SRC and ERK Regulate the Turnover of Cytoskeletal Keratin Filaments
Abstract
1. Introduction
2. Results
2.1. Experimental Design
2.2. Inhibition of SRC Reduces Keratin Filament Turnover
2.3. Inhibition of FAK Signaling Does Not Affect Keratin Turnover
2.4. Inhibition of PI3K/AKT Signaling Has Only a Minor Effect on Keratin Turnover
2.5. Inhibition of ERK Decreases Keratin Filament Turnover
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Pharmacological Inhibitors
4.3. Microscopy
4.4. Antibodies and Immunoblotting
4.5. Image Analysis and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Windoffer, R.; Woll, S.; Strnad, P.; Leube, R.E. Identification of novel principles of keratin filament network turnover in living cells. Mol. Biol. Cell 2004, 15, 2436–2448. [Google Scholar] [CrossRef]
- Kolsch, A.; Windoffer, R.; Wurflinger, T.; Aach, T.; Leube, R.E. The keratin-filament cycle of assembly and disassembly. J. Cell Sci. 2010, 123 Pt 13, 2266–2272. [Google Scholar] [CrossRef] [PubMed]
- Windoffer, R.; Beil, M.; Magin, T.M.; Leube, R.E. Cytoskeleton in motion: The dynamics of keratin intermediate filaments in epithelia. J. Cell Biol. 2011, 194, 669–678. [Google Scholar] [CrossRef]
- Leube, R.E.; Moch, M.; Kolsch, A.; Windoffer, R. “Panta rhei”: Perpetual cycling of the keratin cytoskeleton. Bioarchitecture 2011, 1, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.H.; Yoon, M.; Moir, R.D.; Khuon, S.; Flitney, F.W.; Goldman, R.D. Insights into the dynamic properties of keratin intermediate filaments in living epithelial cells. J. Cell Biol. 2001, 153, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Sawant, M.; Schwarz, N.; Windoffer, R.; Magin, T.M.; Krieger, J.; Mucke, N.; Obara, B.; Jankowski, V.; Jankowski, J.; Wally, V.; et al. Threonine 150 Phosphorylation of Keratin 5 Is Linked to Epidermolysis Bullosa Simplex and Regulates Filament Assembly and Cell Viability. J. Investig. Dermatol. 2018, 138, 627–636. [Google Scholar] [CrossRef]
- Snider, N.T.; Omary, M.B. Post-translational modifications of intermediate filament proteins: Mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2014, 15, 163–177. [Google Scholar] [CrossRef]
- Nishimura, Y.; Kasahara, K.; Inagaki, M. Intermediate filaments and IF-associated proteins: From cell architecture to cell proliferation. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2019, 95, 479–493. [Google Scholar] [CrossRef]
- Woll, S.; Windoffer, R.; Leube, R.E. p38 MAPK-dependent shaping of the keratin cytoskeleton in cultured cells. J. Cell Biol. 2007, 177, 795–807. [Google Scholar] [CrossRef]
- Feng, X.; Coulombe, P.A. Complementary roles of specific cysteines in keratin 14 toward the assembly, organization, and dynamics of intermediate filaments in skin keratinocytes. J. Biol. Chem. 2015, 290, 22507–22519. [Google Scholar] [CrossRef]
- Osmanagic-Myers, S.; Gregor, M.; Walko, G.; Burgstaller, G.; Reipert, S.; Wiche, G. Plectin-controlled keratin cytoarchitecture affects MAP kinases involved in cellular stress response and migration. J. Cell Biol. 2006, 174, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Moch, M.; Windoffer, R.; Schwarz, N.; Pohl, R.; Omenzetter, A.; Schnakenberg, U.; Herb, F.; Chaisaowong, K.; Merhof, D.; Ramms, L.; et al. Effects of Plectin Depletion on Keratin Network Dynamics and Organization. PLoS ONE 2016, 11, e0149106. [Google Scholar] [CrossRef] [PubMed]
