Abstract
FLOWERING LOCUS T (FT) is a key integrator of flowering pathways. White lupin, a grain legume, encodes four FT homologs: LalbFTa1, LalbFTa2, LalbFTc1, and LalbFTc2. Widespread distribution of white lupin implies diverse phenological adaptations to contrasting ecosystems. Recent studies highlighted associations between FT indels and flowering regulation. Therefore, we surveyed the global white lupin collection for the presence of such indels and potential links to phenology. A panel of 626 white lupin genotypes, representing several European and African agro-climates, was phenotyped under a long-day photoperiod in a two-year study, showing up to 80 days of flowering time difference between early landraces from Eastern Mediterranean and late accessions from France, Madeira, the Canaries, Greece, Italy, and the Azores. As many as seventeen indel variants were identified for LalbFTc1, twelve for LalbFTa2, nine for LalbFTa1, and four for LalbFTc2, yielding roughly three hundred allelic combinations. Significant correlations with phenology were confirmed for one LalbFTa1 indel and twelve LalbFTc1 indels. A large, highly correlated LalbFTc1 indel was revealed to be conserved among all domesticated Old World lupins, carrying all FTc1-promoter candidate binding sites of the same major floral repressor, AGAMOUS-LIKE 15. A small LalbFTa1 indel, providing additional contribution to earliness, showed homology between white and yellow lupins. LalbFTc1 indel-based PCR markers revealed high discriminatory power towards early (PR_42a and PR_71b) or late (PR_58c, PR_36b, PR_80, and PR_60b) flowering.
1. Introduction
White lupin (Lupinus albus L.) is a grain legume with a relatively long history of cultivation, dating back to ancient Greece and Egypt [1,2,3,4]. This species, currently growing in a wide range of environments from East Africa and Western Asia to Madeira and the Azores, originates putatively from the Eastern Mediterranean region, where wild greacus-type accessions can still be found [5,6]. Since ancient times, white lupin has been partially domesticated and distributed across numerous countries, including Italy, France, Spain, Ethiopia, Germany, Switzerland, Poland, and Australia [7,8,9,10,11,12]. As a low-alkaloid crop (min. 0.02%) with high grain protein (up to 38%) and moderate seed oil content (10–13%), characterized by a favorable ratio of omega-6 to omega-3 acids, white lupin has been recognized as valuable for human consumption [13,14,15,16,17]. White lupin evolved in the temperate climate of the Mediterranean Basin, and as a result, it developed natural adaptations to this environment. One such adaptation is responsiveness to vernalization during the vegetative growth phase, which allows synchronization of flowering time with seasonal weather patterns to enhance the chances of successful reproduction. This mechanism has been observed in numerous white lupin landraces across many countries. For instance, a typical vernalization-responsive white lupin “winter” ecotype achieves flowering competence after prolonged cold exposure of autumn-germinated plants [18,19]. Moreover, as a natural adaptation to summer seed dispersal and subsequent germination before the onset of winter, vernalization-responsive white lupin accessions have developed a degree of frost tolerance and cold hardening ability [20,21,22]. Aside from the winter ecotype, the global germplasm pool of white lupin also includes representatives of a vernalization-independent “spring” ecotype, which is characterized by rapid flowering and low frost tolerance, along with a variety of intermediate forms that show moderate responsiveness to vernalization [18,22,23,24,25,26,27,28]. Breeders have exploited both ecotypes to develop cultivars aligned with the local agro-climatic conditions. The winter ecotype is sown in autumn in regions with mild winters, such as the Mediterranean Basin and Western Europe. In contrast, the spring ecotype is sown after the frost period ends in colder regions, which include central and eastern parts of Europe, the western part of Asia, and the northern territories of North America [7,29,30,31]. In France, the winter ecotype is also effectively intercropped with triticale, significantly suppressing weed growth and increasing total grain yield [32]. In Australia, autumn sowing of white lupin is carried out using vernalization-independent spring accessions to guarantee early flowering [8,33]. In Greece, efforts to reselect early-flowering white lupin germplasm have recently been undertaken [34].
Current studies revealed quantitative control of flowering time in white lupin, evidenced both by linkage mapping of quantitative trait loci (QTLs) in a recombinant inbred line (RIL) population and by a genome-wide association study (GWAS) in a germplasm diversity panel carrying landraces from various eco-geographical locations [10,27,28,35]. In addition to several single-nucleotide polymorphisms (SNPs) and presence/absence variants (PAVs) from different regions of the genome, significant associations with white lupin phenology were also revealed for the newly identified insertion–deletion polymorphism in the promoter region of the FLOWERING LOCUS T homolog, the LalbFTc1 gene (Lalb_Chr14g0364281) [27,35,36]. Similar observations were made for two other Old World domesticated lupin species, narrow-leafed lupin (L. angustifolius) and yellow lupin (L. luteus), up to ~6 kbp upstream of the start codon [37,38,39,40,41,42,43,44]. Moreover, in yellow lupin, besides the LlutFTc1 promoter deletion associated with early phenology and vernalization independence, indels in two other homologs, the LlutFTa1a and LlutFTc2 genes, revealed remarkable associations with flowering time [37]. A question has arisen regarding the level of evolutionary and functional conservation among FT indels in the white lupin genome compared to other cultivated Old World lupin species.
To answer this question, we developed a PCR-based marker array spanning the promoter regions and selected introns of all four FT homologs in the white lupin genome. We implemented this array to assess the newly established large germplasm diversity panel, which comprises 626 genotypes from 32 countries and territories, representing all regions where white lupin currently grows. In parallel, this germplasm panel was subjected to a two-year controlled-environment phenotyping for plant phenology phases. Then, the identified indel polymorphisms in all FT homologs were correlated with the observed white lupin phenology. The shared microsynteny of three Old World lupin species was analyzed in genomic regions where significantly correlated FT indels occur.
2. Results
2.1. Over 80 Days of Flowering Time Difference Was Observed Between Early and Late White Lupin Lines
Data on phenological phases were obtained for all studied genotypes, with a total of 626 lines (Supplementary Tables S1 and S2) in 2020 and 2021. In the first year of observations, the mean number of days from sowing to floral bud emergence ranged from 36.0 to 111.7, from sowing to the start of flowering between 41.3 and 127.0, and from sowing to the end of flowering from 62.0 to 129.0. In the second year, these values were similar, ranging from 33.0 to 114.7 days to floral bud emergence, 40.7 to 121.0 days to the start of flowering, and 54.0 to 128.0 days to the end of flowering. The earliest genotypes included several landraces from Turkey, Syria, and Jordan that developed floral buds 34 to 37 days after sowing and outperformed all domesticated materials in earliness. The earliest cultivars were Butan from Poland (with a mean value of 37.5 days from sowing to floral bud emergence), Lotos from Russia (38.6 days), and Terre from South Africa (38.7 days). In general, early-flowering domesticated materials originated from Russia, Israel, Chile, Hungary, Poland, Greece, Belarus, Germany, Ukraine, and Italy. On the other hand, the latest lines were winter-type cultivars from France—Adam, Aster, and Luxe (104.1–112.8 days)—that developed floral buds about 4–8 weeks later than late-flowering landraces from Madeira (58.7 days), the Canaries (59.6 days), Greece (63.7 days), Italy (64.6 days), and the Azores (76.5 days). High correlation coefficients between years were revealed, reaching 0.97 for days from sowing to floral bud emergence, 0.96 for days from sowing to the start of flowering, and 0.95 for days from sowing to the end of flowering. Slightly decreasing correlation coefficients for subsequent developmental phases may indicate the increasing influence of environmental conditions on the phenology of plants during their growth in the greenhouse. The automated DNA isolation protocol yielded an average concentration of 810.7 ± 329.9 ng/µL DNA (Supplementary Table S3).
2.2. Nine Indels Were Identified in the LalbFTa1 Gene Region, Including One Significantly Correlated with White Lupin Phenology
The white lupin pangenome [45] was screened for the coverage of short-read sequences provided for 39 white lupin varieties (https://www.whitelupin.fr/, accessed on 16 July 2025). Eight candidate structural variants in the LalbFTa1 gene promoter were identified, along with one in the LalbFTa1 third intron. Based on short-read mapping, three lines were selected for sequence alignment: Amiga (Lalb_Chr02 reference), LD37, and P27174. The 13,417 bp long alignment (spanning promoter, all introns and 3′UTR) further confirmed all variants except LD37 deletion of 46 bp (DOI: 10.5281/zenodo.15224868) (Supplementary Figure S1). The alignment included the Lalb_Chr02 sequence and genome sequence contigs: LD37_contig_002595, containing four deletions, three insertions, and one inversion; P27174-4_contig_014309, carrying one deletion; and P27174-4_contig_000651, similar to the reference Amiga sequence. Besides these indels, 160 SNP loci were identified in the white lupin pangenome (Supplementary Table S4).
Markers from the recently published PCR array [27] (Supplementary Table S5) recognized all indel variants (Supplementary Table S6) except three. These include the following: an 8 bp insertion amplified by a PR_10 marker, with a product size difference too small to provide reliable determination on standard agarose gels; a 59 bp insertion not amplified by the PR_05a/PR_05b marker due to primer binding in a 635 bp deletion; and the previously mentioned 46 bp deletion in LD37. To clarify, a PR_13 marker targeting the 46 bp deletion was scored as polymorphic due to an additional shorter polymorphic product visible on a gel; however, it may represent a repetitive element because the main product was identical in all studied lines. The sequence annotated in the pangenome as the 46 bp deletion is a repetitive element present in eighteen LD37 contigs.
To supplement the PR_05a/b PCR marker, which targeted four structural variants and produced complex patterns of PAVs and length polymorphisms, a pair of new PCR markers (PR_74 and PR_75) was designed to fine-tune the scoring accuracy. Similarly, a 59 bp insertion localized at −5649 bp was supplied with a newly developed PR_76 PCR marker. Agarose gel electrophoregrams showing polymorphism of PCR-based markers targeting white lupin LalbFTa1 indels are provided in Supplementary Figure S2.
The screening of a white lupin germplasm testing panel, which included 190 genotypes, along with twenty PCR markers that span the entire promoter and third intron of the LalbFTa1 gene, revealed that ten of the markers were monomorphic and were discarded from further analysis (Table 1, Supplementary Table S7). The remaining ten markers revealed minor allele frequency (MAF) values between 1.1% to 48.9%, with only four below 3% (markers PR_03, PR_04, PR_05b, and QTL11). All polymorphic markers were analyzed across 626 genotypes, yielding similar MAF values to those observed in the germplasm testing panel (Table 1, Supplementary Table S8).

