382. [Google Scholar] [CrossRef] [PubMed]
- Carapeto, A.P.; Marcuello, C.; Faísca, P.F.N.; Rodrigues, M.S. Morphological and Biophysical Study of S100A9 Protein Fibrils by Atomic Force Microscopy Imaging and Nanomechanical Analysis. Biomolecules 2024, 14, 1091. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Ayton, S.; Bush, A.I. The Essential Elements of Alzheimer’s Disease. J. Biol. Chem. 2021, 296, 100105. [Google Scholar] [CrossRef]
- Yang, H.; Yang, M.; Guan, H.; Liu, Z.; Zhao, S.; Takeuchi, S.; Yanagisawa, D.; Tooyama, I. Mitochondrial Ferritin in Neurodegenerative Diseases. Neurosci. Res. 2013, 77, 1–7. [Google Scholar] [CrossRef]
- Lu, Z.; Nie, G.; Li, Y.; Soe-Lin, S.; Tao, Y.; Cao, Y.; Zhang, Z.; Liu, N.; Ponka, P.; Zhao, B. Overexpression of Mitochondrial Ferritin Sensitizes Cells to Oxidative Stress via an Iron-Mediated Mechanism. Antioxid. Redox Signal. 2009, 11, 1791–1803. [Google Scholar] [CrossRef]
- Gordon, N. Friedreich’s Ataxia and Iron Metabolism. Brain Dev. 2000, 22, 465–468. [Google Scholar] [CrossRef]
- Das, N.; Raymick, J.; Sarkar, S. Role of Metals in Alzheimer’s Disease. Metab. Brain Dis. 2021, 36, 1627–1639. [Google Scholar] [CrossRef]
- Yang, A.; Luan, J.; Xu, M.; Du, L.; Lv, K.; Hu, P.; Shu, N.; Yuan, Z.; Shmuel, A.; Ma, G. Regional Brain Iron Correlates with Transcriptional and Cellular Signatures in Alzheimer’s Disease. Alzheimers Dement. J. Alzheimers Assoc. 2025, 21, e14459. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, C.; Pan, R.; Tang, B.; Li, C.; Liu, J.; Tao, W.; Zhang, X.; Yang, T.; Yan, Y.; et al. Cognitive Implications and Associated Transcriptomic Signatures of Distinct Regional Iron Depositions in Cerebral Small Vessel Disease. Alzheimers Dement. J. Alzheimers Assoc. 2025, 21, e70196. [Google Scholar] [CrossRef]
- Xu, J.; Shen, R.; Qian, M.; Zhou, Z.; Xie, B.; Jiang, Y.; Yu, Y.; Dong, W. Ferroptosis in Alzheimer’s Disease: The Regulatory Role of Glial Cells. J. Integr. Neurosci. 2025, 24, 25845. [Google Scholar] [CrossRef] [PubMed]
- Soladogun, A.S.; Zhang, L. The Neural Palette of Heme: Altered Heme Homeostasis Underlies Defective Neurotransmission, Increased Oxidative Stress, and Disease Pathogenesis. Antioxidants 2024, 13, 1441. [Google Scholar] [CrossRef] [PubMed]
- Dey, C.; Pal, P.; Samanta, S.; Ghosh Dey, S. Heme-Aβ Compound 0: A Common Intermediate in ROS Generation and Peroxidase Activity. Dalton Trans. 2025, 54, 7263–7271. [Google Scholar] [CrossRef] [PubMed]