- Laly, A.C.; Sliogeryte, K.; Pundel, O.J.; Ross, R.; Keeling, M.C.; Avisetti, D.; Waseem, A.; Gavara, N.; Connelly, J.T. The keratin network of intermediate filaments regulates keratinocyte rigidity sensing and nuclear mechanotransduction. Sci. Adv. 2021, 7, eabd6187. [Google Scholar] [CrossRef]
- Guo, Y.; Redmond, C.J.; Leacock, K.A.; Brovkina, M.V.; Ji, S.; Jaskula-Ranga, V.; Coulombe, P.A. Keratin 14-dependent disulfides regulate epidermal homeostasis and barrier function via 14-3-3sigma and YAP1. Elife 2020, 9, e53165. [Google Scholar] [CrossRef]
- Liao, J.; Lowthert, L.A.; Ku, N.O.; Fernandez, R.; Omary, M.B. Dynamics of human keratin 18 phosphorylation: Polarized distribution of phosphorylated keratins in simple epithelial tissues. J. Cell Biol. 1995, 131, 1291–1301. [Google Scholar] [CrossRef]
- Sawant, M.S.; Leube, R.E. Consequences of Keratin Phosphorylation for Cytoskeletal Organization and Epithelial Functions. Int. Rev. Cell Mol. Biol. 2017, 330, 171–225. [Google Scholar] [PubMed]
- Ku, N.O.; Omary, M.B. Phosphorylation of human keratin 8 in vivo at conserved head domain serine 23 and at epidermal growth factor-stimulated tail domain serine 431. J. Biol. Chem. 1997, 272, 7556–7564. [Google Scholar] [CrossRef]
- Moch, M.; Herberich, G.; Aach, T.; Leube, R.E.; Windoffer, R. Measuring the regulation of keratin filament network dynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 10664–10669. [Google Scholar] [CrossRef]
- Toivola, D.M.; Zhou, Q.; English, L.S.; Omary, M.B. Type II keratins are phosphorylated on a unique motif during stress and mitosis in tissues and cultured cells. Mol. Biol. Cell 2002, 13, 1857–1870. [Google Scholar] [CrossRef]
- Zhou, Q.; Snider, N.T.; Liao, J.; Li, D.H.; Hong, A.; Ku, N.O.; Cartwright, C.A.; Omary, M.B. Characterization of in vivo keratin 19 phosphorylation on tyrosine-391. PLoS ONE 2010, 5, e13538. [Google Scholar] [CrossRef]
- Ku, N.O.; Azhar, S.; Omary, M.B. Keratin 8 phosphorylation by p38 kinase regulates cellular keratin filament reorganization: Modulation by a keratin 1-like disease causing mutation. J. Biol. Chem. 2002, 277, 10775–10782. [Google Scholar] [CrossRef] [PubMed]
- Menon, M.B.; Schwermann, J.; Singh, A.K.; Franz-Wachtel, M.; Pabst, O.; Seidler, U.; Omary, M.B.; Kotlyarov, A.; Gaestel, M. p38 MAP kinase and MAPKAP kinases MK2/3 cooperatively phosphorylate epithelial keratins. J. Biol. Chem. 2010, 285, 33242–33251. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Zhou, X.; Liao, J.; Omary, M.B. Pervanadate-mediated tyrosine phosphorylation of keratins 8 and 19 via a p38 mitogen-activated protein kinase-dependent pathway. J. Cell Sci. 1999, 112 Pt 13, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Stepulak, A.; Holmstrom, T.H.; Omary, M.B.; Eriksson, J.E. The intermediate filament protein keratin 8 is a novel cytoplasmic substrate for c-Jun N-terminal kinase. J. Biol. Chem. 2002, 277, 10767–10774. [Google Scholar] [CrossRef]
- Xia, B.; Zhang, H.; Yang, M.; Du, S.; Wei, J.; Ding, L. Leukamenin E Induces K8/18 Phosphorylation and Blocks the Assembly of Keratin Filament Networks Through ERK Activation. Int. J. Mol. Sci. 2020, 21, 3164. [Google Scholar] [CrossRef]