Table 1.
PCR validation of indel polymorphism for the LalbFTa1 (Lalb_Chr02g0156991) gene and minor allele frequency (MAF) values in the white lupin germplasm testing panel (190 genotypes) and full germplasm array (626 genotypes).
Correlation analysis highlighted statistically significant associations with white lupin phenology solely for the PR_09 marker (Supplementary Figure S3, Supplementary Table S9), recognizing a 90 bp deletion located 3572 bp upstream of the transcription start site (TSS) of the LalbFTa1 gene (rρ-value ranging from 0.16 to 0.20, Bonferroni-corrected p-value between 2 × 10−5 and 0.0013). A shorter indel sequence was associated with delayed flowering. Interestingly, the position of this indel relative to the TSS in white lupin corresponds to the position of the homologous indel in the yellow lupin LlutFTa1 gene (3848 bp before TSS), which is also significantly associated with plant phenology (Figure 1, Supplementary Table S10) [37]. However, it has the opposite allelic effect, with a shorter indel sequence linked to accelerated flowering.
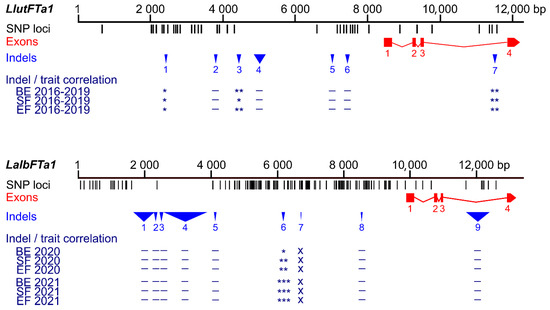
Figure 1.
Comparison of sequence polymorphisms identified in the white lupin LalbFTa1 gene and homologous yellow lupin LluFTa1 gene and correlations of main indels with plant phenology. Black tags visualize SNP and short (≤5 bp) indel loci, whereas red rectangles and blue triangles show exons and longer (≥6 bp) indels, respectively. The Bonferroni-corrected p-value of Spearman’s rank correlation coefficient, calculated for three phenology traits (the number of days to floral bud emergence (BE), start of flowering (SF), and end of flowering (EF)), is shown in the following scheme: ***, p < 0.0001; **, 0.0001 ≤ p < 0.001; *, 0.001 ≤ p ≤ 0.05; –, p > 0.05 (not significant); x, not analyzed. White lupin phenotyping was conducted without pre-sowing vernalization during the 2020 and 2021 growing seasons in a greenhouse located at the Institute of Plant Genetics, Polish Academy of Sciences in Poznań. Phenological and sequence data for yellow lupin were retrieved from [37].
2.3. Twelve Indels Were Identified in the LalbFTa2 Gene Region, but Without Significant Correlation with Plant Phenology
Screening of the white lupin pangenome [45] revealed the presence of twelve potential indels in the LalbFTa2 gene region, nine localized in the promoter, one in the first intron, and two in the third intron. All indels were confirmed by the 11,669 bp long alignment (DOI: 10.5281/zenodo.15224868) (Supplementary Figure S1) of Lalb_Chr21 and genome sequence contigs from lines Clovis (Clovis_contig_006417 carrying one deletion and one insertion) and GR38 (GR38_contig_001073 containing seven insertions and three deletions). Apart from indels, 213 SNP loci were identified in the white lupin pangenome (Supplementary Table S4).
Published PCR markers [27] (Supplementary Table S5) were able to recognize the first five structural variants at the 5′ sequence of the promoter (Supplementary Table S6). Detection of short indels localized at −2681 bp, −1762 bp, −1603 bp, and +700 bp from the TSS was not feasible with simple agarose-based methodology because of a slight length difference of the PCR products between alleles. For the remaining three unsolved indels, located at positions −1194 bp, +1184 bp, and +1900 bp from the TSS, new PCR markers were designed: PR_77, PR_78, and PR_79 (Supplementary Table S5). Among the twenty markers covering the full promoter and the third intron of the LalbFTa2 gene, ten were monomorphic in the white lupin germplasm testing panel of 190 genotypes (Supplementary Table S7). Agarose gel electrophoregrams showing the polymorphism of PCR-based markers targeting white lupin LalbFTa2 gene indels are provided in Supplementary Figure S4.
Polymorphic markers revealed MAF values between 1.3% (PR_79) and 14.5% (PR_16) and were tested on the complete array of 626 genotypes (Table 2, Supplementary Table S8). Then, nine markers revealed MAF values in the 0.5–4.8% range, whereas one (PR_16) was significantly more polymorphic (MAF value 13.8%). All analyzed LalbFTa2 indel markers revealed a lack of significant correlation with the examined traits, with rρ-values between −0.04 and 0.13, as well as p-values between 0.06 and 1 (Supplementary Figure S5, Supplementary Table S9). Visualization of sequence polymorphisms identified in the white lupin LalbFTa2 and the homologous yellow lupin LluFTa2 gene is presented in Figure 2.

Table 2.
PCR validation of indel polymorphism in the LalbFTa2 (Lalb_Chr21g0317021) gene promoter and minor allele frequency (MAF) values in the white lupin germplasm testing panel (190 genotypes) and full germplasm array (626 genotypes).
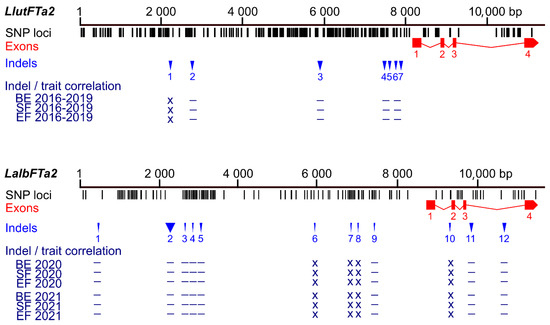
Figure 2.
Comparison of sequence polymorphisms identified in the white lupin LalbFTa2 gene and homologous L. lutes LluFTa2 gene and correlations of main indels with plant phenology. Black tags visualize SNP and short (≤3 bp) indel loci, whereas red rectangles and blue triangles show exons and large (≥4 bp) indels, respectively. The Bonferroni-corrected p-value of Spearman’s rank correlation coefficient, calculated for three phenology traits (the number of days to floral bud emergence (BE), start of flowering (SF), and end of flowering (EF), is shown in the following scheme: ***, p < 0.0001; **, 0.0001 ≤ p < 0.001; *, 0.001 ≤ p ≤ 0.05; –, p > 0.05 (not significant); x, not analyzed (correlations for all analyzed LluFTa2 indel markers were not significant). White lupin phenotyping was conducted without pre-sowing vernalization during the 2020 and 2021 growing seasons in a greenhouse located at the Institute of Plant Genetics, Polish Academy of Sciences in Poznań. Phenological and marker data for yellow lupin were retrieved from [37].
2.4. Seventeen Indel Variants, Including at Least Twelve That Are Significantly Correlated with Plant Phenology, Were Identified in the LalbFTc1 Gene Region
The presence of seventeen indels (including overlapping variants) in the LalbFTc1 gene region was revealed by white lupin pangenome alignment [45], sixteen in the promoter, including nine previously reported [27], and one in the third intron. All indels were confirmed by the 14,708 bp long alignment (DOI: 10.5281/zenodo.15224868) (Supplementary Figure S1) of Lalb_Chr14 and genome sequence contigs from lines P27174 (P27174-4_contig_001786 carrying four deletions and one insertion), GR38 (GR38_contig_001107 with one insertion and three deletions), LD37 (LD37_contig 001630 with four deletions and two insertions), and Sanger-sequenced PCR products obtained for P36a/b and PCR42a/b markers (targeting one insertion and three deletions). Besides indels, 251 SNP loci were identified in the white lupin pangenome (Supplementary Table S4). The PCR markers, including those recently published [27] and those developed in this study (Supplementary Table S5), successfully recognized twelve indel variants and three sets of SNPs located in the binding sites of three primers (Supplementary Table S6).
Large indels, such as the P27174 deletion of 2126 bp, the LD37 deletion of 2388 bp, the GR38 insertion of 3800 bp, the P27174 insertion of 3818 bp, and the LD37 insertion of 3877 bp, were supplemented with several markers, designed to have primer bindings sites flanking indels or anchored in particular deletions. A pair of closely located LAP029B deletions of 7 bp and 28 bp was also supplied with specific primers to facilitate agarose gel-based discrimination of particular variants (markers PR_71a-d). A similar approach was used for an LD37 insertion of 24 bp and an adjacent deletion of 75 bp (marker PR_80).
All analyzed primer pair combinations were polymorphic in the white lupin germplasm testing panel of 190 genotypes (Table 3, Supplementary Table S7). Agarose gel electrophoregrams showing the polymorphism of PCR-based markers targeting white lupin LalbFTc1 gene indels are provided in Supplementary Figure S6. MAF values ranged from 1.1% to 45.3%. A total of 13 primer pairs generated markers with MAF ≥ 10.0. Markers with an MAF value < 1.5 were discarded from complete panel genotyping. Moreover, several polymorphic markers revealed similar segregation patterns, including PR_30 and PR_31 or PR_39, PR_62, PR_67, and PR_70. In these cases, single representative markers (PR_30 and PR_70) were chosen for screening (Supplementary Table S7).

Table 3.
PCR validation of indel polymorphism in the LalbFTc1 (Lalb_Chr14g0364281) gene promoter [27] and minor allele frequency (MAF) values in the white lupin germplasm testing panel (190 genotypes) and full germplasm array (626 genotypes).
As many as 18 markers revealed a significant correlation with white lupin phenological traits (Supplementary Figure S7, Supplementary Table S9). The highest correlation coefficients were identified for markers PR_71d (from −0.43 to −0.40), PR_60c (from 0.31 to 0.35), PR_58c (from −0.34 to −0.31), PR_71a (−0.32), PR_36b (from −0.33 to −0.31), PR_60b (from 0.30 to 0.33), PR_42a (from −0.34 to −0.28), PR_80 (from 0.30 to 0.32), and PR_70 (from −0.31 to −0.28). These markers target LAP029B deletions of 7 bp and 28 bp, LD37 deletion of 2388 bp, LAP022E insertion of 25 bp, LAP022E deletion of 264 bp, LD37 deletion of 70 bp, and LD37 insertion of 18 bp. It is noteworthy that the LD37 deletion of 2388 bp is located at a corresponding position relative to the transcription start site, analogous to the Ku, Jul, and Pal indels in the LanFTc1 promoter that facilitate vernalization-independent early flowering in narrow-leafed lupin, as well as a major indel in the LlutFTc1 promoter, which is associated with vernalization independence in yellow lupin (Figure 3, Supplementary Tables S10 and S11) [27,37,38,41].