- Novack, G.V.; Galeano, P.; Defelipe, L.A.; Campanelli, L.; Campuzano, K.S.; Rotondaro, C.; Castaño, E.M.; Do Carmo, S.; Cuello, A.C.; García-Alai, M.M.; et al. The Supramolecular Architecture of Mitochondrial Complex I in the Rat Brain Is Altered by Alzheimer’s-Like Cerebral Amyloidosis. J. Neurochem. 2025, 169, e70017. [Google Scholar] [CrossRef]
- Mahapatra, G.; Gao, Z.; Bateman, J.R.; Lockhart, S.N.; Bergstrom, J.; DeWitt, A.R.; Piloso, J.E.; Kramer, P.A.; Gonzalez-Armenta, J.L.; Amick, K.A.; et al. Blood-Based Bioenergetic Profiling Reveals Differences in Mitochondrial Function Associated with Cognitive Performance and Alzheimer’s Disease. Alzheimers Dement. J. Alzheimers Assoc. 2023, 19, 1466–1478. [Google Scholar] [CrossRef]
- Bennett, J.P.; Keeney, P.M. Alzheimer’s and Parkinson’s Brain Tissues Have Reduced Expression of Genes for mtDNA OXPHOS Proteins, Mitobiogenesis Regulator PGC-1α Protein and mtRNA Stabilizing Protein LRPPRC (LRP130). Mitochondrion 2020, 53, 154–157. [Google Scholar] [CrossRef]
- Hartl, D.; Schuldt, V.; Forler, S.; Zabel, C.; Klose, J.; Rohe, M. Presymptomatic Alterations in Energy Metabolism and Oxidative Stress in the APP23 Mouse Model of Alzheimer Disease. J. Proteome Res. 2012, 11, 3295–3304. [Google Scholar] [CrossRef]
- Sharma, V.; Sharma, P.; Singh, T.G. Ferroptosis and Alzheimer’s: Unveiling New Avenues for the Treatment and Prevention. Metab. Brain Dis. 2025, 40, 167. [Google Scholar] [CrossRef]
- Mezzanotte, M.; Stanga, S. Brain Iron Dyshomeostasis and Ferroptosis in Alzheimer’s Disease Pathophysiology: Two Faces of the Same Coin. Aging Dis. 2024, 1–26. [Google Scholar] [CrossRef]
- Chandrasekaran, V.P.; Viswanathan, R.; Venkitasubramanian, T.A. Glutamine Synthetase, Glutaminase and Phosphodiesterase Activities in Brain under Hypoxia: In Vitro Effect of Cortisol, GABA and Serotonin on Glutamine Synthetase. Environ. Physiol. Biochem. 1975, 5, 373–377. [Google Scholar]
- Vernone, A.; Ricca, C.; Merlo, D.; Pescarmona, G.; Silvagno, F. The Analysis of Glutamate and Glutamine Frequencies in Human Proteins as Marker of Tissue Oxygenation. R. Soc. Open Sci. 2019, 6, 181891. [Google Scholar] [CrossRef]
- Bertram, L.; McQueen, M.B.; Mullin, K.; Blacker, D.; Tanzi, R.E. Systematic Meta-Analyses of Alzheimer Disease Genetic Association Studies: The AlzGene Database. Nat. Genet. 2007, 39, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Sunkin, S.M.; Ng, L.; Lau, C.; Dolbeare, T.; Gilbert, T.L.; Thompson, C.L.; Hawrylycz, M.; Dang, C. Allen Brain Atlas: An Integrated Spatio-Temporal Portal for Exploring the Central Nervous System. Nucleic Acids Res. 2013, 41, D996–D1008. