Potential miRNAs as Diagnostic Biomarkers for Differentiating Disease States in Ulcerative Colitis: A Systematic Review
Abstract
1. Introduction
2. Methods
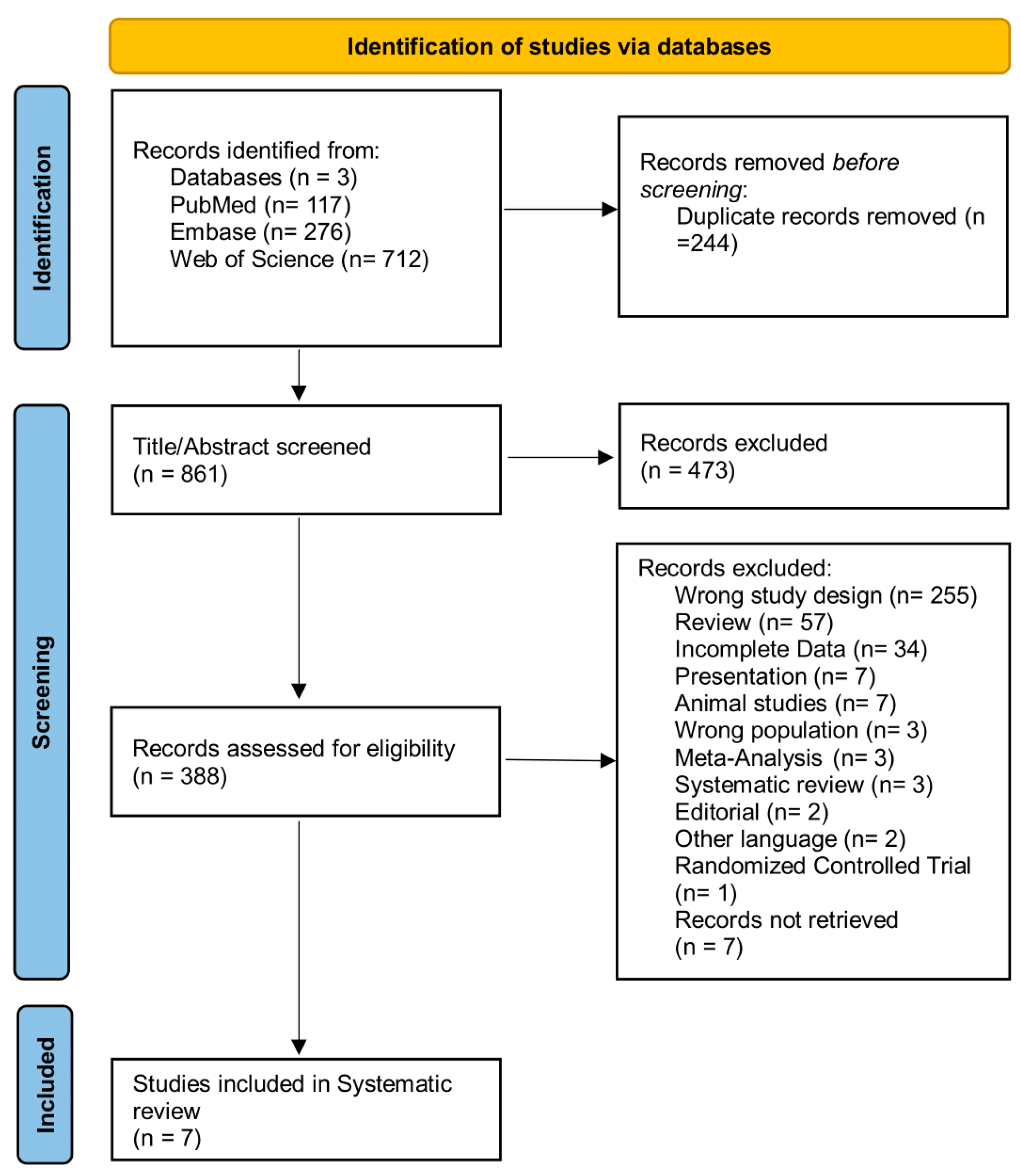
2.1. Search Strategy and Eligibility Criteria
2.2. Quality Assessment and Critical Appraisal
2.3. Data Extraction
2.4. Data Synthesis
3. Results
3.1. Study Characteristics and Methodological Diversity
3.2. miRNA Expression Patterns and Cross-Study Consistency
3.3. Statistical Analysis and Effect Size Evaluation
4. Discussion
Clinical Implementation Considerations and Diagnostic Utility
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feuerstein, J.D.; Moss, A.C.; Farraye, F.A. Ulcerative Colitis. Mayo Clin. Proc. 2019, 94, 1357–1373. [Google Scholar] [CrossRef] [PubMed]
- Gros, B.; Kaplan, G.G. Ulcerative Colitis in Adults: A Review. JAMA 2023, 330, 951–965. [Google Scholar] [CrossRef] [PubMed]
- Harbord, M.; Eliakim, R.; Bettenworth, D.; Karmiris, K.; Katsanos, K.; Kopylov, U.; Kucharzik, T.; Molnár, T.; Raine, T.; Sebastian, S.; et al. Third European Evidence-Based Consensus on Diagnosis and Management of Ulcerative Colitis. Part 2: Current Management. J. Crohns Colitis 2017, 11, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Kornbluth, A.; Sachar, D.B. Ulcerative Colitis Practice Guidelines in Adults: American College of Gastroenterology, Practice Parameters Committee. Am. J. Gastroenterol. 2010, 105, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Rubin, D.T.; Ananthakrishnan, A.N.; Siegel, C.A.; Sauer, B.G.; Long, M.D. ACG Clinical Guideline: Ulcerative Colitis in Adults. Am. J. Gastroenterol. 2019, 114, 384–413. [Google Scholar] [CrossRef] [PubMed]
- Gajendran, M.; Loganathan, P.; Jimenez, G.; Catinella, A.P.; Ng, N.; Umapathy, C.; Ziade, N.; Hashash, J.G. A Comprehensive Review and Update on Ulcerative Colitis. Dis. Mon. 2019, 65, 100851. [Google Scholar] [CrossRef] [PubMed]
- Desai, D.; Dhoble, P. Rapidly Changing Epidemiology of Inflammatory Bowel Disease: Time to Gear up for the Challenge before It Is Too Late. Indian J. Gastroenterol. 2024, 43, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Gorospe, J.; Windsor, J.; Hracs, L.; Coward, S.; Buie, M.; Quan, J.; Caplan, L.; Markovinovic, A.; Cummings, M.; Goddard, Q.; et al. Trends in inflammatory bowel disease incidence and prevalence across epidemiologic stages: A global systematic review with meta-analysis. Inflamm. Bowel Dis. 2024, 30, S00. [Google Scholar] [CrossRef]
- Pasvol, T.J.; Horsfall, L.; Bloom, S.; Segal, A.W.; Sabin, C.; Field, N.; Rait, G. Incidence and Prevalence of Inflammatory Bowel Disease in UK Primary Care: A Population-Based Cohort Study. BMJ Open 2020, 10, e036584. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, M.; Chung, H.; Xu, Y.; Qiu, H. Trends in the Prevalence and Incidence of Ulcerative Colitis in Japan and the US. Int. J. Colorectal Dis. 2023, 38, 135. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Wang, H.; Ye, S.; Guan, J.; Tan, W.; Cheng, S.; Wei, G.; Wu, W.; Wu, F.; Zhou, Y. Up-Regulation of microRNA-126 May Contribute to Pathogenesis of Ulcerative Colitis via Regulating NF-kappaB Inhibitor IκBα. PLoS ONE 2012, 7, e52782. [Google Scholar] [CrossRef] [PubMed]