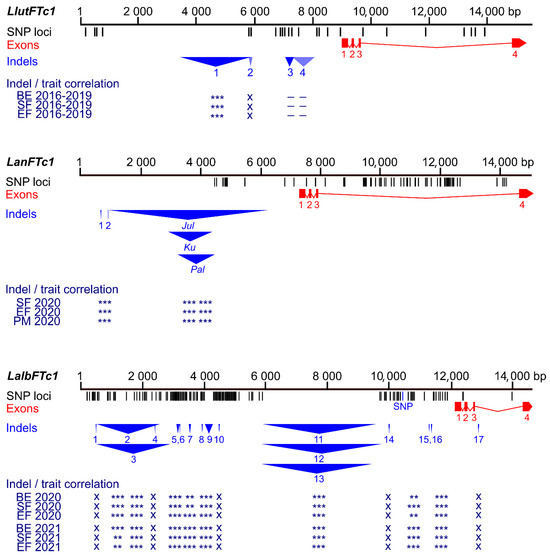
Figure 3.
Comparison of sequence polymorphisms identified in the white lupin LalbFTc1 gene and homologous yellow lupin LluFTc1 and narrow-leafed lupin LanFTc1 genes with indicated correlations of main indels with plant phenology. Black tags visualize SNP and short (≤3 bp) indel loci whereas red rectangles and blue triangles show exons and large (≥4 bp) indels, respectively. The Bonferroni-corrected p-value of Spearman’s rank correlation coefficient, calculated for phenology traits (the number of days to floral bud emergence (BE), start of flowering (SF), end of flowering (EF), and pod maturity (PM)), is shown in the following scheme: ***, p < 0.0001; **, 0.0001 ≤ p < 0.001; *, 0.001 ≤ p ≤ 0.05; –, p > 0.05 (not significant); x, not analyzed. White lupin phenotyping was conducted without pre-sowing vernalization during the 2020 and 2021 growing seasons in a greenhouse located at the Institute of Plant Genetics, Polish Academy of Sciences in Poznań. Phenological and marker data for yellow lupin were retrieved from [37], whereas those for narrow-leafed lupin were retrieved from [39,41], including genotyping performed in this study.
The presence of LAP022E and LAP029B indels was evidenced by sequencing of PCR products obtained for markers PR_36a/b and PR42a/b, whereas the presence of two closely located LD37 indels was shown by sequencing of PR_80 PCR products (DOI 10.5281/zenodo.15223455). Taking into consideration the geographic distribution of analyzed white lupin accessions, two highly associated alleles represented by markers PR_58c and PR_36b were identified predominantly in the Azores, the Canaries, and Greece (Supplementary Tables S1 and S8).
2.5. No Indel Was Found in the LalbFTc2 Gene Promoter, While Four Indels Were Identified in the Introns, Albeit Without Significant Correlation with Phenology
White lupin pangenome alignment [45] revealed no indel in the LalbFTc2 gene promoter. Nevertheless, it highlighted the presence of four indels located between particular exons: one in the second intron and three in the third intron (Supplementary Table S6). The first one, identified in the white lupin line GRC5262B, was visualized by short sequence reads and a GRC5262B_contig_007970 misassembly at approximate positions spanning a length of 209 bp in the reference genome. The region was also marked by the gap between the ends of Dieta_contig_007291 and Dieta_contig_008195, as well as between P27174_contig_011400 and P27174_contig_003576 in the 13,133 bp long alignment (Supplementary Figure S1), including the Lalb_Chr09 sequence (DOI 10.5281/zenodo.15224868). Apart from these indels, 389 SNP loci were identified in the pangenome (Supplementary Table S4).
The screening of the white lupin germplasm testing panel of 190 genotypes using the PR_64 marker targeting the GRC5262B missassembly resulted in a presence/absence variant, suggesting that a large insertion influences PCR amplification. Three other indels (13 bp, 1463 bp, and 8 bp) were localized in the third intron and were identified in alignment with Dieta_contig_007291. The marker PR_73 flanking these indels confirmed the expected PCR product length polymorphism. Apart from the indels, PCR screening with LalbFTc2 gene promoter primers (Supplementary Table S5) revealed one presence/absence variant (marker PR_57), indicating a novel sequence polymorphism absent in the pangenome alignment (Supplementary Tables S6 and S7). Nevertheless, it was rare, identified only in a few white lupin accessions (Table 4). Agarose gel electrophoregrams showing the polymorphism of PCR-based markers targeting white lupin LalbFTc2 gene indels are provided in Supplementary Figure S8.

Table 4.
PCR validation of indel polymorphism in the LalbFTc2 (Lalb_Chr09g0331851) gene promoter and minor allele frequency (MAF) values in the white lupin germplasm testing panel (190 genotypes) and full germplasm array (626 genotypes).
All polymorphic PCR markers (PR_57, PR_64, PR_73) were analyzed in the complete white lupin germplasm panel, revealing MAF values of 0.9%, 2.2%, and 17.4%, respectively (Supplementary Table S8). Although the location of a third LalbFTc2 indel corresponds to the LlutFTc2 indel, which is associated with photoperiodic responsiveness in yellow lupin [37], the correlation of the corresponding PR_73 marker with white lupin phenology was negligible, with rρ-values of 0.09–0.12 and non-significant p-values (Supplementary Figure S9, Supplementary Table S8).
Non-significant correlations were also found in PR_64 and PR_57 markers, representing the first indel and the presence/absence variant, respectively. Interestingly, the PR_57 marker turned out to be highly selective towards winter-type cultivar Aster (represented by lines LAP119c and LAP119d), with only three false-positive homozygotes and one heterozygote in the complete array of 626 genotypes (99.4% accuracy). A comparison of sequence polymorphisms identified in the white lupin LalbFTc2 gene and the homologous yellow lupin LluFTc2 gene and the correlations of main indels with plant phenology is presented in Figure 4.
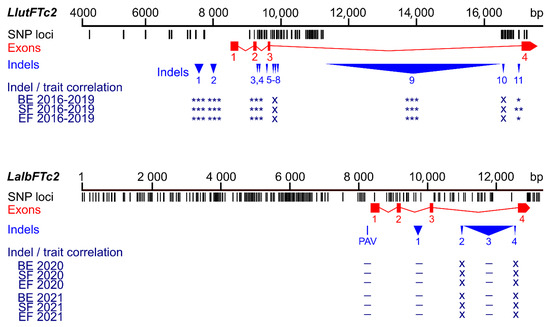
Figure 4.
Comparison of sequence polymorphisms identified in the white lupin LalbFTc2 gene and homologous yellow lupin LluFTc2 gene and correlations of main indels and a presence/absence variant (PAV) with plant phenology. Black tags visualize SNP and short (≤5 bp) indel loci, whereas red rectangles and blue triangles show exons and large (≥6 bp) indels, respectively. p-value of Spearman’s rank correlation coefficient with Bonferroni correction, calculated for three phenology traits (the number of days to floral bud emergence (BE), start of flowering (SF), and end of flowering (EF)), is shown in the following scheme: ***, p < 0.0001; **, 0.0001 ≤ p < 0.001; *, 0.001 ≤ p ≤ 0.05; –, p > 0.05 (not significant); x, not analyzed. White lupin phenotyping was conducted without pre-sowing vernalization during the 2020 and 2021 growing seasons in a greenhouse located at the Institute of Plant Genetics, Polish Academy of Sciences in Poznań. Phenological and marker data for yellow lupin were retrieved from [37].
2.6. LalbFTc1 Promoter Indels Carry Potential Binding Sites for Transcription Factors Involved in Flowering Control
As LalbFTc1 indels revealed the most significant correlations with white lupin phenology (Figure 5), in silico analysis of transcription factor binding sites was performed. An alignment of 14,708 bp carrying all indels recognized in the LalbFTc1 gene region (DOI 10.5281/zenodo.15224868) was submitted to the PlantPAN for annotation of transcription factor binding sites targeting the region upstream of the LalbFTc1 TSS (located at 12,009 bp in the alignment). This analysis revealed that many transcription factors (115) have all their LalbFTc1 promoter binding sites localized exclusively in polymorphic regions (indels or SNPs) (Supplementary Table S12). This set included 18 elements from flowering regulatory pathways, which had potential binding sites, sometimes even in two or more indels. Thus, the overlapping section of 2126 and 2388 indels revealed binding sites for PHYA-INDUCED MOTIF (SORLIP2AT), ABSCISIC ACID RESPONSIVE ELEMENT-BINDING FACTOR 1 or 4 (ABF1 or ABF4), FLOWERING BHLH 4 (FBH4), GATA TRANSCRIPTION FACTOR 25 (GATA25), AGAMOUS-LIKE 15 (AGL15), and INDETERMINATE(ID)-DOMAIN 7 (IDD7), while the remaining part of the 2388 indel contained another AGL15 site. A 264 bp deletion contained an additional IDD7 site, whereas the largest 3800–3877 bp indels included additional binding sites for SORLIP2AT, GATA25, and AGL15, as well as unique loci for DWARF AND DELAYED FLOWERING 2 (DDF2), NAC DOMAIN CONTAINING PROTEIN 50 (NAC050), ELONGATED HYPOCOTYL 5 (HY5), AGAMOUS-LIKE 71 (AGL71), and TGA4/OCTOPINE SYNTHASE (OCS)-ELEMENT-BINDING FACTOR 4 (OBF4). Moreover, LAP022E carried binding sites of MYB30 and MYB73 at SNP loci. The list of transcription factors from flowering regulatory pathways that revealed LalbFTc1 binding sites exclusively in the polymorphic regions of the promoter is provided in Table 5, whereas their putative roles are presented in the Discussion Section.
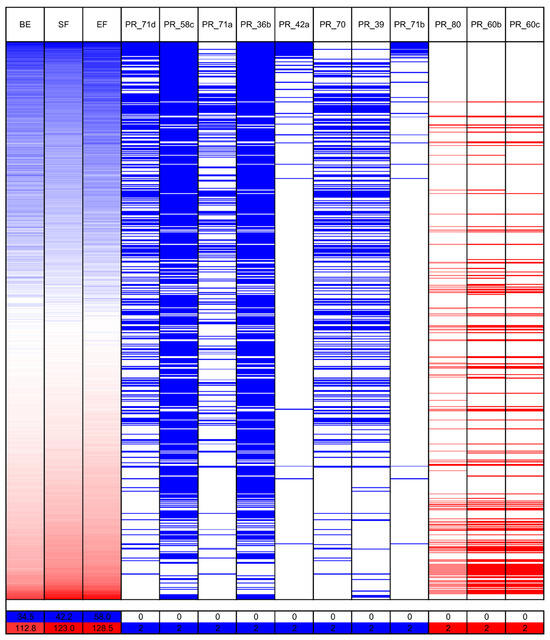
Figure 5.