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Tan, L.; Yu, J.-T. The Role of PICALM in Alzheimer’s Disease. Mol. Neurobiol. 2015, 52, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Y.; Chen, Y.; Huang, X.; Zhou, F.; Wang, W.; Xian, B.; Zhang, X.; Masliah, E.; Chen, Q.; et al. Appoptosin Is a Novel Pro-Apoptotic Protein and Mediates Cell Death in Neurodegeneration. J. Neurosci. Off. J. Soc. Neurosci. 2012, 32, 15565–15576. [Google Scholar] [CrossRef]
- Alqahtani, S.M.; Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Alexiou, A.; Fawzy, M.N.; Papadakis, M.; Al-Botaty, B.M.; Alruwaili, M.; Batiha, G.E.-S. The Neuroprotective Role of Humanin in Alzheimer’s Disease: The Molecular Effects. Eur. J. Pharmacol. 2025, 998, 177510. [Google Scholar] [CrossRef]
- Huang, J.; Chen, S.; Hu, L.; Niu, H.; Sun, Q.; Li, W.; Tan, G.; Li, J.; Jin, L.; Lyu, J.; et al. Mitoferrin-1 Is Involved in the Progression of Alzheimer’s Disease Through Targeting Mitochondrial Iron Metabolism in a Caenorhabditis Elegans Model of Alzheimer’s Disease. Neuroscience 2018, 385, 90–101. [Google Scholar] [CrossRef]
- LeVine, S.M.; Tsau, S.; Gunewardena, S. Exploring Whether Iron Sequestration within the CNS of Patients with Alzheimer’s Disease Causes a Functional Iron Deficiency That Advances Neurodegeneration. Brain Sci. 2023, 13, 511. [Google Scholar] [CrossRef]
- Agrawal, S.; Fox, J.; Thyagarajan, B.; Fox, J.H. Brain Mitochondrial Iron Accumulates in Huntington’s Disease, Mediates Mitochondrial Dysfunction, and Can Be Removed Pharmacologically. Free Radic. Biol. Med. 2018, 120, 317–329. [Google Scholar] [CrossRef]
- Hawrylycz, M.J.; Lein, E.S.; Guillozet-Bongaarts, A.L.; Shen, E.H.; Ng, L.; Miller, J.A.; van de Lagemaat, L.N.; Smith, K.A.; Ebbert, A.; Riley, Z.L.; et al. An Anatomically Comprehensive Atlas of the Adult Human Brain Transcriptome. Nature 2012, 489, 391–399. [Google Scholar] [CrossRef]
- Zhou, Z.D.; Chan, C.H.S.; Ma, Q.H.; Xu, X.H.; Xiao, Z.C.; Tan, E.K. The Roles of Amyloid Precursor Protein (APP) in Neurogenesis: Implications to Pathogenesis and Therapy of Alzheimer Disease. Cell Adhes. Migr. 2011, 5, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Al-Kuraishy, H.M.; Sulaiman, G.M.; Mohammed, H.A.; Al-Gareeb, A.I.; Albuhadily, A.K.; Ali, A.A.; Abu-Alghayth, M.H. Beyond Amyloid Plaque, Targeting α-Synuclein in Alzheimer Disease: The Battle Continues. Ageing Res. Rev. 2025, 105, 102684. [Google Scholar] [CrossRef] [PubMed]