- Strnad, P.; Windoffer, R.; Leube, R.E. In vivo detection of cytokeratin filament network breakdown in cells treated with the phosphatase inhibitor okadaic acid. Cell Tissue Res. 2001, 306, 277–293. [Google Scholar] [CrossRef]
- Strnad, P.; Windoffer, R.; Leube, R.E. Induction of rapid and reversible cytokeratin filament network remodeling by inhibition of tyrosine phosphatases. J. Cell Sci. 2002, 115 Pt 21, 4133–4148. [Google Scholar] [CrossRef]
- Strnad, P.; Windoffer, R.; Leube, R.E. Light-induced resistance of the keratin network to the filament-disrupting tyrosine phosphatase inhibitor orthovanadate. J. Investig. Dermatol. 2003, 120, 198–203. [Google Scholar] [CrossRef][Green Version]
- Windoffer, R.; Leube, R.E. Detection of cytokeratin dynamics by time-lapse fluorescence microscopy in living cells. J. Cell Sci. 1999, 112 Pt 24, 4521–4534. [Google Scholar] [CrossRef]
- Leube, R.E.; Bader, B.L.; Bosch, F.X.; Zimbelmann, R.; Achtstaetter, T.; Franke, W.W. Molecular characterization and expression of the stratification-related cytokeratins 4 and 15. J. Cell Biol. 1988, 106, 1249–1261. [Google Scholar] [CrossRef]
- Ullrich, A.; Coussens, L.; Hayflick, J.S.; Dull, T.J.; Gray, A.; Tam, A.W.; Lee, J.; Yarden, Y.; Libermann, T.A.; Schlessinger, J.; et al. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature 1984, 309, 418–425. [Google Scholar] [CrossRef]
- Gadella, T.W., Jr.; Jovin, T.M. Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J. Cell Biol. 1995, 129, 1543–1558. [Google Scholar] [CrossRef]
- Ratajczyk, S.; Drexler, C.; Windoffer, R.; Leube, R.E.; Fuchs, P. A Ca2+-Mediated Switch of Epiplakin from a Diffuse to Keratin-Bound State Affects Keratin Dynamics. Cells 2022, 11, 3077. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.; Moch, M.; Neckernuss, T.; Paschke, S.; Herrmann, H.; Marti, O. Both monovalent cations and plectin are potent modulators of mechanical properties of keratin K8/K18 networks. Soft Matter 2016, 12, 6964–6974. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, W.; Schuster, M.; Moch, M.; Windoffer, R.; Steinberg, G.; Staiger, C.J.; Panstruga, R. Alloxan Disintegrates the Plant Cytoskeleton and Suppresses mlo-Mediated Powdery Mildew Resistance. Plant Cell Physiol. 2020, 61, 505–518. [Google Scholar] [CrossRef]
- Schwarz, N.; Moch, M.; Windoffer, R.; Leube, R.E. Multidimensional Monitoring of Keratin Intermediate Filaments in Cultured Cells and Tissues. Methods Enzymol. 2016, 568, 59–83. [Google Scholar] [PubMed]
- Vultur, A.; Buettner, R.; Kowolik, C.; Liang, W.; Smith, D.; Boschelli, F.; Jove, R. SKI-606 (bosutinib), a novel Src kinase inhibitor, suppresses migration and invasion of human breast cancer cells. Mol. Cancer Ther. 2008, 7, 1185–1194. [Google Scholar] [CrossRef]
- Bain, J.; McLauchlan, H.; Elliott, M.; Cohen, P. The specificities of protein kinase inhibitors: An update. Biochem. J. 2003, 371 Pt 1, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Shen, N.; Guo, T.; Wang, J.; Xie, C.; Cao, Y.; Liu, L.; Yan, Y.; Meng, S.; Du, S. SPTLC2 drives an EGFR-FAK-HBEGF signaling axis to promote ovarian cancer progression. Oncogene 2024, 44, 679–693. [Google Scholar] [CrossRef]
- Mitra, S.K.; Schlaepfer, D.D. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 2006, 18, 516–523. [Google Scholar] [CrossRef]