Allelic composition heatmap of PCR markers that revealed the highest correlations with white lupin phenology: (the number of days to floral bud emergence (BE), start of flowering (SF), and end of flowering (EF)). The bar below the heatmap indicates the color legend of phenological observations and PCR marker alleles recorded for 626 white lupin genotypes. White lupin phenotyping was conducted without pre-sowing vernalization during the 2020 and 2021 growing seasons in a greenhouse located at the Institute of Plant Genetics, Polish Academy of Sciences in Poznań.

Table 5.
The list of flowering-related transcription factors that have binding sites only in the polymorphic regions of the LalbFTc1 promoter alignment.
3. Discussion
3.1. Conserved Function of FTc1 Promoter Indels in Domesticated Old World Lupin Species
In the present study, significant correlations were identified between the number of days from sowing to reaching a specific growth phase (bud emergence, start of flowering, and end of flowering) and indel polymorphism in the regulatory regions of the LalbFTc1 gene and, to a lesser extent, the LalbFTa1 gene. Interestingly, the positions of partially overlapping LalbFTc1 indels of 2126 bp and 2388 bp associated with early flowering of white lupin correspond to the positions of partially overlapping LanFTc1 indels of 1423 bp, 5162 bp, and 1208 bp underlying Ku, Jul, and Pal, conferring early flowering of narrow-leafed lupin, as well as the LlutFTc1 indel of 2227 bp associated with early flowering of yellow lupin [27,37,38,41,44]. Additionally, the position of a LalbFTa1 indel of 90 bp significantly correlated with flowering time in white lupin and converged with the localization of the LlutFTa1 indel of 58 bp, which is significantly associated with plant phenology in yellow lupin [37]. However, the other two white lupin LalbFTa1 indels, which are localized at similar positions as the two significantly associated LlutFTa1 indels, did not show such associations in the analyzed panel of 626 white lupin genotypes. One of these indels, represented by a QTL11 marker, is located in the third intron of the LalbFTa1 gene. This indel is perfectly co-segregated with one of just a few major QTLs for flowering time in a mapping population derived from a cross between domesticated early-flowering Kiev Mutant and late-flowering Ethiopian landrace P27174 [10,35]. In the current study, the Ethiopian LalbFTa1 allele was found in nine accessions at the homozygous phase and six as heterozygotes. Homozygous lines included two Ethiopian landraces, one Italian landrace (LAP098a), and one Polish breeding line (95441, Wat × L. graecus Boiss.). Three of these lines were early-flowering, whereas the remaining ones revealed late flowering time, like the P27174 landrace. The breakdown of the genotype–phenotype association for this locus in two lines (95133 and 95441) carrying the late QTL11 allele in a homozygous state may result from the presence, in the genomes of these lines, of several LalbFTc1 indels related to earliness, evidenced by the corresponding PR_36b, PR_39, PR_41, PR_42b, PR_58c, PR_60a, PR_60b, PR_60c, PR_70, PR_71a, PR_71b, PR_71c, and PR_71d marker scores. Haplotype analysis of LalbFTc1 promoter indels could improve the correlation study; however, the white lupin genome was recently revealed to have very rapid LD decay [11,27,46,47], and we observed this phenomenon also around FT promoters in our germplasm collection. Therefore, we decided to focus on particular indels rather than on co-segregating markers. Some level of recombination occurred even between two closely located markers, separated by just a few hundred base pairs. Our recent white lupin GWAS highlighted significant association of LalbFTc1 indels with plant phenology and vernalization responsiveness; however, that study was focused on non-domesticated germplasm [27]. In the present study, we supplemented a large panel of landraces with a substantial number of materials representing more than 40 years of white lupin breeding programs in Europe. As this analysis does not control for kinship or relatedness at other genetic loci, it cannot be ruled out that the true genetic basis for the observed phenological variation is conferred by other correlated genetic markers. This issue may be resolved by genome complexity reduction-based sequencing of developed genetic diversity panel and association analysis including both PCR-based and genome-wide markers. Such an approach is currently being implemented.
Interestingly, the flowering time in the narrow-leafed lupin is controlled by a single major gene (LanFTc1) with very few minor QTLs. In contrast, in white and yellow lupins, this trait showed significant quantitative variation, suggesting an involvement of several genes from flowering regulatory pathways [27,28,41,43,48,49,50].
3.2. AGL15 Is the Most Likely the Main Transcription Factor in Maintaining the Late-Flowering Phenotype of White Lupin
The common indel-specific transcription factor identified for FTc1 homologs in three domesticated Old World lupin species is AGL15 [37,41]. AGL15 is a known floral repressor that binds directly to the FT gene promoter at sites partially overlapping with those bound by the floral repressors SHORT VEGETATIVE PHASE (SVP) and FLOWERING LOCUS C (FLC) [51,52,53,54,55]. Moreover, AGL15 is also involved in epigenetic silencing of FT chromatin to prevent precocious flowering in response to long days in Arabidopsis [56]. The white lupin genome, like many other legumes, lacks the FLC gene [28,35,57]; however, it putatively contains AGL15 homologs, as BLAST 2.16.0 alignment of the Glycine max AGL15 coding sequence (AY370659.1) to the white lupin CDS database [58] matched three loci (Lalb_Chr19g0140231, Lalb_Chr01g0000501, and Lalb_Chr04g0263791) with high sequence identity (about 75%) and significant e-values (between 5.2 × 10−128 and 1.9 × 10−133). Therefore, it is possible that AGL15 is a major FT repressor in white lupin, and the elimination of all AGL15 binding sites from the LalbFTc1 promoter releases this gene from negative control and triggers flowering.
Apart from AGL15, a few other candidate transcription factors were highlighted, including ABF1, DDF2, FBH4, NAC050, GATA25, HY5, AGL71, OBF4, IDD7, MYB30, and MYB73. ABF1 is a drought-inducible transcription factor that delays flowering in rice [59]. DDF2 is closely related to DDF1, which confers a dwarf and late-flowering phenotype in Arabidopsis [60]. FBH4 accelerates Arabidopsis flowering in low-nitrogen conditions by transcriptional activation of the direct target CONSTANS (CO) and downstream florigen (FT) genes [61,62]. NAC050 is involved in the repression of floral integrator genes, including FT, providing a late-flowering phenotype of Arabidopsis [63,64]. GATA25 acts as an expression activator and accelerates the flowering time of Arabidopsis, directly binding to the promoters of several flowering pathway genes, including FT [65]. HY5 acts as an epigenetic repressor of flowering time in Arabidopsis [66]. Moreover, overexpression of poplar HY5 in Arabidopsis also results in prolonged vegetative growth and delayed flowering [67]. AGL71 promotes flowering through a gibberellin-dependent pathway [68] and OBF4 interacts with CO and binds to the FT promoter [69], whereas IDD7 is responsible for the inhibition of multiple traits, including plant growth and development, including the transition from the vegetative to generative phase [70]. MYB30 is a CO-independent FT activator accelerating flowering irrespective of the length of photoperiod [71], whereas MYB73 is an FT activator negatively targeted by HETEROCHROMATIN PROTEIN 1 (LHP1) [72]. Taking into consideration the direction of phenotypic effects of indel allelic phases and transcription factors, the best repressors for 2126 bp and 2388 bp deletions, which accelerate flowering, are AGL15 and IDD7. In contrast, for 3800–3877 bp insertions that delay flowering, AGL15, DDF2, and HY5 are the best repressors.
3.3. Novel Perspectives for White Lupin Breeding Towards Spring Sowing in a Changing Climate
The present study highlighted the vast diversity of white lupin germplasm in plant phenology, revealing the presence of numerous very early, vernalization-independent landraces, predominantly from Turkey and Syria, which flowered earlier than current cultivars and breeding lines developed for spring-sown cultivation in Central and Eastern Europe. This finding is coherent with previous reports on the earliness of Turkish white lupin germplasm [28,73]. Indeed, population structure analysis based on DArT-seq markers revealed that very early Turkish, Jordanian, and Syrian landraces are genetically distinct from other potential donors of earliness from Egypt, Kenya, Israel, and Sudan, as well as from French spring cultivars [27]. It would be highly advantageous for white lupin breeders to have several different genetic sources of essential traits, as these will help to avoid the critical domestication bottleneck effect. Such a bottleneck unintentionally occurred in another Old World lupin species, narrow-leafed lupin, when only single donors of major traits were available during major domestication efforts targeting earliness, sweetness, seed coat permeability, and anthracnose resistance [74,75,76,77]. Furthermore, white lupin breeders have already reported substantial challenges in incorporating a key agronomic trait, specifically anthracnose resistance, into standard (Ukrainian) sources of early flowering [8,33,78]. The presence of diverse genetic donors of earliness offers greater flexibility in selecting components for crossbreeding. There is a range of heritable traits that could be transferred between particular genotypes, including, besides those mentioned, anthracnose resistance [11,12,50,79,80,81,82], drought tolerance [46,83], calcareous soil adaptation [47,84,85,86], low alkaloid content [15,87,88,89,90], and resistance to Diaporthe toxica and Pleiochaeta setosa causing Phomopsis blight and Pleiochaeta root rot diseases [91,92], as well as yield-related traits, such as pod length, number of seeds in a pod, thousand grain weight, and number of seeds per plant [93,94,95]. Numerous white lupin genotypes in the present study exhibited intermediate phenology, representing a wide range of flowering dates. They constitute valuable resources for addressing ongoing climate change by producing adapted cultivars exploiting growing seasons extended in both directions, with earlier sowing and later maturity [96]. Current climate change (global warming) has already resulted in significant advancement of spring phenology and the onset of flowering of plants, reaching a few days per 1 °C of spring warming [97,98,99]. Nevertheless, the patterns are not uniform across species and regions (including even delay of flowering in some cases) and exhibit significant latitudinal gradients [97,100]. Earlier spring sowing phenology seems to be beneficial in the context of yield under the future climate. In contrast, maturity time requires adaptation to the water regime, contrasting regions with water limitations (earlier maturity to avoid terminal water stress), and regions without significant water shortage during grain filling in summer (later maturity to exploit the full range of available growing season) [96]. European farmers already responded to ongoing climate changes by advancing spring activities as a form of agronomic adaptation to observed trends [101]. White lupin breeders should follow them by releasing thermoneutral, anthracnose-resistant spring cultivars with increased drought tolerance and phenology adapted to the lengths of current and future growing seasons at target cultivation sites.