- Yermakova, A.V.; Rollins, J.; Callahan, L.M.; Rogers, J.; O’Banion, M.K. Cyclooxygenase-1 in Human Alzheimer and Control Brain: Quantitative Analysis of Expression by Microglia and CA3 Hippocampal Neurons. J. Neuropathol. Exp. Neurol. 1999, 58, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Ossenkoppele, R.; van der Kant, R.; Hansson, O. Tau Biomarkers in Alzheimer’s Disease: Towards Implementation in Clinical Practice and Trials. Lancet Neurol. 2022, 21, 726–734. [Google Scholar] [CrossRef]
- Timalsina, D.R.; Abichandani, L.; Ambad, R. Role of Oxidative Stress Markers F2-Isoprostanes and Presenilin-1 in Clinical Diagnosed Alzheimer’s Diseases. J. Pharm. Bioallied Sci. 2025, 17, S384–S387. [Google Scholar] [CrossRef]
- Wolfe, M.S. Presenilin, γ-Secretase, and the Search for Pathogenic Triggers of Alzheimer’s Disease. Biochemistry 2025, 64, 1662–1672. [Google Scholar] [CrossRef]
- Satoh, K.; Abe-Dohmae, S.; Yokoyama, S.; St George-Hyslop, P.; Fraser, P.E. ATP-Binding Cassette Transporter A7 (ABCA7) Loss of Function Alters Alzheimer Amyloid Processing. J. Biol. Chem. 2015, 290, 24152–24165. [Google Scholar] [CrossRef]
- Swerdlow, R.H. The Alzheimer’s Disease Mitochondrial Cascade Hypothesis: A Current Overview. J. Alzheimers Dis. JAD 2023, 92, 751–768. [Google Scholar] [CrossRef]
- Manzine, P.R.; de França Bram, J.M.; Barham, E.J.; do Vale, F.d.A.C.; Selistre-de-Araújo, H.S.; Cominetti, M.R.; Iost Pavarini, S.C. ADAM10 as a Biomarker for Alzheimer’s Disease: A Study with Brazilian Elderly. Dement. Geriatr. Cogn. Disord. 2013, 35, 58–66. [Google Scholar] [CrossRef]
- Harigaya, Y.; Shoji, M.; Shirao, T.; Hirai, S. Disappearance of Actin-Binding Protein, Drebrin, from Hippocampal Synapses in Alzheimer’s Disease. J. Neurosci. Res. 1996, 43, 87–92. [Google Scholar] [CrossRef]
- Gao, Z.-X.; Zhang, C.; Lu, J.-C.; Zhao, X.; Qiu, H.; Wang, H.-J. Pathological Methamphetamine Exposure Triggers the Accumulation of Neuropathic Protein Amyloid-β by Inhibiting UCHL1. Neurotoxicology 2021, 86, 19–25. [Google Scholar] [CrossRef]
- Lunnon, K.; Keohane, A.; Pidsley, R.; Newhouse, S.; Riddoch-Contreras, J.; Thubron, E.B.; Devall, M.; Soininen, H.; Kłoszewska, I.; Mecocci, P.; et al. Mitochondrial Genes Are Altered in Blood Early in Alzheimer’s Disease. Neurobiol. Aging 2017, 53, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, Y.; Cai, Y.; Aobulikasimu, A.; Wang, Y.; Hu, C.; Miao, Z.; Shao, Y.; Zhao, M.; Hu, Y.; et al. Endo-Lysosomal Network Disorder Reprograms Energy Metabolism in SorL1-Null Rat Hippocampus. Adv. Sci. Weinh. Baden-Wurtt. Ger. 2024, 11, e2407709. [Google Scholar] [CrossRef] [PubMed]
