Recombinant Expression and Characterization of a Novel Type I Baeyer–Villiger Monooxygenase from a Streptomyces Strain Isolated from the Rhizosphere of the Atacama Desert Lupinus oreophilus
Abstract
1. Introduction
2. Results and Discussion
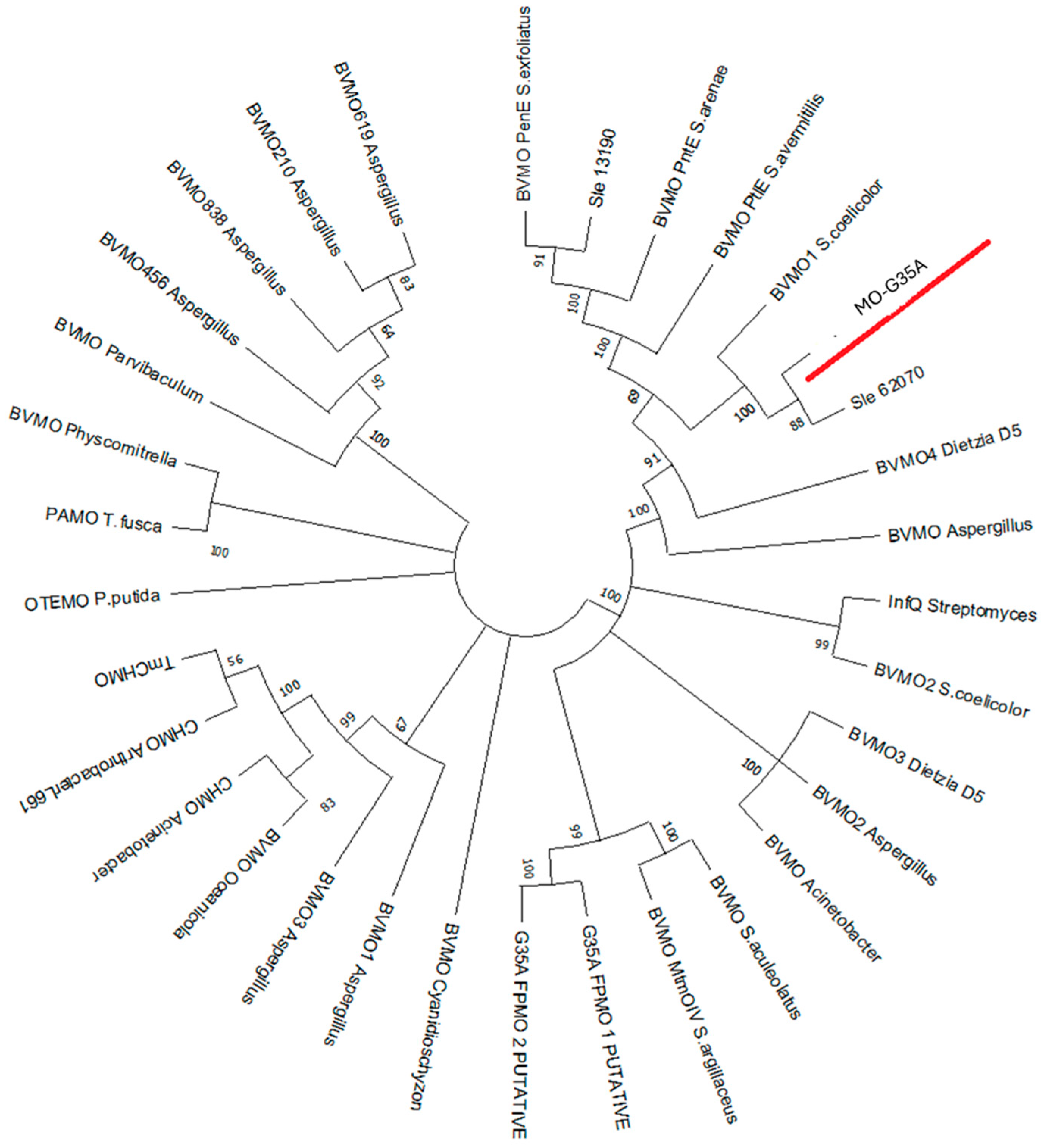
2.1. Identification of a Type I BVMO Using Computational Analysis
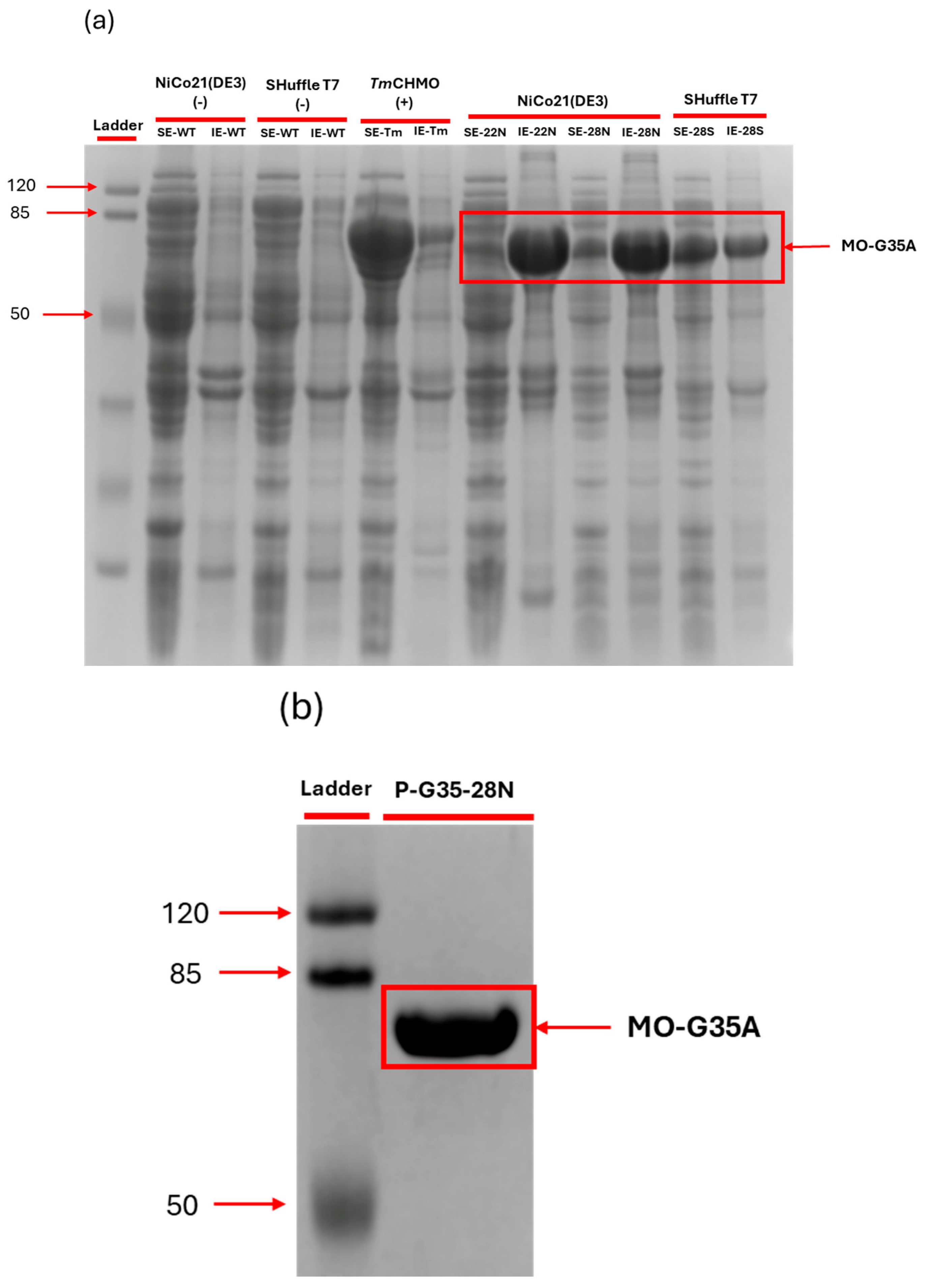
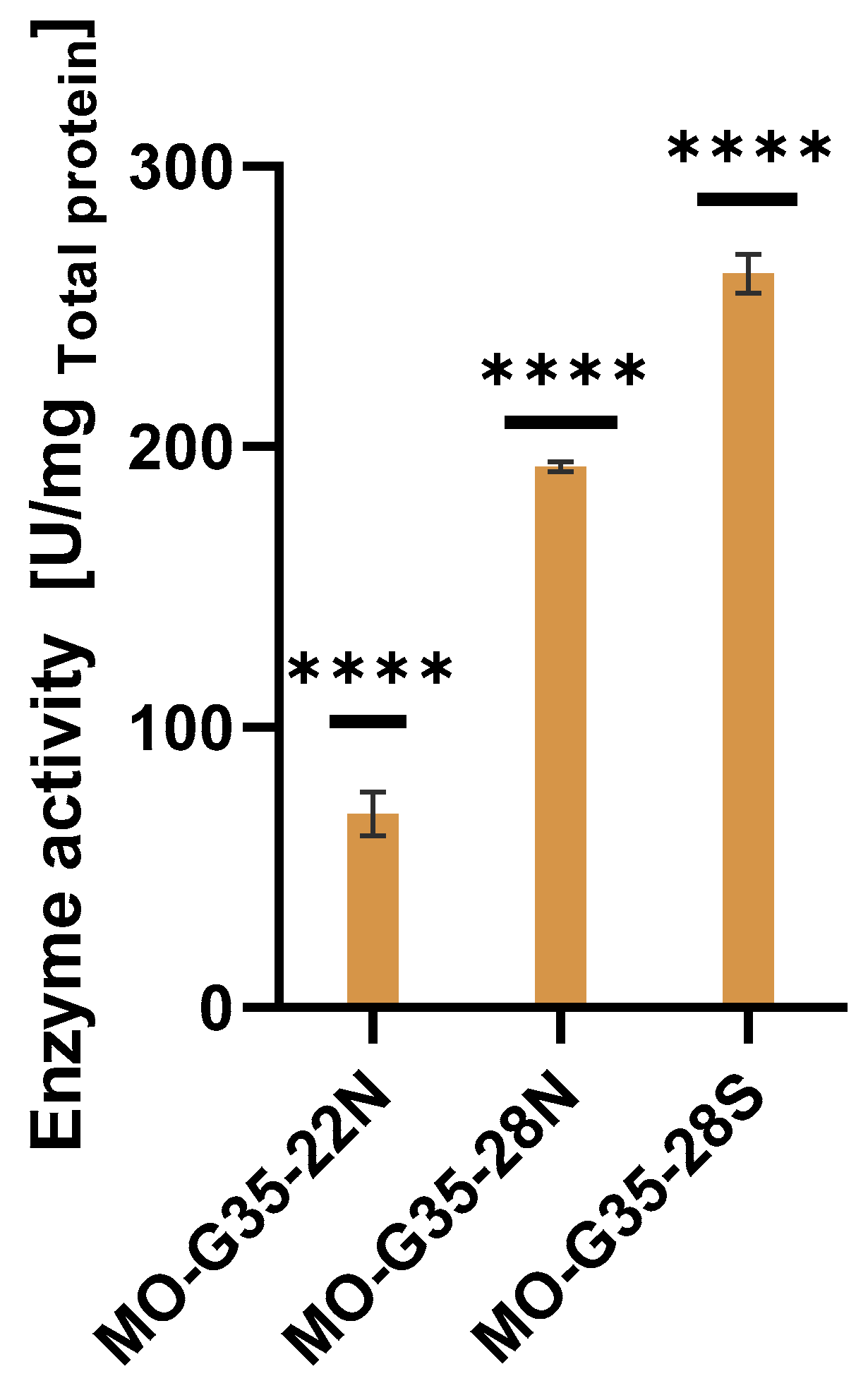
2.2. MO-G35A Expression Systems Comparison
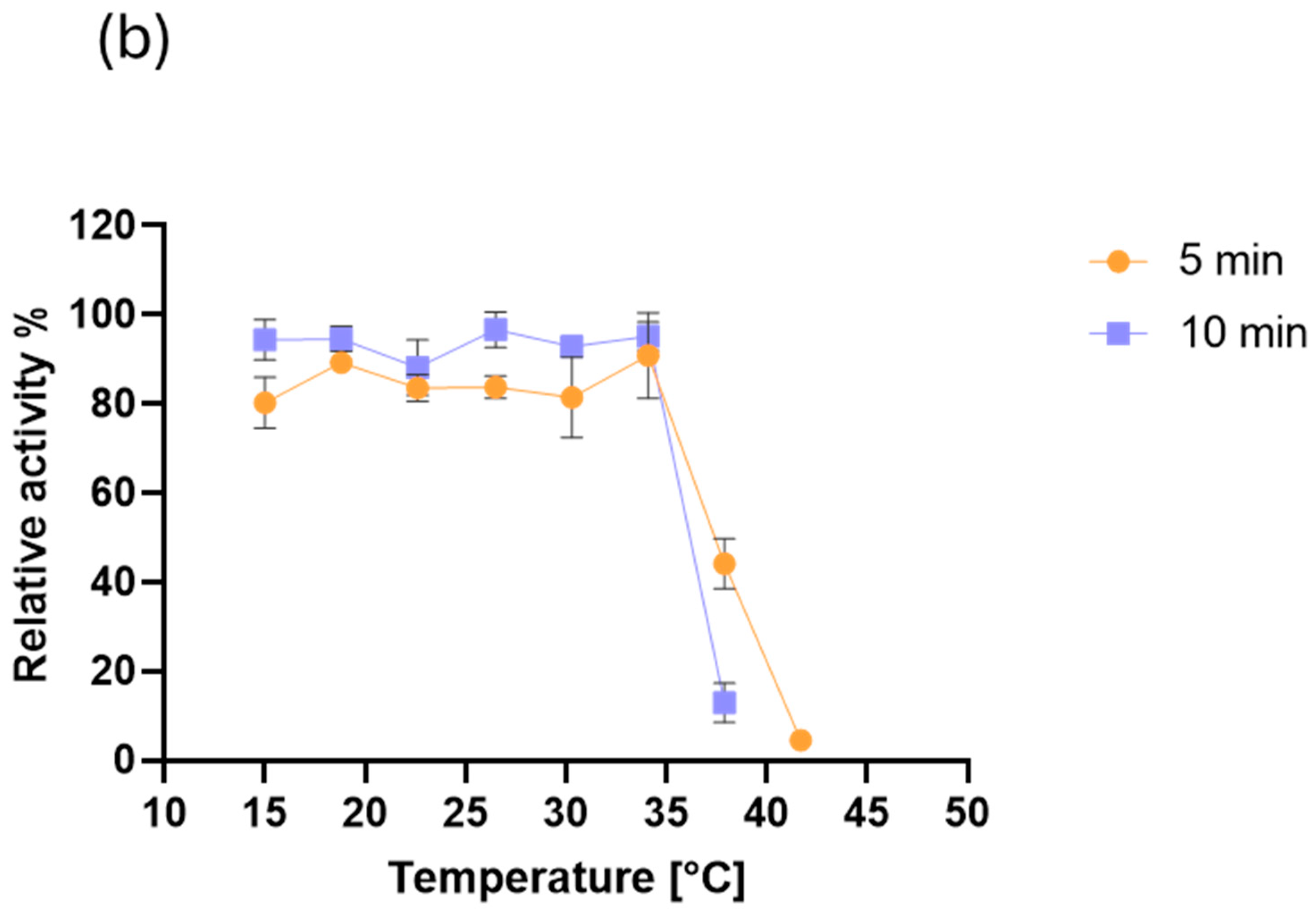
2.3. MO-G35A Biochemical Characterization
3. Materials and Methods
3.1. Chemicals or Reagents
3.2. Bioinformatic Search for Putative Type I BVMOs
3.3. Cloning of MO-G35A
3.4. Expression of MO-G35A
3.5. Purification of Recombinant MO-G35A
3.6. Enzyme Characterization
3.6.1. MO-G35A Activity Assays
3.6.2. Protein Quantification
3.6.3. MO-G35A Kinetic Parameters
3.6.4. pH Effect on MO-G35A Activity
3.6.5. Temperature Effect on MO-G35A Activity
3.6.6. MO-G35A Thermal Stability
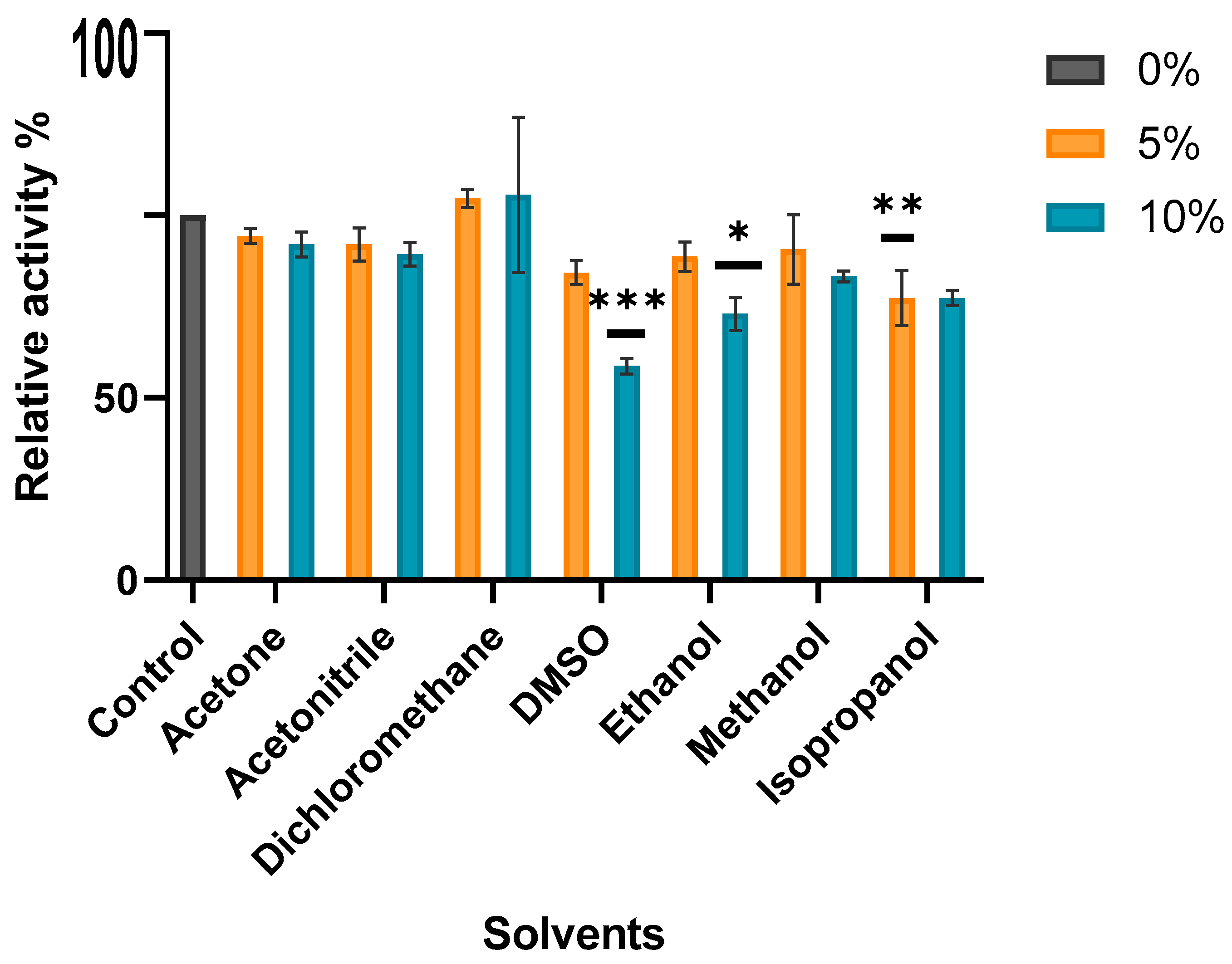
3.6.7. Solvent Effect on MO-G35A Activity
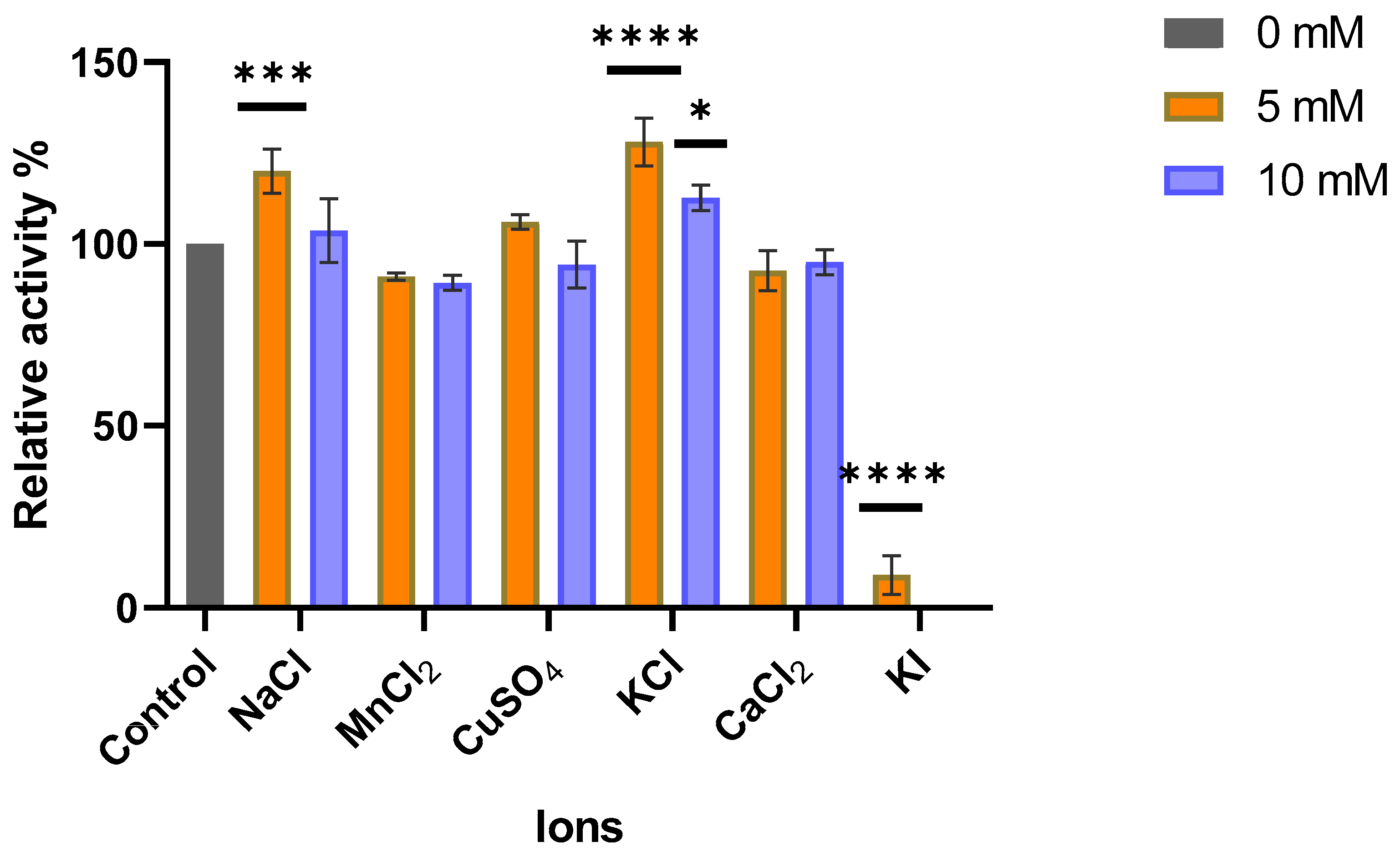
3.6.8. Ions Effect on MO-G35A Activity
3.6.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huijbers, M.M.E.; Montersino, S.; Westphal, A.H.; Tischler, D.; van Berkel, W.J.H. Flavin dependent monooxygenases. Arch. Biochem. Biophys. 2014, 544, 2–17. [Google Scholar] [CrossRef]
- Liu, L.-L.; Liu, H.-F.; Gao, H.-H.; Yang, Z.-Z.; Feng, X.-L.; Gao, J.-M.; Zhao, J.-B. Genome-based analysis of the type II PKS biosynthesis pathway of xanthones in Streptomyces caelestis and their antifungal activity. RSC Adv. 2019, 9, 37376–37383. [Google Scholar] [CrossRef]
- Deng, Y.; Zhou, Q.; Wu, Y.; Chen, X.; Zhong, F. Properties and Mechanisms of Flavin-Dependent Monooxygenases and Their Applications in Natural Product Synthesis. Int. J. Mol. Sci. 2022, 23, 2622. [Google Scholar] [CrossRef]
- van Berkel, W.J.H.; Kamerbeek, N.M.; Fraaije, M.W. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 2006, 124, 670–689. [Google Scholar] [CrossRef] [PubMed]