4. Materials and Methods
4.1. White Lupin Germplasm
The germplasm panel analyzed in this study consisted of 626 genotypes: 313 accessions provided by Poznań Plant Breeding Ltd. (Wiatrowo, Poland) and 313 genotypes selected from 120 accessions based on their phenological diversity within accessions [102] that were provided by the Council for Agricultural Research and Economics (Lodi, Italy). The list of genotypes, including countries/regions of origin, domestication status, and germplasm donors, is provided in Supplementary Table S1. Genotypes originate from 31 countries or territories (i.e., Canaries, Madeira, and Azores) and differ by domestication status: 386 are landraces, 135 wild or primitive accessions, 74 cultivars, 28 breeding lines, and 3 mutants. Domesticated material was selected from major white lupin cultivation countries such as Poland (27 lines), France (22), Germany (13), Ukraine (6), Belarus (6), Russia (5), South Africa (5), Chile (4), Spain (4), Portugal (3), and others, located primarily in the temperate subcontinental climate zone. Landrace collection sites represent a wide range of climates, including tropical and subtropical (Ethiopia), cold semi-arid (Anatolia and Maghreb), dry-summer Mediterranean (sites around the Mediterranean Basin), warm-summer Mediterranean (the Azores and Madeira), and humid temperate (i.e., oceanic, France and Portugal). These regions also diverged in the length of the photoperiod during the white lupin growing season, ranging from approximately 9–10 h in winter sowing in the northern areas of the Mediterranean Basin to 11–12 h in Ethiopia and 12–17 h in spring sowing in other regions of Europe.
4.2. Phenotyping of White Lupin Growth Transition into the Generative Phase
Experiments were conducted without pre-sowing vernalization in a greenhouse located at the Institute of Plant Genetics, Polish Academy of Sciences, Poznań, Poland (52°26′ N 16°54′ E), with an air temperature above 18 °C. Seeds were sown by the end of winter (19 March 2020 and 11 March 2021), and plants were grown under an ambient long-day photoperiod, increasing during plant growth from about 12 h in March to greater than 16 h in June and July. Phenology traits were phenotyped as the number of days from sowing to reaching a specific developmental stage. Floral bud emergence was inspected every second day. The start of flowering was recorded when the first fully colored petals on the main stem appeared, and the end of flowering was registered when half of the petals on the primary inflorescence had faded. All observations were made with a minimum of three and a maximum of ten biological replicates. The experimental design (positions of pots in the greenhouse) is provided in Supplementary Table S2.
4.3. DNA Isolation from the White Lupin Germplasm Panel
Two medium-sized young leaves (about 100–150 mg tissue in total) were collected from 5-week-old plants into 2 mL microcentrifuge tubes (Eppendorf, Hamburg, Germany), immediately frozen in liquid nitrogen and stored in an ultra-low temperature freezer at −70 °C. Homogenization of frozen tissue was performed with two stainless steel beads (ø 5 mm, Qiagen, Hilden, Germany) using TissueLyser II (Qiagen) at 30 rpm for 30 s. The automated isolation system Maxwell® RSC 48 Instrument (Promega, Mannheim, Germany) and the Maxwell® RSC PureFood GMO and Authentication Kit (Promega) [103] were used for DNA isolation without any changes to the standard protocol. DNA concentration and quality (Supplementary Table S3) were measured using a NanoDrop 2000 (ThermoFisher Scientific, Warsaw, Poland). DNA was isolated from 3 plants per genotype separately and subsequently mixed in equal amounts of DNA to provide a final 100 ng/µL concentration (this approach enabled us to detect possible heterogeneity within genotypes).
4.4. PCR-Based Genotyping of Indel Polymorphism in the FT Gene Promoters
PCR-based markers were anchored in the regulatory regions of four FT homologs: LalbFTa1 (Lalb_Chr02g0156991), LalbFTa2 (Lalb_Chr21g0317021), LalbFTc1 (Lalb_Chr14g0364281), and LalbFTc2 (Lalb_Chr09g0331851), spanning by overlapping products the region from 8 kbp upstream of the transcription start site to the first exon of the analyzed genes [27]. Moreover, based on the sequence alignment of the white lupin pangenome sequence [45] and results of preliminary genotyping [27], three additional PCR-based markers (PR_74, PR_75, and PR_76) were designed for LalbFTa1 indels, three (PR_77, PR_78, and PR_79) for LalbFTa2 indels, five (PR_60, PR_62, PR_66, PR_67, and PR_80) for LalbFTc1 indels, and two (PR_64 and PR_73) for LalbFTc2 indels. Sequence alignment was performed using the progressive Mauve algorithm [104], assuming genome collinearity, whereas PCR primers were designed using Primer 3 Plus [105]. Both programs were run in Geneious Prime 2025.0.3 [106]. The list of primer sequences and corresponding white lupin genome coordinates is provided in Supplementary Table S5.
GoTaq G2 Flexi DNA Polymerase (Promega) was used to amplify PCR products. The PCR protocol included initial denaturation (94 °C, 3 min) and 35 cycles composed of three phases—denaturation (94 °C, 30 s), annealing (56–62 °C, 30 s), and elongation (72 °C, 30 s up to 1 kbp, 60 s for longer products)—followed by the final elongation (72 °C, 5 min). The visualization of length differences between alleles was conducted using agarose gel electrophoresis, with the agarose concentration (1–3%) inversely proportional to the size of the expected products. For small indels (shorter than about 50 bp), high-resolution 3:1 agarose (Serva, Heidelberg, Germany) was used, whereas for larger indels, wide-range agarose (Serva) was preferred. Moreover, for comparison of FTc1 allelic effects between lupin species, narrow-leafed lupin germplasm [39] was genotyped with LanFTc1 indel markers [43]. Sanger sequencing of selected PCR products was performed by Genomed (Warsaw, Poland).
4.5. Correlation Analysis
A Kolmogorov–Smirnov test was performed to check the normality of phenotypic data [107]. This test revealed that the datasets for all analyzed phenological traits in both years significantly diverged from a normal distribution. Therefore, a calculation of the Spearman rank correlation was performed. The ranks for phenotypic observations were determined by using the formula =RANK.AVG in Microsoft Excel. Correlations were calculated between rank values and marker scores (0, 1, 2).
To determine if the obtained correlation coefficient was statistically significant, a two-tailed t-test was performed with Bonferroni correction applied by multiplying each calculated p-value by the number of tested markers. To compare specific allelic effects between species, correlation calculations were also conducted for narrow-leafed lupin LanFTc1 indels using phenotypic observations for 126 accessions reported in field studies [39]. This was performed alongside analysis of yellow lupin LlutFTa1, LlutFTa2, LlutFTc1, and LlutFTc2 indels using controlled-environment phenotypic observations and genotypic data published for 111 accessions [37].
4.6. Annotation of Transcription Factor Binding Sites in LalbFTc1 Promoter
To investigate whether the observed sequence polymorphism in the LalbFTc1 promoter may be associated with the presence or absence of binding sites for specific transcription factors from flowering regulatory pathways, we constructed sequence alignments using the reference Amiga sequence and contigs LD37_contig_001630, P27174-4_contig_001786, and GR38_contig_001107 between the PRFTC1F1 primer and the LalbFTc1 transcription start site. Additionally, we sequenced PR_42 PCR products for lines LAP029B and LAP030A between PRFTC1F5 and PRFTc1_R5b primers and sequenced PR_36 PCR products for lines LAP022B and LAP022E between PRFTC1F3 and PRFTc1_R3b primers. Sequences were extracted from the alignments, with gaps representing indel polymorphism and submitted to the Plant Promoter Analysis Navigator (PlantPAN) [108,109] using the Arabidopsis thaliana transcription factors database and by applying a similarity threshold of 0.85. The identified transcription factors were annotated using the genes that control flowering time in Arabidopsis [110]. Additionally, allele-specific transcription factors with potential binding sites identified solely in the polymorphic loci were screened in the literature database for their potential involvement in flowering time regulation.
5. Conclusions
The present study evidenced a high structural variation in promoter sequences of three FT homologs and introns of all four FT genes encoded by the white lupin genome. More than three hundred allelic combinations of FT indels were found in the white lupin germplasm panel by PCR screening, of which one-third could be attributed to the LalbFTc1 gene. The LalbFTc1 promoter was evidenced to carry not only the highest number of indels among all white lupin FT homologs but also the largest ones, up to 3877 bp in length. Moreover, at least twelve LalbFTc1 promoter indels demonstrated significant correlations with plant phenology, and some also showed high discriminatory power towards earliness. A narrow list of indel-specific transcription factors was annotated for the LalbFTc1 promoter, highlighting AGL15 as the most plausible candidate. LalbFTc1 indel-based elimination of all binding sites for important repressive transcription factor(s) from flowering regulatory pathways seems to be a conserved mechanism in Old World lupins, as it was shown to be present in three distinct lineages (white lupin, narrow-leafed lupin, and yellow lupin). Besides contributing to the general knowledge of indel polymorphisms around FT genes and their relations with plant phenology, our study provided support for the ongoing domestication of white lupin as a crop, highlighting key structural mutations that breeders can target to align white lupin growing season requirements with the characteristics of the growing periods experienced in a given location.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms26146858/s1.
Author Contributions
Conceptualization, M.K. and S.R.-B.; methodology, W.B., S.R.-B., and A.S.; validation, W.B., S.R.-B., and A.S.; formal analysis, M.K.; investigation, W.B., S.R.-B., and A.S.; data curation, W.B., S.R.-B., A.S., and M.K.; writing—original draft preparation, M.K.; writing—review and editing, M.K., W.B., and S.R.-B.; visualization, M.K.; supervision, M.K. and S.R.-B.; funding acquisition, S.R.-B. and M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Centre, Poland projects SONATINA3 no. 2019/32/C/NZ9/00055, SONATA17 no. 2021/43/D/NZ9/00293 (white lupin research) and OPUS21 project no. 2021/41/B/NZ9/02226 (yellow lupin analysis). An open access charge was co-financed by Wroclaw University of Environmental and Life Sciences as well as by National Science Centre, Poland.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data generated during this study are included in this published article and its supplementary information files. Moreover, sequence alignments constructed for the LalbFTa1, LalbFTa2, LalbFTc1, and LalbFTc2 genes as well as Sanger sequencing chromatogram alignments for PCR products are available in the Zenodo repository under accession numbers 15223456 (DOI 10.5281/zenodo.15223455) https://zenodo.org/records/15223456 and 15224869 (DOI 10.5281/zenodo.15224868) https://zenodo.org/records/15224869, both records accessed on 3 June 2025.