- Coskun, M.; Bjerrum, J.T.; Seidelin, J.B.; Troelsen, J.T.; Olsen, J.; Nielsen, O.H. MiR-20b, MiR-98, MiR-125b-1*, and Let-7e* as New Potential Diagnostic Biomarkers in Ulcerative Colitis. World J. Gastroenterol. WJG 2013, 19, 4289–4299. [Google Scholar] [CrossRef] [PubMed]
- Van Der Goten, J.; Vanhove, W.; Lemaire, K.; Van Lommel, L.; Machiels, K.; Wollants, W.J.; De Preter, V.; De Hertogh, G.; Ferrante, M.; Van Assche, G.; et al. Integrated MiRNA and MRNA Expression Profiling in Inflamed Colon of Patients with Ulcerative Colitis. PLoS ONE 2014, 9, e116117. [Google Scholar] [CrossRef] [PubMed]
- Schönauen, K.; Le, N.; von Arnim, U.; Schulz, C.; Malfertheiner, P.; Link, A. Circulating and Fecal microRNAs as Biomarkers for Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2018, 24, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Li, X.; Zhu, Y.; Li, N.; Zhang, N.; Niu, M. Faecal MicroRNA as a Biomarker of the Activity and Prognosis of Inflammatory Bowel Diseases. Biochem. Biophys. Res. Commun. 2018, 503, 2443–2450. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, Y.; Li, L.; Yu, Q.; Chao, K.; Zhou, G.; Qiu, Y.; Feng, R.; Huang, S.; He, Y.; et al. Circulating MicroRNA146b-5p Is Superior to C-Reactive Protein as a Novel Biomarker for Monitoring Inflammatory Bowel Disease. Aliment. Pharmacol. Ther. 2019, 49, 733–743. [Google Scholar] [CrossRef] [PubMed]
- El Sabbagh, E.; Shaker, O.; Kassem, A.M.; Khairy, A. Role of Circulating MicroRNA146b-5p and MicroRNA-106a in Diagnosing and Predicting the Severity of Inflammatory Bowel Disease. Egypt. J. Chem. 2023, 66, 545–551. [Google Scholar] [CrossRef]
- Shi, C.; Liang, Y.; Yang, J.; Xia, Y.; Chen, H.; Han, H.; Yang, Y.; Wu, W.; Gao, R.; Qin, H. MicroRNA-21 Knockout Improve the Survival Rate in DSS Induced Fatal Colitis through Protecting against Inflammation and Tissue Injury. PLoS ONE 2013, 8, e66814. [Google Scholar] [CrossRef] [PubMed]
- Polytarchou, C.; Hommes, D.W.; Palumbo, T.; Hatziapostolou, M.; Koutsioumpa, M.; Koukos, G.; van der Meulen-de Jong, A.E.; Oikonomopoulos, A.; van Deen, W.K.; Vorvis, C.; et al. MicroRNA214 Is Associated With Progression of Ulcerative Colitis, and Inhibition Reduces Development of Colitis and Colitis-Associated Cancer in Mice. Gastroenterology 2015, 149, 981–992.e11. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, Y.; Shi, C.; Chen, H.; Zhang, H.; Chen, N.; Zhang, P.; Wang, F.; Yang, J.; Yang, J.; et al. Overexpression of MiR-21 in Patients with Ulcerative Colitis Impairs Intestinal Epithelial Barrier Function through Targeting the Rho GTPase RhoB. Biochem. Biophys. Res. Commun. 2013, 434, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Thorlacius-Ussing, G.; Schnack Nielsen, B.; Andersen, V.; Holmstrøm, K.; Pedersen, A.E. Expression and Localization of MiR-21 and MiR-126 in Mucosal Tissue from Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2017, 23, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Yu, Y.; Tan, S. The Role of the MiR-21-5p-Mediated Inflammatory Pathway in Ulcerative Colitis. Exp. Ther. Med. 2019, 19, 981. [Google Scholar] [CrossRef] [PubMed]