- Boumali, R.; Urli, L.; Naim, M.; Soualmia, F.; Kinugawa, K.; Petropoulos, I.; El Amri, C. Kallikrein-Related Peptidase’s Significance in Alzheimer’s Disease Pathogenesis: A Comprehensive Survey. Biochimie 2024, 226, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Aron, L.; Qiu, C.; Ngian, Z.K.; Liang, M.; Drake, D.; Choi, J.; Fernandez, M.A.; Roche, P.; Bunting, E.L.; Lacey, E.K.; et al. A Neurodegeneration Checkpoint Mediated by REST Protects against the Onset of Alzheimer’s Disease. Nat. Commun. 2023, 14, 7030. [Google Scholar] [CrossRef]
- Dong, W.; Cui, M.-C.; Hu, W.-Z.; Zeng, Q.; Wang, Y.-L.; Zhang, W.; Huang, Y. Genetic and Molecular Evaluation of SQSTM1/P62 on the Neuropathologies of Alzheimer’s Disease. Front. Aging Neurosci. 2022, 14, 829232. [Google Scholar] [CrossRef]
- Liu, F.-J.; Hua, X.-F.; Wang, W.-J. A New Bioinformatics Insight into Human Cancer-Associated Proteins. Oncol. Rep. 2012, 27, 1932–1936. [Google Scholar] [CrossRef]
- Turewicz, M.; Frericks-Zipper, A.; Stepath, M.; Schork, K.; Ramesh, S.; Marcus, K.; Eisenacher, M. BIONDA: A Free Database for a Fast Information on Published Biomarkers. Bioinforma. Adv. 2021, 1, vbab015. [Google Scholar] [CrossRef]
- Pospisil, P.; Iyer, L.K.; Adelstein, S.J.; Kassis, A.I. A Combined Approach to Data Mining of Textual and Structured Data to Identify Cancer-Related Targets. BMC Bioinform. 2006, 7, 354. [Google Scholar] [CrossRef]
- Cavedo, E.; Lista, S.; Khachaturian, Z.; Aisen, P.; Amouyel, P.; Herholz, K.; Jack, C.R.; Sperling, R.; Cummings, J.; Blennow, K.; et al. The Road Ahead to Cure Alzheimer’s Disease: Development of Biological Markers and Neuroimaging Methods for Prevention Trials Across All Stages and Target Populations. J. Prev. Alzheimers Dis. 2014, 1, 181–202. [Google Scholar] [CrossRef]
- Abdukarimov, N.; Kokabi, K.; Kunz, J. Ferroptosis and Iron Homeostasis: Molecular Mechanisms and Neurodegenerative Disease Implications. Antioxidants 2025, 14, 527. [Google Scholar] [CrossRef]
- Ayton, S.; Wang, Y.; Diouf, I.; Schneider, J.A.; Brockman, J.; Morris, M.C.; Bush, A.I. Brain Iron is Associated with Accelerated Cognitive Decline in People with Alzheimer Pathology. Mol. Psychiatry 2020, 25, 2932–2941. [Google Scholar] [CrossRef] [PubMed]