- Sieg, D.J.; Hauck, C.R.; Ilic, D.; Klingbeil, C.K.; Schaefer, E.; Damsky, C.H.; Schlaepfer, D.D. FAK integrates growth-factor and integrin signals to promote cell migration. Nat. Cell Biol. 2000, 2, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Yi, P.; Amazit, L.; LaMarca, H.L.; Ashcroft, F.; Kumar, R.; Mancini, M.A.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. SRC-3Delta4 mediates the interaction of EGFR with FAK to promote cell migration. Mol. Cell 2010, 37, 321–332. [Google Scholar] [CrossRef]
- Slack-Davis, J.K.; Martin, K.H.; Tilghman, R.W.; Iwanicki, M.; Ung, E.J.; Autry, C.; Luzzio, M.J.; Cooper, B.; Kath, J.C.; Roberts, W.G.; et al. Cellular characterization of a novel focal adhesion kinase inhibitor. J. Biol. Chem. 2007, 282, 14845–14852. [Google Scholar] [CrossRef]
- Tse, K.W.; Dang-Lawson, M.; Lee, R.L.; Vong, D.; Bulic, A.; Buckbinder, L.; Gold, M.R. B cell receptor-induced phosphorylation of Pyk2 and focal adhesion kinase involves integrins and the Rap GTPases and is required for B cell spreading. J. Biol. Chem. 2009, 284, 22865–22877. [Google Scholar] [CrossRef] [PubMed]
- Chan, X.Y.; Chang, K.P.; Yang, C.Y.; Liu, C.R.; Hung, C.M.; Huang, C.C.; Liu, H.P.; Wu, C.C. Upregulation of ENAH by a PI3K/AKT/beta-catenin cascade promotes oral cancer cell migration and growth via an ITGB5/Src axis. Cell. Mol. Biol. Lett. 2024, 29, 136. [Google Scholar] [CrossRef]
- Poh, A.R.; Ernst, M. Functional roles of SRC signaling in pancreatic cancer: Recent insights provide novel therapeutic opportunities. Oncogene 2023, 42, 1786–1801. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Kim, S.; Yoon, H.N.; Ku, N.O. Keratin 8/18 Regulate the Akt Signaling Pathway. Int. J. Mol. Sci. 2021, 22, 9227. [Google Scholar] [CrossRef]
- Paramio, J.M.; Segrelles, C.; Ruiz, S.; Jorcano, J.L. Inhibition of protein kinase B (PKB) and PKCzeta mediates keratin K10-induced cell cycle arrest. Mol. Cell. Biol. 2001, 21, 7449–7459. [Google Scholar] [CrossRef]
- Santos, M.; Paramio, J.M.; Bravo, A.; Ramirez, A.; Jorcano, J.L. The expression of keratin k10 in the basal layer of the epidermis inhibits cell proliferation and prevents skin tumorigenesis. J. Biol. Chem. 2002, 277, 19122–19130. [Google Scholar] [CrossRef]
- Kim, S.; Wong, P.; Coulombe, P.A. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature 2006, 441, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Chivu-Economescu, M.; Dragu, D.L.; Necula, L.G.; Matei, L.; Enciu, A.M.; Bleotu, C.; Diaconu, C.C. Knockdown of KRT17 by siRNA induces antitumoral effects on gastric cancer cells. Gastric Cancer 2017, 20, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, S.I.; Zvelebil, M.J.; Shuttleworth, S.J.; Hancox, T.; Saghir, N.; Timms, J.F.; Waterfield, M.D. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem. J. 2007, 404, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kuramitsu, Y.; Baron, B.; Kitagawa, T.; Tokuda, K.; Akada, J.; Maehara, S.I.; Maehara, Y.; Nakamura, K. PI3K inhibitor LY294002, as opposed to wortmannin, enhances AKT phosphorylation in gemcitabine-resistant pancreatic cancer cells. Int. J. Oncol. 2017, 50, 606–612. [Google Scholar] [CrossRef]
- Gilot, D.; Giudicelli, F.; Lagadic-Gossmann, D.; Fardel, O. Akti-1/2, an allosteric inhibitor of Akt 1 and 2, efficiently inhibits CaMKIalpha activity and aryl hydrocarbon receptor pathway. Chem.-Biol. Interact. 2010, 188, 546–552. [Google Scholar] [CrossRef]