- Paul, C.E.; Eggerichs, D.; Westphal, A.H.; Tischler, D.; van Berkel, W.J.H. Flavoprotein monooxygenases: Versatile biocatalysts. Biotechnol. Adv. 2021, 51, 107712. [Google Scholar] [CrossRef] [PubMed]
- Tolmie, C.; Smit, M.S.; Opperman, D.J. Native roles of Baeyer–Villiger monooxygenases in the microbial metabolism of natural compounds. Nat. Prod. Rep. 2019, 36, 326–353. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Choi, G.E.; Gu, D.H.; Seo, P.W.; Kim, J.W.; Park, J.B.; Kim, J.S. Structural basis for the selective addition of an oxygen atom to cyclic ketones by Baeyer-Villiger monooxygenase from Parvibaculum lavamentivorans. Biochem. Biophys. Res. Commun. 2019, 512, 564–570. [Google Scholar] [CrossRef]
- Bosserman, M.A.; Downey, T.; Noinaj, N.; Buchanan, S.K.; Rohr, J. Molecular Insight into Substrate Recognition and Catalysis of Baeyer–Villiger Monooxygenase MtmOIV, the Key Frame-Modifying Enzyme in the Biosynthesis of Anticancer Agent Mithramycin. ACS Chem. Biol. 2013, 8, 2466–2477. [Google Scholar] [CrossRef] [PubMed]
- Pazmiño, D.E.T.; Dudek, H.M.; Fraaije, M.W. Baeyer–Villiger monooxygenases: Recent advances and future challenges. Curr. Opin. Chem. Biol. 2010, 14, 138–144. [Google Scholar] [CrossRef]
- Pazmiño, D.E.T.; Winkler, M.; Glieder, A.; Fraaije, M.W. Monooxygenases as biocatalysts: Classification, mechanistic aspects and biotechnological applications. J. Biotechnol. 2010, 146, 9–24. [Google Scholar] [CrossRef]
- Yu, J.M.; Liu, Y.Y.; Zheng, Y.C.; Li, H.; Zhang, X.Y.; Zheng, G.W.; Li, C.X.; Bai, Y.P.; Xu, J.H. Direct Access to Medium-Chain α,ω-Dicarboxylic Acids by Using a Baeyer–Villiger Monooxygenase of Abnormal Regioselectivity. ChemBioChem 2018, 19, 2049–2054. [Google Scholar] [CrossRef] [PubMed]
- Messiha, H.L.; Ahmed, S.T.; Karuppiah, V.; Suardíaz, R.; Avalos, G.A.A.; Fey, N.; Yeates, S.; Toogood, H.S.; Mulholland, A.J.; Scrutton, N.S. Biocatalytic Routes to Lactone Monomers for Polymer Production. Biochemistry 2018, 57, 1997–2008. [Google Scholar] [CrossRef] [PubMed]