- Ayton, S.; Portbury, S.; Kalinowski, P.; Agarwal, P.; Diouf, I.; Schneider, J.A.; Morris, M.C.; Bush, A.I. Regional Brain Iron Associated with Deterioration in Alzheimer’s Disease: A Large Cohort Study and Theoretical Significance. Alzheimers Dement. J. Alzheimers Assoc. 2021, 17, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Finkelstein, D.I.; Adlard, P.A. Interactions of Metals and Apolipoprotein E in Alzheimer’s Disease. Front. Aging Neurosci. 2014, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Duchateau, L.; Wawrzyniak, N.; Sleegers, K. The ABC’s of Alzheimer Risk Gene ABCA7. Alzheimers Dement. 2024, 20, 3629–3648. [Google Scholar] [CrossRef]
- Choi, J.; Levey, A.I.; Weintraub, S.T.; Rees, H.D.; Gearing, M.; Chin, L.-S.; Li, L. Oxidative Modifications and Down-Regulation of Ubiquitin Carboxyl-Terminal Hydrolase L1 Associated with Idiopathic Parkinson’s and Alzheimer’s Diseases. J. Biol. Chem. 2004, 279, 13256–13264. [Google Scholar] [CrossRef]
- Thonberg, H.; Chiang, H.-H.; Lilius, L.; Forsell, C.; Lindström, A.-K.; Johansson, C.; Björkström, J.; Thordardottir, S.; Sleegers, K.; Van Broeckhoven, C.; et al. Identification and Description of Three Families with Familial Alzheimer Disease That Segregate Variants in the SORL1 Gene. Acta Neuropathol. Commun. 2017, 5, 43. [Google Scholar] [CrossRef]
- Lu, T.; Aron, L.; Zullo, J.; Pan, Y.; Kim, H.; Chen, Y.; Yang, T.-H.; Kim, H.-M.; Drake, D.; Liu, X.S.; et al. REST and Stress Resistance in Ageing and Alzheimer’s Disease. Nature 2014, 507, 448–454. [Google Scholar] [CrossRef]
- Ficiarà, E.; Stura, I.; Guiot, C. Iron Deposition in Brain: Does Aging Matter? Int. J. Mol. Sci. 2022, 23, 10018. [Google Scholar] [CrossRef]
- Ficiarà, E.; Stura, I.; Vernone, A.; Silvagno, F.; Cavalli, R.; Guiot, C. Iron Overload in Brain: Transport Mismatches, Microbleeding Events, and How Nanochelating Therapies May Counteract Their Effects. Int. J. Mol. Sci. 2024, 25, 2337. [Google Scholar] [CrossRef]
- Shen, E.H.; Overly, C.C.; Jones, A.R. The Allen Human Brain Atlas: Comprehensive Gene Expression Mapping of the Human Brain. Trends Neurosci. 2012, 35, 711–714. [Google Scholar] [CrossRef]
- Vernone, A.; Ricca, C.; Pescarmona, G.; Silvagno, F. Chromosome Walking: A Novel Approach to Analyse Amino Acid Content of Human Proteins Ordered by Gene Position. Appl. Sci. 2021, 11, 3511. [Google Scholar] [CrossRef]
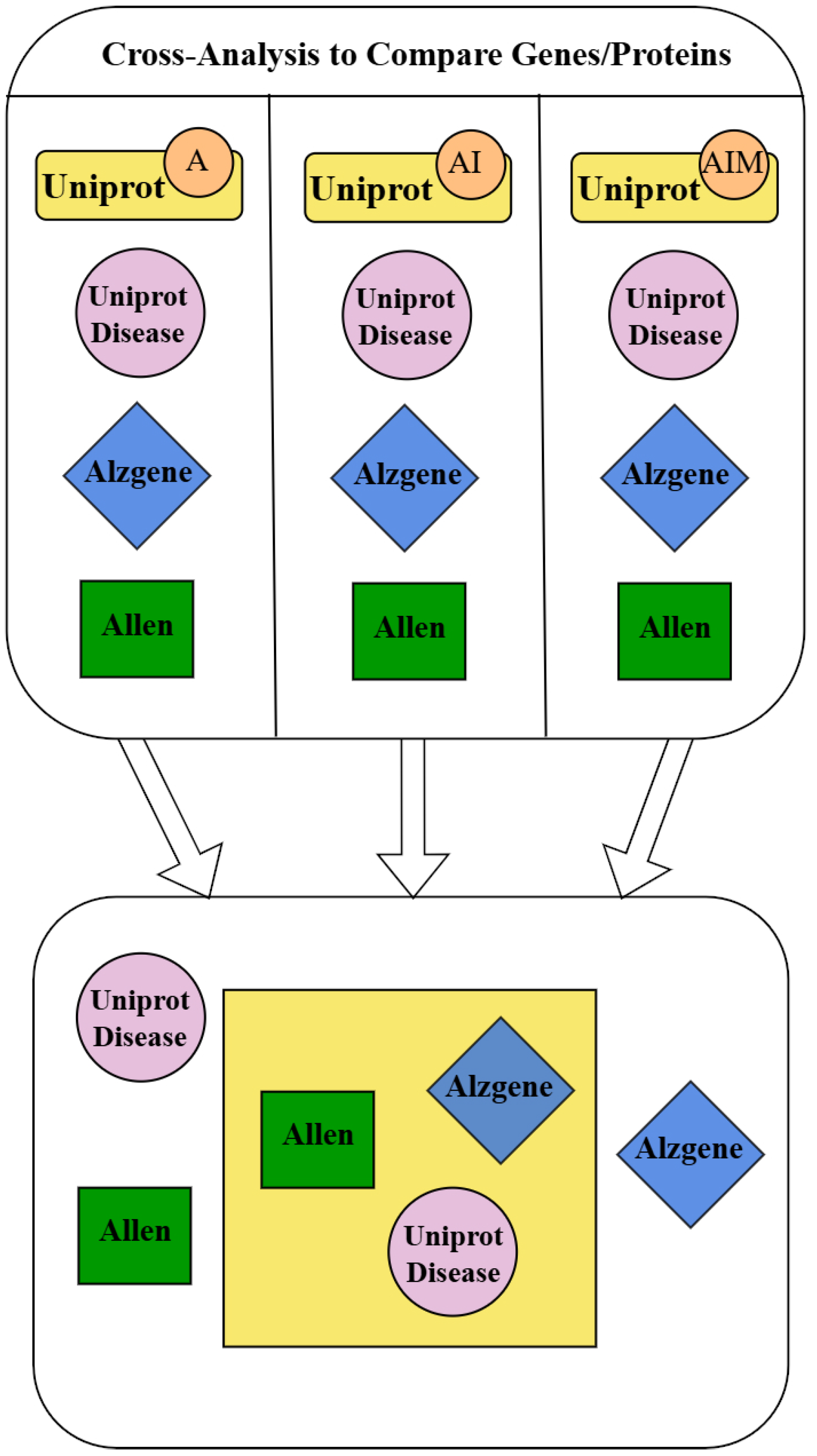
| Overlapping Identification | AIM 1 | AI 2 | A 3 |
|---|---|---|---|
| 5 datasets | APP | 0 | 0 |
| 4 datasets | 0 | APOE | MAPT |
| PSEN1 | |||
| PSEN2 | |||
| ABCA7 | |||
| 3 datasets | PICALM | 0 | ADAM10 |
| SNCA | DBN1 | ||
| UCHL1 | |||