Acknowledgments
The authors would like to acknowledge the National Research Institute for Agriculture, Food and Environment (INRAE) and Paolo Annicchiarico from the Council for Agricultural Research and Economics, Research Centre for Animal Production and Aquaculture, for providing the landrace accessions of white lupin subjected to controlled-environment phenotyping in the study. We thank Jolanta Belter from the Department of Gene Structure and Function, Institute of Plant Genetics, Polish Academy of Sciences, for assistance in greenhouse experiments.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- van der Veen, M. Consumption, Trade and Innovation. Exploring the Botanical Remains from the Roman and Islamic Ports at Quseir a-Qadim, Egypt; Africa Magna: Frankfurt, Germany, 2012; p. 313. [Google Scholar]
- Kurlovich, B.S. Lupins: Geography, Classification, Genetic Resources and Breeding; Kurlovich, B.S., Ed.; Intan: St. Petersburg, Russia, 2002. [Google Scholar]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World, 4th ed.; Oxford University Press: Oxford, UK, 2012; p. 264. [Google Scholar]
- Gladstones, J.S. Lupins as crop plants. Field Crop Abstr. 1970, 23, 26. [Google Scholar]
- Wolko, B.; Clements, J.C.; Naganowska, B.; Nelson, M.N.; Yang, H.A. Lupinus. In Wild Crop Relatives: Genomic and Breeding Resources; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 153–206. [Google Scholar]
- Gladstones, J.S. The Mediterranean white lupin. J. Dep. Agric. West. Aust. Ser. 4 1976, 17, 70–74. [Google Scholar]
- Gresta, F.; Wink, M.; Prins, U.; Abberton, M.T.; Capraro, J.; Scarafoni, A.; Hill, G. Lupins in European cropping systems. In Lupins in European Cropping Systems. Legumes in Cropping Systems; Murphy-Bokern, D., Stoddard, F., Watson, C., Eds.; CABI Publishing: Wallingford, UK, 2017; pp. 88–108. [Google Scholar]
- Adhikari, K.N.; Thomas, G.; Diepeveen, D.; Trethowan, R. Overcoming the barriers of combining early flowering and anthracnose resistance in white lupin (Lupinus albus L.) for the Northern Agricultural Region of Western Australia. Crop Pasture Sci. 2013, 64, 914–921. [Google Scholar] [CrossRef]
- Yeheyis, L.; Mekonnen, W.; Nelson, M.; McNaughton, D.; Tarekegn, A.; Yadelew, Z.; Sanders, H. The search for commercial sweet white lupin (Lupinus albus L.) adaptive to Ethiopian growing condition seems not successful: What should be done? Z. Naturforsch C J. Biosci. 2023, 78, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Książkiewicz, M.; Nazzicari, N.; Yang, H.; Nelson, M.N.; Renshaw, D.; Rychel, S.; Ferrari, B.; Carelli, M.; Tomaszewska, M.; Stawiński, S.; et al. A high-density consensus linkage map of white lupin highlights synteny with narrow-leafed lupin and provides markers tagging key agronomic traits. Sci. Rep. 2017, 7, 15335. [Google Scholar] [CrossRef] [PubMed]
- Alkemade, J.A.; Nazzicari, N.; Messmer, M.M.; Annicchiarico, P.; Ferrari, B.; Voegele, R.T.; Finckh, M.R.; Arncken, C.; Hohmann, P. Genome-wide association study reveals white lupin candidate gene involved in anthracnose resistance. Theor. Appl. Genet. 2022, 135, 1011–1024. [Google Scholar] [CrossRef] [PubMed]
- Alkemade, J.; Messmer, M.; Arncken, C.; Leska, A.; Annicchiarico, P.; Nazzicari, N.; Książkiewicz, M.; Voegele, R.T.; Finckh, M.; Hohmann, P. A high-throughput phenotyping tool to identify field-relevant anthracnose resistance in white lupin. Plant Dis. 2021, 105, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Arnoldi, A.; Greco, S. Nutritional and nutraceutical characteristics of lupin protein. Nutrafoods 2011, 10, 23–29. [Google Scholar] [CrossRef]
- Boschin, G.; D’Agostina, A.; Annicchiarico, P.; Arnoldi, A. The fatty acid composition of the oil from Lupinus albus cv. Luxe as affected by environmental and agricultural factors. Eur. Food Res. Technol. 2007, 225, 769–776. [Google Scholar] [CrossRef]
- Kroc, M.; Rybiński, W.; Wilczura, P.; Kamel, K.A.; Kaczmarek, Z.; Barzyk, P.; Święcicki, W. Quantitative and qualitative analysis of alkaloids composition in the seeds of a white lupin (Lupinus albus L.) collection. Genet. Resour. Crop Evol. 2017, 64, 1853–1860. [Google Scholar] [CrossRef]
- Pereira, A.; Ramos, F.; Sanches Silva, A. Lupin (Lupinus albus L.) seeds: Balancing the good and the bad and addressing future challenges. Molecules 2022, 27, 8557. [Google Scholar] [CrossRef] [PubMed]
- Prusinski, J. White lupin (Lupinus albus L.)—Nutritional and health values in human nutrition—A review. Czech J. Food Sci. 2017, 35, 95–105. [Google Scholar] [CrossRef]
- Adhikari, K.N.; Buirchell, B.J.; Sweetingham, M.W. Length of vernalization period affects flowering time in three lupin species. Plant Breed. 2012, 131, 631–636. [Google Scholar] [CrossRef]
- Huyghe, C. Winter growth of autumn-sown white lupin (Lupinus albus L.) main apex growth model. Ann. Bot. 1991, 67, 429–434. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Iannucci, A. Winter survival of pea, faba bean and white lupin cultivars across contrasting Italian locations and sowing times, and implications for selection. J. Agric. Sci. 2007, 145, 611–622. [Google Scholar] [CrossRef]
- Shield, I.F.; Scott, T.; Stevenson, H.J.; Leach, J.E.; Todd, A.D. The causes of over-winter plant losses of autumn-sown white lupins (Lupinus albus) in different regions of the UK over three seasons. J. Agric. Sci. 2000, 135, 173–183. [Google Scholar] [CrossRef]
- Huyghe, C.; Papineau, J. Winter development of autumn sown white lupin: Agronomic and breeding consequences. Agronomie 1990, 10, 709–716. [Google Scholar] [CrossRef]
- Rahman, M.; Gladstones, J. Control of lupin flower initiation by vernalization, photoperiod and temperature under controlled environment. Aust. J. Exp. Agric. 1972, 12, 638–645. [Google Scholar] [CrossRef]
- Clapham, W.M.; Willcott, J.B. Thermosensitivity in Spring White Lupin. Ann. Bot. 1995, 76, 349–357. [Google Scholar] [CrossRef]
- Putnam, D.H.; Simmons, S.R.; Hardman, L.L. Vernalization and Seeding Date Effects on Yield and Yield Components of White Lupin. Crop Sci. 1993, 33, 1076–1083. [Google Scholar] [CrossRef]
- Rahman, M.; Gladstones, J. Effects of temperature and photoperiod on flowering and yield components of lupin genotypes in the field. Aust. J. Exp. Agric. 1974, 14, 205–213. [Google Scholar] [CrossRef]
- Rychel-Bielska, S.; Bielski, W.; Surma, A.; Annicchiarico, P.; Belter, J.; Kozak, B.; Galek, R.; Harzic, N.; Książkiewicz, M. A GWAS study highlights significant associations between a series of indels in a FLOWERING LOCUS T gene promoter and flowering time in white lupin (Lupinus albus L.). BMC Plant Biol. 2024, 24, 722. [Google Scholar] [CrossRef] [PubMed]
- Rychel-Bielska, S.; Surma, A.; Bielski, W.; Kozak, B.; Galek, R.; Książkiewicz, M. Quantitative control of early flowering in white lupin (Lupinus albus L.). Int. J. Mol. Sci. 2021, 22, 3856. [Google Scholar] [CrossRef] [PubMed]
- López-Bellido, L.; Fuentes, M.; Lhamby, J.C.B.; Castillo, J.E. Growth and yield of white lupin (Lupinus albus) under Mediterranean conditions: Effect of sowing date. Field Crop. Res. 1994, 36, 87–94. [Google Scholar] [CrossRef]
- Yakovenko, G.L.; Lukashevisn, M.I.; Ageeva, P.A.; Novik, N.V.; Zakharova, M.V. Status and prospects of breeding of cultivated species of Lupin in Russia. IOP Conf. Ser. Earth Environ. Sci. 2021, 663, 012014. [Google Scholar] [CrossRef]
- Faluyi, M.A.; Zhou, X.M.; Zhang, F.; Leibovitch, S.; Migner, P.; Smith, D.L. Seed quality of sweet white lupin (Lupinus albus) and management practice in eastern Canada. Eur. J. Agron. 2000, 13, 27–37. [Google Scholar] [CrossRef]
- Carton, N.; Naudin, C.; Piva, G.; Corre-Hellou, G. Intercropping Winter Lupin and Triticale Increases Weed Suppression and Total Yield. Agriculture 2020, 10, 316. [Google Scholar] [CrossRef]
- Adhikari, K.; Buirchell, B.; Yan, G.; Sweetingham, M. Two complementary dominant genes control flowering time in albus lupin (Lupinus albus L.). Plant Breed. 2011, 130, 496–499. [Google Scholar] [CrossRef]
- Zafeiriou, I.; Polidoros, A.N.; Baira, E.; Kasiotis, K.M.; Machera, K.; Mylona, P.V. Mediterranean White Lupin Landraces as a Valuable Genetic Reserve for Breeding. Plants 2021, 10, 2403. [Google Scholar] [CrossRef] [PubMed]
- Rychel, S.; Książkiewicz, M.; Tomaszewska, M.; Bielski, W.; Wolko, B. FLOWERING LOCUS T, GIGANTEA, SEPALLATA and FRIGIDA homologs are candidate genes involved in white lupin (Lupinus albus L.) early flowering. Mol. Breed. 2019, 39, 43. [Google Scholar] [CrossRef]
- Surma, A.; Książkiewicz, M.; Bielski, W.; Kozak, B.; Galek, R.; Rychel-Bielska, S. Development and validation of PCR marker array for molecular selection towards spring, vernalization-independent and winter, vernalization-responsive ecotypes of white lupin (Lupinus albus L.). Sci. Rep. 2025, 15, 2659. [Google Scholar] [CrossRef] [PubMed]