- Fink, M.J.; Rudroff, F.; Mihovilovic, M.D. Baeyer–Villiger monooxygenases in aroma compound synthesis. Bioorg Med. Chem. Lett. 2011, 21, 6135–6138. [Google Scholar] [CrossRef]
- Alphand, V.; Carrea, G.; Wohlgemuth, R.; Furstoss, R.; Woodley, J.M. Towards large-scale synthetic applications of Baeyer-Villiger monooxygenases. Trends Biotechnol. 2003, 21, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Zhu, J.; Wu, Y.Q.; Zhang, Y.; Xia, J.Y.; Zhao, Q.; Lin, G.Q.; Yu, H.L.; Xu, J.H. Enzymatic Preparation of the Chiral (S)-Sulfoxide Drug Esomeprazole at Pilot-Scale Levels. Org. Process Res. Dev. 2020, 24, 1124–1130. [Google Scholar] [CrossRef]
- Matsui, T.; Dekishima, Y.; Ueda, M. Biotechnological production of chiral organic sulfoxides: Current state and perspectives. Appl. Microbiol. Biotechnol. 2014, 98, 7699–7706. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Kamerbeek, N.M.; Heidekamp, A.J.; Fortin, R.; Janssen, D.B. The Prodrug Activator EtaA from Mycobacterium tuberculosis Is a Baeyer-Villiger Monooxygenase. J. Biol. Chem. 2004, 279, 3354–3360. [Google Scholar] [CrossRef]
- Leisch, H.; Morley, K.; Lau, P.C.K. Baeyer−Villiger Monooxygenases: More Than Just Green Chemistry. Chem. Rev. 2011, 111, 4165–4222. [Google Scholar] [CrossRef]
- Willetts, A. Bicyclo[3.2.0]carbocyclic Molecules and Redox Biotransformations: The Evolution of Closed-Loop Artificial Linear Biocatalytic Cascades and Related Redox-Neutral Systems. Molecules 2023, 28, 7249. [Google Scholar] [CrossRef]
- Coker, J.A. Extremophiles and biotechnology: Current uses and prospects. F1000Research 2016, 5, 396. [Google Scholar] [CrossRef]
- Gran-Scheuch, A.; Trajkovic, M.; Parra, L.; Fraaije, M.W. Mining the Genome of Streptomyces leeuwenhoekii: Two New Type I Baeyer–Villiger Monooxygenases From Atacama Desert. Front. Microbiol. 2018, 9, 1609. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Wu, J.; Heuts, D.P.H.M.; van Hellemond, E.W.; Spelberg, J.H.L.; Janssen, D.B. Discovery of a thermostable Baeyer–Villiger monooxygenase by genome mining. Appl. Microbiol. Biotechnol. 2005, 66, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Romero, E.; Castellanos, J.R.G.; Mattevi, A.; Fraaije, M.W. Characterization and Crystal Structure of a Robust Cyclohexanone Monooxygenase. Angew. Chem. Int. Ed. 2016, 55, 15852–15855. [Google Scholar] [CrossRef]
