CRISPR-Cas Gene Editing Technology in Potato
Abstract
1. Introduction
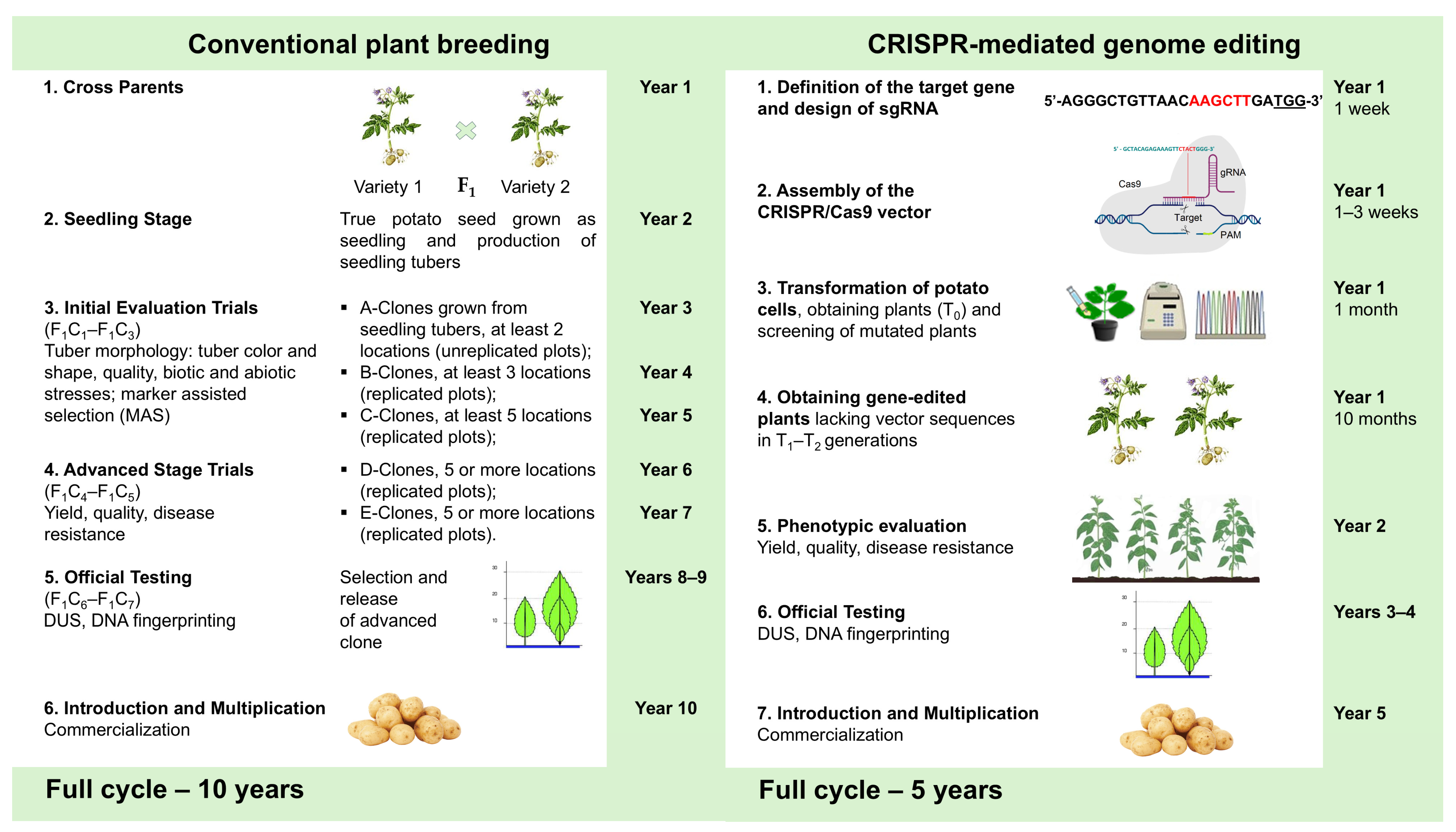
2. Using CRISPR-Cas Gene Editing Technology to Improve Potato Breeding Process
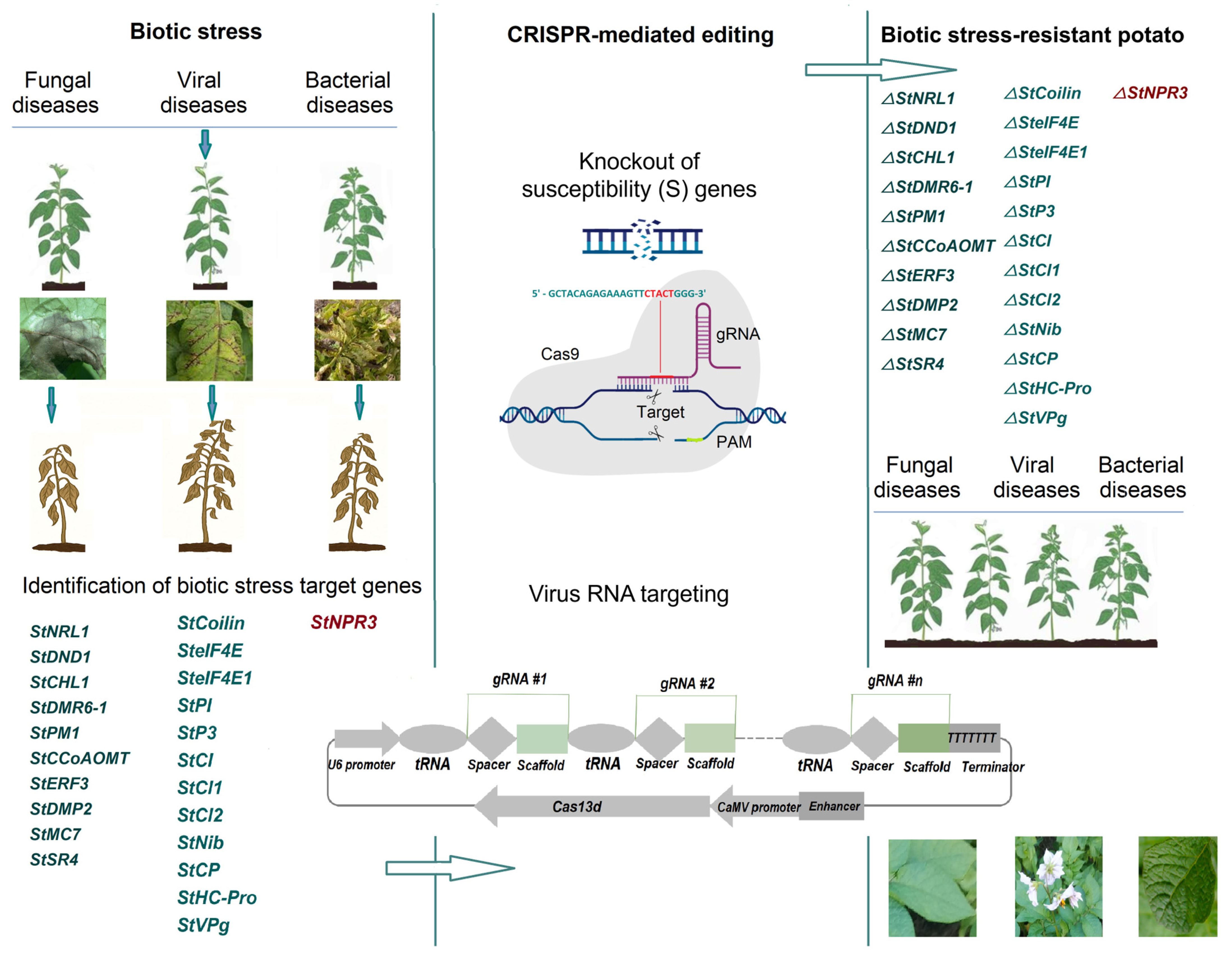
3. Using CRISPR-Cas Gene Editing Technology to Increase Disease Resistance in Potato
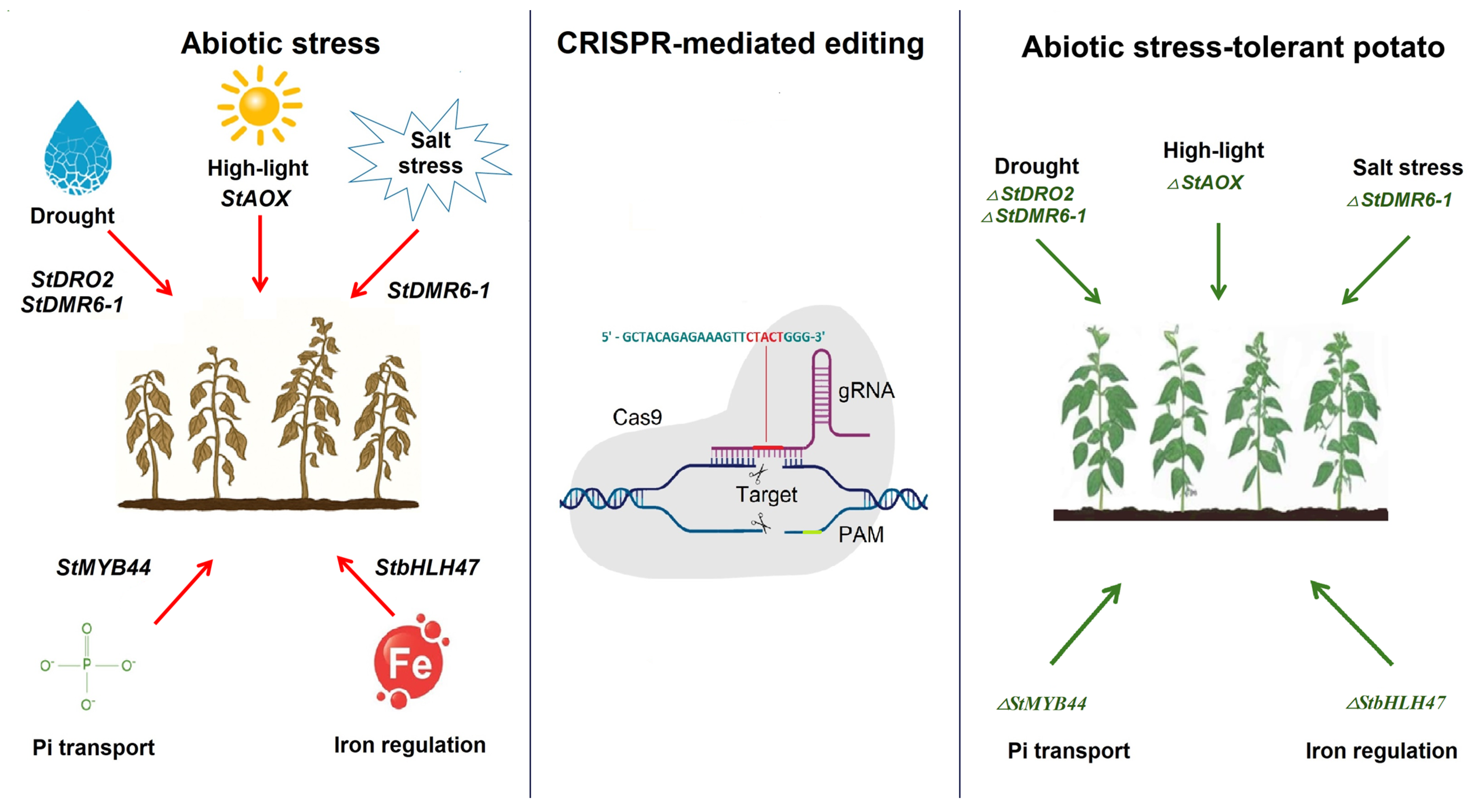
4. Using CRISPR-Cas Gene Editing Technology to Increase Abiotic Tolerance in Potato
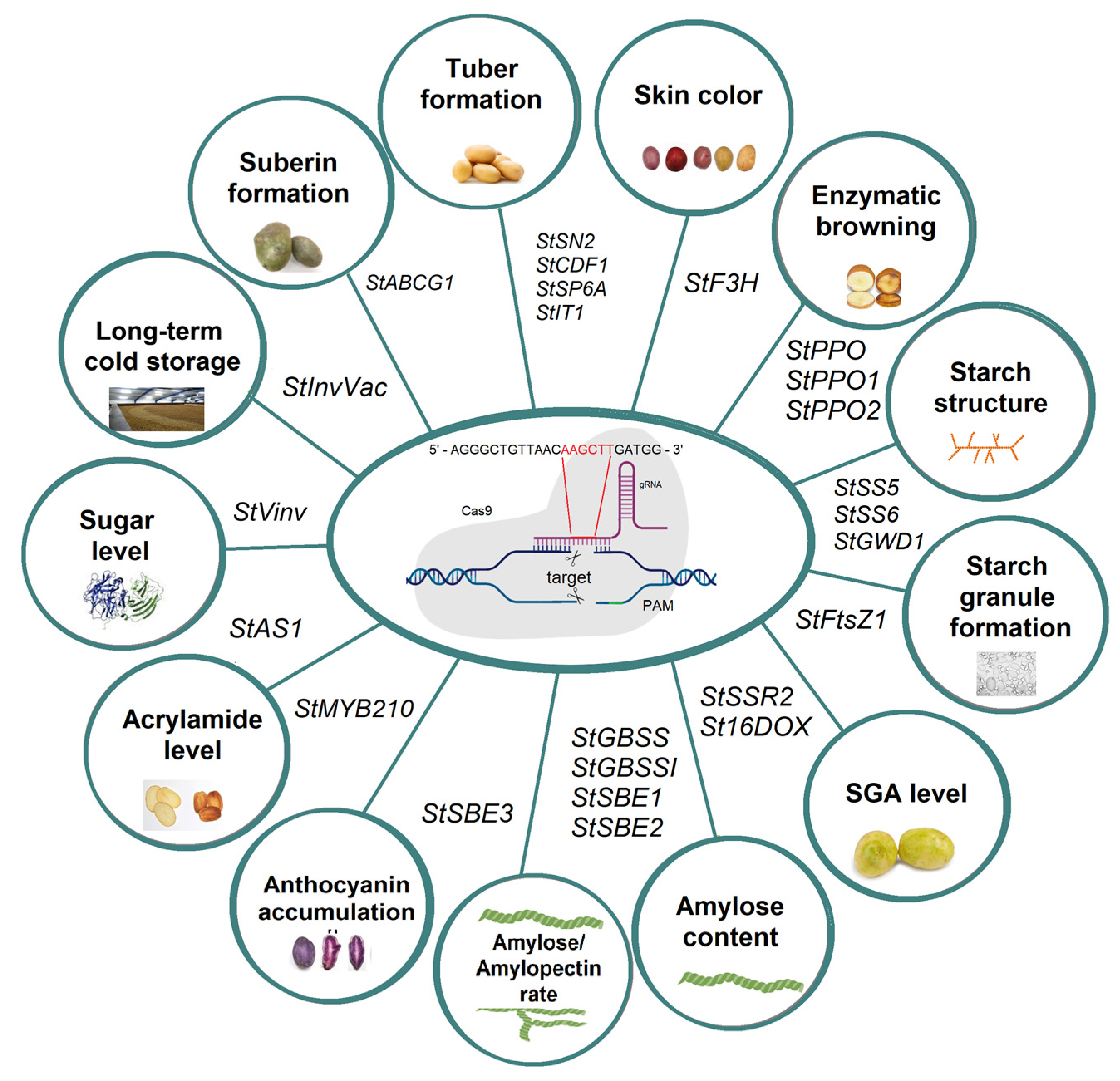
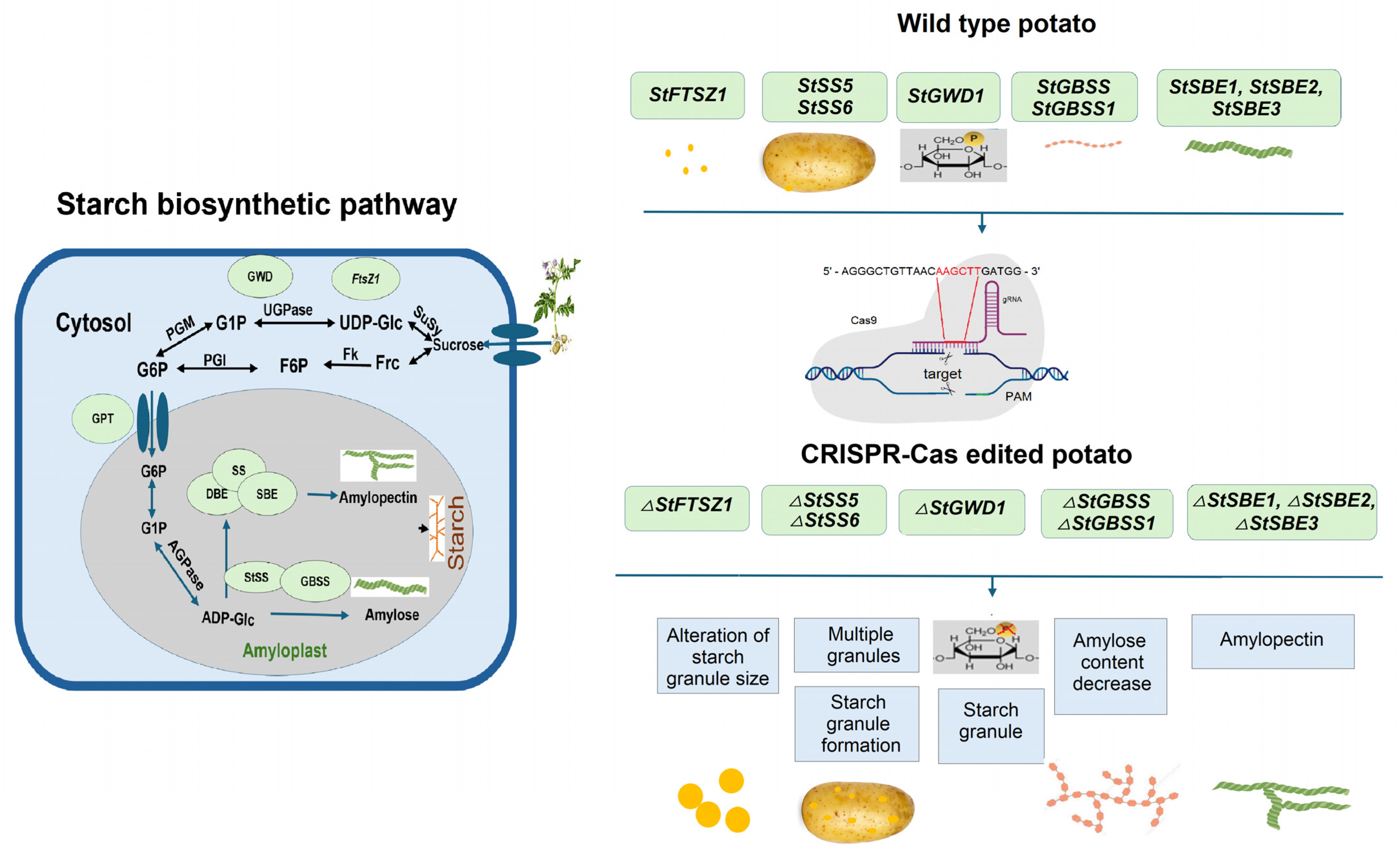
5. Using CRISPR-Cas Gene Editing Technology for Potato Quality Improvement
| Target Gene | Delivery Method | Type of Editing | Improvement | References |
|---|---|---|---|---|
| StSN2 | Agrobacterium- mediated transformation | Knockout | Tuber formation | [95] |
| StCDF1 promoter | Agrobacterium- mediated transformation | Knockout | Potato tuberization | [96] |
| StSP6A | Agrobacterium- mediated transformation | Knockout | Induction and formation of potato tubers | [97] |
| StIT1 | Agrobacterium- mediated transformation | Knockout | Tuber initiation | [99] |
| StF3H | Agrobacterium- mediated transformation | Knockout | Skin color change | [98] |
| StPPO | Agrobacterium- mediated transformation | Knockout | Reduced enzymatic browning | [104] |
| StPPO1, StPPO2 | Agrobactierum- and geminivirus-based transformation | Knockout | Reduced enzymatic browning | [41] |
| StPPO2 | Protoplast transfection with RNPs | Knockout | Reduced enzymatic browning | [101,102] |
| StFtsZ1 | Agrobacterium- mediated transformation | Knockout | Increase in starch granule size | [109] |
| StSS5 | Agrobacterium- mediated transformation | Knockout | Starch granule formation and tuber development | [110] |
| StSS6 | Agrobacterium- mediated transformation | Knockout | Starch structure | [111] |
| StGWD1 | Agrobacterium- mediated transformation | Knockout | Amylose content | [113] |
| StSSR2 | Agrobacterium- mediated transformation | Knockout | Decreasing level of SGA | [30] |
| St16DOX | Agrobacterium- mediated transformation | Knockout | Decreasing level of SGA | [114] |
| StGBSS | Agrobacterium- mediated transformation | Knockout | Amylose content decrease | [115] |
| StGBSS | PEG-mediated protoplast transfection | Knockout | Amylose content decrease | [36,116] |
| StGBSS | PEG-mediated protoplast transfection | Knockout | Amylose content decrease | [35] |
| StGBSSI | Agrobacterium- mediated transformation | Knockout | Amylose content decrease | [117] |
| StGBSSI | Agrobacterium- mediated transformation | Knockout | Amylose content decrease | [118] |
| StGBSSI | Agrobacterium- mediated transformation | Base edit | Amylose content decrease | [119] |
| StSBE1, StSBE2 | Agrobacterium- mediated transformation | Knockout | Amylose content decrease | [120] |
| StSBE1, StSBE2 | Agrobacterium-mediated transformation and by PEG-mediated delivery into protoplasts | Knockout | Decrease in branching frequency of amylopectin | [121,122] |
| StSBE3 | Agrobacterium- mediated transformation | Knockout | Amylose/amylopectin content decrease | [123] |
| StMYB210 | Agrobacterium- mediated transformation | Knockout | Regulation of anthocyanin accumulation in tuber flesh | [125] |
| StVinv, StAS1 | RNP-particle bombardment and Agrobacterium- mediated transformation | Knockout | Reduced browning and acrylamide | [38] |
| StVinv | Agrobacterium- mediated transformation | Knockout | Optimize level of sugars | [29] |
| StVinv | Agrobacterium- mediated transformation | Knockout | Optimize level of sugars | [130] |
| StVinv | Agrobacterium- mediated transformation | Knockout | Root development | [129] |
| StInvVac | Protoplast transfection | Knockout | Long-term cold storage and bruising resistance | [126] |
| StABCG1 | Agrobacterium- mediated transformation | Knockout | Reduced suberin formation | [131] |
6. Challenges and Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRISPR | clustered regularly interspaced short palindromic repeats |
| Cas | CRISPR-associated protein |
| MAS | marker-assisted selection |
| GE | gene editing |
| ZFN | zinc finger nuclease |
| TALEN | transcription activator-like effector nuclease |
| (s)gRNA | (Single) Guide RNA (Ribonucleic Acid) |
| CRISPRa | CRISPR-mediated transcriptional activation |
| CRISPRi | CRISPR interference |
| BE | base editing |
| PE | prime editing |
| DSB | double-strand break |
| HR | homologous recombination |
| NHEJ | non-homologous end joining |
| HDR | homology-directed repair |
| PEG-mediated | polyethylene glycol-mediated |
| RNAi | RNA interference |
| StNRL1 | Solanum tuberosum NPH3/RPT2-like1 |
| MLO | Mildew Locus O |
| SWEET | Sugars Will Eventually be Exported Transporter |
| StDMR6 | S. tuberosum downy mildew resistance 6 |
| StbHLH47 | S. tuberosum basic helix–loop–helix 47 |
| StDRO2 | S. tuberosum deeper rooting 2 |
| RSA | root system architecture |
| StLike3 | S. tuberosum FMO GS-OX-Like3 |
| StDND1 | S. tuberosum defense no death 1 |
| StCHL1 | S. tuberosum CIB1/HBI1-like1 |
| StPM1 | S. tuberosum PLASMA MEMBRANE PROTEIN 1 |
| AWPM-19 | ABA-induced wheat plasma membrane polypeptide-19 |
| StCCoAOMT | S. tuberosum Caffeoyl-CoA O-methyltransferase |
| StERFs | S. tuberosum ethylene response factors |
| StDMP2 | S. tuberosum DOMAIN OF UNKNOWN FUNCTION 679 membrane protein 2 |
| ER | endoplasmic reticulum |
| StPRRs | S. tuberosum pattern recognition receptors |
| StNDR1 | S. tuberosum non-race-specific disease resistance 1 |
| MC | metacaspase |
| PCD | programmed cell death |
| StMC7 | S. tuberosum metacaspase 7 |
| SR1 | Signal Response 1 |
| StSR4 | S. tuberosum Signal Responsive 4 |
| CAMTA3 | calmodulin (CaM)-binding transcription activator 3 |
| Clso | Candidatus Liberibacter solanacearum |
| StNPR(3) | S. tuberosum non-expressor of pathogenesis-related (3) |
| PVY | potato virus Y |
| SteIF | S. tuberosum eukaryotic translation initiation factor |
| nCBP | New Cap-Binding Protein |
| Cas13a | CRISPR-associated protein “a” |