- Plewiński, P.; Rychel-Bielska, S.; Kozak, B.; Maureira-Butler, I.J.; Iqbal, M.M.; Nelson, M.N.; Książkiewicz, M. FLOWERING LOCUS T indel variants confer vernalization-independent and photoperiod-insensitive flowering of yellow lupin (Lupinus luteus L.). Hortic. Res. 2022, 9, uhac180. [Google Scholar] [CrossRef] [PubMed]
- Rychel-Bielska, S.; Plewiński, P.; Kozak, B.; Galek, R.; Książkiewicz, M. Photoperiod and vernalization control of flowering-related genes: A case study of the narrow-leafed lupin (Lupinus angustifolius L.). Front. Plant Sci. 2020, 11, 572135. [Google Scholar] [CrossRef] [PubMed]
- Plewiński, P.; Ćwiek-Kupczyńska, H.; Rudy, E.; Bielski, W.; Rychel-Bielska, S.; Stawiński, S.; Barzyk, P.; Krajewski, P.; Naganowska, B.; Wolko, B.; et al. Innovative transcriptome-based genotyping highlights environmentally responsive genes for phenology, growth and yield in a non-model grain legume. Plant Cell Environ. 2020, 43, 2680–2698. [Google Scholar] [CrossRef] [PubMed]
- Plewiński, P.; Książkiewicz, M.; Rychel-Bielska, S.; Rudy, E.; Wolko, B. Candidate domestication-related genes revealed by expression quantitative trait loci mapping of narrow-leafed lupin (Lupinus angustifolius L.). Int. J. Mol. Sci. 2019, 20, 5670. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.N.; Książkiewicz, M.; Rychel, S.; Besharat, N.; Taylor, C.M.; Wyrwa, K.; Jost, R.; Erskine, W.; Cowling, W.A.; Berger, J.D.; et al. The loss of vernalization requirement in narrow-leafed lupin is associated with a deletion in the promoter and de-repressed expression of a Flowering Locus T (FT) homologue. New Phytol. 2017, 213, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Książkiewicz, M.; Rychel, S.; Nelson, M.N.; Wyrwa, K.; Naganowska, B.; Wolko, B. Expansion of the phosphatidylethanolamine binding protein family in legumes: A case study of Lupinus angustifolius L. FLOWERING LOCUS T homologs, LanFTc1 and LanFTc2. BMC Genom. 2016, 17, 820. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.M.; van der Zanden, J.; Saradadevi, R.; Berger, J.D.; Kamphuis, L.G.; Pradhan, A.; Sharma, D.; Nelson, M.N.; Cowling, W.A. A multiplex PCR marker distinguishes between a series of four LanFTc1 alleles regulating flowering time in narrow-leafed lupin (Lupinus angustifolius). Plant Breed. 2021, 140, 1090–1101. [Google Scholar] [CrossRef]
- Taylor, C.M.; Kamphuis, L.G.; Zhang, W.; Garg, G.; Berger, J.D.; Mousavi-Derazmahalleh, M.; Bayer, P.E.; Edwards, D.; Singh, K.B.; Cowling, W.A.; et al. INDEL variation in the regulatory region of the major flowering time gene LanFTc1 is associated with vernalization response and flowering time in narrow-leafed lupin (Lupinus angustifolius L.). Plant Cell Environ. 2019, 42, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Hufnagel, B.; Soriano, A.; Taylor, J.; Divol, F.; Kroc, M.; Sanders, H.; Yeheyis, L.; Nelson, M.; Péret, B. Pangenome of white lupin provides insights into the diversity of the species. Plant Biotechnol. J. 2021, 19, 2532–2543. [Google Scholar] [CrossRef] [PubMed]
- Pecetti, L.; Annicchiarico, P.; Crosta, M.; Notario, T.; Ferrari, B.; Nazzicari, N. White lupin drought tolerance: Genetic variation, trait genetic architecture, and genome-enabled prediction. Int. J. Mol. Sci. 2023, 24, 2351. [Google Scholar] [CrossRef] [PubMed]
- Annicchiarico, P.; de Buck, A.J.; Vlachostergios, D.N.; Heupink, D.; Koskosidis, A.; Nazzicari, N.; Crosta, M. White Lupin Adaptation to Moderately Calcareous Soils: Phenotypic Variation and Genome-Enabled Prediction. Plants 2023, 12, 1139. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.M.; Garg, G.; Berger, J.D.; Ribalta, F.M.; Croser, J.S.; Singh, K.B.; Cowling, W.A.; Kamphuis, L.G.; Nelson, M.N. A Trimethylguanosine Synthase1-like (TGS1) homologue is implicated in vernalisation and flowering time control. Theor. Appl. Genet. 2021, 134, 3411–3426. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.M.; Huynh, M.; Udall, J.A.; Kilian, A.; Adhikari, K.N.; Berger, J.D.; Erskine, W.; Nelson, M.N. The first genetic map for yellow lupin enables genetic dissection of adaptation traits in an orphan grain legume crop. BMC Genet. 2019, 20, 68. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.T.T.; Ellwood, S.R.; Adhikari, K.; Nelson, M.N.; Oliver, R.P. The first genetic and comparative map of white lupin (Lupinus albus L.): Identification of QTLs for anthracnose resistance and flowering time, and a locus for alkaloid content. DNA Res. 2007, 14, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, D.E.; Wang, C.-T.; Zheng, Y.; Adamczyk, B.J.; Singhal, R.; Hall, P.K.; Perry, S.E. The MADS-domain factors AGAMOUS-LIKE15 and AGAMOUS-LIKE18, along with SHORT VEGETATIVE PHASE and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol. 2014, 165, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.J.; Lehti-Shiu, M.D.; Fernandez, D.E. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007, 50, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.L.; Madrigal, P.; Tsuda, K.; Rawat, V.; Richter, R.; Romera-Branchat, M.; Fornara, F.; Schneeberger, K.; Krajewski, P.; Coupland, G. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015, 16, 31. [Google Scholar] [CrossRef] [PubMed]
- Bouché, F.; Woods, D.P.; Amasino, R.M. Winter Memory throughout the Plant Kingdom: Different Paths to Flowering. Plant Physiol. 2017, 173, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Kröber, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wang, Y.; He, Y. Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol. 2013, 11, e1001649. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Foucher, F.; Ferrandiz, C.; Macknight, R.; Navarro, C.; Morin, J.; Vardy, M.E.; Ellis, N.; Beltran, J.P.; Rameau, C.; et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005, 137, 1420–1434. [Google Scholar] [CrossRef] [PubMed]
- Hufnagel, B.; Marques, A.; Soriano, A.; Marquès, L.; Divol, F.; Doumas, P.; Sallet, E.; Mancinotti, D.; Carrere, S.; Marande, W.; et al. High-quality genome sequence of white lupin provides insight into soil exploration and seed quality. Nat. Commun. 2020, 11, 492. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Zhao, T.; Gomez, A.; Li, C.; Yu, C.; Li, H.; Lin, J.; Yang, Y.; Liu, B.; et al. A Drought-Inducible Transcription Factor Delays Reproductive Timing in Rice. Plant Physiol. 2016, 171, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 2004, 37, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Sanagi, M.; Aoyama, S.; Kubo, A.; Lu, Y.; Sato, Y.; Ito, S.; Abe, M.; Mitsuda, N.; Ohme-Takagi, M.; Kiba, T.; et al. Low nitrogen conditions accelerate flowering by modulating the phosphorylation state of FLOWERING BHLH 4 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2022942118. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, B.; Kang, Y.; Cui, X.; Liu, A.; Deleris, A.; Greenberg, M.V.C.; Cui, X.; Qiu, Q.; Lu, F.; et al. C-terminal domains of histone demethylase JMJ14 interact with a pair of NAC transcription factors to mediate specific chromatin association. Cell Discov. 2015, 1, 15003. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.Q.; Ma, Z.Y.; Huang, H.W.; Mo, H.; Zhao, T.T.; Li, L.; Cai, T.; Chen, S.; Ma, L.; He, X.J. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 2015, 43, 1469–1484. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lee, J.; Kim, B.; Shin, J.; Kang, T.-A.; Kim, W.-C. GATA25, a novel regulator, accelerates the flowering time of Arabidopsis thaliana. Appl. Biol. Chem. 2022, 65, 28. [Google Scholar] [CrossRef]
- Chu, L.; Yang, C.; Zhuang, F.; Gao, Y.; Luo, M. The HDA9-HY5 module epigenetically regulates flowering time in Arabidopsis thaliana. J. Cell. Physiol. 2022, 237, 2961–2968. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chen, Z.; Feng, Q.; Long, T.; Ding, J.; Shu, P.; Deng, H.; Yu, P.; Tan, W.; Liu, S.; et al. ELONGATED HYPOCOTYL 5a modulates FLOWERING LOCUS T2 and gibberellin levels to control dormancy and bud break in poplar. Plant Cell 2024, 36, 1963–1984. [Google Scholar] [CrossRef] [PubMed]
- Dorca-Fornell, C.; Gregis, V.; Grandi, V.; Coupland, G.; Colombo, L.; Kater, M.M. The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 2011, 67, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Young Hun, S.; Na Young, S.; Su Young, S.; Hye Jin, K.; Dae-Jin, Y.; Chae Oh, L.; Sang Yeol, L.; Kyu Young, K.; Jong Chan, H. Isolation of CONSTANS as a TGA4/OBF4 Interacting Protein. Mol. Cells 2008, 25, 559–565. [Google Scholar] [CrossRef]
- Sun, B.; Fan, Y.; Duan, H.; Liu, X.; Chen, Y.; Shang, G.; Liu, Y.; Yang, H.; Qu, C.; Li, J.; et al. Genome-wide characterization of Brassica napus INDETERMINATE DOMAIN genes reveals a negative role for BnA08.IDD7 in plant development. Ind. Crop. Prod. 2022, 175, 114263. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Adrian, J.; Gissot, L.; Coupland, G.; Yu, D.; Turck, F. Elevated levels of MYB30 in the phloem accelerate flowering in Arabidopsis through the regulation of FLOWERING LOCUS T. PLoS ONE 2014, 9, e89799. [Google Scholar] [CrossRef] [PubMed]