- Azua-Bustos, A.; González-Silva, C.; Fairén, A.G. The Atacama Desert in Northern Chile as an Analog Model of Mars. Front. Astron. Space Sci. 2022, 8, 810426. [Google Scholar] [CrossRef]
- Navarro-González, R.; Rainey, F.A.; Molina, P.; Bagaley, D.R.; Hollen, B.J.; de la Rosa, J.; Small, A.M.; Quinn, R.C.; Grunthaner, F.J.; Cáceres, L.; et al. Mars-Like Soils in the Atacama Desert, Chile, and the Dry Limit of Microbial Life. Science 2003, 302, 1018–1021. [Google Scholar] [CrossRef]
- Bull, A.T.; Asenjo, J.A.; Goodfellow, M.; Gómez-Silva, B. The Atacama Desert: Technical Resources and the Growing Importance of Novel Microbial Diversity. Annu. Rev. Microbiol. 2016, 70, 215–234. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Robinson, C.K.; Barnum, T.; Fricke, W.F.; Davila, A.F.; Jedynak, B.; McKay, C.P.; DiRuggiero, J. Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome 2013, 1, 28. [Google Scholar] [CrossRef]
- Rateb, M.E.; Houssen, W.E.; Arnold, M.; Abdelrahman, M.H.; Deng, H.; Harrison, W.T.A.; Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Ferguson, G.; et al. Chaxamycins A–D, Bioactive Ansamycins from a Hyper-arid Desert Streptomyces sp. J. Nat. Prod. 2011, 74, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, M.S.A.; Philippon, T.; Asenjo, J.A.; Bull, A.T.; Goodfellow, M.; Ebel, R.; Jaspars, M.; Rateb, M.E. Asenjonamides A–C, antibacterial metabolites isolated from Streptomyces asenjonii strain KNN 42.f from an extreme-hyper arid Atacama Desert soil. J. Antibiot 2018, 71, 425–431. [Google Scholar] [CrossRef]
- Rateb, M.E.; Ebel, R.; Jaspars, M. Natural product diversity of actinobacteria in the Atacama Desert. Antonie Van Leeuwenhoek 2018, 111, 1467–1477. [Google Scholar] [CrossRef]
- Amaresan, N.; Kumar, K.; Naik, J.H.; Bapatla, K.G.; Mishra, R.K. Streptomyces in Plant Growth Promotion. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2018; pp. 125–135. [Google Scholar] [CrossRef]