| NIb | Nuclear Inclusion protein b |
| CP | Coat Protein |
| Cas13d | CRISPR-associated protein “d” |
| PTG | Polycistronic tRNA-gRNA |
| LshCas13a | Leptotrichia shahii Cas13a |
| EPSPS | 5-enolpyruvylshikimate-3-phosphate synthase |
| GVR | geminivirus replicon |
| StALS | S. tuberosum Acetolactate Synthase |
| PVS | potato virus S |
| PVX | potato virus X |
| PLRV | potato leafroll virus |
| StAOX | S. tuberosum alternative oxidase |
| ROS | reactive oxygen species |
| Pi | phosphate |
| RNA-Seq | RNA sequencing |
| StMYB44 | S. tuberosum MYB 44 transcription factor in S. tuberosum |
| StVinv, StInvVac | S. tuberosum vacuolar invertase |
| StSN2 | S. tuberosum Snakin-2 |
| ABA | abscisic acid |
| STP1 | Sugar Transporter Protein 1 |
| StCDF1 | S. tuberosum cycling dof factor 1 |
| StSP6A | S. tuberosum self-pruning 6A |
| StPHO1 | S. tuberosum phosphate transporter 1 |
| S-RNAse | S-locus RNAse |
| SLF | S-locus F-box |
| Sli | S-locus inhibitor |
| SaCas9 | Staphylococcus aureus CRISPR-Cas9 |
| GBSS | granule-bound starch synthase |
| StGBSS | S. tuberosum granule-bound starch synthase |
| StGBSS(I) | S. tuberosum granule-bound starch synthase (I) |
| INVINH1 | invertase inhibitor 1 |
| StPDS | S. tuberosum phytoendesaturase |
| StER | S. tuberosum Erecta |
| B33 | also named Patatin class I promoter |
| Suc, SUT | sucrose transporter |
| AmA1 | Amaranth Albumin 1 |
| StF3H | S. tuberosum Flavanone 3-hydroxylase |
| StPPO | S. tuberosum Polyphenol Oxidase |
| pGEF(-U) | Fluorescent Editor Geminivirus-Based Plasmid (Universal) |
| GTPase | Guanosine Triphosphate hydrolase |
| StFtsZ1 | S. tuberosum Filamentous temperature sensitive 1 |
| SNP | Single Nucleotide Polymorphism |
| SGAs | steroidal glycoalkaloids |
| StSSR2 | S. tuberosum sterol side chain reductase 2 |
| 2OGD | 2-oxoglutarate-dependent dioxygenase |
| St16DOX | S. tuberosum 16-hydroxylase |
| StSS5 | S. tuberosum starch synthease 5 |
| StSS6 | S. tuberosum starch synthease 6 |
| StGWD1 | S. tuberosum α-glucan water dikinase 1 |
| StMYB210 | S. tuberosum R2R3 MYB transcription factor genes |
| pre-tRNA | precursor of transfer RNA |
| CBE | cytidine base editor |
| StSBE | S. tuberosum Starch Branching Enzyme |
| RNP | ribonucleoprotein |
| CL | chain length |
| StIAA | S. tuberosum auxin/indole-3-acetic acid |
| StABCG1 | S. tuberosum suberin transporter |
| StAS1 | S. tuberosum asparagine synthetase 1 |
| TPS | true potato seed |
| IR | Intergenic Region |
| SSN | single-strand nick |
References
- Anders, S.; Cowling, W.; Pareek, A.; Gupta, K.J.; Singla-Pareek, S.L.; Foyer, C.H. Gaining Acceptance of Novel Plant Breeding Technologies. Trends Plant Sci. 2021, 26, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Sapakhova, Z.; Abilda, Z.; Toishimanov, M.; Daurov, D.; Daurova, A.; Raissova, N.; Sidorik, A.; Kanat, R.; Zhambakin, K.; Shamekova, M. Early Generation Selection of Potato Breeding Lines. Horticulturae 2024, 10, 1121. [Google Scholar] [CrossRef]
- Daurov, D.; Daurova, A.; Sapakhova, Z.; Kanat, R.; Akhmetzhanova, D.; Abilda, Z.; Toishimanov, M.; Raissova, N.; Otynshiyev, M.; Zhambakin, K.; et al. The Impact of the Growth Regulators and Cultivation Conditions of Temporary Immersion Systems (TISs) on the Morphological Characteristics of Potato Explants and Microtubers. Agronomy 2024, 14, 1782. [Google Scholar] [CrossRef]
- Asakaviciute, R.; Zelya, A.; Kacergius, A.; Andriychuk, T.; Zelya, G.; Skoreyko, A.; Razukas, A. Assessment of Potato Varieties of Lithuanian Breeding Resistance Potato Wart Causative Agents and Late Blight. Sci. Rep. 2025, 15, 5915. [Google Scholar] [CrossRef]
- Campos, H.; Ortiz, O. The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind; Springer International Publishing: Cham, Switzerland, 2019; ISBN 9783030286835. [Google Scholar]
- Ortiz, R.; Reslow, F.; Huicho, J.; Vetukuri, R.; Crossa, J. Adaptability, Stability, and Productivity of Potato Breeding Clones and Cultivars at High Latitudes in Europe. Discov. Life 2024, 54, 13. [Google Scholar] [CrossRef]
- Getachew Bekele, B. Review on Genetics and Breeding of Potato (Solanum tuberosum L.). Glob. Sci. J. 2022, 10, 1205–1221. [Google Scholar]
- Carputo, D.; Aversano, R.; Frusciante, L. Breeding Potato for Quality Traits. Acta Hortic. 2005, 684, 55–64. [Google Scholar] [CrossRef]
- Nacheva, E.; Pevicharova, G. Potato Breeding Lines for Processing. Genet. Breed. 2008, 37, 77–84. [Google Scholar]
- Zhang, S.; Wang, X.; Kinay, P.; Dau, Q. Climate Change Impacts on Potato Storage. Foods 2024, 13, 1119. [Google Scholar] [CrossRef]
- Berindean, I.V.; Taoutaou, A.; Rida, S.; Ona, A.D.; Stefan, M.F.; Costin, A.; Racz, I.; Muntean, L. Modern Breeding Strategies and Tools for Durable Late Blight Resistance in Potato. Plants 2024, 13, 1711. [Google Scholar] [CrossRef]
- Naess, S.K.; Bradeen, J.M.; Wielgus, S.M.; Haberlach, G.T.; McGrath, J.M.; Helgeson, J.P. Resistance to late blight in Solanum bulbocastanum is mapped to chromosome 8. Theor. Appl. Genet. 2000, 101, 697–704. [Google Scholar] [CrossRef]
- Tek, A.L.; Stevenson, W.R.; Helgeson, J.P.; Jiang, J. Transfer of Tuber Soft Rot and Early Blight Resistances from Solanum Brevidens into Cultivated Potato. Theor. Appl. Genet. 2004, 109, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.; Li, J.; Dong, J.; Wu, J.; Wang, H.; Zuo, Y.; Cai, X.; Song, B. Molecular Marker-Assisted Selection for Frost Tolerance in a Diallel Population of Potato. Cells 2023, 12, 1226. [Google Scholar] [CrossRef]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; Wang, J.; et al. Genome Sequence and Analysis of the Tuber Crop Potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Tussipkan, D.; Manabayeva, S.A. Employing CRISPR/Cas Technology for the Improvement of Potato and Other Tuber Crops. Front. Plant Sci. 2021, 12, 747476. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Buckseth, T.; Challam, C.; Zinta, R.; Bhatia, N.; Dalamu, D.; Naik, S.; Poonia, A.K.; Singh, R.K.; Luthra, S.K.; et al. CRISPR/Cas Genome Editing in Potato: Current Status and Future Perspectives. Front. Genet. 2022, 13, 827808. [Google Scholar] [CrossRef]