- Chae, J.; Han, S.J.; Karthik, S.; Kim, H.J.; Kim, J.H.; Yun, H.R.; Chung, Y.S.; Sung, S.; Heo, J.B. LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) partially inhibits the transcriptional activation of FT by MYB73 and regulates flowering in Arabidopsis. Plant J. 2024, 120, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Le Sech, L.; Huyghe, C. Diallel analysis in white lupin: Consequences for breeding. Agronomie 1991, 11, 719–726. [Google Scholar] [CrossRef][Green Version]
- Cowling, W.A. Genetic diversity in narrow-leafed lupin breeding after the domestication bottleneck. In The Lupin Genome; Singh, K.B., Kamphuis, L.G., Nelson, M.N., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–17. [Google Scholar]
- Mousavi-Derazmahalleh, M.; Bayer, P.E.; Nevado, B.; Hurgobin, B.; Filatov, D.; Kilian, A.; Kamphuis, L.G.; Singh, K.B.; Berger, J.D.; Hane, J.K.; et al. Exploring the genetic and adaptive diversity of a pan-Mediterranean crop wild relative: Narrow-leafed lupin. Theor. Appl. Genet. 2018, 131, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.D.; Buirchell, B.J.; Luckett, D.J.; Nelson, M.N. Domestication bottlenecks limit genetic diversity and constrain adaptation in narrow-leafed lupin (Lupinus angustifolius L.). Theor. Appl. Genet. 2012, 124, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Stefanova, K.T.; Buirchell, B. Multiplicative mixed models for genetic gain assessment in lupin breeding. Crop Sci. 2010, 50, 880–891. [Google Scholar] [CrossRef]
- Adhikari, K.N.; Buirchell, B.J.; Thomas, G.J.; Sweetingham, M.W.; Yang, H. Identification of anthracnose resistance in Lupinus albus L. and its transfer from landraces to modern cultivars. Crop Pasture Sci. 2009, 60, 472–479. [Google Scholar] [CrossRef]
- Schwertfirm, G.; Schneider, M.; Haase, F.; Riedel, C.; Lazzaro, M.; Ruge-Wehling, B.; Schweizer, G. Genome-wide association study revealed significant SNPs for anthracnose resistance, seed alkaloids and protein content in white lupin. Theor. Appl. Genet. 2024, 137, 155. [Google Scholar] [CrossRef] [PubMed]
- Rychel-Bielska, S.; Nazzicari, N.; Plewiński, P.; Bielski, W.; Annicchiarico, P.; Książkiewicz, M. Development of PCR-based markers and whole-genome selection model for anthracnose resistance in white lupin (Lupinus albus L.). J. Appl. Genet. 2020, 61, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Jacob, I.; Feuerstein, U.; Heinz, M.; Schott, M.; Urbatzka, P. Evaluation of new breeding lines of white lupin with improved resistance to anthracnose. Euphytica 2017, 213, 236. [Google Scholar] [CrossRef]
- Ruge-Wehling, B.; Dieterich, R.; Thiele, C.; Eickmeyer, F.; Wehling, P. Resistance to anthracnose in narrow-leafed lupin (Lupinus angustifolius L.): Sources of resistanceand development of molecular markers. J. Für Kult. 2009, 61, 62–65. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Romani, M.; Pecetti, L. White lupin (Lupinus albus) variation for adaptation to severe drought stress. Plant Breed. 2018, 137, 782–789. [Google Scholar] [CrossRef]
- Christiansen, J.L.; Raza, S.; Jørnsgård, B.; Mahmoud, S.A.; Ortiz, R. Potential of landrace germplasm for genetic enhancement of white lupin in Egypt. Genet. Resour. Crop Evol. 2000, 47, 425–430. [Google Scholar] [CrossRef]
- Raza, S.; Abdel-Wahab, A.; JØrnsgård, B.; Christiansen, J.L. Calcium tolerance and ion uptake of Egyptian lupin landraces on calcareous soils. Afr. Crop Sci. J. 2001, 9, 393–400. [Google Scholar] [CrossRef]
- Kerley, S.J.; Norgaard, C.; Leach, J.E.; Christiansen, J.L.; Huyghe, C.; Römer, P. The development of potential screens based on shoot calcium and iron concentrations for the evaluation of tolerance in Egyptian genotypes of white lupin (Lupinus albus L.) to limed soils. Ann. Bot. 2002, 89, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Mancinotti, D.; Czepiel, K.; Taylor, J.L.; Golshadi Galehshahi, H.; Møller, L.A.; Jensen, M.K.; Motawia, M.S.; Hufnagel, B.; Soriano, A.; Yeheyis, L.; et al. The causal mutation leading to sweetness in modern white lupin cultivars. Sci. Adv. 2023, 9, eadg8866. [Google Scholar] [CrossRef] [PubMed]
- Rychel, S.; Książkiewicz, M. Development of gene-based molecular markers tagging low alkaloid pauper locus in white lupin (Lupinus albus L.). J. Appl. Genet. 2019, 60, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Renshaw, D.; Luckett, D.; Clements, J.; Yan, G.; Adhikari, K.; Buirchell, B.; Sweetingham, M.; Yang, H. Development of a sequence-specific PCR marker linked to the gene “pauper” conferring low-alkaloids in white lupin (Lupinus albus L.) for marker assisted selection. Mol. Breed. 2009, 23, 153–161. [Google Scholar] [CrossRef]
- Harrison, J.E.M.; Williams, W. Genetical control of alkaloids in Lupinus albus. Euphytica 1982, 31, 357–364. [Google Scholar] [CrossRef]
- Raman, R.; Vipin, C.A.; Luckett, D.J.; Cowley, R.B.; Ash, G.J.; Harper, J.D.I.; Kilian, A.; Raman, H. Localisation of loci involved in resistance to Diaporthe toxica and Pleiochaeta setosa in white lupin (Lupinus albus L.). Open J. Genet. 2014, 4, 210–226. [Google Scholar] [CrossRef]
- Cowley, R.; Luckett, D.J.; Ash, G.J.; Harper, J.D.I.; Vipin, C.A.; Raman, H.; Ellwood, S. Identification of QTLs associated with resistance to Phomopsis pod blight (Diaporthe toxica) in Lupinus albus. Breed. Sci. 2014, 64, 83–89. [Google Scholar] [CrossRef] [PubMed]
- González-Andrés, F.; Casquero, P.A.; San-Pedro, C.; Hernández-Sánchez, E. Diversity in White Lupin (Lupinus albus L.) Landraces from Northwest Iberian plateau. Genet. Resour. Crop Evol. 2007, 54, 27–44. [Google Scholar] [CrossRef]
- Georgieva, N.A.; Kosev, V. Phenotypic Variability of White Lupine (Lupinus albus L.) Germplasm. Not. Sci. Biol. 2017, 9, 397–403. [Google Scholar] [CrossRef][Green Version]
- Mikić, A.; Mihailović, V. Significance of genetic resources of cool season annual legumes: III: Locally cultivated and maintained crop landraces. Ratar. Povrt. 2014, 51, 190–203. [Google Scholar] [CrossRef]
- Minoli, S.; Jägermeyr, J.; Asseng, S.; Urfels, A.; Müller, C. Global crop yields can be lifted by timely adaptation of growing periods to climate change. Nat. Commun. 2022, 13, 7079. [Google Scholar] [CrossRef] [PubMed]
- Tourville, J.C.; Murray, G.L.D.; Nelson, S.J. Distinct latitudinal patterns of shifting spring phenology across the Appalachian Trail Corridor. Ecology 2024, 105, e4403. [Google Scholar] [CrossRef] [PubMed]
- Pareja-Bonilla, D.; Arista, M.; Morellato, L.P.C.; Ortiz, P.L. Better soon than never: Climate change induces strong phenological reassembly in the flowering of a Mediterranean shrub community. Ann. Bot. 2023, 135, 239–254. [Google Scholar] [CrossRef] [PubMed]
- Geissler, C.; Davidson, A.; Niesenbaum, R.A. The influence of climate warming on flowering phenology in relation to historical annual and seasonal temperatures and plant functional traits. PeerJ 2023, 11, e15188. [Google Scholar] [CrossRef] [PubMed]
- Austin, M.W.; Smith, A.B.; Olsen, K.M.; Hoch, P.C.; Krakos, K.N.; Schmocker, S.P.; Miller-Struttmann, N.E. Climate change increases flowering duration, driving phenological reassembly and elevated co-flowering richness. New Phytol. 2024, 243, 2486–2500. [Google Scholar] [CrossRef] [PubMed]
- Menzel, A.; Yuan, Y.; Matiu, M.; Sparks, T.; Scheifinger, H.; Gehrig, R.; Estrella, N. Climate change fingerprints in recent European plant phenology. Glob. Change Biol. 2020, 26, 2599–2612. [Google Scholar] [CrossRef] [PubMed]
- Annicchiarico, P.; Harzic, N.; Carroni, A.M. Adaptation, diversity, and exploitation of global white lupin (Lupinus albus L.) landrace genetic resources. Field Crop. Res. 2010, 119, 114–124. [Google Scholar] [CrossRef]
- McGaughey, K.D.; Yilmaz-Swenson, T.; Elsayed, N.M.; Cruz, D.A.; Rodriguez, R.R.; Kritzer, M.D.; Peterchev, A.V.; Gray, M.; Lewis, S.R.; Roach, J.; et al. Comparative evaluation of a new magnetic bead-based DNA extraction method from fecal samples for downstream next-generation 16S rRNA gene sequencing. PLoS ONE 2018, 13, e0202858. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Massey Jr, F.J. The Kolmogorov-Smirnov Test for Goodness of Fit. J. Am. Stat. Assoc. 1951, 46, 68–78. [Google Scholar] [CrossRef]
- Chow, C.-N.; Yang, C.-W.; Wu, N.-Y.; Wang, H.-T.; Tseng, K.-C.; Chiu, Y.-H.; Lee, T.-Y.; Chang, W.-C. PlantPAN 4.0: Updated database for identifying conserved non-coding sequences and exploring dynamic transcriptional regulation in plant promoters. Nucleic Acids Res. 2024, 52, D1569–D1578. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.N.; Lee, T.Y.; Hung, Y.C.; Li, G.Z.; Tseng, K.C.; Liu, Y.H.; Kuo, P.L.; Zheng, H.Q.; Chang, W.C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, D1155–D1163. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, A.; Richter, R. Genetic and molecular basis of floral induction in Arabidopsis thaliana. J. Exp. Bot. 2020, 71, 2490–2504. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).