- Guan, Y.; Chen, X. Recent Applications of Flavin-Dependent Monooxygenases in Biosynthesis, Pharmaceutical Development, and Environmental Science. Catalysts 2023, 13, 1495. [Google Scholar] [CrossRef]
- Ahmad, I.; Nawaz, N.; Darwesh, N.M.; Rahman, S.; Mustafa, M.Z.; Khan, S.B.; Patching, S.G. Overcoming challenges for amplified expression of recombinant proteins using Escherichia coli. Protein Expr. Purif. 2018, 144, 12–18. [Google Scholar] [CrossRef]
- Astakala, R.V.; Preet, G.; Milne, B.F.; Tibyangye, J.; Razmilic, V.; Castro, J.F.; Asenjo, J.A.; Andrews, B.; Ebel, R.; Jaspars, M. Mutactimycin AP, a New Mutactimycin Isolated from an Actinobacteria from the Atacama Desert. Molecules 2022, 27, 7185. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kobayashi, K.; Hasegawa, Y.; Iwaki, H. Cloning, expression, and characterization of Baeyer–Villiger monooxygenases from eukaryotic Exophiala jeanselmei strain KUFI-6N. Biosci. Biotechnol. Biochem. 2021, 85, 1675–1685. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Kim, D.H.; Kim, S.J.; Kim, J.H.; Bae, K.H.; Lee, C.H. The Analysis and Application of a Recombinant Monooxygenase Library as a Biocatalyst for the Baeyer-Villiger Reaction. J. Microbiol. Biotechnol. 2007, 17, 1083–1089. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Eppink, M.H.M.; Van Berkel, W.J.H.; Schreuder, H.A. Identification of a novel conserved sequence motif in flavoprotein hydroxylases with a putative dual function in FAD/NAD(P)H binding. Protein Sci. 1997, 6, 2454–2458. [Google Scholar] [CrossRef] [PubMed]
- Fraaije, M.W.; Kamerbeek, N.M.; van Berkel, W.J.H.; Janssen, D.B. Identification of a Baeyer–Villiger monooxygenase sequence motif. FEBS Lett. 2002, 518, 43–47. [Google Scholar] [CrossRef]
- Riebel, A.; Dudek, H.M.; de Gonzalo, G.; Stepniak, P.; Rychlewski, L.; Fraaije, M.W. Expanding the set of rhodococcal Baeyer–Villiger monooxygenases by high-throughput cloning, expression and substrate screening. Appl. Microbiol. Biotechnol. 2012, 95, 1479–1489. [Google Scholar] [CrossRef]