- Kaur, N.; Qadir, M.; Francis, D.V.; Alok, A.; Tiwari, S.; Ahmed, Z.F.R. CRISPR/Cas9: A Sustainable Technology to Enhance Climate Resilience in Major Staple Crops. Front. Genome Ed. 2025, 7, 1533197. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, U.K.; Öztürk, Z.N.; Gökçe, A.F. Drought Stress: Review and Recommendations; Springer Nature: Cham, Switzerland, 2025; ISBN 9783031806100. [Google Scholar]
- Butler, N.M.; Atkins, P.A.; Voytas, D.F.; Douches, D.S. Generation and Inheritance of Targeted Mutations in Potato (Solanum tuberosum L.) Using the CRISPR/Cas System. PLoS ONE 2015, 10, e0144591. [Google Scholar] [CrossRef] [PubMed]
- Heidersbach, A.J.; Dorighi, K.M.; Gomez, J.A.; Jacobi, A.M.; Haley, B. A Versatile, High-Efficiency Platform for CRISPR-Based Gene Activation. Nat. Commun. 2023, 14, 902. [Google Scholar] [CrossRef]
- Larson, M.H.; Gilbert, L.A.; Wang, X.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR Interference (CRISPRi) for Sequence-Specific Control of Gene Expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef]
- Brito, L.F.; Schultenkämper, K.; Passaglia, L.M.P.; Wendisch, V.F. CRISPR Interference-Based Gene Repression in the Plant Growth Promoter Paenibacillus Sonchi Genomovar Riograndensis SBR5. Appl. Microbiol. Biotechnol. 2020, 104, 5095–5106. [Google Scholar] [CrossRef]
- Azameti, M.K.; Dauda, W.P. Base Editing in Plants: Applications, Challenges, and Future Prospects. Front. Plant Sci. 2021, 12, 664997. [Google Scholar] [CrossRef]
- Vu, T.V.; Nguyen, N.T.; Kim, J.; Hong, J.C.; Kim, J.Y. Prime Editing: Mechanism Insight and Recent Applications in Plants. Plant Biotechnol. J. 2024, 22, 19–36. [Google Scholar] [CrossRef]
- Rozov, S.M.; Permyakova, N.V.; Deineko, E.V. The Problem of the Low Rates of CRISPR/Cas9-Mediated Knock-Ins in Plants: Approaches and Solutions. Int. J. Mol. Sci. 2019, 20, 3371. [Google Scholar] [CrossRef]
- Jeggo, P.A.; Martin, S.G.; Williams, K.J.; Prise, K.M. Advances in Radiation Biology—Highlights from the 16th ICRR special feature: Introductory editorial. Br. J. Radiol. 2020, 93, 20209006. [Google Scholar] [CrossRef] [PubMed]
- Clasen, B.M.; Stoddard, T.J.; Luo, S.; Demorest, Z.L.; Li, J.; Cedrone, F.; Tibebu, R.; Davison, S.; Ray, E.E.; Daulhac, A.; et al. Improving Cold Storage and Processing Traits in Potato through Targeted Gene Knockout. Plant Biotechnol. J. 2016, 14, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Yasmeen, A.; Shakoor, S.; Azam, S.; Bakhsh, A.; Shahid, N.; Latif, A.; Shahid, A.A.; Husnain, T.; Rao, A.Q. CRISPR/Cas-Mediated Knockdown of Vacuolar Invertase Gene Expression Lowers the Cold-Induced Sweetening in Potatoes. Planta 2022, 256, 107. [Google Scholar] [CrossRef]
- Zheng, Z.; Ye, G.; Zhou, Y.; Pu, X.; Wang, S.; Wang, J. Editing Sterol Side Chain Reductase 2 Gene (StSSR2) via CRISPR/Cas9 Reduces the Total Steroidal Glycoalkaloids in Potato. All. Life 2021, 14, 401–413. [Google Scholar] [CrossRef]
- Kavuri, N.R.; Ramasamy, M.; Qi, Y.; Mandadi, K. Applications of CRISPR/Cas13-Based RNA Editing in Plants. Cells 2022, 11, 2665. [Google Scholar] [CrossRef]
- Sood, S.; Dipta, B.; Mangal, V.; Thakur, A.K.; Dutt, S.; Sharma, N.; Kumar, V.; Singh, B. CRISPR/Cas-Mediated Genome Editing for Improving Key Traits in Potato (Solanum tuberosum L.). J. Plant Growth Regul. 2024, 44, 529–542. [Google Scholar] [CrossRef]
- Sutula, M.; Tussipkan, D.; Kali, B.; Manabayeva, S. Molecular Mechanisms Underlying Defense Responses of Potato (Solanum tuberosum L.) to Environmental Stress and CRISPR/Cas-Mediated Engineering of Stress Tolerance. Plants 2025, 14, 1983. [Google Scholar] [CrossRef]
- Metje-Sprink, J.; Menz, J.; Modrzejewski, D.; Sprink, T. DNA-Free Genome Editing: Past, Present and Future. Front. Plant Sci. 2019, 9, 1957. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.S.; Ohlsson, P.; Gonzalez, M.N.; Samuelsson, M.; Hofvander, P. Genome Editing in Potato via CRISPR-Cas9 Ribonucleoprotein Delivery. Physiol. Plant 2018, 164, 378–384. [Google Scholar] [CrossRef]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.-S.; Samuelsson, M.; Hofvander, P. Efficient Targeted Multiallelic Mutagenesis in Tetraploid Potato (Solanum tuberosum) by Transient CRISPR-Cas9 Expression in Protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Kim, S.W.; Lee, S.; Koo, Y. Optimized Protocols for Protoplast Isolation, Transfection, and Regeneration in the Solanum Genus for the CRISPR/Cas-Mediated Transgene-Free Genome Editing. Appl. Biol. Chem. 2024, 67, 21. [Google Scholar] [CrossRef]
- Ly, D.N.P.; Iqbal, S.; Fosu-Nyarko, J.; Milroy, S.; Jones, M.G.K. Multiplex CRISPR-Cas9 Gene-Editing Can Deliver Potato Cultivars with Reduced Browning and Acrylamide. Plants 2023, 12, 379. [Google Scholar] [CrossRef]
- Shamekova, M.; Mendoza, M.R.; Hsieh, Y.-C.; Lindbo, J.; Omarov, R.T.; Scholthof, H.B. Tombusvirus-Based Vector Systems to Permit over-Expression of Genes or That Serve as Sensors of Antiviral RNA Silencing in Plants. Virology 2014, 452–453, 159–165. [Google Scholar] [CrossRef]
- Mahmood, M.A.; Naqvi, R.Z.; Rahman, S.U.; Amin, I.; Mansoor, S. Plant Virus-Derived Vectors for Plant Genome Engineering. Viruses 2023, 15, 531. [Google Scholar] [CrossRef]
- Acha, G.; Vergara, R.; Muñoz, M.; Mora, R.; Aguirre, C.; Muñoz, M.; Kalazich, J.; Prieto, H. A Traceable DNA-Replicon Derived Vector to Speed up Gene Editing in Potato: Interrupting Genes Related to Undesirable Postharvest Tuber Traits as an Example. Plants 2021, 10, 1882. [Google Scholar] [CrossRef]
- Bradshaw, J.E. A Brief History of the Impact of Potato Genetics on the Breeding of Tetraploid Potato Cultivars for Tuber Propagation. Potato Res. 2022, 65, 461–501. [Google Scholar] [CrossRef]
- Ye, M.; Peng, Z.; Tang, D.; Yang, Z.; Li, D.; Xu, Y.; Zhang, C.; Huang, S. Generation of Self-Compatible Diploid Potato by Knockout of S-RNase. Nat. Plants 2018, 4, 651–654. [Google Scholar] [CrossRef]