- Guglielmi, G.; Crucitta, S.; Bertani, L.; Ruglioni, M.; Svizzero, G.B.; Ceccarelli, L.; Del Re, M.; Danesi, R.; Costa, F.; Fogli, S. Expression of Circulating Let-7e and MiR-126 May Predict Clinical Remission in Patients With Crohn’s Disease Treated With Anti-TNF-α Biologics. Inflamm. Bowel Dis. 2024, 30, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.G.; He, Q.; Guo, S.Q.; Shi, Z.Z. Reduced Serum MIR-98 Predicts Unfavorable Clinical Outcome of Colorectal Cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8345–8353. [Google Scholar] [CrossRef] [PubMed]
- Casado-Bedmar, M.; Roy, M.; Berthet, L.; Hugot, J.P.; Yang, C.; Manceau, H.; Peoc’h, K.; Chassaing, B.; Merlin, D.; Viennois, E. Fecal Let-7b and MiR-21 Directly Modulate the Intestinal Microbiota, Driving Chronic Inflammation. Gut Microbes 2024, 16, 2394249. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, X.; Wu, Q.; Song, J.; Wang, L.; Li, G. MiR-125b Inhibits Goblet Cell Differentiation in Allergic Airway Inflammation by Targeting SPDEF. Eur. J. Pharmacol. 2016, 782, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Valmiki, S.; Ahuja, V.; Puri, N.; Paul, J. MiR-125b and MiR-223 Contribute to Inflammation by Targeting the Key Molecules of NFκB Pathway. Front. Med. 2020, 6, 494658. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.Y.; Wang, Z.; Wang, Z.H.; Jiang, X.G.; Lu, W.H. Inhibition of MiR-155 Alleviates Sepsis-Induced Inflammation and Intestinal Barrier Dysfunction by Inactivating NF-ΚB Signaling. Int. Immunopharmacol. 2021, 90, 107218. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zuo, D.; Liu, X.; Fan, H.; Chen, Q.; Deng, S.; Shou, Z.; Tang, Q.; Yang, J.; Nan, Z.; et al. MiR-155 Contributes to Th17 Cells Differentiation in Dextran Sulfate Sodium (DSS)-Induced Colitis Mice via Jarid2. Biochem. Biophys. Res. Commun. 2017, 488, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, F.; Li, H.; Fan, H.; Wu, H.; Dong, Y.; Chu, S.; Tan, C.; Wang, Q.; He, H.; et al. MiR-155 Contributes to Intestinal Barrier Dysfunction in DSS-Induced Mice Colitis via Targeting HIF-1α/TFF-3 Axis. Aging 2020, 12, 14966. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Deng, S.; Yang, J.; Shou, Z.; Wei, C.; Zhang, L.; Zhu, F.; Gao, F.; Liu, X.; Liu, Y.; et al. Antagomir of MiR-31-5p Modulates Macrophage Polarization via the AMPK/SIRT1/NLRP3 Signaling Pathway to Protect against DSS-Induced Colitis in Mice. Aging 2024, 16, 5336–5353. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Tabuchi, T.; Minami, Y.; Takahashi, Y.; Itoh, T.; Nakamura, M. Expression of Let-7i Is Associated with Toll-like Receptor 4 Signal in Coronary Artery Disease: Effect of Statins on Let-7i and Toll-like Receptor 4 Signal. Immunobiology 2012, 217, 533–539. [Google Scholar] [CrossRef] [PubMed]