- Riebel, A.; de Gonzalo, G.; Fraaije, M.W. Expanding the biocatalytic toolbox of flavoprotein monooxygenases from Rhodococcus jostii RHA1. J. Mol. Catal. B Enzym. 2013, 88, 20–25. [Google Scholar] [CrossRef]
- Aalbers, F.S.; Fraaije, M.W. Coupled reactions by coupled enzymes: Alcohol to lactone cascade with alcohol dehydrogenase–cyclohexanone monooxygenase fusions. Appl. Microbiol. Biotechnol. 2017, 101, 7557–7565. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Aalbers, F.S.; Tang, W.; Röllig, R.; Fraaije, M.W.; Kara, S. Convergent Cascade Catalyzed by Monooxygenase–Alcohol Dehydrogenase Fusion Applied in Organic Media. ChemBioChem 2019, 20, 1653–1658. [Google Scholar] [CrossRef]
- Purwani, N.N.; Martin, C.; Savino, S.; Fraaije, M.W. Modular Assembly of Phosphite Dehydrogenase and Phenylacetone Monooxygenase for Tuning Cofactor Regeneration. Biomolecules 2021, 11, 905. [Google Scholar] [CrossRef] [PubMed]
- Mokhonov, V.V.; Vasilenko, E.A.; Gorshkova, E.N.; Astrakhantseva, I.V.; Novikov, D.V.; Novikov, V.V. SlyD-deficient Escherichia coli strains: A highway to contaminant-free protein extraction. Biochem. Biophys. Res. Commun. 2018, 499, 967–972. [Google Scholar] [CrossRef]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 2012, 11, 753. [Google Scholar] [CrossRef]
- Ren, G.; Ke, N.; Berkmen, M. Use of the SHuffle Strains in Production of Proteins. Curr. Protoc. Protein Sci. 2016, 85, 5.26.1–5.26.21. [Google Scholar] [CrossRef]
- Studier, F.W. T7 Expression Systems for Inducible Production of Proteins from Cloned Genes in E. coli. Curr. Protoc. Mol. Biol. 2018, 124, e63. [Google Scholar] [CrossRef]
- Craig, D.B.; Dombkowski, A.A. Disulfide by Design 2.0: A web-based tool for disulfide engineering in proteins. BMC Bioinform. 2013, 14, 346. [Google Scholar] [CrossRef]
- Chen, J.; Song, J.; Zhang, S.; Wang, Y.; Cui, D.; Wang, C. Chaperone Activity of DsbC. J. Biol. Chem. 1999, 274, 19601–19605. [Google Scholar] [CrossRef]