- Taylor, M. Routes to Genetic Gain in Potato. Nat. Plants 2018, 4, 631–632. [Google Scholar] [CrossRef] [PubMed]
- Enciso-Rodriguez, F.; Manrique-Carpintero, N.C.; Nadakuduti, S.S.; Buell, C.R.; Zarka, D.; Douches, D. Overcoming Self-Incompatibility in Diploid Potato Using CRISPR-Cas9. Front. Plant Sci. 2019, 10, 376. [Google Scholar] [CrossRef]
- Eggers, E.J.; van der Burgt, A.; van Heusden, S.A.W.; de Vries, M.E.; Visser, R.G.F.; Bachem, C.W.B.; Lindhout, P. Neofunctionalisation of the Sli Gene Leads to Self-Compatibility and Facilitates Precision Breeding in Potato. Nat. Commun. 2021, 12, 4141. [Google Scholar] [CrossRef]
- Zong, Y.; Song, Q.; Li, C.; Jin, S.; Zhang, D.; Wang, Y.; Qiu, J.-L.; Gao, C. Efficient C-to-t Base Editing in Plants Using a Fusion of Ncas9 and Human Apobec3a. Nat. Biotechnol. 2018, 36, 950–953. [Google Scholar] [CrossRef]
- Butler, N.M.; Baltes, N.J.; Voytas, D.F.; Douches, D.S. Geminivirus-Mediated Genome Editing in Potato (Solanum tuberosum L.) Using Sequence-Specific Nucleases. Front. Plant Sci. 2016, 7, 1045. [Google Scholar] [CrossRef]
- Das Dangol, S.; Çalışkan, M.E.; Bakhsh, A. CRISPR-Cas9-Mediated Reduced Expression of Potato Apoplastic Invertase Inhibitor Gene and Analysis of Transformation Efficiency Parameters. Potato Res. 2024, 68, 1699–1715. [Google Scholar] [CrossRef]
- Butler, N.M.; Jansky, S.H.; Jiang, J. First-Generation Genome Editing in Potato Using Hairy Root Transformation. Plant Biotecnol. J. 2020, 18, 2201–2209. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Strickland, L.W.; Hamilton, J.P.; Trusky, J.K.; Fang, C.; Butler, N.M.; Douches, D.S.; Buell, C.R.; Jiang, J. Jan and Mini-Jan, a Model System for Potato Functional Genomics. Plant Biotechnol. J. 2025, 23, 1243–1256. [Google Scholar] [CrossRef]
- Kiryushkin, A.S.; Ilina, E.L.; Guseva, E.D.; Pawlowski, K.; Demchenko, K.N. Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation. Plants 2022, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elsalam, K.A.; Ahmad, A.; Zhang, B. Genome Modified Plants and Microbes in Food and Agriculture CRISPRized Horticulture Crops; Academic Press: London, UK, 2024. [Google Scholar]
- Nourozi, M.; Nazarain-Firouzabadi, F.; Ismaili, A.; Ahmadvand, R.; Poormazaheri, H. CRISPR/Cas StNRL1 Gene Knockout Increases Resistance to Late Blight and Susceptibility to Early Blight in Potato. Front. Plant Sci. 2023, 14, 1278127. [Google Scholar] [CrossRef]
- Daurov, D.; Argynbayeva, A.; Daurova, A.; Zhapar, K.; Sapakhova, Z.; Zhambakin, K.; Shamekova, M. Monitoring the Spread of Potato Virus Diseases in Kazakhstan. Am. J. Potato Res. 2023, 100, 63–70. [Google Scholar] [CrossRef]
- Price, J.A.; Coyne, D.; Blok, V.C.; Jones, J.T. Potato Cyst Nematodes Globodera Rostochiensis and G. Pallida. Mol. Plant Pathol. 2021, 22, 495–507. [Google Scholar] [CrossRef]
- Tessema, G.L.; Seid, H.E. Potato Bacterial Wilt in Ethiopia: History, Current Status, and Future Perspectives. PeerJ 2023, 11, e14661. [Google Scholar] [CrossRef]
- Weng, L.; Tang, Z.; Sardar, M.F.; Yu, Y.; Ai, K.; Liang, S.; Alkahtani, J.; Lyv, D. Unveiling the Frontiers of Potato Disease Research through Bibliometric Analysis. Front. Microbiol. 2024, 15, 1430066. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, M.A.; Statsyuk, N.V.; Demidova, V.N.; Semeniuk, I.N.; Smetanina, T.I.; Ukolova, A.Y.; Vyatchinov, A.A. A Complex Approach to Control Black Dot Disease in Potato. Agronomy 2024, 14, 1373. [Google Scholar] [CrossRef]
- Bagchi, P.; Sawicka, B.; Stamenkovic, Z.; Marković, D.; Bhattacharjee, D. Potato Late Blight Outbreak: A Study on Advanced Classification Models Based on Meteorological Data. Sensors 2024, 24, 7864. [Google Scholar] [CrossRef]
- Ahmad, S.; Wei, X.; Sheng, Z.; Hu, P.; Tang, S. CRISPR/Cas9 for Development of Disease Resistance in Plants: Recent Progress, Limitations and Future Prospects. Brief. Funct. Genom. 2018, 19, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Kieu, N.P.; Lenman, M.; Wang, E.S.; Petersen, B.L.; Andreasson, E. Mutations Introduced in Susceptibility Genes through CRISPR/Cas9 Genome Editing Confer Increased Late Blight Resistance in Potatoes. Sci. Rep. 2021, 11, 4487. [Google Scholar] [CrossRef]
- Kusch, S.; Panstruga, R. Mlo-Based Resistance: An Apparently Universal “Weapon” to Defeat Powdery Mildew Disease. Mol. Plant-Microbe Interact. 2017, 30, 179–189. [Google Scholar] [CrossRef]
- Engelhardt, S.; Stam, R.; Hückelhoven, R. Good Riddance? Breaking Disease Susceptibility in the Era of New Breeding Technologies. Agronomy 2018, 8, 114. [Google Scholar] [CrossRef]
- Sun, K.; Wolters, A.-M.A.; Vossen, J.H.; Rouwet, M.E.; Loonen, A.E.H.M.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Silencing of Six Susceptibility Genes Results in Potato Late Blight Resistance. Transgenic Res. 2016, 25, 731–742. [Google Scholar] [CrossRef]
- Karlsson, M.; Kieu, N.P.; Lenman, M.; Marttila, S.; Resjö, S.; Zahid, M.A.; Andreasson, E. CRISPR/Cas9 Genome Editing of Potato StDMR6-1 Results in Plants Less Affected by Different Stress Conditions. Hortic. Res. 2024, 11, uhae130. [Google Scholar] [CrossRef]
- Bi, W.; Liu, J.; Li, Y.; He, Z.; Chen, Y.; Zhao, T.; Liang, X.; Wang, X.; Meng, X.; Dou, D.; et al. CRISPR/Cas9-Guided Editing of a Novel Susceptibility Gene in Potato Improves Phytophthora Resistance without Growth Penalty. Plant Biotechnol. J. 2024, 22, 4–6. [Google Scholar] [CrossRef]
- Hegde, N.; Joshi, S.; Soni, N.; Kushalappa, A.C. The Caffeoyl-CoA O-Methyltransferase Gene SNP Replacement in Russet Burbank Potato Variety Enhances Late Blight Resistance through Cell Wall Reinforcement. Plant Cell Rep. 2021, 40, 237–254. [Google Scholar] [CrossRef]
- Razzaq, H.A.; Ijaz, S.; Haq, I.U.; Khan, I.A. Functional Inhibition of the StERF3 Gene by Dual Targeting through CRISPR/Cas9 Enhances Resistance to the Late Blight Disease in Solanum tuberosum L. Mol. Biol. Rep. 2022, 12, 11675–11684. [Google Scholar] [CrossRef]