- Wang, L.; Li, S.; Cai, K.; Xiao, Y.; Ye, L. TLR7 Agonists Modulate the Activation of Human Conjunctival Epithelial Cells Induced by IL-1beta via the ERK1/2 Signaling Pathway. Inflammation 2023, 46, 1430–1444. [Google Scholar] [CrossRef]
- Morris, E.J.; Jha, S.; Restaino, C.R.; Dayananth, P.; Zhu, H.; Cooper, A.; Carr, D.; Deng, Y.; Jin, W.; Black, S.; et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013, 3, 742–750. [Google Scholar] [CrossRef]
- Ku, N.O.; Omary, M.B. Keratins turn over by ubiquitination in a phosphorylation-modulated fashion. J. Cell Biol. 2000, 149, 547–552. [Google Scholar] [CrossRef]
- Egu, D.T.; Schmitt, T.; Ernst, N.; Ludwig, R.J.; Fuchs, M.; Hiermaier, M.; Moztarzadeh, S.; Moron, C.S.; Schmidt, E.; Beyersdorfer, V.; et al. EGFR Inhibition by Erlotinib Rescues Desmosome Ultrastructure and Keratin Anchorage and Protects against Pemphigus Vulgaris IgG-Induced Acantholysis in Human Epidermis. J. Investig. Dermatol. 2024, 144, 2440–2452. [Google Scholar] [CrossRef]
- Green, K.J.; Niessen, C.M.; Rubsam, M.; Perez White, B.E.; Broussard, J.A. The Desmosome-Keratin Scaffold Integrates ErbB Family and Mechanical Signaling to Polarize Epidermal Structure and Function. Front. Cell Dev. Biol. 2022, 10, 903696. [Google Scholar] [CrossRef]
- Te Molder, L.; de Pereda, J.M.; Sonnenberg, A. Regulation of hemidesmosome dynamics and cell signaling by integrin alpha6beta4. J. Cell Sci. 2021, 134, jcs259004. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, J. Common and distinct elements in cellular signaling via EGF and FGF receptors. Science 2004, 306, 1506–1507. [Google Scholar] [CrossRef] [PubMed]
- Hynes, N.E.; MacDonald, G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009, 21, 177–184. [Google Scholar] [CrossRef]
- Moustakas, A. Crosstalk between TGF-beta and EGF receptors via direct phosphorylation. J. Cell Biol. 2024, 223, e202403075. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Accornero, P.; Miretti, S.; Starvaggi Cucuzza, L.; Martignani, E.; Baratta, M. Epidermal growth factor and hepatocyte growth factor cooperate to enhance cell proliferation, scatter, and invasion in murine mammary epithelial cells. J. Mol. Endocrinol. 2010, 44, 115–125. [Google Scholar] [CrossRef]
- Osmani, N.; Labouesse, M. Remodeling of keratin-coupled cell adhesion complexes. Curr. Opin. Cell Biol. 2015, 32, 30–38. [Google Scholar] [CrossRef]
- Tehrani, S.; Tomasevic, N.; Weed, S.; Sakowicz, R.; Cooper, J.A. Src phosphorylation of cortactin enhances actin assembly. Proc. Natl. Acad. Sci. USA 2007, 104, 11933–11938. [Google Scholar] [CrossRef]
- Yamada, T.; Aoyama, Y.; Owada, M.K.; Kawakatsu, H.; Kitajima, Y. Scraped-wounding causes activation and association of C-Src tyrosine kinase with microtubules in cultured keratinocytes. Cell Struct. Funct. 2000, 25, 351–359. [Google Scholar] [CrossRef]
- He, Y.; Ren, Y.; Wu, B.; Decourt, B.; Lee, A.C.; Taylor, A.; Suter, D.M. Src and cortactin promote lamellipodia protrusion and filopodia formation and stability in growth cones. Mol. Biol. Cell 2015, 26, 3229–3244. [Google Scholar] [CrossRef]