- Jensen, C.N.; Cartwright, J.; Ward, J.; Hart, S.; Turkenburg, J.P.; Ali, S.T.; Allen, M.J.; Grogan, G. A Flavoprotein Monooxygenase that Catalyses a Baeyer–Villiger Reaction and Thioether Oxidation Using NADH as the Nicotinamide Cofactor. ChemBioChem 2012, 13, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Ceccoli, R.D.; Bianchi, D.A.; Carabajal, M.A.; Rial, D.V. Genome mining reveals new bacterial type I Baeyer-Villiger monooxygenases with (bio)synthetic potential. Mol. Catal. 2020, 486, 110875. [Google Scholar] [CrossRef]
- Niero, M.; Righetto, I.; Beneventi, E.; de Laureto, P.P.; Fraaije, M.W.; Filippini, F.; Bergantino, E. Unique Features of a New Baeyer–Villiger Monooxygenase from a Halophilic Archaeon. Catalysts 2020, 10, 128. [Google Scholar] [CrossRef]
- Fürst, M.J.L.J.; Boonstra, M.; Bandstra, S.; Fraaije, M.W. Stabilization of cyclohexanone monooxygenase by computational and experimental library design. Biotechnol. Bioeng. 2019, 116, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Bisagni, S.; Hatti-Kaul, R.; Mamo, G. Cloning, expression and characterization of a versatile Baeyer-Villiger monooxygenase from Dietzia sp. D5. AMB Express 2014, 4, 23. [Google Scholar] [CrossRef]
- Chánique, A.M.; Polidori, N.; Sovic, L.; Kracher, D.; Assil-Companioni, L.; Galuska, P.; Parra, L.P.; Gruber, K.; Kourist, R. A Cold-Active Flavin-Dependent Monooxygenase from Janthinobacterium svalbardensis Unlocks Applications of Baeyer–Villiger Monooxygenases at Low Temperature. ACS Catal. 2023, 13, 3549–3562. [Google Scholar] [CrossRef]
- Secundo, F.; Fiala, S.; Fraaije, M.W.; de Gonzalo, G.; Meli, M.; Zambianchi, F.; Ottolina, G. Effects of water miscible organic solvents on the activity and conformation of the baeyer–villiger monooxygenases from Thermobifida fusca and Acinetobacter calcoaceticus: A comparative study. Biotechnol. Bioeng. 2011, 108, 491–499. [Google Scholar] [CrossRef]
- Doukyu, N.; Ogino, H. Organic solvent-tolerant enzymes. Biochem. Eng. J. 2010, 48, 270–282. [Google Scholar] [CrossRef]
- Wei, S.; Liu, Y.; Zhou, J.; Xu, G.; Ni, Y. Two enantiocomplementary Baeyer-Villiger monooxygenases newly identified for asymmetric oxyfunctionalization of thioether. Mol. Catal. 2021, 513, 111784. [Google Scholar] [CrossRef]