- Bi, W.; Chen, Y.; Song, Y.; Liu, J.; Zhao, T.; Sun, C.; Qin, J.; Tu, Z.; Li, Y.; Wang, X.; et al. Potato DMP2 Positively Regulates Plant Immunity by Modulating Endoplasmic Reticulum Homeostasis. J. Integr. Plant Biol. 2025, 67, 1568–1581. [Google Scholar] [CrossRef]
- Poudel, B.; Sathe, A.; Bede, J.C.; Kushalappa, A.C. Editing Metacaspase (StMC7) Gene Enhances Late Blight Resistance in Russet Burbank Potato. PLoS ONE 2025, 20, e0325702. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.-B.; Park, S.-J.; Park, J.-S.; Lee, H.-J.; Shin, S.Y.; Lee, S.M.; Choi, G.J.; Kim, S.-G.; Cho, H.S.; Jeon, J.-H.; et al. Editing of StSR4 by Cas9-RNPs Confers Resistance to Phytophthora Infestans in Potato. Front. Plant Sci. 2022, 13, 997888. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, M.; Rajkumar, M.S.; Bedre, R.; Irigoyen, S.; Berg-Falloure, K.; Kolomiets, M.V.; Mandadi, K.K. Genome Editing of NPR3 Confers Potato Resistance to Candidatus liberibacter spp. Plant Biotechnol. J. 2024, 22, 2635–2637. [Google Scholar] [CrossRef]
- Makhotenko, A.V.; Khromov, A.V.; Snigir, E.A.; Makarova, S.S.; Makarov, V.V.; Suprunova, T.P.; Kalinina, N.O.; Taliansky, M.E. Functional Analysis of Coilin in Virus Resistance and Stress Tolerance of Potato Solanum tuberosum Using CRISPR-Cas9 Editing. Dokl. Biochem. Biophys. 2019, 484, 88–91. [Google Scholar] [CrossRef]
- Noureen, A.; Khan, M.Z.; Amin, I.; Zainab, T.; Mansoor, S. CRISPR/Cas9-Mediated Targeting of Susceptibility Factor EIF4E-Enhanced Resistance Against Potato Virus Y. Front. Genet. 2022, 13, 922019. [Google Scholar] [CrossRef]
- Lucioli, A.; Tavazza, R.; Baima, S.; Fatyol, K.; Burgyan, J.; Tavazza, M. CRISPR-Cas9 Targeting of the EIF4E1 Gene Extends the Potato Virus Y Resistance Spectrum of the Solanum tuberosum L. cv. Desirée. Front. Microbiol. 2022, 13, 873930. [Google Scholar] [CrossRef]
- Zhan, X.; Zhang, F.; Zhong, Z.; Chen, R.; Wang, Y.; Chang, L.; Bock, R.; Nie, B.; Zhang, J. Generation of Virus-Resistant Potato Plants by RNA Genome Targeting. Plant Biotechnol. J. 2019, 17, 1814–1822. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Liu, W.; Nie, B.; Zhang, F.; Zhang, J. Cas13d-Mediated Multiplex RNA Targeting Confers a Broad-Spectrum Resistance against RNA Viruses in Potato. Commun. Biol. 2023, 6, 855. [Google Scholar] [CrossRef]
- Noureen, A.; Khan, M.Z.; Amin, I.; Zainab, T.; Ahmad, N.; Haider, S.; Mansoor, S. Broad-Spectrum Resistance against Multiple PVY-Strains by CRSIPR/Cas13 System in Solanum tuberosum Crop. GM Crops Food 2022, 13, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Asensio-S.-Manzanera, M.C.; Santiago-Calvo, Y.; Palomo-Gómez, J.L.; Marquínez-Ramírez, R.; Bastin, S.; García-Méndez, E.M.; Hernández-Suárez, E.; Siverio-de-la-Rosa, F. Survey of Candidatus Liberibacter Solanacearum and Its Associated Vectors in Potato Crop in Spain. Insects 2022, 13, 964. [Google Scholar] [CrossRef]
- Gutierrez Sanchez, P.A.; Babujee, L.; Jaramillo Mesa, H.; Arcibal, E.; Gannon, M.; Halterman, D.; Jahn, M.; Jiang, J.; Rakotondrafara, A.M. Overexpression of a Modified EIF4E Regulates Potato Virus y Resistance at the Transcriptional Level in Potato. BMC Genom. 2020, 21, 18. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Mehmood, M.A.; Shahid, M.; Fatma, S.; Khan, A.; Ali, S. Prospects for Potato Genome Editing to Engineer Resistance against Viruses and Cold-Induced Sweetening. GM Crops Food 2020, 11, 185–205. [Google Scholar] [CrossRef]
- Nasr-Eldin, M.; Messiha, N.; Othman, B.; Megahed, A.; Elhalag, K. Induction of Potato Systemic Resistance against the Potato Virus Y (PVYNTN), Using Crude Filtrates of Streptomyces Spp. under Greenhouse Conditions. Egypt. J. Biol. Pest Control 2019, 29, 62. [Google Scholar] [CrossRef]
- Rajput, M.; Choudhary, K.; Kumar, M.; Vivekanand, V.; Chawade, A.; Ortiz, R.; Pareek, N. Rna Interference and Crispr/Cas Gene Editing for Crop Improvement: Paradigm Shift towards Sustainable Agriculture. Plants 2021, 10, 1914. [Google Scholar] [CrossRef]
- Cao, Y.; Zhou, H.; Zhou, X.; Li, F. Control of Plant Viruses by CRISPR/Cas System-Mediated Adaptive Immunity. Front. Microbiol. 2020, 11, 593700. [Google Scholar] [CrossRef]
- Bakhsh, A.; Hussain, T.; Rahamkulov, I.; Demirel, U.; Çalışkan, M.E. Transgenic Potato Lines Expressing CP4-EPSP Synthase Exhibit Resistance against Glyphosate. Plant Cell Tissue Organ. Cult. 2020, 140, 23–34. [Google Scholar] [CrossRef]
- Kanat, R.; Shamekova, M.; Sapakhova, Z.; Toishimanov, M.; Daurov, D.; Raissova, N.; Abilda, Z.; Daurova, A.; Zhambakin, K. Gene Expression Analysis for Drought Tolerance in Early Stage of Potato Plant Development. Biology 2024, 13, 857. [Google Scholar] [CrossRef] [PubMed]
- Sapakhova, Z.; Raissova, N.; Daurov, D.; Zhapar, K.; Daurova, A.; Zhigailov, A.; Zhambakin, K.; Shamekova, M. Sweet Potato as a Key Crop for Food Security under the Conditions of Global Climate Change: A Review. Plants 2023, 12, 2516. [Google Scholar] [CrossRef] [PubMed]
- Choudry, M.W.; Riaz, R.; Nawaz, P.; Ashraf, M.; Ijaz, B.; Bakhsh, A. CRISPR-Cas9 Mediated Understanding of Plants’ Abiotic Stress-Responsive Genes to Combat Changing Climatic Patterns. Funct. Integr. Genom. 2024, 24, 132. [Google Scholar] [CrossRef]
- Wang, P.; Wu, X.; Li, N.; Nie, H.; Ma, Y.; Wu, J.; Zhang, Z.; Ma, Y. The StbHLH47 Transcription Factor Negatively Regulates Drought Tolerance in Potato (Solanum tuberosum L.). BMC Plant Biol. 2025, 25, 14. [Google Scholar] [CrossRef]
- Chauhan, H.; Alok, A.; Aiana; Upadhyay, S.K.; Pandey, A.; Singh, K. CRISPR/Cas9 Edited StbHLH47 Lines Exhibit Altered Expression Profiling of Iron Regulating Genes and Increased Iron Content in Solanum tuberosum. Curr. Plant Biol. 2024, 38, 100354. [Google Scholar] [CrossRef]
- Zhao, J.; Yao, B.; Peng, Z.; Yang, X.; Li, K.; Zhang, X.; Zhu, H.; Zhou, X.; Wang, M.; Jiang, L.; et al. Splicing Defect of StDRO2 Intron 1 Promotes Potato Root Growth by Disturbing Auxin Transport to Adapt to Drought Stress. Hortic. Plant J. 2024, 11, 706–720. [Google Scholar] [CrossRef]