- Thomas, S.M.; Brugge, J.S. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 1997, 13, 513–609. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Meng, S.; Mei, L.; Zhao, Z.J.; Jove, R.; Wu, J. Roles of Gab1 and SHP2 in paxillin tyrosine dephosphorylation and Src activation in response to epidermal growth factor. J. Biol. Chem. 2004, 279, 8497–8505. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.W.; McLean, G.W.; Wyke, A.W.; Paraskeva, C.; Parkinson, E.K.; Frame, M.C.; Brunton, V.G. The catalytic activity of the Src family kinases is required to disrupt cadherin-dependent cell-cell contacts. Mol. Biol. Cell 2000, 11, 51–64. [Google Scholar] [CrossRef]
- Li, L.; Okura, M.; Imamoto, A. Focal adhesions require catalytic activity of Src family kinases to mediate integrin-matrix adhesion. Mol. Cell. Biol. 2002, 22, 1203–1217. [Google Scholar] [CrossRef]
- Palovuori, R.; Sormunen, R.; Eskelinen, S. SRC-induced disintegration of adherens junctions of madin-darby canine kidney cells is dependent on endocytosis of cadherin and antagonized by Tiam-1. Lab. Investig. 2003, 83, 1901–1915. [Google Scholar] [CrossRef]
- Westhoff, M.A.; Serrels, B.; Fincham, V.J.; Frame, M.C.; Carragher, N.O. SRC-mediated phosphorylation of focal adhesion kinase couples actin and adhesion dynamics to survival signaling. Mol. Cell. Biol. 2004, 24, 8113–8133. [Google Scholar] [CrossRef] [PubMed]
- Destaing, O.; Sanjay, A.; Itzstein, C.; Horne, W.C.; Toomre, D.; De Camilli, P.; Baron, R. The tyrosine kinase activity of c-Src regulates actin dynamics and organization of podosomes in osteoclasts. Mol. Biol. Cell 2008, 19, 394–404. [Google Scholar] [CrossRef]
- Kuga, T.; Hoshino, M.; Nakayama, Y.; Kasahara, K.; Ikeda, K.; Obata, Y.; Takahashi, A.; Higashiyama, Y.; Fukumoto, Y.; Yamaguchi, N. Role of Src-family kinases in formation of the cortical actin cap at the dorsal cell surface. Exp. Cell Res. 2008, 314, 2040–2054. [Google Scholar] [CrossRef]
- Rotty, J.D.; Coulombe, P.A. A wound-induced keratin inhibits Src activity during keratinocyte migration and tissue repair. J. Cell Biol. 2012, 197, 381–389. [Google Scholar] [CrossRef]
- Saha, S.K.; Kim, K.; Yang, G.M.; Choi, H.Y.; Cho, S.G. Cytokeratin 19 (KRT19) has a Role in the Reprogramming of Cancer Stem Cell-Like Cells to Less Aggressive and More Drug-Sensitive Cells. Int. J. Mol. Sci. 2018, 19, 1423. [Google Scholar] [CrossRef]
- Kaplan, K.B.; Bibbins, K.B.; Swedlow, J.R.; Arnaud, M.; Morgan, D.O.; Varmus, H.E. Association of the amino-terminal half of c-Src with focal adhesions alters their properties and is regulated by phosphorylation of tyrosine 527. EMBO J. 1994, 13, 4745–4756. [Google Scholar] [CrossRef] [PubMed]
- Maa, M.C.; Leu, T.H. Vanadate-dependent FAK activation is accomplished by the sustained FAK Tyr-576/577 phosphorylation. Biochem. Biophys. Res. Commun. 1998, 251, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Windoffer, R.; Kolsch, A.; Woll, S.; Leube, R.E. Focal adhesions are hotspots for keratin filament precursor formation. J. Cell Biol. 2006, 173, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Leube, R.E.; Moch, M.; Windoffer, R. Intermediate filaments and the regulation of focal adhesion. Curr. Opin. Cell Biol. 2015, 32, 13–20. [Google Scholar] [CrossRef]
- Dave, J.M.; Kang, H.; Abbey, C.A.; Maxwell, S.A.; Bayless, K.J. Proteomic profiling of endothelial invasion revealed receptor for activated C kinase 1 (RACK1) complexed with vimentin to regulate focal adhesion kinase (FAK). J. Biol. Chem. 2013, 288, 30720–30733. [Google Scholar] [CrossRef]