| MT-ND1 | |||
| SORL1 | |||
| KLK6 | |||
| REST | |||
| SQSTM1 |
| Gene Name (Primary) | Entry Name 1 | Protein Name | E/Q | Dataset 2 | GO 3 Term | Role in AD |
|---|---|---|---|---|---|---|
| PICALM | PICAL_HUMAN | PICALM | 0.862 | 3 | I | [24] |
| SLC25A38 | S2538_HUMAN | Appoptosin | 0.571 | 2 | M | [25] |
| APP | A4_HUMAN | APP | 2.556 | 5 | A | [31] |
| SNCA | SYUA_HUMAN | Alpha-synuclein | 3.000 | 3 | I,M | [32] |
| PTGS1 | PGH1_HUMAN | COX-1 | 1.333 | 2 | - | [33] |
| MAPT | TAU_HUMAN | MAPT | 1.788 | 4 | M | [34] |
| PSEN1 | PSN1_HUMAN | Presenilin-1 | 1.684 | 4 | M | [35] |
| PSEN2 | PSN2_HUMAN | Presenilin-2 | 3.000 | 4 | M | [36] |
| ABCA7 | ABCA7_HUMAN | ABCA7 | 1.384 | 3 | - | [37] |
| APOE | APOE_HUMAN | Apolipoprotein E | 1.250 | 4 | - | [38] |
| ADAM10 | ADA10_HUMAN | ADAM10 | 1.294 | 3 | - | [39] |
| DBN1 | DREB_HUMAN | Drebrin | 2.676 | 3 | - | [40] |
| UCHL1 | UCHL1_HUMAN | UCHL1 | 1.833 | 3 | - | [41] |
| MT-ND1 | NU1M_HUMAN | MT-ND1 | 1.833 | 3 | M | [42] |
| SORL1 | SORL_HUMAN | SorL1 | 1.595 | 3 | - | [43] |
| KLK6 | KLK6_HUMAN | Kallikrein-6 | 0.786 | 3 | - | [44] |
| REST | REST_HUMAN | REST | 2.125 | 3 | - | [45] |
| SQSTM1 | SQSTM_HUMAN | Sequestosome-1 | 3.417 | 3 | M | [46] |
| SLC25A37 | MFRN1_HUMAN | Mitoferrin-1 | 0.765 | 1 | - | [27] |
| SLC25A28 | MFRN2_HUMAN | Mitoferrin-2 | 1.063 | 1 | - | [28] |
| FXN | FRDA_HUMAN | Frataxin | 1.429 | 1 | - | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vernone, A.; Stura, I.; Guiot, C.; D’Agata, F.; Silvagno, F. Data Mining and Biochemical Profiling Reveal Novel Biomarker Candidates in Alzheimer’s Disease. Int. J. Mol. Sci. 2025, 26, 7536. https://doi.org/10.3390/ijms26157536
Vernone A, Stura I, Guiot C, D’Agata F, Silvagno F. Data Mining and Biochemical Profiling Reveal Novel Biomarker Candidates in Alzheimer’s Disease. International Journal of Molecular Sciences. 2025; 26(15):7536. https://doi.org/10.3390/ijms26157536
Chicago/Turabian StyleVernone, Annamaria, Ilaria Stura, Caterina Guiot, Federico D’Agata, and Francesca Silvagno. 2025. "Data Mining and Biochemical Profiling Reveal Novel Biomarker Candidates in Alzheimer’s Disease" International Journal of Molecular Sciences 26, no. 15: 7536. https://doi.org/10.3390/ijms26157536
APA StyleVernone, A., Stura, I., Guiot, C., D’Agata, F., & Silvagno, F. (2025). Data Mining and Biochemical Profiling Reveal Novel Biomarker Candidates in Alzheimer’s Disease. International Journal of Molecular Sciences, 26(15), 7536. https://doi.org/10.3390/ijms26157536