- Hua, D.; Ma, M.; Ge, G.; Suleman, M.; Li, H. The Role of Cyanide-Resistant Respiration in Solanum tuberosum L. against High. Light. Stress. Plant Biol. 2020, 22, 425–432. [Google Scholar] [CrossRef]
- Zhou, X.; Zha, M.; Huang, J.; Li, L.; Imran, M.; Zhang, C. StMYB44 Negatively Regulates Phosphate Transport by Suppressing Expression of PHOSPHATE1 in Potato. J. Exp. Bot. 2017, 68, 1265–1281. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Y.; Li, L.; Yan, L.; Wang, X.; Wang, Q.; Lai, X. StSN2 Enhances Tuber Formation in Potato via Upregulating of the ABA Signaling Pathway. Front. Plant Sci. 2025, 16, 1566237. [Google Scholar] [CrossRef]
- Wan, M.; Xie, H.; Guo, H.; Jing, S.; Zeng, D.; Li, B.; Zhu, B.; Zeng, Z. Developing a Pipeline for Identification, Characterization and Molecular Editing of Cis-Regulatory Elements: A Case Study in Potato. aBIOTECH 2024, 6, 91–96. [Google Scholar] [CrossRef]
- Lei, C.; Ye, M.; Li, C.; Gong, M. H2O2 Participates in the Induction and Formation of Potato Tubers by Activating Tuberization-Related Signal Transduction Pathways. Agronomy 2023, 13, 1398. [Google Scholar] [CrossRef]
- Wulff-Vester, A.; Andersson, M.; Brurberg, M.B.; Hofvander, P.; Alsheikh, M.; Harwood, W.; Hvoslef-Eide, T. Colour Change in Potato (Solanum tuberosum L.) Tubers by Disruption of the Anthocyanin Pathway via Ribonucleoprotein Complex Delivery of the CRISPR/Cas9 System. Plant Cell Tissue Organ. Cult. 2024, 157, 25. [Google Scholar] [CrossRef]
- Tang, D.; Jia, Y.; Zhang, J.; Li, H.; Cheng, L.; Wang, P.; Bao, Z.; Liu, Z.; Feng, S.; Zhu, X.; et al. Genome Evolution and Diversity of Wild and Cultivated Potatoes. Nature 2022, 606, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Meng, L.; Zhang, G.; Yang, X.; Pang, B.; Cheng, J.; He, B.; Sun, F. Unraveling Crop Enzymatic Browning through Integrated Omics. Front. Plant Sci. 2024, 15, 1342639. [Google Scholar] [CrossRef]
- González, M.N.; Massa, G.A.; Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.S.; Storani, L.; Décima Oneto, C.A.; Hofvander, P.; Feingold, S.E. Reduced Enzymatic Browning in Potato Tubers by Specific Editing of a Polyphenol Oxidase Gene via Ribonucleoprotein Complexes Delivery of the CRISPR/Cas9 System. Front. Plant Sci. 2020, 10, 1649. [Google Scholar] [CrossRef]
- González, M.N.; Massa, G.A.; Andersson, M.; Décima Oneto, C.A.; Turesson, H.; Storani, L.; Olsson, N.; Fält, A.S.; Hofvander, P.; Feingold, S.E. Comparative Potato Genome Editing: Agrobacterium Tumefaciens-Mediated Transformation and Protoplasts Transfection Delivery of CRISPR/Cas9 Components Directed to StPPO2 Gene. Plant Cell Tissue Organ. Cult. 2021, 145, 291–305. [Google Scholar] [CrossRef]
- Martínez-Prada, M.d.M.; Curtin, S.J.; Gutiérrez-González, J.J. Potato Improvement through Genetic Engineering. GM Crops Food 2021, 12, 479–496. [Google Scholar] [CrossRef]
- Jayakody, T.B.; Zarka, D.; Cho, K.H.; Jensen, J.; Sikora, S.; Buell, C.R.; Douches, D.S.; Nadakuduti, S.S. Genome-Wide Evaluation of Gene Editing Outcomes Using CRISPR/Cas9 in Seed Propagated Camelina Sativa and Vegetatively Propagated Solanum tuberosum. Front. Plant Sci. 2024, 15, 1496861. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Manghwar, H.; Sun, L.; Wang, P.; Wang, G.; Sheng, H.; Zhang, J.; Liu, H.; Qin, L.; Rui, H.; et al. Whole Genome Sequencing Reveals Rare Off-Target Mutations and Considerable Inherent Genetic or/and Somaclonal Variations in CRISPR/Cas9-Edited Cotton Plants. Plant Biotechnol. J. 2019, 17, 858–868. [Google Scholar] [CrossRef]
- Wang, X.; Tu, M.; Wang, Y.; Yin, W.; Zhang, Y.; Wu, H.; Gu, Y.; Li, Z.; Xi, Z.; Wang, X. Whole-Genome Sequencing Reveals Rare off-Target Mutations in CRISPR/Cas9-Edited Grapevine. Hortic. Res. 2021, 8, 114. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 System Produces Specific and Homozygous Targeted Gene Editing in Rice in One Generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Chincinska, I.A.; Miklaszewska, M.; Sołtys-Kalina, D. Recent Advances and Challenges in Potato Improvement Using CRISPR/Cas Genome Editing. Planta 2023, 257, 25. [Google Scholar] [CrossRef]
- Pfotenhauer, A.C.; Occhialini, A.; Harbison, S.A.; Li, L.; Piatek, A.A.; Luckett, C.R.; Yang, Y.; Stewart, C.N.; Lenaghan, S.C. Genome-Editing of FtsZ1 for Alteration of Starch Granule Size in Potato Tubers. Plants 2023, 12, 1878. [Google Scholar] [CrossRef]
- Helle, S.; Bray, F.; Verbeke, J.; Devassine, S.; Courseaux, A.; Facon, M.; Tokarski, C.; Rolando, C.; Szydlowski, N. Proteome Analysis of Potato Starch Reveals the Presence of New Starch Metabolic Proteins as Well as Multiple Protease Inhibitors. Front. Plant Sci. 2018, 9, 746. [Google Scholar] [CrossRef]
- Sevestre, F.; Facon, M.; Wattebled, F.; Szydlowski, N. Facilitating Gene Editing in Potato: A Single-Nucleotide Polymorphism (SNP) Map of the Solanum tuberosum L. cv. Desiree genome. Sci. Rep. 2020, 10, 2045. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Song, L.; Yi, X.; Ni, X.; Cui, X.; Duan, S.; Jiang, R.; Lyu, D.; Wang, J.; Hu, B.; et al. Potato STARCH SYNTHEASE 5 Is Critical for Simple Starch Granule Initiation in Amyloplasts and Tuber Development. Plant J. 2025, 122, e70206. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, M.; Ito, K.; Hamada, K.; Takeuchi, A.; Asano, K.; Noda, T.; Watanabe, A.; Hokura, A.; Teramura, H.; Takahashi, F.; et al. Peculiar Properties of Tuber Starch in a Potato Mutant Lacking the α-Glucan Water Dikinase 1 Gene GWD1 Created by Targeted Mutagenesis Using the CRISPR/DMac3-Cas9 System. Plant Biotechnol. 2023, 40, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Nakayasu, M.; Akiyama, R.; Lee, H.J.; Osakabe, K.; Osakabe, Y.; Watanabe, B.; Sugimoto, Y.; Umemoto, N.; Saito, K.; Muranaka, T.; et al. Generation of α-Solanine-Free Hairy Roots of Potato by CRISPR/Cas9 Mediated Genome Editing of the St16DOX Gene. Plant Physiol. Biochem. 2018, 131, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Abeuova, L.; Kali, B.; Tussipkan, D.; Akhmetollayeva, A.; Ramankulov, Y.; Manabayeva, S. CRISPR/Cas9-Mediated Multiple Guide RNA-Targeted Mutagenesis in the Potato. Transgenic Res. 2023, 32, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Veillet, F.; Chauvin, L.; Kermarrec, M.-P.; Sevestre, F.; Merrer, M.; Terret, Z.; Szydlowski, N.; Devaux, P.; Gallois, J.-L.; Chauvin, J.-E. The Solanum tuberosum GBSSI Gene: A Target for Assessing Gene and Base Editing in Tetraploid Potato. Plant Cell Rep. 2019, 38, 1065–1080. [Google Scholar] [CrossRef]