- Gregor, M.; Osmanagic-Myers, S.; Burgstaller, G.; Wolfram, M.; Fischer, I.; Walko, G.; Resch, G.P.; Jorgl, A.; Herrmann, H.; Wiche, G. Mechanosensing through focal adhesion-anchored intermediate filaments. FASEB J. 2014, 28, 715–729. [Google Scholar] [CrossRef]
- Havel, L.S.; Kline, E.R.; Salgueiro, A.M.; Marcus, A.I. Vimentin regulates lung cancer cell adhesion through a VAV2-Rac1 pathway to control focal adhesion kinase activity. Oncogene 2015, 34, 1979–1990. [Google Scholar] [CrossRef]
- Zholudeva, A.O.; Potapov, N.S.; Kozlova, E.A.; Lomakina, M.E.; Alexandrova, A.Y. Impairment of Assembly of the Vimentin Intermediate Filaments Leads to Suppression of Formation and Maturation of Focal Contacts and Alteration of the Type of Cellular Protrusions. Biochem. (Mosc.) 2024, 89, 184–195. [Google Scholar] [CrossRef]
- Pan, Y.; Jing, R.; Pitre, A.; Williams, B.J.; Skalli, O. Intermediate filament protein synemin contributes to the migratory properties of astrocytoma cells by influencing the dynamics of the actin cytoskeleton. FASEB J. 2008, 22, 3196–3206. [Google Scholar] [CrossRef]
- Sun, N.; Huiatt, T.W.; Paulin, D.; Li, Z.; Robson, R.M. Synemin interacts with the LIM domain protein zyxin and is essential for cell adhesion and migration. Exp. Cell Res. 2010, 316, 491–505. [Google Scholar] [CrossRef]
- Bordeleau, F.; Galarneau, L.; Gilbert, S.; Loranger, A.; Marceau, N. Keratin 8/18 modulation of protein kinase C-mediated integrin-dependent adhesion and migration of liver epithelial cells. Mol. Biol. Cell 2010, 21, 1698–1713. [Google Scholar] [CrossRef] [PubMed]
- Braicu, C.; Buse, M.; Busuioc, C.; Drula, R.; Gulei, D.; Raduly, L.; Rusu, A.; Irimie, A.; Atanasov, A.G.; Slaby, O.; et al. A Comprehensive Review on MAPK: A Promising Therapeutic Target in Cancer. Cancers 2019, 11, 1618. [Google Scholar] [CrossRef] [PubMed]
- Osaki, L.H.; Gama, P. MAPKs and signal transduction in the control of gastrointestinal epithelial cell proliferation and differentiation. Int. J. Mol. Sci. 2013, 14, 10143–10161. [Google Scholar] [CrossRef]
- Ganguly, P.; Macleod, T.; Wong, C.; Harland, M.; McGonagle, D. Revisiting p38 Mitogen-Activated Protein Kinases (MAPK) in Inflammatory Arthritis: A Narrative of the Emergence of MAPK-Activated Protein Kinase Inhibitors (MK2i). Pharmaceuticals 2023, 16, 1286. [Google Scholar] [CrossRef]
- Huang, C.; Jacobson, K.; Schaller, M.D. MAP kinases and cell migration. J. Cell Sci. 2004, 117 Pt 20, 4619–4628. [Google Scholar] [CrossRef]
- Waschke, J.; Spindler, V. Desmosomes and extradesmosomal adhesive signaling contacts in pemphigus. Med. Res. Rev. 2014, 34, 1127–1145. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, P.; Hu, P.; Liu, Z.; Diaz, L.A.; Enghild, J.J.; Chua, M.P.; Rubenstein, D.S. Desmosome signaling. Inhibition of p38MAPK prevents pemphigus vulgaris IgG-induced cytoskeleton reorganization. J. Biol. Chem. 2005, 280, 23778–23784. [Google Scholar] [CrossRef]
- Perez Verdaguer, M.; Zhang, T.; Paulo, J.A.; Gygi, S.; Watkins, S.C.; Sakurai, H.; Sorkin, A. Mechanism of p38 MAPK-induced EGFR endocytosis and its crosstalk with ligand-induced pathways. J. Cell Biol. 2021, 220, e202102005. [Google Scholar] [CrossRef]