| Database | Keywords | Results |
|---|---|---|
| PubMed | (“microRNA” OR “miRNA” OR “mir” OR “microRNAs” OR “miRNAs”) | 222,472 |
| (“biomarker” OR “biomarkers” OR “diagnostic marker” OR “molecular marker”) | 927,326 | |
| (“ulcerative colitis” OR “active ulcerative colitis” OR “inactive ulcerative colitis” OR “remission ulcerative colitis” OR “flare ulcerative colitis” OR “quiescent ulcerative colitis”) | 66,276 | |
| #1 AND #2 AND #3 | 117 | |
| Web of Science | microRNA OR miRNA OR mir OR microRNAs OR miRNAs AND biomarker OR biomarkers OR diagnostic marker OR molecular marker AND ulcerative colitis OR active ulcerative colitis OR inactive ulcerative colitis OR remission ulcerative colitis OR flare ulcerative colitis OR quiescent ulcerative colitis | 712 |
| Embase | microRNA OR miRNA OR mir OR microRNAs OR miRNAs AND biomarker OR biomarkers OR diagnostic marker OR molecular marker AND ulcerative colitis OR active ulcerative colitis OR inactive ulcerative colitis OR remission ulcerative colitis OR flare ulcerative colitis OR quiescent ulcerative colitis | 276 |
| Parameters | Inclusion Criteria | Exclusion Criteria |
|---|---|---|
| Population | Adult individuals with a confirmed diagnosis of ulcerative colitis and differentiation based on disease state | Studies about adult individuals with non-UC conditions (e.g., Crohn’s disease, general IBD, etc.) |
| Intervention | miRNAs as diagnostic tool to assess UC disease state | Studies that use any other biomolecules as biomarkers |
| Comparison | N/A | N/A |
| Outcome | miRNAs as diagnostic biomarker for differentiating active and inactive UC | No significant change in miRNA expression level capable of differentiating active UC from inactive UC |
| NOS Score | ||||
|---|---|---|---|---|
| Author, Year | Study Design | Selection | Comparability | Outcome/Exposure |
| [12] | Case–Control | 


 | 
 | |
| [13] | Cohort | 


 |  | |
| [14] | Case–Control | 


 |  | 
 |
| [15] | Cohort | 


 |  | |
| [16] | Case–Control | 


 | 
 | |
| [17] | Cohort | 


 |  | 
 |
| [18] | Case–Control | 


 | 
 | |
| Author and Year | Study Design | Sample Type | Sample Size (aUC/iUC/HC) | Gender (M/F) | miRNA Measurement |
|---|---|---|---|---|---|
| [12] | Case–control | Pinch biopsies | 12/10/15 | 5/7, 4/6, 7/8 | qRT-PCR |
| [13] | Cohort | Pinch biopsies | 20/19/20 | 9/11, 6/13, 10/10 | miRNA microarray, qPCR |
| [14] | Case–control | Colonic mucosal biopsies | 10/7/10 | 6/4, 4/3, 5/5 | miRNA microarray, qRT-PCR |
| [15] | Cohort | Serum and fecal samples | 10/8/35 | 4/6, 4/4, 14/21 | qPCR |
| [16] | Case–control | Fecal samples | 41/25/66 | NR | miRNA microarray, qPCR |
| [17] | Cohort | Serum samples | 55/45/41 | NR | qRT-PCR |
| [18] | Case–control | Blood samples | 33/2/30 | NR | qPCR |