- Lehuedé, L.; Córdova, A.; Rodríguez, S.; Astudillo-Castro, C.; Vera, C.; Salazar, O. A sequential bioconversion-purification process to produce high purity xylo-oligosaccharides (XOS) from Nothophagus pumilio sawdust wastes. Ind. Crops Prod. 2024, 214, 118531. [Google Scholar] [CrossRef]





| Strain | Plasmid | Host | Reference |
|---|---|---|---|
| MO-G35A-p22N | pET-22b | E. coli NiCo21(DE3) | This work |
| MO-G35A-p28N | pET-28a | E. coli NiCo21(DE3) | This work |
| MO-G35A-p28S | pET-28a | E. coli Shuffle T7 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, C.; Rodríguez, S.; Reyes-Godoy, J.P.; Razmilic, V.; Martínez, I. Recombinant Expression and Characterization of a Novel Type I Baeyer–Villiger Monooxygenase from a Streptomyces Strain Isolated from the Rhizosphere of the Atacama Desert Lupinus oreophilus. Int. J. Mol. Sci. 2025, 26, 5940. https://doi.org/10.3390/ijms26135940
González C, Rodríguez S, Reyes-Godoy JP, Razmilic V, Martínez I. Recombinant Expression and Characterization of a Novel Type I Baeyer–Villiger Monooxygenase from a Streptomyces Strain Isolated from the Rhizosphere of the Atacama Desert Lupinus oreophilus. International Journal of Molecular Sciences. 2025; 26(13):5940. https://doi.org/10.3390/ijms26135940
Chicago/Turabian StyleGonzález, Carolina, Sebastián Rodríguez, José Pablo Reyes-Godoy, Valeria Razmilic, and Irene Martínez. 2025. "Recombinant Expression and Characterization of a Novel Type I Baeyer–Villiger Monooxygenase from a Streptomyces Strain Isolated from the Rhizosphere of the Atacama Desert Lupinus oreophilus" International Journal of Molecular Sciences 26, no. 13: 5940. https://doi.org/10.3390/ijms26135940
APA StyleGonzález, C., Rodríguez, S., Reyes-Godoy, J. P., Razmilic, V., & Martínez, I. (2025). Recombinant Expression and Characterization of a Novel Type I Baeyer–Villiger Monooxygenase from a Streptomyces Strain Isolated from the Rhizosphere of the Atacama Desert Lupinus oreophilus. International Journal of Molecular Sciences, 26(13), 5940. https://doi.org/10.3390/ijms26135940