- Toinga-Villafuerte, S.; Vales, M.I.; Awika, J.M.; Rathore, K.S. CRISPR/Cas9-Mediated Mutagenesis of the Granule-Bound Starch Synthase Gene in the Potato Variety Yukon Gold to Obtain Amylose-Free Starch in Tubers. Int. J. Mol. Sci. 2022, 23, 4640. [Google Scholar] [CrossRef] [PubMed]
- Johansen, I.E.; Liu, Y.; Jørgensen, B.; Bennett, E.P.; Andreasson, E.; Nielsen, K.L.; Blennow, A.; Petersen, B.L. High Efficacy Full Allelic CRISPR/Cas9 Gene Editing in Tetraploid Potato. Sci. Rep. 2019, 9, 17715. [Google Scholar] [CrossRef]
- Kusano, H.; Ohnuma, M.; Mutsuro-Aoki, H.; Asahi, T.; Ichinosawa, D.; Onodera, H.; Asano, K.; Noda, T.; Horie, T.; Fukumoto, K.; et al. Establishment of a Modified CRISPR/Cas9 System with Increased Mutagenesis Frequency Using the Translational Enhancer DMac3 and Multiple Guide RNAs in Potato. Sci. Rep. 2018, 8, 13753. [Google Scholar] [CrossRef]
- Zhao, X.; Jayarathna, S.; Turesson, H.; Fält, A.S.; Nestor, G.; González, M.N.; Olsson, N.; Beganovic, M.; Hofvander, P.; Andersson, R.; et al. Amylose Starch with No Detectable Branching Developed through DNA-Free CRISPR-Cas9 Mediated Mutagenesis of Two Starch Branching Enzymes in Potato. Sci. Rep. 2021, 11, 4311. [Google Scholar] [CrossRef]
- Tuncel, A.; Corbin, K.R.; Ahn-Jarvis, J.; Harris, S.; Hawkins, E.; Smedley, M.A.; Harwood, W.; Warren, F.J.; Patron, N.J.; Smith, A.M. Cas9-Mediated Mutagenesis of Potato Starch-Branching Enzymes Generates a Range of Tuber Starch Phenotypes. Plant Biotechnol. J. 2019, 17, 2259–2271. [Google Scholar] [CrossRef]
- Tuncel, A.; Qi, Y. CRISPR/Cas Mediated Genome Editing in Potato: Past Achievements and Future Directions. Plant Sci. 2022, 325, 111474. [Google Scholar] [CrossRef]
- Takeuchi, A.; Ohnuma, M.; Teramura, H.; Asano, K.; Noda, T.; Kusano, H.; Tamura, K.; Shimada, H. Creation of a Potato Mutant Lacking the Starch Branching Enzyme Gene Stsbe3 That Was Generated by Genome Editing Using the CRISPR/DMac3-Cas9 System. Plant Biotechnol. 2021, 38, 345–353. [Google Scholar] [CrossRef]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.P.; Guyon-Debast, A.; Chauvin, J.E.; Nogué, F.; Mazier, M. Transgene-Free Genome Editing in Tomato and Potato Plants Using Agrobacterium-Mediated Delivery of a CRISPR/Cas9 Cytidine Base Editor. Int. J. Mol. Sci. 2019, 20, 402. [Google Scholar] [CrossRef]
- Du, H.; Zhai, Z.; Pu, J.; Liang, J.; Wang, R.; Zhang, Z.; Wang, P.; Zhu, Y.; Huang, L.; Li, D.; et al. Two Tandem R2R3 MYB Transcription Factor Genes Cooperatively Regulate Anthocyanin Accumulation in Potato Tuber Flesh. Plant Biotechnol. J. 2025, 23, 1521–1534. [Google Scholar] [CrossRef]
- Massa, G.A.; Decima Oneto, C.A.; González, M.N.; Hornum, A.P.; Arizmendi, A.; Sucar, S.; Divito, S.B.; Feingold, S.E. CRISPR/Cas9-Mediated Development of Potato Varieties with Long-Term Cold Storage and Bruising Resistance. Biology 2025, 14, 445. [Google Scholar] [CrossRef]
- Wiberley-Bradford, A.E.; Bethke, P.C. Suppression of the Vacuolar Invertase Gene Delays Senescent Sweetening in Chipping Potatoes. J. Sci. Food Agric. 2018, 98, 354–360. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, A.; Butler, N.M.; Zeng, Z.; Xin, H.; Wang, L.; Lv, Z.; Eshel, D.; Douches, D.S.; Jiang, J. Molecular Dissection of an Intronic Enhancer Governing Cold-Induced Expression of the Vacuolar Invertase Gene in Potato. Plant Cell 2024, 36, 1985–1999. [Google Scholar] [CrossRef]
- Yasmeen, A.; Bakhsh, A.; Ajmal, S.; Muhammad, M.; Sadaqat, S.; Awais, M.; Azam, S.; Latif, A.; Shahid, N.; Rao, A.Q. CRISPR/Cas9-Mediated Genome Editing in Diploid and Tetraploid Potatoes. Acta Physiol. Plant 2023, 45, 32. [Google Scholar] [CrossRef]
- Sattar, T.; Yaseen, S.; Waheed, U.; Farid, H.N.; Khan, Z. CRISPR/Cas9-Mediated Inhibition of Vacuolar Invertase (VInv) Gene in Potato. Pak. J. Biochem. Biotechnol. 2023, 4, 23–34. [Google Scholar] [CrossRef]
- Perino, E.H.B.; Smolka, U.; Gorzolka, K.; Grützner, R.; Marillonnet, S.; Vahabi, K.; Rosahl, S. The Suberin Transporter StABCG1 Is Required for Barrier Formation in Potato Leaves. Sci. Rep. 2025, 15, 7930. [Google Scholar] [CrossRef] [PubMed]
- Corte, L.E.-D.; Mahmoud, L.M.; Moraes, T.S.; Mou, Z.; Grosser, J.W.; Dutt, M. Development of Improved Fruit, Vegetable, and Ornamental Crops Using the CRISPR/Cas9 Genome Editing Technique. Plants 2019, 8, 601. [Google Scholar] [CrossRef]
- Mansour, S.A.; Belal, M.H.; Abou-Arab, A.A.K.; Ashour, H.M.; Gad, M.F. Evaluation of Some Pollutant Levels in Conventionally and Organically Farmed Potato Tubers and Their Risks to Human Health. Food Chem. Toxicol. 2009, 47, 615–624. [Google Scholar] [CrossRef]
- Dong, H.; Huang, Y.; Wang, K. The Development of Herbicide Resistance Crop Plants Using Crispr/Cas9-Mediated Gene Editing. Genes 2021, 12, 912. [Google Scholar] [CrossRef]
- Faizal, A.; Nugroho, S.; Sembada, A.A.; Theda, Y.; Komariyah, T.; Esyanti, R.R. Genome Editing in Future Crop Protection: Utilizing CRISPR/Cas9 to Improve Crop Resistance against Diseases, Pests, and Weeds. Discov. Agric. 2024, 2, 104. [Google Scholar] [CrossRef]
- Part 205—National Organic Program. 2025. Available online: https://www.ecfr.gov/current/title-7/part-205 (accessed on 4 July 2025).
- European Commission. Study on the Status of New Genomic Techniques Under Union Law and in Light of the Court of Justice Ruling in Case C-528/16. SWD(2021) 92 Final. 2021. Available online: https://food.ec.europa.eu/system/files/2021-04/gmo_mod-bio_ngt_eu-study.pdf (accessed on 4 July 2025).