| Author (Year) | miRNAs Studied | Disease Duration (Years) Mean (Range) | Disease Activity Method | Key Findings |
|---|---|---|---|---|
| [12] | miR-21, miR-126, miR-375 | 1.8 (0.5–4) aUC, 4 (2–6) iUC | UCDAI: 0.9 (0–2) iUC, 8.91 (8–10) aUC | miR-126 (↑ 18-fold, p < 0.05) in aUC vs. HC. miR-21 (↑ 14.7-fold, p < 0.05) in aUC vs. HC. No significant changes in iUC vs. HC. |
| [13] | miR-20b, -99a, -203, -26b, -98, -125b-1, let-7e | 15/5 aUC, 8/11 iUC | Mayo: 0 (0–1) iUC, 6 (2–12) aUC | miR-20b (↑ p < 0.05) in aUC vs. HC. let-7e (↑ p < 0.05) in iUC vs. HC. miR-98 (↑ p < 0.05) in iUC vs. aUC and HC. miR-125b-1 (↑ p < 0.01) in aUC vs. HC. |
| [14] | miR-21-5p, miR-31-5p, miR-146a-5p, miR-155-5p, miR-650, miR-196b-5p, miR-200c-3p, miR-375, miR-200b-3p, miR-422a | 7.1 (0.6–20.1) aUC, 6.6 (4.0–14.4) iUC | Mayo: 0 (±0.5) iUC, 8 (±1.5) aUC | miR-21-5p, miR-31-5p, miR-146a-5p, miR-155-5p, miR-650, miR-375 (↑) in aUC vs. HC. miR-196b-5p, miR-196b-3p, miR-200c-3p (↓) in aUC vs. HC. |
| [15] | miR-16, miR-21, miR-155, miR-223 | NR | Mayo: ≤5.27 iUC, >5.27 aUC | miR-21, miR-223 (↑) in iUC vs. HC serum. miR-21, miR-155 (↓) in aUC vs. iUC serum. miR-16, miR-155, miR-223 (↓) in aUC vs. HC feces. miR-155 (↓) in aUC and iUC vs. HC feces. |
| [16] | miR-199a, miR-223-3p, miR-1226, miR-548ab, miR-515-5p | 6.5 aUC, 11.5 iUC | Modified Mayo: ≤2 iUC, >2 aUC | miR-515, miR-548ab, miR-1226 (↓) in aUC vs. HC and iUC vs. HC. miR-199a-5p, miR-223-3p (↑) in aUC vs. HC and iUC. |
| [17] | miR-197-5p, miR-603, miR-145-3p, miR-574-3p, miR-34a-5p, miR-323a-3p, miR-141-3p, miR-146b-5p, miR-193b-3p, miR-31-5p, miR-27a, miR-27b, miR-944, miR-204-3p, miR-206, miR-24-1-5p, miR-135b-5p | NR | Mayo: ≤2 iUC, >2 aUC | miR-146b-5p (↑) in aUC vs. iUC and HC. |
| [18] | miR-106a, miR-146b | NR | Mayo: No specific cut values | No significant change between aUC and iUC. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, A.U.; Chacon-Millan, P.; Stiuso, P. Potential miRNAs as Diagnostic Biomarkers for Differentiating Disease States in Ulcerative Colitis: A Systematic Review. Int. J. Mol. Sci. 2025, 26, 6822. https://doi.org/10.3390/ijms26146822
Khan AU, Chacon-Millan P, Stiuso P. Potential miRNAs as Diagnostic Biomarkers for Differentiating Disease States in Ulcerative Colitis: A Systematic Review. International Journal of Molecular Sciences. 2025; 26(14):6822. https://doi.org/10.3390/ijms26146822
Chicago/Turabian StyleKhan, Atta Ullah, Pilar Chacon-Millan, and Paola Stiuso. 2025. "Potential miRNAs as Diagnostic Biomarkers for Differentiating Disease States in Ulcerative Colitis: A Systematic Review" International Journal of Molecular Sciences 26, no. 14: 6822. https://doi.org/10.3390/ijms26146822
APA StyleKhan, A. U., Chacon-Millan, P., & Stiuso, P. (2025). Potential miRNAs as Diagnostic Biomarkers for Differentiating Disease States in Ulcerative Colitis: A Systematic Review. International Journal of Molecular Sciences, 26(14), 6822. https://doi.org/10.3390/ijms26146822