- Park, M.K.; Lee, H.J.; Shin, J.; Noh, M.; Kim, S.Y.; Lee, C.H. Novel participation of transglutaminase-2 through c-Jun N-terminal kinase activation in sphingosylphosphorylcholine-induced keratin reorganization of PANC-1 cells. Biochim. Biophys. Acta 2011, 1811, 1021–1029. [Google Scholar] [CrossRef]
- Russell, D.; Ross, H.; Lane, E.B. ERK involvement in resistance to apoptosis in keratinocytes with mutant keratin. J. Investig. Dermatol. 2010, 130, 671–681. [Google Scholar] [CrossRef]
- Pan, X.; Kane, L.A.; Van Eyk, J.E.; Coulombe, P.A. Type I keratin 17 protein is phosphorylated on serine 44 by p90 ribosomal protein S6 kinase 1 (RSK1) in a growth- and stress-dependent fashion. J. Biol. Chem. 2011, 286, 42403–42413. [Google Scholar] [CrossRef] [PubMed]
- Omary, M.B.; Ku, N.O.; Liao, J.; Price, D. Keratin modifications and solubility properties in epithelial cells and in vitro. Sub-Cell. Biochem. 1998, 31, 105–140. [Google Scholar]
- Zhou, X.; Liao, J.; Hu, L.; Feng, L.; Omary, M.B. Characterization of the major physiologic phosphorylation site of human keratin 19 and its role in filament organization. J. Biol. Chem. 1999, 274, 12861–12866. [Google Scholar] [CrossRef] [PubMed]
- Moch, M.; Schwarz, N.; Windoffer, R.; Leube, R.E. The keratin-desmosome scaffold: Pivotal role of desmosomes for keratin network morphogenesis. Cell Mol. Life Sci. 2020, 77, 543–558. [Google Scholar] [CrossRef]
- Romashin, D.D.; Tolstova, T.V.; Varshaver, A.M.; Kozhin, P.M.; Rusanov, A.L.; Luzgina, N.G. Keratins 6, 16, and 17 in Health and Disease: A Summary of Recent Findings. Curr. Issues Mol. Biol. 2024, 46, 8627–8641. [Google Scholar] [CrossRef]
- Langhofer, M.; Hopkinson, S.B.; Jones, J.C. The matrix secreted by 804G cells contains laminin-related components that participate in hemidesmosome assembly in vitro. J. Cell Sci. 1993, 105 Pt 3, 753–764. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
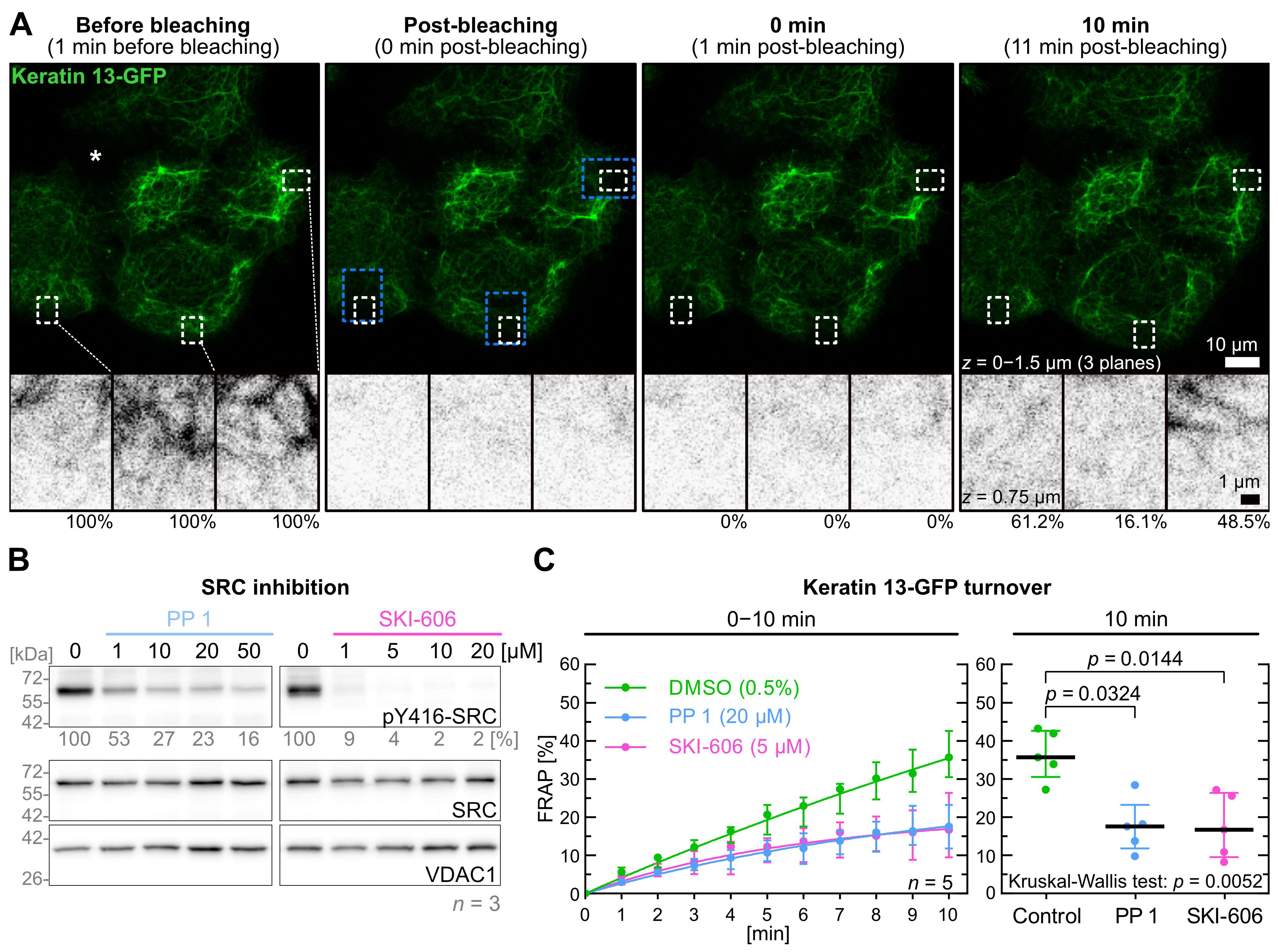

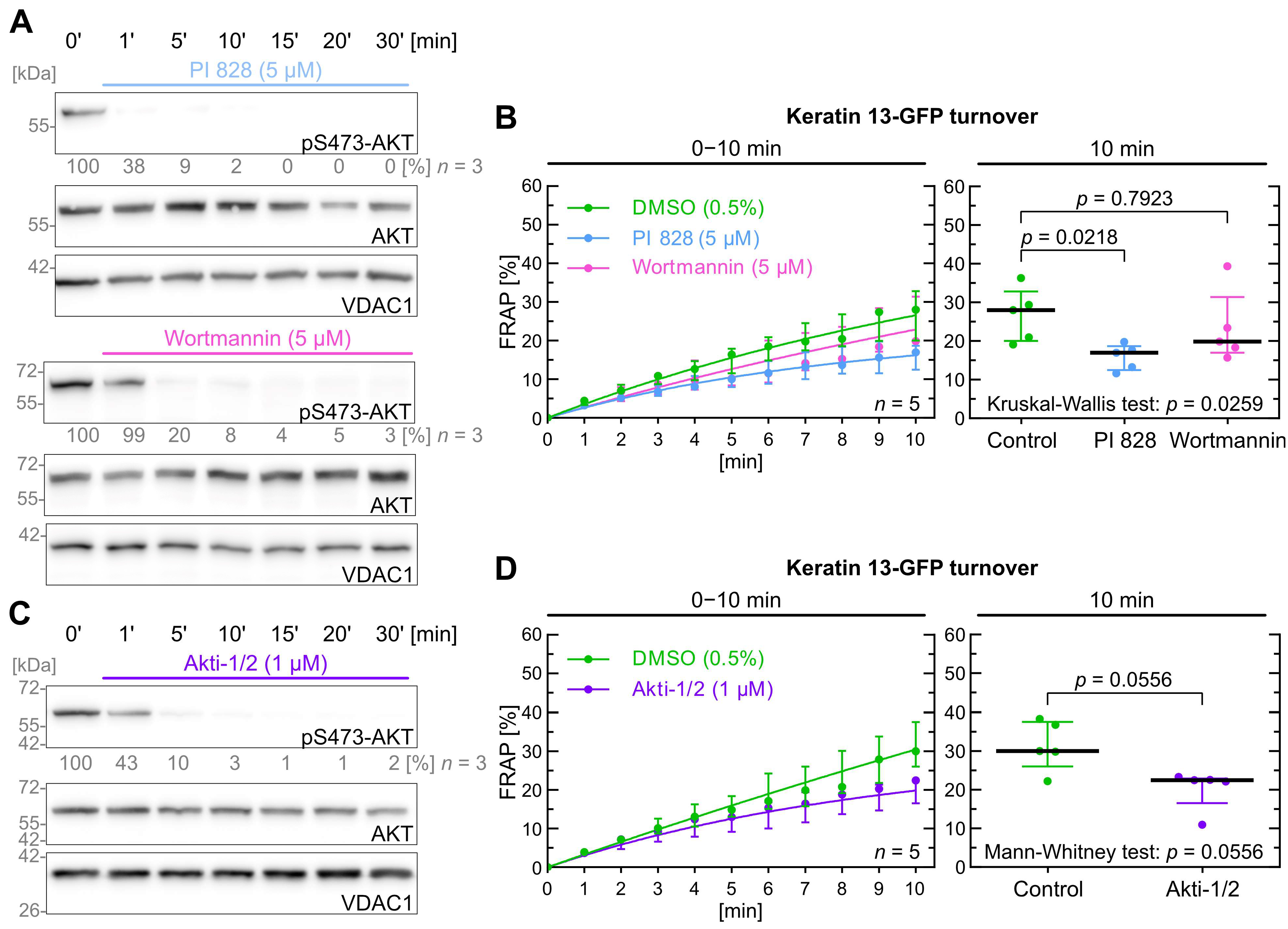
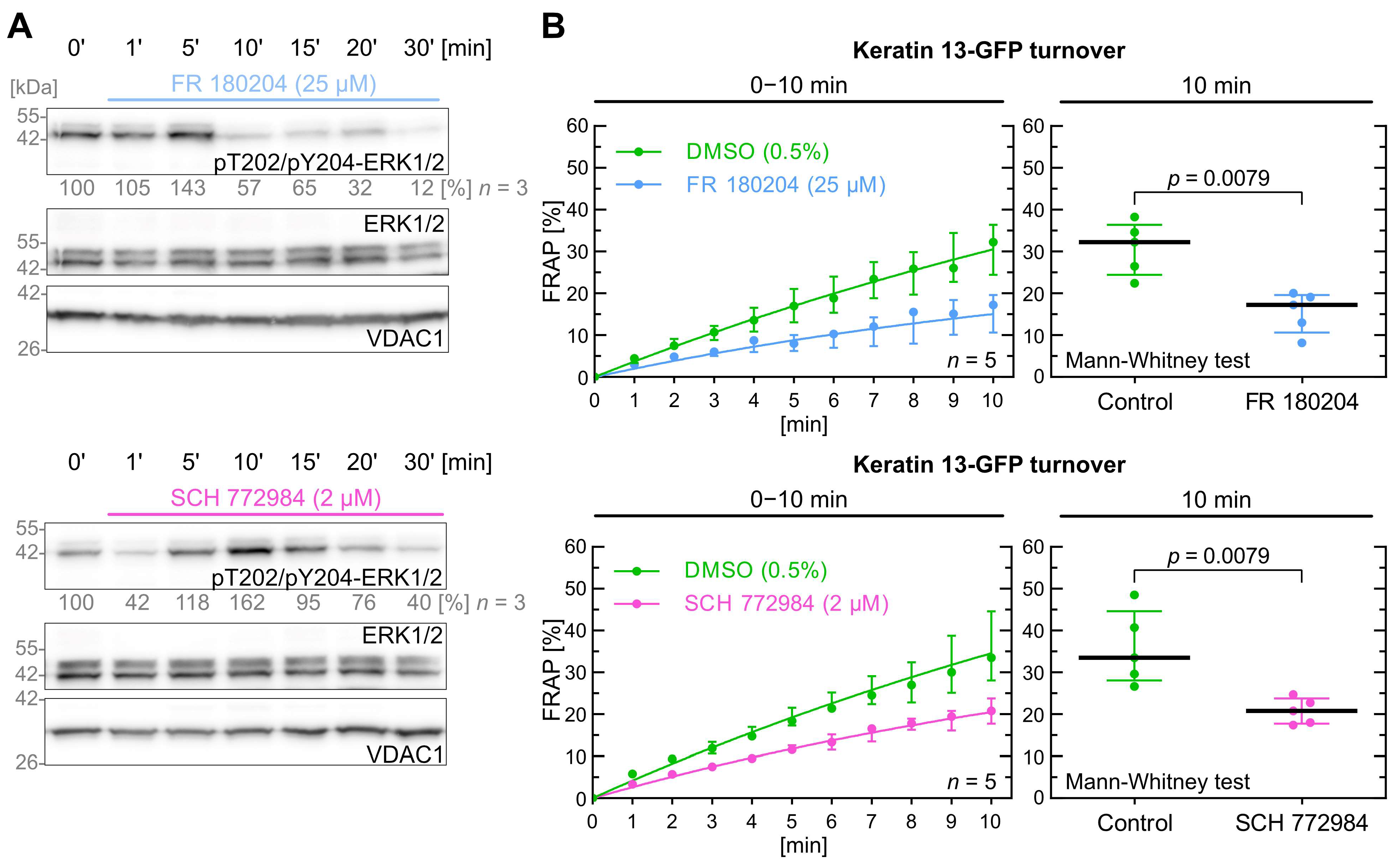
| Kinase | Inhibitor | Inhibitor [µM] | Keratin Turnover 1 |
|---|---|---|---|
| SRC | PP 1 | 20 | ↓ 51% (p = 0.032) |
| SKI-606 | 5 | ↓ 53% (p = 0.014) | |
| ERK1/2 | FR 180204 | 25 | ↓ 47% (p = 0.008) |
| SCH 772984 | 2 | ↓ 37% (p = 0.008) | |
| FAK | PF-573228 | 2 | ↑ 6% (p > 0.999) |
| PF-431396 | 0.5 | ↑ 10% (p > 0.999) | |
| PI3K | PI 828 | 5 | ↓ 39% (p = 0.022) |
| Wortmannin | 5 | ↓ 29% (p = 0.792) | |
| AKT | Akti-1/2 | 1 | ↓ 25% (p = 0.056) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moch, M.; Leube, R.E. SRC and ERK Regulate the Turnover of Cytoskeletal Keratin Filaments. Int. J. Mol. Sci. 2025, 26, 5476. https://doi.org/10.3390/ijms26125476
Moch M, Leube RE. SRC and ERK Regulate the Turnover of Cytoskeletal Keratin Filaments. International Journal of Molecular Sciences. 2025; 26(12):5476. https://doi.org/10.3390/ijms26125476
Chicago/Turabian StyleMoch, Marcin, and Rudolf E. Leube. 2025. "SRC and ERK Regulate the Turnover of Cytoskeletal Keratin Filaments" International Journal of Molecular Sciences 26, no. 12: 5476. https://doi.org/10.3390/ijms26125476
APA StyleMoch, M., & Leube, R. E. (2025). SRC and ERK Regulate the Turnover of Cytoskeletal Keratin Filaments. International Journal of Molecular Sciences, 26(12), 5476. https://doi.org/10.3390/ijms26125476