- Molitorisová, A.; Clemens, S.; Fresco, L.; Hubar-Kołodziejczyk, A.; Tosun, J.; Niggli, U.; Qaim, M.; Visser, R.G.F.; Weber, A.P.M.; Wesseler, J.; et al. New Genomic Techniques in Organic Production: Considerations for Science-Based, Effective, and Acceptable EU Regulation. Cell Rep. Sustain. 2025, 2, 100405. [Google Scholar] [CrossRef]
- Cardi, T.; Murovec, J.; Bakhsh, A.; Boniecka, J.; Bruegmann, T.; Bull, S.E.; Eeckhaut, T.; Fladung, M.; Galovic, V.; Linkiewicz, A.; et al. CRISPR/Cas-Mediated Plant Genome Editing: Outstanding Challenges a Decade after Implementation. Trends Plant Sci. 2023, 28, 1144–1165. [Google Scholar] [CrossRef] [PubMed]
- Puchta, H. Regulation of Gene-Edited Plants in Europe: From the Valley of Tears into the Shining Sun? aBIOTECH 2024, 5, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Hussain, A.; Manghwar, H.; Xie, K.; Xie, S.; Zhao, S.; Larkin, R.M.; Qing, P.; Jin, S.; Ding, F. Genome Editing with the CRISPR-Cas System: An Art, Ethics and Global Regulatory Perspective. Plant Biotechnol. J. 2020, 18, 1651–1669. [Google Scholar] [CrossRef]
- Waltz, E. Gene-edited CRISPR mushroom escapes US regulation. Nature 2016, 532, 293. [Google Scholar] [CrossRef] [PubMed]
- Lema, M.A. Regulatory Aspects of Gene Editing in Argentina. Transgenic Res. 2019, 28, 147–150. [Google Scholar] [CrossRef] [PubMed]
- da Cunha, N.B.; Silva Junior, J.J.d.; Araújo, A.M.M.; de Souza, L.R.; Leite, M.L.; Medina, G.d.S.; Rodriguez, G.R.; dos Anjos, R.M.; Rodrigues, J.C.M.; Costa, F.F.; et al. Updates on the Regulatory Framework of Edited Organisms in Brazil: A Molecular Revolution in Brazilian Agribusiness. Genes 2025, 16, 553. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.A. The Global Advance of Genome-Edited Plants to the Market: The Key Role of Chile in Its Development. Plants 2024, 13, 3597. [Google Scholar] [CrossRef]
- Gatica-Arias, A. The regulatory current status of plant breeding technologies in some Latin American and the Caribbean countries. PCTOC 2020, 141, 229–242. [Google Scholar] [CrossRef]
- Mou, T.; Song, Q.; Liu, Y.; Song, J. Initiating the commercialization of genetically modified staple crops in China: Domestic biotechnological advancements, regulatory milestones, and governance frameworks. GM Crops Food 2025, 16, 450–481. [Google Scholar] [CrossRef] [PubMed]
- USDA. Biotechnology and Other New Production Technologies, Biotechnology and Other New Production Technologies Addendum, Biotechnology-Plants and Animals, Cloning; USDA: Washington, DC, USA, 2025. [Google Scholar]
- Kim, K.; Kim, C.K.; Kim, W.-C. The Strategy of Knock-in with Homology-Directed Genome Editing in the Model Ornamental Plant Petunia Using CRISPR/Cas9 Ribonucleoprotein Complex. Sci. Hortic. 2024, 326, 112714. [Google Scholar] [CrossRef]
- Zhang, Z.; Hua, L.; Gupta, A.; Tricoli, D.; Edwards, K.J.; Yang, B.; Li, W. Development of an Agrobacterium-Delivered CRISPR/Cas9 System for Wheat Genome Editing. Plant Biotechnol. J. 2019, 17, 1623–1635. [Google Scholar] [CrossRef]
- Gan, W.C.; Ling, A.P.K. CRISPR/Cas9 in Plant Biotechnology: Applications and Challenges. Biotechnologia 2022, 103, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.-L.; Dong, L.; Wang, Z.-P.; Zhang, H.-Y.; Han, C.-Y.; Liu, B.; Wang, X.-C.; Chen, Q.-J. A CRISPR/Cas9 Toolkit for Multiplex Genome Editing in Plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.W.; Qi, Y. CRISPR/Cas9 for Plant Genome Editing: Accomplishments, Problems and Prospects. Plant Cell Rep. 2016, 35, 1417–1427. [Google Scholar] [CrossRef]
- Zhang, S.; Shen, J.; Li, D.; Cheng, Y. Strategies in the Delivery of Cas9 Ribonucleoprotein for CRISPR/Cas9 Genome Editing. Theranostics 2020, 11, 614–648. [Google Scholar] [CrossRef]
- Jiang, W.; Bush, J.; Sheen, J. A Versatile and Efficient Plant Protoplast Platform for Genome Editing by Cas9 RNPs. Front. Genome Ed. 2021, 3, 719190. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, X.; Liu, H.; Li, Z. Highly Efficient DNA-Free Plant Genome Editing Using Virally Delivered CRISPR–Cas9. Nat. Plants 2020, 6, 773–779. [Google Scholar] [CrossRef]
- Chiong, K.T.; Cody, W.B.; Scholthof, H.B. RNA Silencing Suppressor-Influenced Performance of a Virus Vector Delivering Both Guide RNA and Cas9 for CRISPR Gene Editing. Sci. Rep. 2021, 11, 6769. [Google Scholar] [CrossRef]
- Bradshaw, J.E. Breeding Diploid F 1 Hybrid Potatoes for Propagation from Botanical Seed (TPS): Comparisons with Theory and Other Crops. Plants 2022, 2022, 1121. [Google Scholar] [CrossRef]
- Sood, S.; Bhardwaj, V.; Mangal, V.; Kardile, H.; Dipta, B.; Kumar, A.; Singh, B.; Siddappa, S.; Sharma, A.K.; Dalamu, A.K.; et al. Development of near Homozygous Lines for Diploid Hybrid TPS Breeding in Potatoes. Heliyon 2024, 10, e31507. [Google Scholar] [CrossRef]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Egorova, A.A.; Chalaya, N.A.; Fomin, I.N.; Barchuk, A.I.; Gerasimova, S.V. De Novo Domestication Concept for Potato Germplasm Enhancement. Agronomy 2022, 12, 462. [Google Scholar] [CrossRef]
- Jansky, S.H.; Charkowski, A.O.; Douches, D.S.; Gusmini, G.; Richael, C.; Bethke, P.C.; Spooner, D.M.; Novy, R.G.; De Jong, H.; De Jong, W.S.; et al. Reinventing Potato as a Diploid Inbred Line-Based Crop. Crop Sci. 2016, 56, 1412–1422. [Google Scholar] [CrossRef]
- Nadakuduti, S.S.; Buell, C.R.; Voytas, D.F.; Starker, C.G.; Douches, D.S. Genome Editing for Crop Improvement—Applications in Clonally Propagated Polyploids with a Focus on Potato (Solanum tuberosum L.). Front. Plant Sci. 2018, 9, 1607. [Google Scholar] [CrossRef]
| Target Gene | Delivery Method | Type of Editing | Improvement | References |
|---|---|---|---|---|
| StS-RNase | Agrobacterium- mediated transformation | Knockout | Pollen tube growth | [45,46,47,48] |
| StSli | Agrobacterium- mediated transformation | Knockout | Overcoming self-pollen rejection | [46] |
| StTaALS | Agrobacterium- mediated transformation | Base editing | Fundamental research | [47] |
| StALS1 | Agrobacterium- mediated transformation | Knockout | Herbicide resistance | [20] |
| StALS1 | Agrobacterium- and geminivirus-mediated GE | Knockout | Herbicide tolerance | [48] |
| StScINVINH1 | Agrobacterium- mediated transformation | Knockout | Callus induction and regeneration | [49] |
| StPDS | Agrobacterium tumefaciens-based transformation | Knockout | Broadens the number of genotypes for transformation and reduces chimerism | [50] |
| StDwarf, StEr | Agrobacterium- mediated transformation | Knockout | Plant growth | [51] |
| Target Gene | Delivery Method | Type of Editing | Improvement | References |
|---|---|---|---|---|
| StNRL1 | Agrobacterium- mediated transformation | Knockout | Late blight resistance | [54] |
| StDND1, StCHL1, StDMR6-1 | Agrobacterium- mediated transformation | Knockout | Late blight resistance | [62] |
| StDMR6-1 | Agrobacterium- mediated transformation | Knockout | Late blight, early blight, and common scab | [66] |
| StPM1 | Agrobacterium- mediated transformation | Knockout | Late blight resistance | [67] |
| StCCoAOMT | Agrobacterium- mediated transformation | Knockout | Late blight resistance | [68] |
| StERF3 | Agrobacterium- mediated transformation | Knockout | Late blight resistance | [69] |
| StDMP2 | Agrobacterium- mediated transformation | Knockout | Late blight resistance | [70] |
| StMC7 | Agrobacterium-mediated transformation | Knockout | Late blight resistance | [71] |
| StSR4 | RNP-mediated CRISPR/Cas9 GE | Knockout | Late blight resistance | [72] |
| StNPR3 | Agrobacterium-mediated transformation | Knockout | Resistance to Clso | [73] |
| StCoilin gene | RNP-mediated CRISPR/Cas9 GE | Knockout | Potato virus Y | [74] |
| SteIF4E | Agrobacterium-mediated transformation | Knockout | Potato virus Y | [75] |
| SteIF4E1 | Protoplast transfection | Knockout | Potato virus Y | [76] |
| StP3, StCI, StNib, StCP | Agrobacterium-mediated transformation | Virus RNA targeting | Potato virus Y | [77] |
| StPI, StHC-Pro, StP3, StCl1, StCl2, and StVPg | Agrobacterium-mediated transformation | Virus RNA targeting | Potato virus Y | [78] |
| StCP gene | Agrobacterium-mediated transformation | Virus RNA targeting | Multiplex virus resistance PVY, PVS, PVX, or PLRV | [79] |
| Target Gene | Delivery Method | Type of Editing | Improvement | References |
|---|---|---|---|---|
| StDMR6-1 | Agrobacterium- mediated transformation | Knockout | Drought and salt tolerance | [66] |
| StDRO2 | Agrobacterium- mediated transformation | Nonsense mutation | Drought tolerance | [92] |
| StAOX | Agrobacterium- mediated transformation | Knockout | High-light tolerance | [93] |
| StMYB44 | Agrobacterium- mediated transformation | Knockout | Pi transport regulation | [94] |
| StbHLH47 | Agrobacterium- mediated transformation | Knockout | Decreased ferric chelate reductase (FCR) activity | [91] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapakhova, Z.; Kanat, R.; Choi, K.; Daurov, D.; Daurova, A.; Zhambakin, K.; Shamekova, M. CRISPR-Cas Gene Editing Technology in Potato. Int. J. Mol. Sci. 2025, 26, 7496. https://doi.org/10.3390/ijms26157496
Sapakhova Z, Kanat R, Choi K, Daurov D, Daurova A, Zhambakin K, Shamekova M. CRISPR-Cas Gene Editing Technology in Potato. International Journal of Molecular Sciences. 2025; 26(15):7496. https://doi.org/10.3390/ijms26157496
Chicago/Turabian StyleSapakhova, Zagipa, Rakhim Kanat, Khanylbek Choi, Dias Daurov, Ainash Daurova, Kabyl Zhambakin, and Malika Shamekova. 2025. "CRISPR-Cas Gene Editing Technology in Potato" International Journal of Molecular Sciences 26, no. 15: 7496. https://doi.org/10.3390/ijms26157496
APA StyleSapakhova, Z., Kanat, R., Choi, K., Daurov, D., Daurova, A., Zhambakin, K., & Shamekova, M. (2025). CRISPR-Cas Gene Editing Technology in Potato. International Journal of Molecular Sciences, 26(15), 7496. https://doi.org/10.3390/ijms26157496






